Abstract
The clinical outcome of Helicobacter pylori infections is determined by multiple host-pathogen interactions that may develop to chronic gastritis, and sometimes peptic ulcers or gastric cancer. Highly virulent strains encode a type IV secretion system (T4SS) that delivers the effector protein CagA into gastric epithelial cells. Translocated CagA undergoes tyrosine phosphorylation at EPIYA-sequence motifs, called A, B and C in Western-type strains, by members of the oncogenic Src and Abl host kinases. Phosphorylated EPIYA-motifs mediate interactions of CagA with host signaling factors – in particular various SH2-domain containing human proteins – thereby hijacking multiple downstream signaling cascades. Observations of tyrosine-phosphorylated CagA are mainly based on the use of commercial phosphotyrosine antibodies, which originally were selected to detect phosphotyrosines in mammalian proteins. Systematic studies of phosphorylated EPIYA-motif detection by the different antibodies would be very useful, but are not yet available. To address this issue, we synthesized phospho- and non-phosphopeptides representing each predominant Western CagA EPIYA-motif, and determined the recognition patterns of seven different phosphotyrosine antibodies in Western blots, and also performed infection studies with diverse representative Western H. pylori strains. Our results show that a total of 9–11 amino acids containing the phosphorylated EPIYA-motifs are necessary and sufficient for specific detection by these antibodies, but revealed great variability in sequence recognition. Three of the antibodies recognized phosphorylated EPIYA-motifs A, B and C similarly well; whereas preferential binding to phosphorylated motif A and motifs A and C was found with two and one antibodies, respectively, and the seventh anti-phosphotyrosine antibody did not recognize any phosphorylated EPIYA-motif. Controls showed that none of the antibodies recognized the corresponding non-phospho CagA peptides, and that all of them recognized phosphotyrosines in mammalian proteins. These data are valuable in judicious application of commercial anti-phosphotyrosine antibodies and in characterization of CagA phosphorylation during infection and disease development.
Introduction
Posttranslational modification of proteins by kinases is important in many cell signaling processes. Although phosphorylation of some serine, threonine and histidine residues occurs both in prokaryotes and eukaryotes, tyrosine phosphorylation is believed generally to be mostly restricted to higher organisms, where it plays important roles in signal transduction and developmental regulation [1]. In fact, typical tyrosine kinase genes have been reported in only very few sequenced bacterial genomes [2], [3]. However, there are numerous reports of effector proteins from pathogenic bacteria that undergo tyrosine phosphorylation after translocation into eukaryotic host cells, which is a remarkable example of convergent evolution, not just descent from a common ancestor [2], [4]–[6]. In various cases, effector protein phosphotyrosines together with some flanking residues act as recognition motifs for eukaryotic signaling factors. They recruit in particular cellular binding partners that contain SH2 (Src homology 2) domains, but not PTB (phosphotyrosine binding) domains, and thereby target and subvert eukaryotic signal transduction pathways in ways that benefit the pathogen [2], [4]. This virulence strategy is well-established for six different bacterial pathogens: enteropathogenic Escherichia coli (EPEC), Helicobacter pylori, Chlamydia trachomatis, Bartonella henselae, Anaplasma phagocytophilum and Ehrlichia chaffeensis [7]–[13].
The virulence factor CagA of the gastric pathogen H. pylori provides a prime example of such tyrosine phosphorylatable effector proteins [8], [14]–[18]. CagA is delivered to host cells via a type IV secretion system (T4SS), a complex syringe-like pilus device whose synthesis is encoded in the cag pathogenicity island and induced on contact with target cells [19]–[22]. A hallmark of cultured AGS gastric epithelial cells infected by CagA-producing H. pylori strains is the development of the so-called “hummingbird” or “elongation” cell phenotype [8], [23], [24]. This in vitro phenotype is likely to reflect several in vivo signaling processes that control immune responses, wound healing, metastasis and invasive growth of cancer cells [25], [26]. CagA interacts with more than 20 host cell proteins, variously in phosphorylation-dependent and phosphorylation-independent manners [5]. Sequence analysis and site-directed mutagenesis identified a series of EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs near the CagA carboxy-terminus as phosphorylation sites and showed that phospho-CagA is essential for AGS cell elongation [23], [24], [27]–[30]. Four specific EPIYA-motifs termed A, B, C and D have been identified based primarily on relative positions in CagA and adjoining amino acid sequences despite some diversity in flanking sequences and even in EPIYA-motifs themselves [5], [6], [31]-[33]. Whereas EPIYA-A and EPIYA-B motifs are found in all CagA proteins, EPIYA-C is mainly found in strains of African and Indo-European ancestry, whereas CagA from most East-Asian strains contain the more potent EPIYA-D motif in place of EPIYA-C. Although most CagA proteins contain only three EPIYA-motifs, some strains have additional EPIYA-copies [5], [6], due to recombination between repetitions in flanking DNA sequences [31], [34]–[44]. Two-dimensional gel electrophoresis (2-DE) of phospho-CagA proteins from infections of AGS or MKN-28 cells by Western H. pylori strains with 3-4 EPIYA-motifs has shown that only one or two tyrosines (but not three) can be phosphorylated per CagA molecule [45], [46].
The host tyrosine kinases active on these CagA EPIYA-motifs were identified as members of the Src [24], [47] and Abl [48], [49] families. We found that c-Src only phosphorylated EPIYA-C or EPIYA-D, while c-Abl phosphorylated EPIYA-A, EPIYA-B, EPIYA-C, and EPIYA-D. Further analysis revealed that none of the phosphorylated EPIYA-motifs alone was sufficient for inducing AGS cell elongation [46]. Site-directed mutagenesis has shown that the preferred combination of phosphorylated EPIYA-motifs in Western strains was EPIYA-A and EPIYA-C, either across two CagA molecules or simultaneously on one [46]. These studies thus identified a tightly regulated hierarchic phosphorylation model for CagA starting at EPIYA-C/D, followed by phosphorylation at EPIYA-A or EPIYA-B. However, the observation that CagA can undergo tyrosine phosphorylation in host cells is mainly based on the use of commercial α-phosphotyrosine antibodies in Western blots [8], [14]–[17]. These antibodies had been selected years ago to specifically detect a broad range of phosphorylated tyrosine residues in mammalian proteins. Three of these phosphotyrosine-specific antibodies have been shown to exhibit a similar phosphotyrosine-binding preference in mammalian proteins, preferably with a proline residue at position +3 and a leucine at position -1 [50]. However, proline and leucine residues are not present at this position in the corresponding phosphorylation sites of CagA [2], [5], [6]. In addition, systematic studies on the specific recognition patterns of phosphotyrosine motifs in the delivered bacterial effectors such as CagA by a large number of different antibodies are not yet available. Therefore, we addressed this important question and synthesized phospho- and non-phospho peptides of each CagA EPIYA-motif from Western strains to investigate the recognition specificity by seven commercially available phosphotyrosine antibodies. We also performed infection studies with H. pylori to investigate the recognition patterns of phosphorylated CagA upon delivery into host target cells.
Materials and Methods
Bacterial strains and culture conditions
All wild-type H. pylori strains were typical type-I isolates expressing CagA. The generation of an isogenic ΔcagA mutant has been described [17]. H. pylori was grown in thin layers on horse serum GC agar plates supplemented with vancomycin (10 µg/mL), nystatin (1 µg/mL), and trimethoprim (5 µg/mL) as described previously [51], [52]. All antibiotics were obtained from Sigma-Aldrich (St. Louis, MO, USA). Bacteria were grown at 37°C for 2 days in an anaerobic jar containing a Campygen gas mix of 5% O2, 10% CO2, and 85% N2 (Oxoid, Wesel, Germany) [53], [54].
Synthesis of phospho- and non-phospho CagA peptides
The C-STEPIYAKVNK (EPIYA-A), C-STEPI(pY)AKVNK (phospho-EPIYA-A), C-PEEPIYTQVAK (EPIYA-B), C-PEEPI(pY)TQVAK (phospho-EPIYA-B), C-SPEPIYATIDD (EPIYA-C) and C-SPEPI(pY)ATIDD (phospho-EPIYA-C) sequences were synthesized by Jerini AG (Berlin, Germany) and the C-TEPI(pY)AKVN, C-EPI(pY)AKV and C-PI(pY)AK peptides by Biosyntan GmbH (Berlin, Germany). These 11-mer peptides were chosen to compare the three EPIYA-motifs because α-phosphotyrosine antibodies typically recognize short phosphopeptides [35], [50], [55], [56]. Commonly, 11-mer peptides are also used for immunizations to generate phospho-specific antibodies, which then recognize the corresponding phosphopeptides bound to affinity columns and in ELISA (Biogenes, Berlin, Germany). All above EPIYA peptides were purified by HPLC, and full-length synthesis as well as purity of each peptide was confirmed by mass spectrometry by Jerini AG and Biosyntan AG. The peptides were resolved at a concentration of 1 mg/mL in DMSO and stored at −20°C.
Cloning and purification of recombinant CagA
A cagA gene fragment of 891 bp (corresponding to amino acid positions 890-1,186 in the C-terminus of CagA from H. pylori strain 26695) including the EPIYA-motifs A, B and C was synthesized by Geneart (Regensburg, Germany). This cagA fragment was then subcloned into the pGEX-2T vector using the restriction enzymes BamHI and EcoRI. The resultant construct was transformed into the E. coli strain BL21. Protein expression was carried out in 800 mL of pre-warmed LB medium, which were inoculated with 8 mL of an overnight culture. When cells reached an OD600 nm of 0.4, protein expression was induced with 1 mM IPTG. After 4 hours, the cells were harvested and the pellet was stored at −20°C. Prior to cell disruption by sonication, the pellet was suspended in 30 mL PBS (phosphate buffered saline) supplemented with 1 mM DTT. To remove cell debris, the sample was centrifuged at 25,000 rpm for 50 min. Subsequently, the supernatant was loaded onto a 1 mL GSTrap HP column (GE Healthcare, Munich, Germany) and bound target protein was eluted with 50 mM Tris-HCl pH 8.0 containing 10 mM reduced Glutathione and 1 mM DTT. Fractions were analyzed by SDS-PAGE and Western blotting. Selected CagA-positive fractions were pooled and further purified using HiLoad 16/60 Superdex 75 (GE Healthcare) equilibrated with 20 mM Tris-HCl pH 8.0, 150 mM NaCl and 1 mM DTT. Peak fractions were analyzed by SDS-PAGE and Western blot. Fractions containing the purified CagA fragment showed no detectable impurities by other proteins.
In vitro phosphorylation of CagA with recombinant c-Abl kinase
1010 cells of H. pylori strain 26695 expressing wild-type CagA (or isogenic ΔcagA mutant as control) were harvested in 1 mL of kinase buffer as described previously [47]. A total of two units of recombinant human c-Abl tyrosine kinase (NEB GmbH, Frankfurt/M., Germany) and 1 µmol/L of ATP were mixed with 30 µL H. pylori lysate or 10 µL recombinant CagA and incubated for 30 min at 30°C as described previously [49]. The reactions were terminated by a 5 min denaturing step at 95°C.
Dotblot analysis
Twenty µg of each CagA peptide or 30 µl of the in vitro kinase reaction products described above were mixed in 1 mL of blotting buffer (192 mM Glycin; 20 mM Tris-HCl, pH 8,4; 0.1% SDS; 20% Methanol). These peptide samples were spotted onto Immobilon-P membrane (Merck Millipore, Darmstadt, Germany) using the BioDot SF apparatus (Bio-Rad, Munich, Germany). The resulting Dotblots were dried and subjected to antibody detection as described below for Western blots.
Eukaryotic cell culture and elongation phenotype quantitation assays
Human adherent gastric adenocarcinoma epithelial cells (AGS, ATCC CRL-1730) were grown in 6-well plates containing RPMI 1640 medium (Invitrogen) supplemented with 25 mM HEPES buffer and 10% heat-inactivated FBS (Biochrom, Berlin, Germany) for 2 days to approximately 70% confluence [57], [58]. Cells were serum-deprived overnight and infected with H. pylori at a MOI of 50 for 6 hours. After infection, the cells were harvested in ice-cold PBS containing 1 mmol/L Na3VO4 (Sigma-Aldrich). Elongated AGS cells in each experiment were quantitated in 10 different 0.25-mm2 fields using an Olympus IX50 phase contrast microscope. All experiments were performed in triplicate and subjected to statistical analysis as described below.
SDS-PAGE and immunoblot analysis
AGS cell pellets with attached bacteria were mixed with equal amounts of 2 x SDS-PAGE buffer and boiled for 5 minutes. Proteins were separated by SDS-PAGE on 6% polyacrylamide gels and blotted onto PVDF membranes (Immobilon-P, Merck Millipore). Membranes were blocked in TBST with 3% BSA or 5% skim milk for 1 hour at room temperature. Membranes were incubated with the seven α-phosphotyrosine antibodies (Table 1) or mouse monoclonal α-CagA antibody (Austral Biologicals, San Ramon, CA, USA) according to the instructions of the manufacturer. Phosphorylated and non-phosphorylated CagA proteins were detected using horseradish peroxidase–conjugated anti-mouse or anti-rabbit polyvalent sheep immunoglobulin secondary antibodies in the ECL Plus chemoluminescence Western blot system of GE Healthcare [59]–[62].
Table 1. Characteristics of commercial α-phosphotyrosine antibodies used in this study.
| Antibody Name | Short namea | Origin/isotype | Used antigen(s) | Given information on antibody specificity | Dilution used | Order number | Company |
| p-Tyr (PY99) | α-PY-99 | mouse-mAB-IgG2b | np | no cross reactivity with Phospho-Ser or Phospho-Thr | 1∶1,000 | sc-7020 | Santa Cruz Biotechnology |
| Anti-phosphotyrosine (clone PY20) | α-PY-20 (BD) | mouse-mAB-IgG2b | np | np | 1∶500 | 61000 | BD Biosciences |
| p-Tyr (PY20) | α-PY-20 (SC) | mouse-mAB-IgG2b | np | np | 1∶1,000 | sc-508 | Santa Cruz Biotechnology |
| Phospho-Tyrosine Mouse mAB (P-Tyr-100) | α-PY-100 | mouse-mAB-IgG1 | mix of phospho-tyrosine peptides (np) | no cross reactivity with Phospho-Ser or Phospho-Thr | 1∶1,000 | 9411 | Cell Signaling |
| Mouse anti-phospho-tyrosine (clone PY69) | α-PY-69 | mouse-mAB-IgG2a | np | np | 1∶1,000 | 610430 | BD Biosciences |
| Phospho-Tyrosine Mouse mAB (P-Tyr-102) | α-PY-102 | mouse-mAB-IgG2 | mix of phospho-tyrosine peptides (np) | no cross reactivity with Phospho-Ser or Phospho-Thr | 1∶1,000 | 9416 | Cell Signaling |
| p-Tyr (PY350) | α-PY-350 | rabbit-pAB-IgG | np | no cross reactivity with Phospho-Ser or Phospho-Thr | 1∶500 | sc-18182 | Santa Cruz Biotechnology |
Abbreviations: IgG (immunoglobulin G); mAB (monoclonal antibody); np (information not provided by the company); pAB (polyclonal antibody); p-Tyr, PY (phosphotyrosine); Ser (serine); Thr (threonine); Tyr (tyrosine).
short name used in this study.
Quantification of Dotblot and Western blot signals
Spot or band intensities on blots probed with the different α-phosphotyrosine antibodies were quantitated with the Lumi-Imager F1 (Roche Diagnostics, Mannheim, Germany). Densitometric measurement of signal intensities revealed the percentage of phosphorylation per sample [63]. The strongest spot on Dotblots was set at 100% as indicated in the corresponding Figures.
Statistical analysis
All data were evaluated using Student t-test with SigmaStat statistical software (version 2.0). All error bars shown in figures and those quoted following the +/− signs represent standard deviations.
Results
Strong recognition of 9-mer and 11-mer CagA phosphopeptides by α-phosphotyrosine antibodies
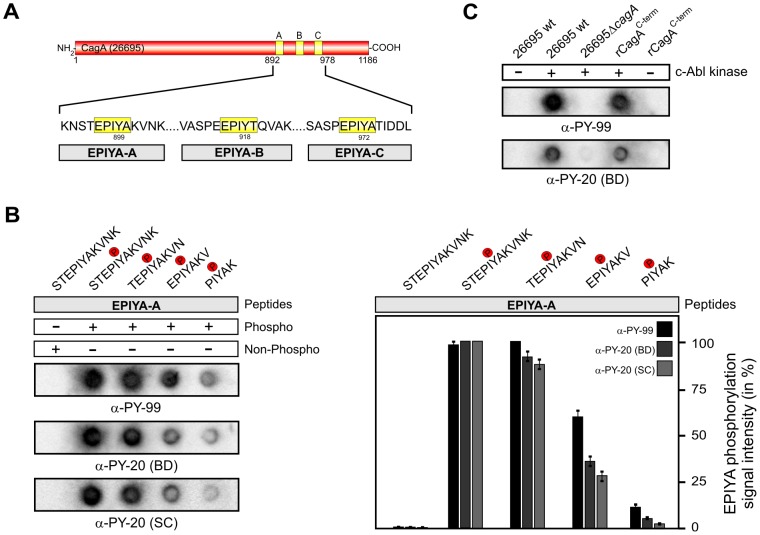
The majority of CagA proteins in Western type clinical isolates contain three EPIYA-motifs, such as that of the first fully sequenced H. pylori strain 26695 (Figure 1A). Commercial phosphotyrosine antibodies typically recognize short mammalian phosphopeptides; in some cases even five amino acids were previously shown to be sufficient for recognition [35], [50], [55], [56]. To systematically analyze the recognition capabilities of phosphorylated CagA EPIYA-motifs by α-phosphotyrosine antibodies we first synthesized a series of peptides derived from the EPIYA-A motif exhibiting the phosphotyrosine residue in the middle +/− five, four, three or two flanking amino acids, including the STEPIYAKVNK (11-mer), TEPIYAKVN (9-mer), EPIYAKV (7-mer) and PIYAK (5-mer) sequences as indicated (Figure 1B, top). Twenty µg of each EPIYA-peptide was immobilized per spot on PVDF membranes using the Dotblot method followed by probing with the α-phosphotyrosine antibodies α-PY99 and α-PY20 (BD). The results show that both antibodies recognized 11-mers and 9-mers with similar strong intensity, while recognition of 7-mers and 5-mers was significantly reduced (Figures 1B). An 11-mer of the corresponding non-phospho-EPIYA peptide did not produce signals in a parallel control experiment (Figure 1B). In further tests, the efficient Dotblot approach detected full-length CagA in bacterial lysates or recombinant C-terminal CagA when phosphorylated by c-Abl in in vitro kinase reactions (Figure 1C). This experiment also confirmed that the α-phosphotyrosine antibodies do not cross-react with non-phospho CagA proteins in control reactions without c-Abl kinase (Figure 1C and data not shown), as expected.
Figure 1. Short EPIYA-phosphopeptides of H. pylori CagA are sufficient for detection by α-phosphotyrosine antibodies.
(A) Typical Western CagA proteins of H. pylori such as that of strain 26695 [76] contain the EPIYA-A, EPIYA-B, and EPIYA-C segments as indicated. These motifs represent tyrosine phosphorylation sites, which can be phosphorylated by c-Abl and c-Src host kinases. (B) The indicated phospho- and non-phospho peptides of the EPIYA-A motif were synthesized and immobilized on PVDF membranes using a Dotblot apparatus. All Dotblots were probed with the indicated commercial phosphotyrosine antibodies and exposed as described in the Material & Methods section. Quantified spot intensities of the Dotblots from three independent experiments are shown to the right. Signal intensities were measured densitometrically with the Lumi-Imager F1 and revealed the percentage of phosphorylation signal per sample. The strongest spot on every Dotblot was set at 100% for each of the different α-phosphotyrosine antibodies as indicated. The results show that 11-mer and 9-mer phosphopeptides are sufficient for strong recognition by the antibodies. (C) Control Dotblot analyses used products of in vitro kinase reactions of c-Abl with either bacterial lysates (from H. pylori wild-type strain 26695 and isogenic ΔcagA mutant) or a purified recombinant CagA C-terminal fragment. Phosphorylated CagA proteins can be also detected by this Dotblot method using seven phosphotyrosine antibodies (Table 1), while the non-phosphorylated CagA forms cannot.
Recognition of phosphopeptides from EPIYA-A, -B and -C by α-phosphotyrosine antibodies
After validating the Dotblot approach using phospho-CagA peptides functions, we synthesized 11-mer phospho- and non-phosphopeptides of EPIYA-B (PEEPIYTQVAK) and EPIYA-C (SPEPIYATIDD) motifs as indicated (Figure 2A, top) and probed them with a set of seven different commercially available α-phosphotyrosine antibodies (Table 1). The results confirmed that these antibodies do not cross-react with the corresponding non-phospho CagA peptides (Figure 2A), and showed, interestingly, that all antibodies except α-PY350 predominantly recognized the EPIYA-A phosphopeptide, which did not recognize any of the CagA-derived phosphopeptides. Some remarkable differences, however, were seen in the abilities of the antibodies to react with the EPIYA-B and EPIYA-C phosphopeptides. In particular, three antibodies (α-PY99, α-PY20-BD, α-PY20-SC) recognized all three phosphorylated EPIYA-A, EPIYA-B and EPIYA-C peptides with similar strong signals, while two others (α-PY100, α-PY69) recognized the phosphorylated EPIYA-A peptide preferentially, and phospho-EPIYA-B and phospho-EPIYA-C more weakly (Figure 2A). A sixth antibody (α-PY102) recognized phosphorylated EPIYA-A strongly and EPIYA-C very weakly, while the seventh antibody, α-PY350, did not recognize any phosphorylated EPIYA-peptide, as noted above (Figure 2A). This inability to detect signals with the α-PY350 antibody was unaffected by binding five-fold more peptide (0.1 mg) to membranes or use of twice as much antibody (data not shown), even though we confirmed that our batch of α-PY350 effectively recognizes phosphorylated host proteins, as discussed below. The quantification of Dotblot data from three independent experiments is presented in Figure 2B. Taken together, the results revealed enormous variability in the capability of seven α-phosphotyrosine antibodies to recognize phosphorylated EPIYA-motifs A, B and/or C.
Figure 2. Variable recognition of synthetic 11-mer CagA phospho-peptides by seven commercial α-phosphotyrosine antibodies.
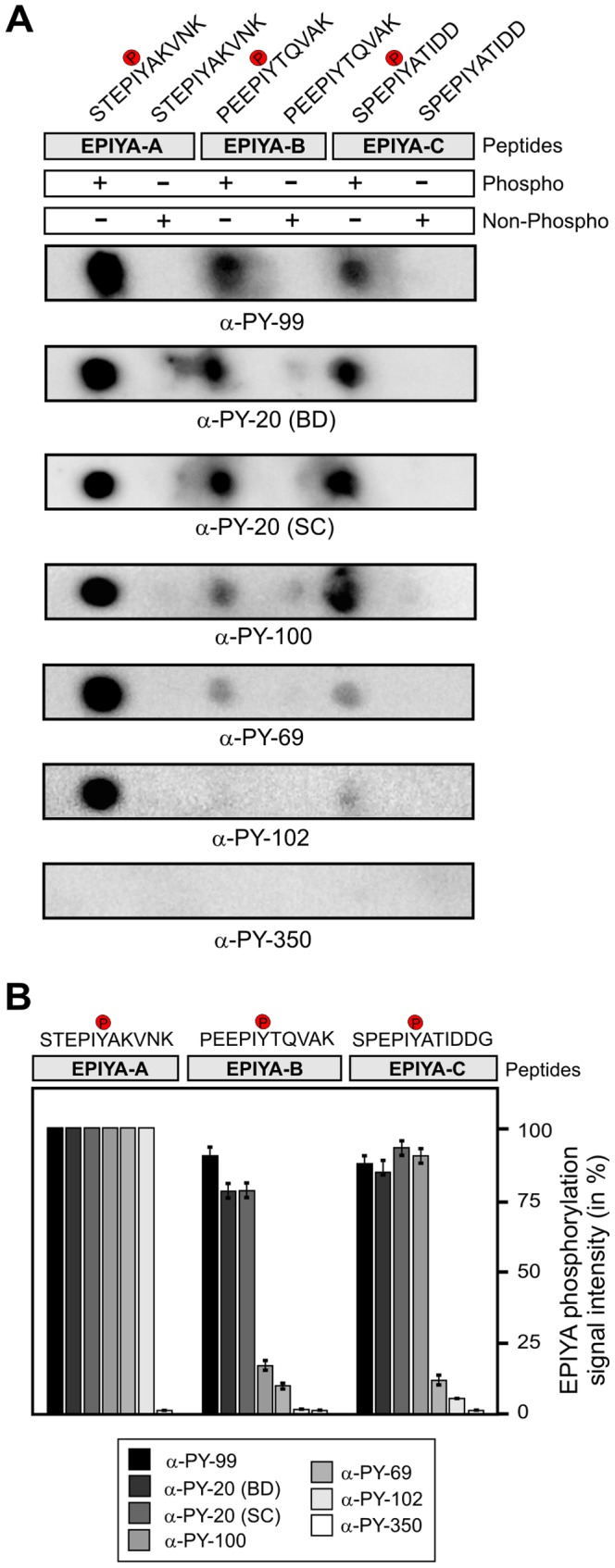
(A) Dotblot analysis of the indicated phospho- and non-phospho peptides derived from single EPIYA-motifs A, B and C. All Dotblots were probed with the indicated commercial phosphotyrosine antibodies and exposed as described in the Material & Methods section. (B) Quantification of spot intensities on Dotblots. Signal intensities were measured densitometrically with the Lumi-Imager F1 and revealed the percentage of phosphorylation signal per sample. The strongest spot on every Dotblot was set at 100% for each of the different α-phosphotyrosine antibodies as indicated. Quantitation results are shown for three independent experiments.
Recognition of phosphorylated CagA protein during infection of AGS cells
Next, we aimed to investigate recognition patterns of phosphorylated CagA proteins during infection. For this purpose, we selected eight representative H. pylori strains from different continents, including Africa, Europe, Asia, North America and South America (Table 2). Sequence alignment of CagA carboxy-terminal regions establishes that all three EPIYA-motifs A, B and C are present, although with some heterogeneity in flanking amino acid sequences (Figure 3). AGS cells were infected for 6 hours with these H. pylori strains and the elongation phenotype of infected cells was monitored over time, to indicate successful CagA delivery and phosphorylation [64]–[66]. About 60−70% of AGS cells exhibited the elongation phenotype after infection with each H. pylori strain, suggesting that high amounts of phospho-CagA are produced (Figure 4). Next, protein lysates were prepared from infected AGS cells and analyzed with the different antibodies. First, the samples were probed with a monoclonal α-CagA antibody recognizing both non-phosphorylated and phosphorylated forms of CagA, to ensure that similar amounts of CagA protein were loaded in each lane (Figure 5, top). The various CagA variants exhibit different band sizes between 130−150 kDa as expected from the different strains (Table 2). The samples were then probed with the seven α-phosphotyrosine antibodies. The results confirmed that all antibodies recognized host cell proteins (Figure 5, asterisks), whereas phospho-CagA was recognized by only some of the antibodies (Figure 5, arrows). The quantification data for phospho-CagA from three independent experiments are presented in Table 3. Two antibodies (α-PY99 and α-PY20-BD) produced strong phospho-CagA bands from all eight strains with little background in the 125−150 kDa region (Figure 5). This indicates that sufficient detectable phospho-CagA was generated during infection and confirmed the elongation phenotype data for each strain noted above. Interestingly, phospho-CagA from two strains (26695 and Sat464) was strongly recognized by six of the seven α-phosphotyrosine antibodies. Antibodies α-PY20-SC, α-PY100 and α-PY102, which recognized all three phospho-EPIYAs in the above Dotblots gave mixed results. Although these three antibodies produced strong bands with phospho-CagA from H. pylori strains 26695 and Sat464, they produced only weak signals with phospho-CagA from strains P310, Lit75, B8, 2002-370 and Oki61 (Figure 5). In addition, it should be noted that the phospho-CagA patterns among these three antibodies were not identical. For example, phospho-CagA from strain Gam94/24 was strongly recognized by α-PY20-SC, but only weakly by α-PY100 and not by α-PY102. The seventh antibody, α-PY350, did not recognize phosphorylated CagA from any H. pylori strain tested, in agreement with results obtained using EPIYA phosphopeptides, although it did recognize major 125, 150 and 170 kDa host phosphoproteins (Figure 5, bottom). Antibody α-PY69 also appears to be not very useful for studying CagA phosphorylation in AGS cells because of the presence of a cross-reacting host phosphoprotein at about 140 kDa, which is in the size range of various phospho-CagA bands (Figure 5).
Table 2. Characteristics of H. pylori strains and encoded CagA proteins used in this study.
| H. pylori strain | Origin | Pathology | CagA EPIYA type | Accession Number | Reference |
| 26695 | UK | Gastritis | ABC | NP_207343.1 | [76] |
| P310 | Germany | Gastric cancer | ABC | AAY18598 | [80] |
| Lit75 | Lithuania | Non-atrophic gastritis | ABC | YP_005772684 | D. Kersulyte and D. Berg (unpublished) |
| B8 | USA | Peptic ulcer | ABC | FN598874.1 | [81] |
| 2002−370 | Mexico | Non-atrophic gastritis | ABC | JN390443 | [46] |
| Sat464 | Peruvian Amazon | Not evaluated directly [Non-atrophic gastritis assumed] | ABC | YP_005768457 | D. Kersulyte and D. Berg (unpublished) |
| Gam94/24 | Gambia | Not evaluated directly [Non-atrophic gastritis assumed] | ABC | YP_005780282 | D. Kersulyte and D. Berg (unpublished) |
| Oki61 | Japan | Duodenal ulcer | ABC | AB725848 | [82] |
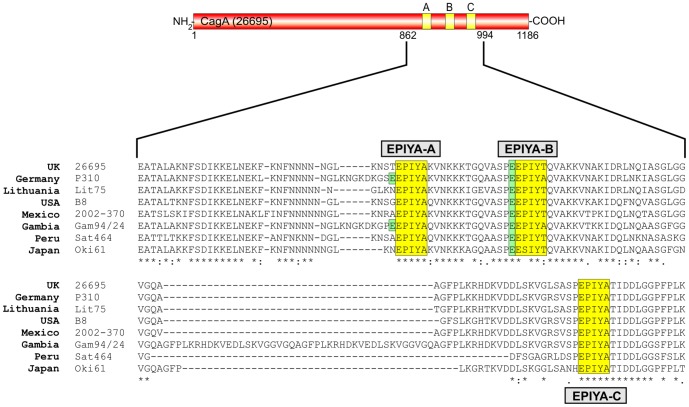
Figure 3. Sequence comparison of the EPIYA-motifs in CagA proteins from different clinical H. pylori strains used in the study.
Western CagA proteins of Helicobacter pylori vary in the carboxy-terminal phosphorylation sites EPIYA-A, EPIYA-B, and EPIYA-C depending on their geographical origin. These EPIYA-repeats serve as tyrosine phosphorylation sites of CagA and can be targeted by c-Abl and c-Src kinases [24], [30], . Multiple EPIYA-segments A, B and C are shaded with yellow and variations in the flanking regions were found among clinical H. pylori isolates from different continents as indicated. One striking feature of EPIYA-B (and in some strains in EPIYA-A) is the presence of a negatively charged glutamate residue in the -4 position (shaded with green), which is highly conserved in EPIYA-B among the different H. pylori strains and may affect the binding capabilities of phosphotyrosine antibodies as discussed in the text. The CagA protein sequences were obtained from databases (Table 2) and sequence alignment was done using the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Figure 4. AGS cell elongation induced during infection with different clinical H. pylori strains.
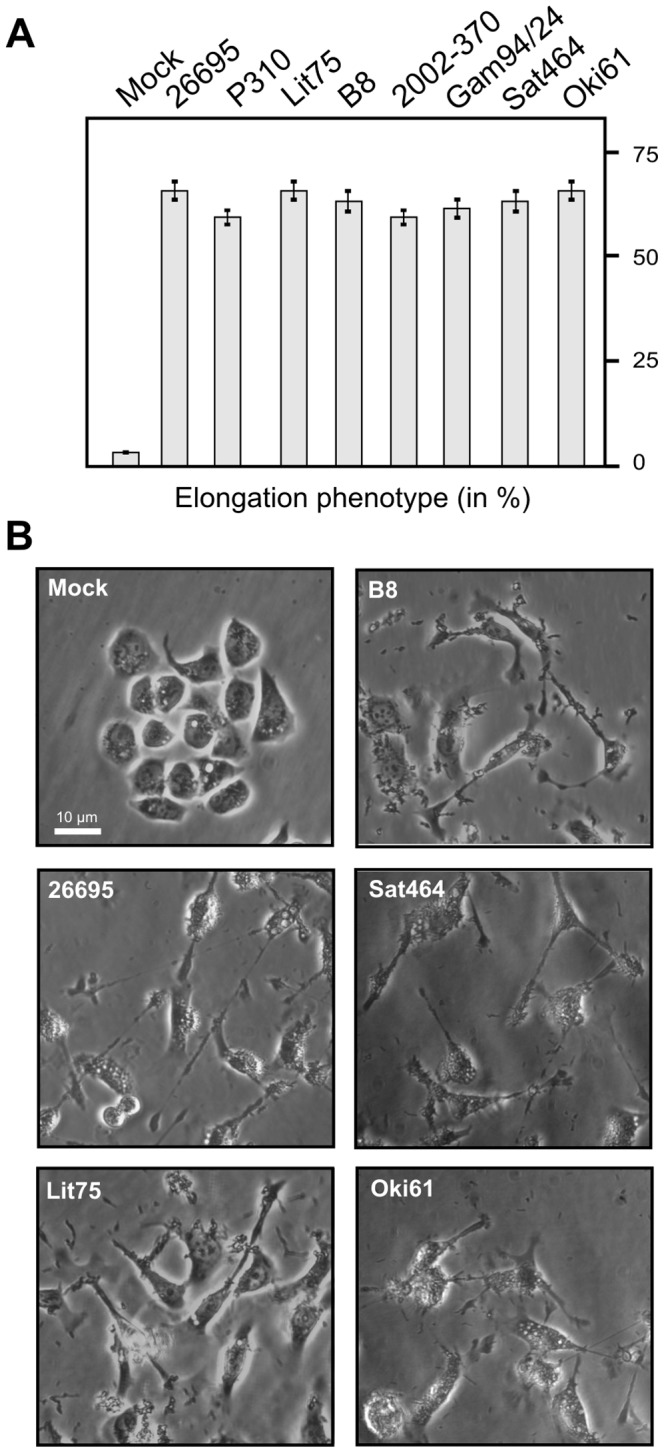
AGS cells were infected for 6-expressing H. pylori strains as indicated. (A) The number of elongated cells in each experiment was quantitated in triplicate in 10 different 0.25-mm2 fields [77]–[79]. (B) Representative phase contrast micrographs of AGS cells infected with the different strains as indicated.
Figure 5. Role of EPIYA motifs in CagA phosphorylation during H. pylori infection was investigated with seven different α-phosphotyrosine antibodies.
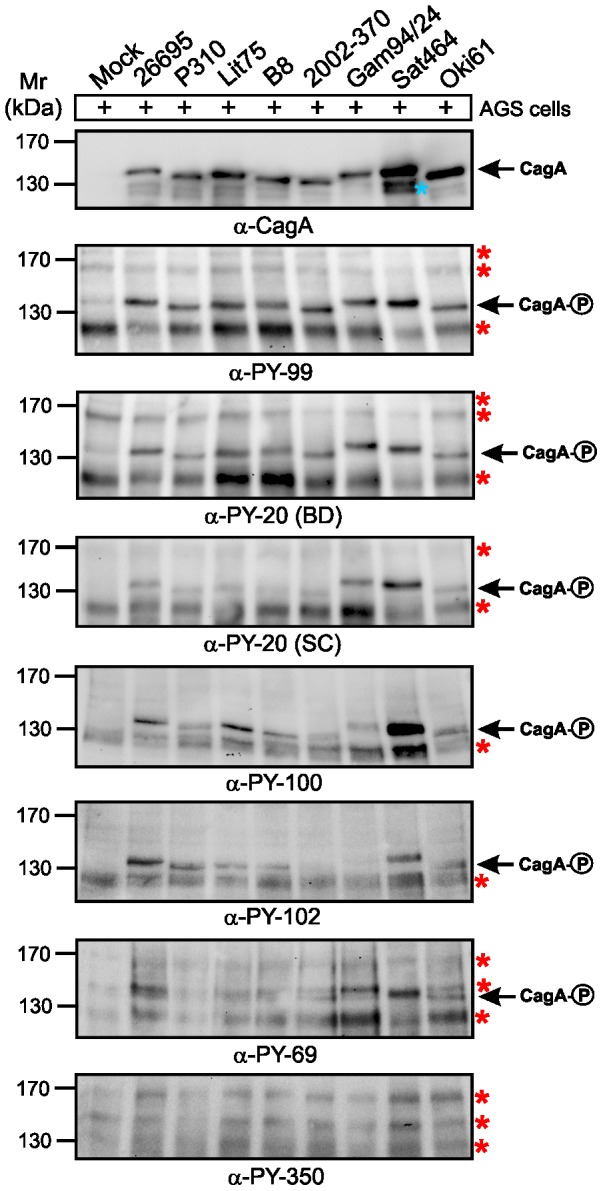
AGS cells were infected for 6-expressing H. pylori strains as indicated. The samples in Figure 4 were harvested after photographing. Phosphorylation of CagA was examined using the indicated α–phosphotyrosine antibodies. Loading of equal amounts of CagA from each sample was confirmed by probing with a monoclonal α-CagA antibody. A larger section of the ∼120−180 kDa range is shown and contains the phospho-CagA bands of different sizes (arrows) as well as a set of tyrosine-phosphorylated host cell proteins (red asterisks). The blue asterisk indicates a putative N-terminal fragment of CagA which sometimes appears on SDS-PAGE gels [23].
Table 3. Recognition of EPIYA-phosphopeptides and phosphorylated CagA proteins by commercial α-phosphotyrosine antibodies.
| Phospho-antibody name | Recognition of phosphopeptides | Phosphorylation signal intensity of translocated CagA by H. pylori strains | |||||||||
| EPIYA-A | EPIYA-B | EPIYA-C | 26695 | P310 | Lit75 | B8 | 2002-370 | Gam94/24 | Sat464 | Oki61 | |
| α-PY-99 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| α-PY-20 (BD) | +++ | ++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| α-PY-20 (SC) | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | ++ |
| α-PY-100 | +++ | + | ++ | ++ | ++ | ++ | ++ | + | ++ | +++ | ++ |
| α-PY-69 | +++ | + | + | +++ | +/− | +/− | +/− | +/− | ++ | +++ | ++ |
| α-PY-102 | +++ | − | + | +++ | ++ | ++ | ++ | − | − | ++ | ++ |
| α-PY-350 | − | − | − | − | − | − | − | − | − | − | − |
Abbreviations used: PY (phosphotyrosine); EPIYA motif (glutamic acid-proline-isoleucine-tyrosine-alanine phosphorylation motif in CagA),
Antibody recognition: +++ (strong signal); ++ (moderate signal); + (weak signal); − (no signal).
Phospho-EPIYA recognition patterns are influenced both by length and amino acid sequence
The binding specificities of the α-PY20 and α-PY100 antibodies have been experimentally characterized using microarrays of phosphopeptide libraries derived from mammalian proteins [50]. This knowledge allowed to investigate the sequence features responsible for differences in binding affinity observed for the EPIYA-B phosphopeptide in more detail. This peptide is recognized by α-PY100 with lower affinity, whereas the other phosphopeptides (EPIYA-A and EPIYA-C) are recognized by α-PY20 and α-PY100 with similar affinity (Figure 1, Table 3). One striking feature of phospho-EPIYA-B is the presence of a negatively charged glutamate residue in the -4 position, which is highly conserved in CagA among different H. pylori strains (Figure 3, shaded with green). The peptide array data from Tinti and co-workers [50] indicates that the presence of a glutamate at this position negatively affects α-PY100 binding, but not α-PY20 binding. Thus, the differences in α-PY20 and α-PY100 binding of phospho-EPIYA-B in our experiments are in line with data from mammalian phosphoproteins and can most likely be attributed to this sequence position. In addition, phosphorylated CagA from different H. pylori strains during infection is generally detected better by α-PY20 than by α-PY100 (Figure 5 and Table 3). These differences in α-PY100 binding most likely result from strain-specific sequence variations in the vicinity of the EPIYA-A motif, which is generally less well-conserved than in the vicinity of the EPIYA-motifs B and C (Figure 3). Phospho-CagA produced by the H. pylori strains P310 and Gam94/24 is bound by α-PY100 with relatively low affinity (Figure 5), which can be attributed at least in part to a glutamate at the -4 position of the EPIYA-A motif (Figure 3, shaded with green). Further inspection of the overall data in Table 3 suggests that additional features also affect α-PY100 binding specificity, as evidenced by the low affinity of phospho-CagA from H. pylori strains B8 and 2002-370 produced during infection. These features are likely determined by residues flanking the EPIYA core motif, as suggested by the significantly weaker binding of the 5-mer and 7-mer peptides described above.
Discussion
The CagA protein and its EPIYA-motifs are known for long time as important virulence markers of H. pylori [5], [6], [18], [23], [24], [27]–[34], [67], [68]. These EPIYA-repeat motifs were originally described in 1993 by the group of Antonello Covacci [69]. Even before these sequences were identified to be the sites of CagA tyrosine phosphorylation [23], certain variations within the EPIYA-region at the sequence level have been reported and associated with gastric disease [70], [71]. After the discovery of CagA tyrosine phosphorylation about 15 years ago [18], intensive efforts were undertaken to identify the involved host cell kinases [5]. Mammals encode about 90 protein tyrosine kinases [72], which phosphorylate their mammalian substrates with specificity depending on amino acid sequences next to the targeted tyrosine residue [73]. The EPIYA-motifs in CagA commonly have isoleucine at the –1 position and the small amino acid alanine at the +1 position, which is similar to the phosphorylation consensus motif EEIYG/E of the host kinase c-Src [47]. Indeed, several lines of evidence have then demonstrated that c-Src and also c-Abl family kinases mediate CagA phosphorylation in vitro and in vivo [24], [46]–[49], [74]. However, progress in this research field has been hampered by a lack of standardized commercial EPIYA-specific phospho-antibodies and lack of knowledge about which phospho-EPIYAs are recognized by the set of available α-phosphotyrosine antibodies (Table 1). Systematic examination of which phosphotyrosine residues in the three different EPIYA-motifs are recognized by these various antibodies has not yet been reported. Thus, despite the years of research, detailed, clear-cut conclusions about CagA phosphorylation patterns in clinical strains have not been possible. Here, we investigated for the first time the recognition specificity of seven commercially available α-phosphotyrosine antibodies with regard to CagA EPIYA-motifs A, B and C in Western H. pylori strains. We found that these antibodies exhibit a remarkable variety in recognition of the various phosphorylated EPIYA-motifs. These results shed light on the usefulness of these antibodies in research and provide valuable new insights for future studies on CagA phosphorylation sites and downstream signaling.
It is well known that the α-phosphotyrosine antibodies were originally developed for mammalian proteins and typically recognize short amino acid stretches containing the phosphorylated tyrosine residue, including synthetic phospho-peptides [35], [50], [55], [56]. We therefore proposed that phospho-peptides derived from the CagA EPIYA-motifs would be useful for studying the recognition capabilities by seven commercial antibodies. Using this strategy we found that 9-mers and 11-mers of EPIYA-phosphopeptides are necessary and sufficient for strong antibody recognition. In addition, three α-phosphotyrosine antibodies (α-PY99, α-PY20-BD and α-PY20-SC) recognized all three 11-mer phospho-EPIYA peptides (A, B and C) with similar and very strong affinity, confirming that this approach works for peptides derived from bacterial effector proteins in addition to mammalian peptides. Overall, this observation nicely correlated with the pronounced recognition of phospho-CagA in cell lysates produced after infection with eight different H. pylori strains (Table 3). Another antibody (α-PY100) reacted preferentially with phospho-EPIYA peptides A and C, and also produced acceptable phospho-CagA patterns by Western blotting of extracts from infected cells. In addition, antibody α-PY102 strongly recognized phospho-EPIYA peptide A and very weakly phospho-EPIYA peptide C, but reacted only with six of eight phospho-CagAs in infected cells. Although α-PY69 also recognized phospho-EPIYA-A preferentially (with weak signals for B and C), it also strongly reacted with host cell proteins in the 125–140 kDa range and is therefore not useful for studying CagA phosphorylation during infection. Importantly, the antibodies α-PY99, α-PY20-BD, α-PY20-SC, α-PY100 and α-PY102 did not react with AGS host cell proteins in the 130–150 kDa range, another important criterion that makes them useful for H. pylori studies. These results allow us to recommend the use of up to five α-phosphotyrosine antibodies for studies of infection by H. pylori (α-PY99, α-PY20-BD and α-PY20-SC, and if needed, also α-PY100 and α-PY69) based on their ability to recognise a broad spectrum of different phospho-CagAs, and thereby to clarify what EPIYAs are phosphorylated.
Most knowledge about phosphotyrosine-based protein-protein interactions is derived from use of α-phosphotyrosine antibodies to investigate mammalian signaling factors [1]. Recent studies have applied the microarray technology to characterize the substrate specificity of widely used α-phosphotyrosine antibodies (including α-PY100 and α-PY20) in human phosphopeptides [50]. The conclusions that were drawn from this analysis are: (i) the antibodies exhibit a similar phosphotyrosine binding specificity whilst at the same time showing specific binding preference depending on some flanking amino acids; and (ii) the similar phosphotyrosine binding specificity is rather broad although proline residues are preferred at position +3 and leucine at position –1 [50]. In addition, the investigations by Tinti and co-workers indicated that the presence of a negatively charged residue such as glutamate at the –4 position specifically affects interaction with α-PY100, but not with α-PY20 [50]. Interestingly, EPIYA-B has a glutamate residue at the –4 position (and sometimes at EPIYA-A), which is highly conserved in CagA among the different H. pylori strains (Figure 3). However, inspection of our overall data in Table 3 suggests that additional features also affect α-PY100 binding specificity, as evidenced by the low affinity to phospho-CagA from strains B8 and 2002-370. Both H. pylori strains differ at several sequence positions in the vicinity of EPIYA-motif A rendering it difficult to clearly correlate antibody affinity with a single sequence position. This also suggests the existence of other structural features, like secondary structure of the motif and its vicinity, which may additionally affect α-phosphotyrosine antibody binding specificity.
How could one apply this knowledge of binding preferences of α-phosphotyrosine antibodies for certain EPIYA-motifs? The use of α-phosphotyrosine antibodies by probing one-dimensional SDS-PAGE blots of lysates from H. pylori infected cells provides a useful first picture of CagA phosphorylation events [75]. However, this picture is not completely useful because increased phospho-CagA signal intensities that can arise over time could result either from increased amounts of translocated CagA molecules undergoing phosphorylation at a specific site, from increased phosphorylation of multiple sites per CagA molecule, or both – possibilities that cannot be distinguished on 1-DE gels. Using 2-DE gel separation of different CagA protein spots, we have recently shown that CagA can be simultaneously phosphorylated either on one or two EPIYAs per molecule [46]. This suggests the appearance of multiple differentially phosphorylated CagA protein species in host cells, each with different functions, and thus could explain how CagA achieves signaling to many different host binding partners [46]. To investigate this hypothesis further and to obtain better tools for clinical applications, it will be desirable in the near future to generate phospho-specific α-CagA antibodies for each EPIYA-motif. In this context, the α-PY69 and α-PY102 antibodies may already be useful because they predominantly detect phospho-EPIYA-A as shown here. A very few studies have reported generation of phospho-specific α-CagA antibodies designed for one specific EPIYA-motif, although to our knowledge these are not commercially available. A first study showed phosphorylation of EPIYA-motif B after infection and after transfection of CagA from strain NCTC11637 [28]; and another study reported on specific phosphorylation of EPIYA-C in CagA from strains 26695 and P12, including respective phenylalanine substitution controls [22]. In addition, the generation of a phosphospecific antibody against EPIYA-C motif in CagA from strain NCTC11637 was described, but this antibody also recognized the phosphorylated CagA forms of strains GC401 and G501, which lack the EPIYA-motif C, a result suggesting that this antibody may be rather unspecific, able to recognize other phosphorylation sites [30]. Thus, the production of more reliable EPIYA-site specific phospho-antibodies could yield highly valuable research tools.
Our studies focused on the EPIYA-motifs A, B and C of Western H. pylori strains. However, it will be also important to investigate the phosphorylation of the generally more potent CagA proteins East-Asian strains with their EPIYA-A, B and D motifs, even though the East-Asian 11-mer EPIYA-D sequence (SPEPIYATIDF) is similar to the Western EPIYA-C sequence (SPEPIYATIDD) [46]. We recently showed that c-Src kinase only phosphorylates CagA EPIYA-C and EPIYA-D motifs, not EPIYA-A and EPIYA-B, using Western blots and the α-PY99 antibody [46]. This result suggests that phospho-EPIYA-D maybe recognized by many or all antibodies that recognize phospho-EPIYA-C. However, this has not yet been tested experimentally, and more studies are certainly warranted to understand the CagA phosphorylation sites in greater detail. A detailed analysis of the various East-Asian EPIYAs is currently underway in our laboratories. One additional option would be to use the phosphopeptide microarray technology to characterize all known individual phospho-EPIYA-motifs and associated amino acid polymorphisms, as was described for human proteins [50], and to test for antibody recognition and host effector protein binding. This would help to better understand the role of single EPIYA-motifs for CagA function and possibly allow correlations and risk predictions for the development of diverse gastric diseases in the future. In addition to CagA, further studies should also focus on the investigation of EPIYA-like phosphorylation sites and downstream signaling used by other bacterial effector proteins from pathogens such as EPEC, Chlamydia, Bartonella, Anaplasma and Ehrlichia species, which collectively represents a fascinating new research area [5], [7]–[13].
Acknowledgments
We thank Dr. Martin Grieβl (FAU Erlangen, Germany) for technical advice in CagA purification, Dr. Javier Torres (Unidad de Investigacion en Enfermedades Infecciosas UMAE Pediatria, IMSS, Mexico) for providing H. pylori strain 2002-370 and Dr. Gabriele Rieder (University Salzburg, Austria) for providing strain B8.
Funding Statement
The work of SB and HS is supported through DFG grants (projects B10 and A2 of CRC-796). The research in DB's group has been supported by US National Institutes of Health Grants (R21 AI078237 and R21 AI088337). YY is supported by US National Institutes of Health Grant R01DK62813. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hunter T (2009) Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol 21: 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Backert S, Selbach M (2005) Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol 13: 476–484. [DOI] [PubMed] [Google Scholar]
- 3. Cousin C, Derouiche A, Shi L, Pagot Y, Poncet S, et al. (2013) Protein-serine/threonine/tyrosine kinases in bacterial signaling and regulation. FEMS Microbiol Lett 346: 11–19. [DOI] [PubMed] [Google Scholar]
- 4. Selbach M, Paul FE, Brandt S, Guye P, Daumke O, et al. (2009) Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe 5: 397–403. [DOI] [PubMed] [Google Scholar]
- 5. Backert S, Tegtmeyer N, Selbach M (2010) The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter 15: 163–176. [DOI] [PubMed] [Google Scholar]
- 6. Hayashi T, Morohashi H, Hatakeyama M (2013) Bacterial EPIYA effectors—where do they come from? What are they? Where are they going? Cell Microbiol 15: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, et al. (1997) Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91: 511–520. [DOI] [PubMed] [Google Scholar]
- 8. Segal ED, Cha J, Lo J, Falkow S, Tompkins LS (1999) Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. . Proc Natl Acad Sci U S A 96: 14559–14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, et al. (2004) A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A 101: 10166–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schulein R, Guye P, Rhomberg TA, Schmid MC, Schröder G, et al. (2005) A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A 102: 856–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ijdo JW, Carlson AC, Kennedy EL (2007) Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell Microbiol 9: 1284–1296. [DOI] [PubMed] [Google Scholar]
- 12. Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y (2007) Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol 9: 2644–57. [DOI] [PubMed] [Google Scholar]
- 13. Rikihisa Y, Lin M (2010) Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol 13: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, et al. (2000) Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 15. Stein M, Rappuoli R, Covacci A (2000) Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci U S A 97: 1263–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Asahi M, Azuma T, Ito S, Ito Y, Suto H, et al. (2000) Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med 191: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Backert S, Ziska E, Brinkmann V, Zimny-Arndt U, Fauconnier A, et al. (2000) Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell Microbiol 2: 155–164. [DOI] [PubMed] [Google Scholar]
- 18. Covacci A, Rappuoli R (2000) Tyrosine-phosphorylated bacterial proteins: Trojan horses for the host cell. J Exp Med 191: 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rohde M, Puels A, Buhrdorf R, Fischer W, Haas R (2003) A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49: 218–234. [DOI] [PubMed] [Google Scholar]
- 20. Tanaka J, Suzuki T, Mimuro H, Sasakawa C (2003) Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol 5: 395–404. [DOI] [PubMed] [Google Scholar]
- 21. Backert S, Meyer TF (2006) Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol 9: 207–217. [DOI] [PubMed] [Google Scholar]
- 22. Kwok T, Zabler D, Urman S, Rohde M, Hartig R, et al. (2007) Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449: 862–866. [DOI] [PubMed] [Google Scholar]
- 23. Backert S, Moese S, Selbach M, Brinkmann V, Meyer TF (2001) Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol Microbiol 42: 631–644. [DOI] [PubMed] [Google Scholar]
- 24. Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, et al. (2002) c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol 43: 971–980. [DOI] [PubMed] [Google Scholar]
- 25. Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, et al. (2003) Cell migration: integrating signals from front to back. Science 302: 1704–1709. [DOI] [PubMed] [Google Scholar]
- 26. Schneider S, Weydig C, Wessler S (2008) Targeting focal adhesions: Helicobacter pylori-host communication in cell migration. Cell Commun Signal 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, et al. (2002) SHP2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295: 683–686. [DOI] [PubMed] [Google Scholar]
- 28. Puels M, Fischer W, Haas R (2002) Activation of Helicobacter pylori CagA by tyrosine phosphorylation is essential for dephosphorylation of host cell proteins in gastric epithelial cells. Mol Microbiol 43: 962–969. [DOI] [PubMed] [Google Scholar]
- 29. Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R, et al. (2002) Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol Cell 10: 745–755. [DOI] [PubMed] [Google Scholar]
- 30. Asahi M, Tanaka Y, Izumi T, Ito Y, Naiki H, et al. (2003) Helicobacter pylori CagA containing ITAM-like sequences localized to lipid rafts negatively regulates VacA-induced signaling in vivo . Helicobacter 8: 1–14. [DOI] [PubMed] [Google Scholar]
- 31. Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X (2009) A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori . PLoS One 4: e7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Backert S, Selbach M (2008) Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol 10: 1573–1581. [DOI] [PubMed] [Google Scholar]
- 33. Yamaoka Y (2010) Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol 7: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aras RA, Lee Y, Kim SK, Israel D, Peek RM Jr, et al. (2003) Natural variation in populations of persistently colonizing bacteria affect human host cell phenotype. J Infect Dis 188: 486–496. [DOI] [PubMed] [Google Scholar]
- 35. Kim M, Shin DS, Kim J, Lee YS (2010) Substrate screening of protein kinases: detection methods and combinatorial peptide libraries. Biopolymers 94: 753–762. [DOI] [PubMed] [Google Scholar]
- 36. Argent RH, Zhang Y, Atherton J (2005) Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J Clin Microbiol 43: 791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, et al. (2006) Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 130: 1181–1190. [DOI] [PubMed] [Google Scholar]
- 38. Panayotopoulou EG, Sgouras DN, Papadakos K, Kalliaropoulos A, Papatheodoridis G, et al. (2007) Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. J Clin Microbiol 45: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, et al. (2008) Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135: 91–99. [DOI] [PubMed] [Google Scholar]
- 40. Schmidt HM, Goh KL, Fock KM, Hilmi I, Dhamodaran S, et al. (2009) Distinct cagA EPIYA motifs are associated with ethnic diversity in Malaysia and Singapore. Helicobacter 14: 256–263. [DOI] [PubMed] [Google Scholar]
- 41. Miura M, Ohnishi N, Tanaka S, Yanagiya K, Hatakeyama M (2009) Differential oncogenic potential of geographically distinct Helicobacter pylori CagA isoforms in mice. Int J Cancer 125: 2497–2504. [DOI] [PubMed] [Google Scholar]
- 42. Truong BX, Mai VT, Tanaka H, Ly le T, Thong TM, et al. (2009) Diverse characteristics of the CagA gene of Helicobacter pylori strains collected from patients from southern vietnam with gastric cancer and peptic ulcer. J Clin Microbiol 47: 4021–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones KR, Joo YM, Jang S, Yoo YJ, Lee HS, et al. (2009) Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J Clin Microbiol 47: 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Furuta Y, Yahara K, Hatakeyama M, Kobayashi I (2011) Evolution of cagA oncogene of Helicobacter pylori through recombination. PLoS ONE 6: e23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Backert S, Müller EC, Jungblut PR, Meyer TF (2001) Tyrosine phosphorylation patterns and size modification of the Helicobacter pylori CagA protein after translocation into gastric epithelial cells. Proteomics 1: 608–617. [DOI] [PubMed] [Google Scholar]
- 46. Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, et al. (2012) c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest 122: 1553–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Selbach M, Moese S, Hauck CR, Meyer TF, Backert S (2002) Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. . J Biol Chem 277: 6775–6778. [DOI] [PubMed] [Google Scholar]
- 48. Poppe M, Feller SM, Römer G, Wessler S (2007) Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene 26: 3462–3472. [DOI] [PubMed] [Google Scholar]
- 49. Tammer I, Brandt S, Hartig R, König W, Backert S (2007) Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology 132: 1309–1319. [DOI] [PubMed] [Google Scholar]
- 50. Tinti M, Nardozza AP, Ferrari E, Sacco F, Corallino S, et al. (2012) The 4G10, pY20 and p-TYR-100 antibody specificity: profiling by peptide microarrays. N Biotechnol 29: 571–517. [DOI] [PubMed] [Google Scholar]
- 51. Kumar Pachathundikandi S, Brandt S, Madassery J, Backert S (2011) Induction of TLR-2 and TLR-5 expression by Helicobacter pylori switches cagPAI-dependent signalling leading to the secretion of IL-8 and TNF-α. PLoS One 6: e19614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiedemann T, Hofbaur S, Tegtmeyer N, Huber S, Sewald N, et al. (2012) Helicobacter pylori CagL dependent induction of gastrin expression via a novel αvβ5-integrin-integrin linked kinase signalling complex. Gut 61: 986–996. [DOI] [PubMed] [Google Scholar]
- 53. Hirsch C, Tegtmeyer N, Rohde M, Rowland M, Oyarzabal OA, et al. (2012) Live Helicobacter pylori in the root canal of endodontic-infected deciduous teeth. J Gastroenterol 47: 936–940. [DOI] [PubMed] [Google Scholar]
- 54. Tegtmeyer N, Rivas Traverso F, Rohde M, Oyarzabal OA, Lehn N, et al. (2013) Electron microscopic, genetic and protein expression analyses of Helicobacter acinonychis strains from a Bengal tiger. PLoS One 8: e71220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blaydes JP, Vojtesek B, Bloomberg GB, Hupp TR (2000) The development and use of phospho-specific antibodies to study protein phosphorylation. Methods Mol Biol 99: 177–189. [DOI] [PubMed] [Google Scholar]
- 56. Houseman BT, Huh JH, Kron SJ, Mrksich M (2002) Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol 20: 270–274. [DOI] [PubMed] [Google Scholar]
- 57. Conradi J, Tegtmeyer N, Woźna M, Wissbrock M, Michalek C, et al. (2012) An RGD helper sequence in CagL of Helicobacter pylori assists in interactions with integrins and injection of CagA. Front Cell Infect Microbiol 2: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hoy B, Geppert T, Boehm M, Reisen F, Plattner P, et al. (2012) Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J Biol Chem 287: 10115–10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tegtmeyer N, Hartig R, Delahay RM, Rohde M, Brandt S, et al. (2010) A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. J Biol Chem 285: 23515–23526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boehm M, Krause-Gruszczynska M, Rohde M, Tegtmeyer N, Takahashi S, et al. (2011) Major host factors involved in epithelial cell invasion of Campylobacter jejuni: role of fibronectin, integrin beta1, FAK, Tiam-1, and DOCK180 in activating Rho GTPase Rac1. Front Cell Infect Microbiol 1: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boehm M, Hoy B, Rohde M, Tegtmeyer N, Bæk KT, et al. (2012) Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Traverso FR, Bohr UR, Oyarzabal OA, Rohde M, Clarici A, et al. (2010) Morphologic, genetic, and biochemical characterization of Helicobacter magdeburgensis, a novel species isolated from the intestine of laboratory mice. Helicobacter 15: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tegtmeyer N, Wittelsberger R, Hartig R, Wessler S, Martinez-Quiles N, et al. (2011) Serine phosphorylation of cortactin controls focal adhesion kinase activity and cell scattering induced by Helicobacter pylori . Cell Host Microbe 9: 520–531. [DOI] [PubMed] [Google Scholar]
- 64. Backert S, Schwarz T, Miehlke S, Kirsch C, Sommer C, et al. (2004) Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. Infect Immun 72: 1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brandt S, Wessler S, Hartig R, Backert S (2009) Helicobacter pylori activates protein kinase C delta to control Raf in MAP kinase signalling: role in AGS epithelial cell scattering and elongation. Cell Motil Cytoskeleton 66: 874–892. [DOI] [PubMed] [Google Scholar]
- 66. Tegtmeyer N, Zabler D, Schmidt D, Hartig R, Brandt S, et al. (2009) Importance of EGF receptor, HER2/Neu and Erk1/2 kinase signalling for host cell elongation and scattering induced by the Helicobacter pylori CagA protein: antagonistic effects of the vacuolating cytotoxin VacA. Cell Microbiol 11: 488–505. [DOI] [PubMed] [Google Scholar]
- 67. Backert S, Clyne M, Tegtmeyer N (2011) Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori . Cell Commun Signal 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smolka AJ, Backert S (2012) How Helicobacter pylori infection controls gastric acid secretion. J Gastroenterol 47: 609–618. [DOI] [PubMed] [Google Scholar]
- 69. Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, et al. (1993) Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. . Proc Natl Acad Sci U S A. 90: 5791–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR (1998) Variants of the 3' region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. . J Clin Microbiol. 36: 2258–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yamaoka Y, El-Zimaity HM, Gutierrez O, Figura N, Kim JG, et al. (1999) Relationship between the cagA 3' repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. . Gastroenterol. 117: 342–349. [DOI] [PubMed] [Google Scholar]
- 72. Robinson DR, Wu YM, Lin SF (2000) The protein tyrosine kinase family of the human genome. Oncogene 19: 5548–5557. [DOI] [PubMed] [Google Scholar]
- 73. Songyang Z, Carraway KL3rd, Eck MJ, Harrison SC, Feldman RA, et al (1995) Catalytic specificity of protein-tyrosine kinases is critical for selective signaling. Nature 373: 536–539. [DOI] [PubMed] [Google Scholar]
- 74. Posselt G, Backert S, Wessler S (2013) The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun Signal 11: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tegtmeyer N, Backert S (2011) Role of Abl and Src family kinases in actin-cytoskeletal rearrangements induced by the Helicobacter pylori CagA protein. Eur J Cell Biol 90: 880–890. [DOI] [PubMed] [Google Scholar]
- 76. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature 388: 539–547. [DOI] [PubMed] [Google Scholar]
- 77. Moese S, Selbach M, Kwok T, Brinkmann V, König W, et al. (2004) Helicobacter pylori induces AGS cell motility and elongation via independent signaling pathways. Infect Immun 72: 3646–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Moese S, Selbach M, Brinkmann V, Karlas A, Haimovich B, et al. (2007) The Helicobacter pylori CagA protein disrupts matrix adhesion of gastric epithelial cells by dephosphorylation of vinculin. Cell. Microbiol 9: 1148–1161. [DOI] [PubMed] [Google Scholar]
- 79. Hoy B, Löwer M, Weydig C, Carra G, Tegtmeyer N, et al. (2010) Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 11: 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brandt S, Kwok T, Hartig R, König W, Backert S (2005) NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci U S A 102: 9300–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Farnbacher M, Jahn T, Willrod D, Daniel R, Haas R, et al. (2010) Sequencing, annotation, and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genomics 11: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Matsunari O, Shiota S, Suzuki R, Watada M, Kinjo N, et al. (2012) Association between Helicobacter pylori virulence factors and gastroduodenal diseases in Okinawa, Japan. J Clin Microbiol 50: 876–583. [DOI] [PMC free article] [PubMed] [Google Scholar]