Abstract
Nitric oxide (NO) is a bioactive molecule involved in numerous biological events that has been reported to display both pro-oxidant and antioxidant properties in plants. Several reports exist which demonstrate the protective action of sodium nitroprusside (SNP), a widely used NO donor, which acts as a signal molecule in plants responsible for the expression regulation of many antioxidant enzymes. This study attempts to provide a novel insight into the effect of application of low (100 μΜ) and high (2.5 mM) concentrations of SNP on the nitrosative status and nitrate metabolism of mature (40 d) and senescing (65 d) Medicago truncatula plants. Higher concentrations of SNP resulted in increased NO content, cellular damage levels and reactive oxygen species (ROS) concentration, further induced in older tissues. Senescing M. truncatula plants demonstrated greater sensitivity to SNP-induced oxidative and nitrosative damage, suggesting a developmental stage-dependent suppression in the plant’s capacity to cope with free oxygen and nitrogen radicals. In addition, measurements of the activity of nitrate reductase (NR), a key enzyme involved in the generation of NO in plants, indicated a differential regulation in a dose and time-dependent manner. Furthermore, expression levels of NO-responsive genes (NR, nitrate/nitrite transporters) involved in nitrogen assimilation and NO production revealed significant induction of NR and nitrate transporter during long-term 2.5 mM SNP application in mature plants and overall gene suppression in senescing plants, supporting the differential nitrosative response of M. truncatula plants treated with different concentrations of SNP.
Keywords: sodium nitroprusside, nitrosative stress, oxidative stress, antioxidants, Medicago truncatula, hydrogen peroxide, nitric oxide, nitrate reductase
Introduction
Over the past years the free radical nitric oxide (NO) has emerged as a signal molecule in many important physiological processes in higher plants in a similar fashion to reactive oxygen species (ROS).1 It is also apparent that NO, along with ROS, is involved in plants’ responses to a multitude of environmental stimuli such as salinity, drought, high light intensity and mechanical wounding.2,3 Interestingly, the existence of a cross-talk between ROS and NO is well documented and has been recently reviewed.4 Several excellent reviews exist which provide latest insights into this multifaceted molecule (see refs. 5 and 6). NO is biologically active at a concentration of 1 nmol/l and participates in signaling cascades that drive plant growth and developmental processes.7 Current knowledge of the role of NO in plants is still rather limited by the relative lack of mutants with altered NO production as well as molecules that can sense and transduce NO signals.8
NO donors are compounds that produce NO when applied to biological systems and are able to either mimic an endogenous NO-related response or substitute for an endogenous NO deficiency.9 The most commonly used one is sodium nitroprusside (SNP), an NO+ donor.10 According to Zandonadi et al.,11 the reason SNP is so widely used is due to its relatively low cost and well-documented application as NO donor, while Floryszak-Wieczorek et al.9 state that SNP gives a continuous, long-lasting NO production compared with other NO donors which is often desirable.
The term reactive nitrogen species (RNS) has been formulated to designate NO and the NO-derived molecules such as nitrogen dioxide (·NO2), peroxynitrite (ONOO−), S-nitrosothiols (RSNOs) and S–nitrosoglutathione (GSNO).12 Nitrosative stress is induced by pathophysiological levels of NO and S-nitrosothiols, resulting from the nitrosylation of critical protein cysteine (Cys) thiols (S-nitrosylation) and metal co-factors.13 Although NO is characterized by its inherent toxic nature and is known to potentially be damaging to cells depending on its concentration and on the situation,14 the NO-triggered defense responses are now widely recognized.15 NO can also have a direct, protective effect against abiotic stress factors, as it alleviates the deleterious effects of ROS in establishing stress tolerance responses,15 partly by increasing the activity of antioxidant enzymes.16
Nitrate reductase (NR) is the first enzyme in the nitrate assimilation pathway, reducing nitrate into nitrite.17 Cytosolic NR is also rapidly emerging as one of the main sources of NO in plants under aerobic conditions.18 Plants have several mechanisms to regulate the activity, level and location of NR in response to a range of environmental conditions and chemicals.19 The signals that alter NR activity are tightly regulated at the transcriptional and post-translational levels.20,21 In higher plants, NR is rapidly inactivated/activated by phosphorylation/dephosphorylation, respectively, in response to different environmental stimuli and treatments.22
Early studies on nitrate signaling demonstrated that nitrate induces the de novo synthesis of NR.23 Subsequent work also demonstrated that nitrate induces other genes in the nitrate assimilation pathway, namely nitrate transporters (NRTs).24 The uptake of nitrate (NO3−) by plant cells relies on transport systems, usually membrane carriers belonging to either NRT1 or NRT2 transporter families (see ref. 25) or a channel into the vacuole of which the expression has been shown to be enhanced by nitrate.26 In addition to the nitrate uptake system, plants have an inducible nitrate efflux system, requiring both RNA and protein synthesis; however, it has a much slower turnover rate than the uptake system.27 Considering that the physiological concentration of nitrite in cells can be very low (μM range),28, a nitrite transporter might also be of importance in nitrite uptake by the inner envelope of higher plant chloroplasts, in order to prevent the accumulation of toxic nitrite in the cytosol.29
The present study attempts to elucidate the importance of different concentrations of SNP and the NO induced in cellular integrity and ROS/RNS interplay, as well as in NR expression and regulatory activity by investigating the dose-, developmental stage- and time-dependent effect of SNP application in NR activity. It also examines the role of SNP and the NO produced as a signal regulating ΝR gene expression and triggering the mobilization of other genes implicated in N metabolism and transport (i.e., nitrate/nitrite transporters), thus providing novel insights into the complex regulation of NO metabolism and the cross-talk between ROS and RNS in M. truncatula plants.
Results
Physiological characterization of SNP-treated M. truncatula plants
Vacuum infiltration of M. truncatula plants with different concentrations of SNP and subsequent macroscopic observation five days after SNP application revealed no phenotypic differences in 40-d-old plants, whereas 65-d-old plants treated with 2.5 mM SNP showed increased damage levels indicated by wilted, chlorotic leaves in comparison with control and 100 μΜ SNP-infiltrated samples (Fig. 1).

Figure 1. Phenotypic response of Medicago truncatula plants five days after application of varying SNP concentrations by vacuum infiltration. Senescing (65d) plants treated with 2.5 mM SNP show increased damage levels indicated by wilted, chlorotic leaves.
Effect of SNP in photosynthetic pigment content
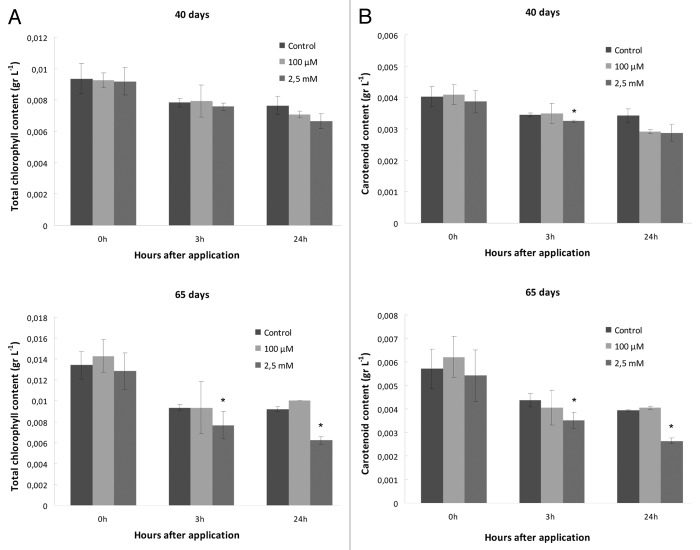
Similar trends were observed in carotenoid and total chlorophyll content: 2.5 mM SNP application resulted in significant reduction in both photosynthetic pigment contents of senescing plants, with a more dramatic effect after 24 h of SNP application (Fig. 2). Carotenoid and total chlorophyll content was also reduced in 40-d-old plants, although the differences were not statistically significant.
Figure 2. Effect of SNP application on photosynthetic pigment content (A) Τotal chlorophyll (α + β) content, (B) Carotenoid content (where upper rows represent measurements made with 40 d plants, while lower rows indicate measurements made with 65 d plants). Asterisks denote statistically different values according to the Tukey pairwise comparison test (p < 0.05). Values are means ± SE (n = 3).
Cellular damage levels and ROS measurements
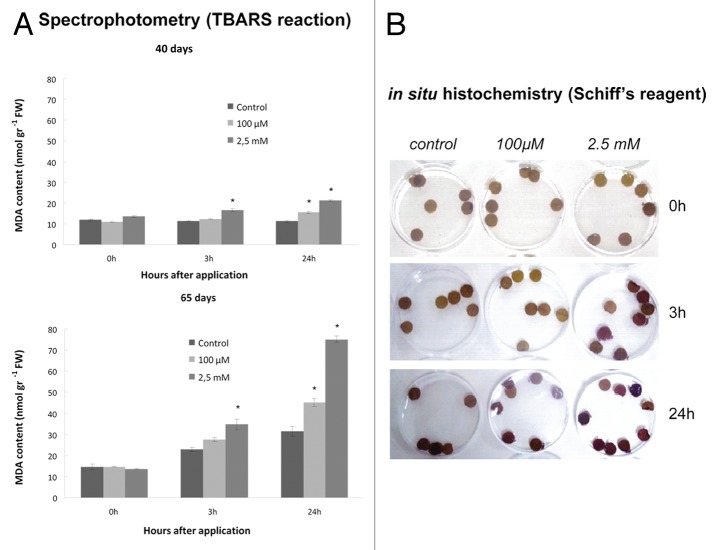
A temporal examination of cellular damage levels was performed by means of spectrophotometric determination of lipid peroxidation, a commonly used marker of oxidative damage. Increasing SNP concentrations resulted in increasing membrane damage, reaching maximum levels at 24 h after application in both mature and senescing plants. The latter demonstrated much higher lipid peroxidation levels in response to SNP application, while control (0 d plants) showed higher constitutive lipid peroxidation (Fig. 3A). In situ histochemical localization of lipid peroxidation with the use of Schiff’s reagent demonstrated similar overall patterns (Fig. 3B).
Figure 3. Effect of SNP application on cellular damage as indicated by lipid peroxidation (MDA content). (A) quantitative analysis by spectrophotometry, where upper row represents measurements made with 40 d plants, while lower row indicates measurements made with 65 d plants. (B) qualitative analysis by in situ histochemistry using Schiff’s reagent. Figure presents 40 d old plants; similar trend was observed with 65 d plants (data not shown). Asterisks denote statistically different values according to the Tukey pairwise comparison test (p < 0.05). Values are means ± SE (n = 3).
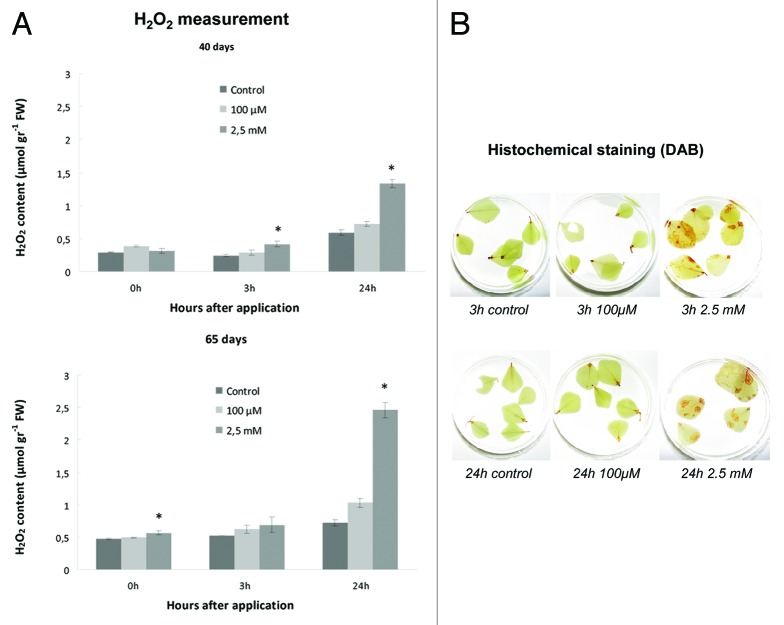
Spectrophotometric determination of cellular reactive oxygen (H2O2) species content in SNP-treated plants showed a gradual increase in H2O2 in both mature and senescing plants, reaching maximal levels at 24 h after 2.5 mM SNP application (Fig. 4A). Overall, higher concentrations of SNP resulted in increased H2O2 concentration, further increasing in older tissues. Hydrogen peroxide content was also increased after 3 h, although the increase was not statistically significant. In situ histochemical localization of hydrogen peroxide with the use of DAB demonstrated similar overall patterns (Fig. 4B).
Figure 4. Effect of SNP application on hydrogen peroxide levels. (A) quantitative analysis by spectrophotometry, where upper row represents measurements made with 40 d plants, while lower row indicates measurements made with 65 d plants. (B) qualitative analysis by in situ histochemistry using DAB reagent. Figure presents 65 d old plants; similar trend was observed with 40 d plants (data not shown). Asterisks denote statistically different values according to the Tukey pairwise comparison test (p < 0.05). Values are means ± SE (n = 3).
Effects of NO content on NR enzymatic activity
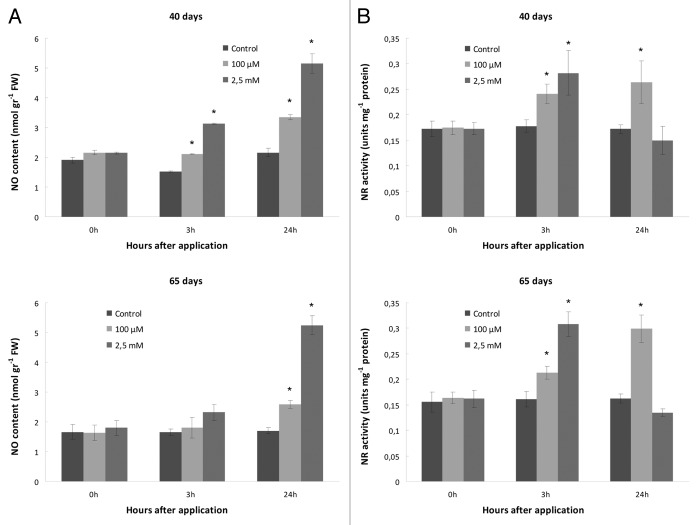
Nitrite-derived nitric oxide content in SNP-treated leaves was measured using the Griess reagent (see “Materials and Methods” section). Similar overall trends were observed in both developmental stages of M. truncatula plants, demonstrating increasing nitrite-derived NO content with increasing SNP concentrations applied. Maximum NO contents were recorded in both mature and senescing plants infiltrated with 2.5 mM SNP after 24 h (Fig. 5A).
Figure 5. Effect of SNP application on nitrite-derived nitric oxide content and NR enzymatic activity. (A) NO measurements made with mature (40 d) and senescing (65 d) plants after lower (100 μΜ) and higher (2.5 mM) SNP application. (B) Measurements of NR enzymatic activity in mature (40 d) and senescing (65 d) plants after lower (100 μΜ) and higher (2.5 mM) SNP application (Upper and lower rows represent measurements made with 40 d and 65 d plants respectively). Asterisks denote statistically different values according to the Tukey pairwise comparison test (p < 0.05). Values are means ± SE (n = 3).
To further elucidate the possible role and mechanism by which SNP regulates NR activity in M. truncatula plants, NR enzymatic activity was measured in mature and senescing plants after low (100 μΜ) and high (2.5 mM) SNP application. At the lower SNP application (100 μΜ), low SNP concentration caused a short-term (3 h) activation of NR activity further increased after the long-term effect of SNP application (24 h) in mature and to a further extent in senescing plants (Fig. 5B). In addition, the short-term application of the higher SNP concentration (3h, 2.5 mM SNP), also caused an increase in NR activity (Fig. 5B).
Although the stimulating effect of NO in NR activity was observed at the lower SNP concentration application, higher concentrations partly inhibited NR activity, as indicated by the lower levels of NR activity after 24 h of 2.5 mM SNP application. This effect was more obvious in senescing plants (65 d) treated with the higher SNP concentration (Fig. 5B).
Expression analysis of genes implicated in NO metabolism after SNP application
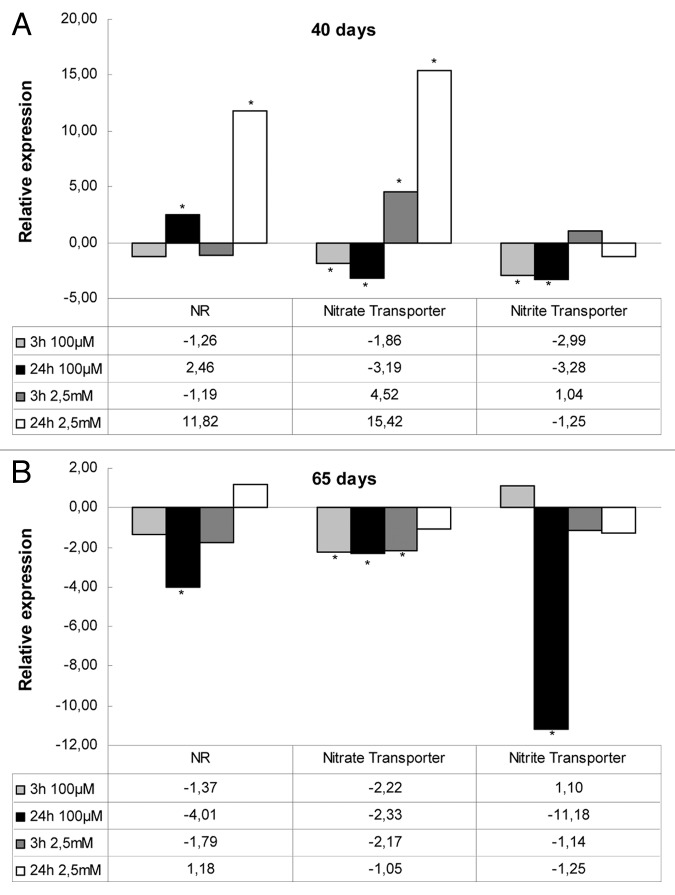
The expression patterns of three key genes (nitrate and nitrite transporters, nitrate reductase) involved in the NO metabolism was assayed by a quantitative real-time RT-PCR approach, revealing differential regulation in leaves of both mature and senescing plants for the genes examined (Fig. 6). Analysis of the expression of genes encoding proteins associated with NO biosynthesis (NR) or transporters (nitrate/nitrite transporter) was performed following 3 h and 24 h of 100 μΜ and 2.5 mM SNP application in mature and senescing plants (Fig. 6).
Figure 6. NO effect on gene expression profiles of NO-responsive genes (NR and nitrate/nitrite transporters). Gene expression analysis was determined by qRT-PCR in leaves of Medicago truncatula Jemalong A17 plants vacuum-infiltrated with 100 μΜ and 2.5 mM SNP at 3/24 h in mature (40 d (A) and senescing (65 d (B) plants. Asterisks denote statistically different values according to pairwise fixed reallocation randomization test (p < 0.05) (n = 3).
A similar trend of suppressed expression of NR as well as nitrite and nitrate transporter genes was observed in senescing (65 d) plants. Contrarily, a significant induction was observed in NR and nitrate transporter gene expression during the long-term effect (24 h) of the higher SNP concentration (2.5 mM) in 40-d-old plants (Fig. 6). Thus, there was a significant induction in transcript abundance of NR and to a lesser extent in nitrate transporter in NO-accumulating cells. By contrast, an overall suppression of the nitrite transporter gene was observed in mature and to a further extent in senescing plants (Fig. 6).
Discussion
Nitric oxide is a key player in several biological cellular processes, acting either as a signaling or as a toxic molecule in plants.1,30 It is also acknowledged as a major component in the establishment of plant symbiosis with nitrogen-fixing bacteria.31 The present study was performed in an attempt to elucidate the effect of application of two different concentrations of SNP, a widely used NO donor, in M. truncatula plants in relation with RNS/ROS signals and cellular NO metabolism. Among the different types of NO donors, SNP was used because of its high efficiency to release NO in plant cells,32 as well as continuous, long-lasting NO production.9 Such a focus was also largely given due to the importance and frequency of SNP use in Plant NO studies (> 600 research articles published during the past 10 years; Source: http://www.scopus.com).
In order to characterize the effect of SNP and NO produced in M. truncatula plants in a concentration-dependent manner, two different SNP concentrations (100 μΜ and 2.5 mΜ) were chosen, based on previous studies.33,34 Moreover, the important role of NO during senescence (see Leshem et al.35) prompted us to investigate the effect of SNP application in Medicago truncatula plants of different developmental stages (40-d-old mature plants and 65-d-old senescing plants), further supported by the investigation of short-term (3 h) and long-term (24 h) SNP application effect.
Carotenoids are low molecular weight compounds that act as non-enzymatic antioxidants primarily quenching singlet oxygen,36 while chlorophyll degradation is a known stress-related marker.37 Phenotypic plant observations in 2.5 mM SNP-treated plants revealed increased visible damage levels and a decline in Car and Chl contents in senescing plants (Fig. 2), while treatment with 100 μM SNP did not affect the plant’s growth and viability (Fig. 1) and the cellular status of the cell. This result is in accordance with Sung and Hong,38 indicating the effect of different NO concentrations in plant development and a similar decline in Car and Chl,37,39 further suggesting that Chl catabolism is highly regulated during development and senescence.39,40 One possible explanation for the observed phenotypic and physiological behavior of the SNP-treated M. truncatula plants may be due to the major induction of ROS / RNS production, according to the increased sensitivity of various plants to oxidative damage under severe stress conditions.30,41 Indeed, higher NO concentration (nitrosative stress) resulted in H2O2 accumulation, accompanied by increased cellular damage levels (accumulation of MDA) in mature plants, further induced in senescing plants (Figs. 3 and 4), thereby indicating its toxic role.42 Contrarily, plants treated with the lower NO concentration resulted in lower H2O2 and MDA content in leaves (Figs. 3 and 4), indicating the protective role of NO by reacting with ROS and consequently inhibiting the detrimental effects of membrane lipid peroxidation.43
Interestingly, NO content is markedly increased in leaves of 2.5 mM SNP-treated mature plants compared with control samples after short-term (3 h) application, unlike H2O2 content which remains at similar levels and is only increased after long-term 2.5 mM SNP application, therefore suggesting that NO induction in leaves appears to precede H2O2 induction (Figs. 4A and 5A). NO has been shown to be involved in the signaling pathway upstream of H2O2 synthesis, thus justifying the observed timing of induction of the two signaling molecules.44
Although NO synthesis in plants is still a matter of debate,45 NO3− may be reduced to NO in M. truncatula plants in a two-step mechanism involving NR and other systems implicated in electron transport chain.46 In an attempt to further clarify the actual mechanism of NR regulation by NO in SNP-treated plants, it was observed that NO accumulation in M. truncatula plants (24 h, 2.5 mM SNP) resulted in a decline in NR activity (Fig. 5B), similar to previous research findings.46,47 This is likely to be the result of NO toxicity.42 It is also possible that higher concentrations of NO result in a negative feed-back regulatory mechanism, thus inhibiting this NO biosynthetic enzyme. However, the significant induction rather than suppression of NR transcripts observed in mature plants treated with 2.5 mM SNP (Fig. 6), suggests that NR activity is regulated by SNP at the post-translational rather than the transcriptional level. Such possibility for post-translational modification of NR activity is further supported by the fact that low concentration of SNP (100 μM) caused a transient increase in NR activity, reaching maximal activation after 24 h (Fig. 5B) in both developmental stages, in contrast with transcript levels that were not induced but were actually suppressed in senescing plants and in transient response mature plants (Fig. 6). Several reports demonstrate the post-translational regulation of NR activity,22,49,50while SNP treatment has been shown to result in massive carbonylation, nitration and S-nitrosylation of citrus proteins.41
The lack of direct correlation between NR gene expression and enzymatic activity, revealing the complexity of the NR regulatory mechanism and the differential gene expression regulation according to the signal,51 could also be partially attributed to the presence of two NR encoding genes in the M. truncatula genome (NR1: TC137636; Mtr.10604.1.S1_at, and NR2: TC130773; Mtr.42446.1.S1_at; see ref. 46), similarly to Arabidopsis thaliana.52 It could be speculated that one or both of the genes is differentially regulated at different environmental/exogenous conditions (i.e., different SNP concentrations) and time points, or that both genes are regulated simultaneously by different metabolic pathways.
Following our observations dealing with the interaction of NO with oxidative events indicative of the plant’s developmental status at both age and biochemical/physiological level, further experiments were performed in order to show whether NO SNP exhibits developmental stage-specific regulation of nitrate/nitrite uptake. In an attempt to examine whether the influx rather than the total pool of nitrate/nitrite is critical for the regulation of NR expression, nitrate and nitrite transporter gene expression analysis was performed. A gene encoding a putative nitrate transporter (UP|Q852P5 [Q852P5], Nitrate transporter, partial [28%]) was chosen based on previous microarray analyses where expression of this transporter was found to be regulated in salt-stressed M. truncatula plants (Filippou and Fotopoulos; data not shown), similar to observations in Arabidopsis.53 Similarly, transcript levels of the nitrate transporter gene were suppressed after 3 h and 24 h of low or high SNP application in senescing plants, showing that NO assimilation may exert a repressive effect on nitrate influx.54 Mature plants, however, demonstrated a different mode of regulation for the higher SNP concentration where the nitrate transporter transcript levels strongly increased in NO-accumulating cells (Fig. 6). It is possible however that the increased sensitivity of senescing plants to nitrosative stress leads to a negative feedback regulation of this nitrate transporter via products of NO3− assimilation (i.e., NO), while it is constitutively expressed in mature plants at higher NO concentrations.55
Focusing on mature plants displaying highest NO concentration (24 h, 2.5 mM SNP), we tried to correlate NR activity with nitrate transporter expression levels, since the nitrate influx rather than the total pool is critical for the regulation of NR expression and activity.56 Furthermore, the regulatory function of NO on NR depending on levels of nitrate supply is also established.34 Transcript abundance of the nitrate transporter gene in NO-accumulating cells (24 h, 2.5 mM SNP; Figure 6) coupled with a significant decline in NR activity (Fig. 5). Similarly, in an NR-deficient line, major induction of the NRT2 mRNA transporter was observed in mature plants leading to NO3− accumulation.17,55 However, NO accumulation was also shown to inhibit NR leaf activity in senescing M. truncatula plants (Fig. 5), although gene expression of the nitrate transporter strongly diminished in leaves after SNP application (Fig. 6), probably due to senescence-induced deregulation of the plant metabolic mechanisms.57
Considering the other different NO concentrations observed (3 h 2.5 mM, 3 h and 24 h 100 μΜ SNP), a rather complex regulation of NR expression/ activity and nitrate transporter expression is observed, implying that feedback regulation may be occurring in both developmental stages of M. truncatula plants, not only by modifying the expression of NR but also possibly via transcriptional or post-translational modification of the nitrate transporter, as a result of the general regulatory role of NO on components of multiple plant metabolic pathways.58
Expression analysis was further elaborated by focusing on the impact of SNP treatment on expression of nitrite transporters, as the effect of nitrite content on NR activity is well established.51 A putative nitrite transporter (T10255 nitrite transport protein, partial [29%]), showing differential regulation in M. truncatula salinity-stressed plants in a similar fashion to the nitrate transporter following a microarray analysis approach (Filippou and Fotopoulos; data not shown), was further analyzed in SNP-treated plants. A general trend of suppressed expression was observed in both developmental stages (Fig. 6), while NO accumulation resulted in a parallel suppression of nitrite transporter transcripts and NR activity in mature plants (Figs. 5 and 6), suggesting a negative feed-back regulatory mechanism for NO, a by-product of nitrite reduction, in excess amounts.59 Plants need to adapt a defense mechanism since NO accumulation might be toxic for the cell leading to cell death,42 thus supporting the inactivation of the nitrite transporter and NR activity observed (Fig. 5 and 6). Furthermore, lower NO concentration (24 h, 100 μΜ SNP) inside the cell appears to be critical, acting as an inducer of NR expression and activity to form more NO. As a result, nitrite transporter expression is suppressed, since there is no necessity to activate the transporter for nitrite influx and further NO generation.
Overall, the results presented in the current study support the notion that SNP and the NO produced can act as either a protective or cytotoxic molecule in a concentration-dependent manner. Senescing M. truncatula plants demonstrated greater sensitivity to NO-induced oxidative and nitrosative damage, as well as repression of both NO3− uptake and assimilatory systems by NO2− and NR activity, suggesting a developmental stage-dependent suppression in the plant's capacity to cope with free oxygen and nitrogen radicals. In addition, our findings underscore an important cross-talk between H2O2 and NO signaling pathways in response to nitrosative stress. However, it should be noted that certain responses could also be potentially attributed, at least partially, to the effect of side products produced by SNP such as cyanide,59,60 although other studies reported no CN- effect using SNP to trigger NO-induced processes.16 Furthermore, the genomic complexity of NR and other NO-responsive genes regulated by SNP and the different expression patterns observed pose the necessity for full transcriptomic analyses that would provide more conclusive evidence. In closing, fully understanding and deciphering the mechanism by which a plant “recognizes” and responds to NO could prove to be remarkably valuable toward the elucidation of the plant’s global stress response and the engineering of tolerant crops.
Material and Methods
Plant material and growth conditions
This study was conducted using M. truncatula genotype Jemalong A17. After scarification, seeds were sown in sterile perlite:sand (3:1) pots and placed at 4 °C for 4 d for stratification. Plants were grown in a growth chamber at 22/16 °C day/night temperatures, at 60–70% RH, with a photosynthetic photon flux density of 100 μmol m2 s−1 and a 16/8 h photoperiod. Experiments were performed in triplicate using pooled samples (each “replicate” sample comprising tissues from a minimum of three independent plants).
SNP application treatment
Mature (40 d) and senescing (65 d) plants were vacuum-infiltrated with low (100 μΜ) and high (2.5 mM) concentrations of SNP and samples were obtained at 3 h (transient effect) and 24 h (long-term effect) as previously described.61 Vacuum-infiltration was chosen as it represents the optimal method for SNP application.62 Control samples were vacuum-infiltrated with dH2O. Supplementary Figure 1 represents a detailed outline of the experimental design followed. Leaf samples were flash frozen in liquid nitrogen and stored at −80°C for subsequent analyses.
Carotenoid and chlorophyll content
Leaf pigments were extracted from 9 mm-diameter leaf discs in dimethyl sulfoxide as described by Richardson et al.63 Carotenoid and chlorophyll concentrations were determined using the equations described by Sims and Gamon.64
Lipid peroxidation assay
The extent of lipid peroxidation was determined from measurement of malondialdehyde (MDA) content resulting from the thiobarbituric acid (TBA) reaction as described by Hodges et al.,65 using an extinction coefficient of 155 mM−1 cm−1.
Hydrogen peroxide and nitric oxide quantification
Hydrogen peroxide was quantified using the KI method, as described by Velikova et al.66 NO content was measured using the Griess reagent in homogenates prepared with Na-acetate buffer (pH 3.6) as described by Zhou et al.67 Nitrite-derived NO content was calculated by comparison to a standard curve of NaNO2.
Nitrate reductase assay
The assay was performed essentially as described,68 with some modifications. The buffer used for preparation of crude extracts contained 100 mM potassium phosphate (pH 7.5), 5 mM (CH3COO)2Mg, 10% (v/v) glycerol, 10% (w/v) polyvinylpyrollidone, 0,1% (v/v) Triton ×-100, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine (prepared fresh) and 1 mM 6-aminocaproic acid. Leaf tissue was extracted in the appropriate buffer using a mortar and pestle and the mixture was thoroughly homogenized. Cell extract was centrifuged at 14,000 × g for 15 min and the clear supernatant was used immediately for measurement of enzyme activity. NR enzyme activity was expressed as specific enzyme activity (units/mg protein).
Histochemical detection of reactive oxygen species and lipid peroxidation
The detection of H2O2 in tissues was performed according to Thordal-Christensen et al.69 with the following modifications. Detached leaves were incubated in 1 mg/ml 3,3′-diaminobenzidine (DAB)-HCl, pH 3.8 in the dark at room temperature for 8–10 h, then chlorophyll was removed by boiling in ethanol (96%, v/v) for 10 min. The assay was based on the instant polymerization of DAB (to form a reddish-brown complex which is stable in most solvents), as soon as it comes into contact with H2O2 in the presence of peroxidases. To determine the specificity of DAB staining, leaves were also stained in the presence of 200 units ml/1 catalase (bovine liver, Sigma-Aldrich) in a control experiment.
Leaf disks were stained with Schiff’s reagent for stress-induced lipid peroxidation.69,70 Leaf disks were stained with Schiff’s reagent (Sigma) for 30 min, and rinsed with 0.5% (w/v) K2S2O5 in 0.05 M HCl, which detects aldehydes originated from lipid peroxides.
RNA isolation, cDNA synthesis and real-time RT-PCR assay
Total RNA was prepared from leaves with the Qiagen RNeasy® Plant Mini Kit (Qiagen) followed by DNase digestion (RNase-free DNase Set; Qiagen). RNA integrity was analyzed spectrophotometrically and by gel electrophoresis. For real-time RT-PCR analyses, 1 µg of total RNA was converted into cDNA using Primescript 1st Strand Synthesis kit according to the manufacturer’s protocol. Subsequently, real-time PCR was performed with Biorad IQ5 (Biorad). The reaction mix contained 4 μl cDNA in RT buffer (diluted 1:5), 0.75 μM of each primer (Table S1) and 1× master mix (SYBR Green Super Mix). Reactions were performed in triplicate and the thermocycler conditions were: 95 °C for 5 min, then 40 cycles of 95 °C for 30 sec, annealing temperature for 30 sec, 72 °C for 30 sec, 80 °C for 2 sec, plate read at 78 °C, followed by 72 °C for 10 min. The annealing temperature of the tested primers is 60 °C and 53 °C for the reference gene (Table S1). Relative quantification of gene expression and statistical analysis of all qRT-PCR data (pairwise fixed reallocation randomization test) were performed using the REST software according to Pfaffl et al.71 Actin 11 gene was used as a housekeeping reference gene.72
Statistical analyses
Statistical analyses of all measurements (excluding qRT-PCR data) were performed using SPSS v.11 (SPSS Inc.,). Biochemical and physiological damage measurements were subjected to ANOVA, and then significant differences between individual means determined using Tukey’s pairwise comparison test at the 5% confidence level.
Supplementary Material
Acknowledgments
We would like to thank Mr. F. Melnikov for excellent technical assistance. This work was supported by Cyprus University of Technology Internal Grant EX032 to VF, who also acknowledges grants-in-aid from COST Action FA0605.
Glossary
Abbreviations:
- NO
nitric oxide
- SNP
sodium nitroprusside
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- NR
nitrate reductase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplementary material may be found here: http://www.landesbioscience.com/journals/psb/article/25479/
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/25479
References
- 1.1. Neill SJ, Desikan R, Hancock J. Nitric oxide signalling in plants. New Phytol. 2003;159:11–35. doi: 10.1046/j.1469-8137.2003.00804.x. [DOI] [PubMed] [Google Scholar]
- 2.Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, et al. Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol. 2008;49:1711–1722. doi: 10.1093/pcp/pcn144. [DOI] [PubMed] [Google Scholar]
- 3.3. Filippou P, Antoniou C, Fotopoulos V. Effect of drought and rewatering on the antioxidant response of Medicago truncatula plants. Plant Signal Behav. 2011;6:270–7. doi: 10.4161/psb.6.2.14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molassiotis A, Fotopoulos V. Oxidative and nitrosative signaling in plants: two branches in the same tree? Plant Signal Behav. 2011;6:210–4. doi: 10.4161/psb.6.2.14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.5. Moreau M, Lindermayr C, Durner J, Klessig DF. NO synthesis and signalling in plants - where do we stand? Physiol Plant. 2010;138:372–83. doi: 10.1111/j.1399-3054.2009.01308.x. [DOI] [PubMed] [Google Scholar]
- 6.Baudouin E. The language of nitric oxide signalling. Plant Biol (Stuttg) 2011;13:233–42. doi: 10.1111/j.1438-8677.2010.00403.x. [DOI] [PubMed] [Google Scholar]
- 7.Beligni MV, Lamattina L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta. 2000;210:215–21. doi: 10.1007/PL00008128. [DOI] [PubMed] [Google Scholar]
- 8.Mur LAJ, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, et al. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013;5 doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floryszak-Wieczorek J, Milczarek G, Arasimowicz M, Ciszewski A. Do nitric oxide donors mimic endogenous NO-related response in plants? Planta. 2006;224:1363–72. doi: 10.1007/s00425-006-0321-1. [DOI] [PubMed] [Google Scholar]
- 10.Ederli L, Reale L, Madeo L, Ferranti F, Gehring C, Fornaciari M, et al. NO release by nitric oxide donors in vitro and in planta. Plant Physiol Biochem. 2009;47:42–8. doi: 10.1016/j.plaphy.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Zandonadi DB, Santos MP, Dobbss LB, Olivares FL, Canellas LP, Binzel ML, et al. Nitric oxide mediates humic acids-induced root development and plasma membrane H+-ATPase activation. Planta. 2010;231:1025–36. doi: 10.1007/s00425-010-1106-0. [DOI] [PubMed] [Google Scholar]
- 12.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine, (4th ed.), Clarendon Press, Oxford, 2007. [Google Scholar]
- 13.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–26. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beligni Ma, Lamattina L. Is nitric oxide toxic or protective? Trends Plant Sci. 1999;4:299–300. doi: 10.1016/S1360-1385(99)01451-X. [DOI] [PubMed] [Google Scholar]
- 15.Qiao W, Fan LM. Nitric oxide signaling in plant responses to abiotic stresses. J Integr Plant Biol. 2008;50:1238–46. doi: 10.1111/j.1744-7909.2008.00759.x. [DOI] [PubMed] [Google Scholar]
- 16.Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, et al. Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot. 2008;59:165–76. doi: 10.1093/jxb/erm293. [DOI] [PubMed] [Google Scholar]
- 17.Fan X, Gordon-Weeks R, Shen Q, Miller AJ. Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J Exp Bot. 2006;57:1333–40. doi: 10.1093/jxb/erj110. [DOI] [PubMed] [Google Scholar]
- 18.Mur LAJ, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, et al. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013;5 doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford NM. Mechanisms for nitric oxide synthesis in plants. J Exp Bot. 2006;57:471–8. doi: 10.1093/jxb/erj050. [DOI] [PubMed] [Google Scholar]
- 20.Campbell WH. Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:277–303. doi: 10.1146/annurev.arplant.50.1.277. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki H, Sakihama Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 2000;468:89–92. doi: 10.1016/S0014-5793(00)01203-5. [DOI] [PubMed] [Google Scholar]
- 22.Lillo C, Lea US, Leydecker MT, Meyer C. Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in constitutive activation of the enzyme in vivo and nitrite accumulation. Plant J. 2003;35:566–73. doi: 10.1046/j.1365-313X.2003.01828.x. [DOI] [PubMed] [Google Scholar]
- 23.Remmler JL, Campbell WH. Regulation of corn leaf nitrate reductase II. synthesis and turnover of the enzyme's activity and protein. Plant Physiol. 1986;80:442–7. doi: 10.1104/pp.80.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stitt M. Nitrate regulation of metabolism and growth. Curr Opin Plant Biol. 1999;2:178–86. doi: 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- 25.Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. Nitrate transport and signalling. J Exp Bot. 2007;58:2297–306. doi: 10.1093/jxb/erm066. [DOI] [PubMed] [Google Scholar]
- 26.De Angeli A, Monachello D, Ephritikhine G, Frachisse JM, Thomine S, Gambale F, et al. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature. 2006;442:939–42. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- 27.Aslam M, Travis RL, Rains DW. Evidence for substrate induction of a nitrate efflux system in barley roots. Plant Physiol. 1996;112:1167–75. doi: 10.1104/pp.112.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot. 2002;53:103–10. doi: 10.1093/jexbot/53.366.103. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura M, Georgescu MN, Takahashi M. A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 2007;48:1022–35. doi: 10.1093/pcp/pcm073. [DOI] [PubMed] [Google Scholar]
- 30.Lamotte O, Courtois C, Barnavon L, Pugin A, Wendehenne D. Nitric oxide in plants: the biosynthesis and cell signalling properties of a fascinating molecule. Planta. 2005;221:1–4. doi: 10.1007/s00425-005-1494-8. [DOI] [PubMed] [Google Scholar]
- 31.del Giudice J, Cam Y, Damiani I, Fung-Chat F, Meilhoc E, Bruand C, et al. Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol. 2011;191:405–17. doi: 10.1111/j.1469-8137.2011.03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar P, Tewari RK, Sharma PN. Sodium nitroprusside-mediated alleviation of iron deficiency and modulation of antioxidant responses in maize plants. AoB Plants. 2010 doi: 10.1093/aobpla/plq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan X, Su X, You Y, Qu H, Li Y, Jiang Y. Effect of nitric oxide on pericarp browning of harvested longan fruit in relation to phenolic metabolism. Food Chem. 2007;104:571–6. doi: 10.1016/j.foodchem.2006.12.007. [DOI] [Google Scholar]
- 34.Jin CW, Du ST, Zhang YS, Lin XY, Tang CX. Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum) Ann Bot. 2009;104:9–17. doi: 10.1093/aob/mcp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leshem YY, Wills RBH, Veng-Va Ku V. Evidence for the function of the free radical gas, nitric oxide (NO*), as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol Biochem. 1998;36:825–33. doi: 10.1016/S0981-9428(99)80020-5. [DOI] [Google Scholar]
- 36.Collins A. Carotenoids and genomic stability. Mutat Res. 2001;475:1–28. doi: 10.1016/S0027-5107(01)00071-9. [DOI] [PubMed] [Google Scholar]
- 37.Singh S, Khan NA, Nazar R, Anjum NA. Photosynthetic traits and activities of antioxidant enzymes in blackgram (Vigna mungo L. Hepper) under cadmium stress. Am J Plant Physiol. 2008;3:25–32. doi: 10.3923/ajpp.2008.25.32. [DOI] [Google Scholar]
- 38.Sung CH, Hong JK. Sodium nitroprusside mediates seedling development and attenuation of oxidative stresses in Chinese cabbage. Plant Biotechnol Rep. 4. 2010:243–251. [Google Scholar]
- 39.Qian HF, Chen W, Li JJ, Wang J, Zhou Z, Liu WP, et al. The effect of exogenous nitric oxide on alleviating herbicide damage in Chlorella vulgaris. Aquat Toxicol. 2009;92:250–7. doi: 10.1016/j.aquatox.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell. 2007;19:1649–64. doi: 10.1105/tpc.106.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanou G, Filippou P, Belghazi M, Job D, Diamantidis G, Fotopoulos V, et al. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 2012;72:585–99. doi: 10.1111/j.1365-313X.2012.05100.x. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro AD. Nitric oxide signaling in plants. Vitam Horm. 2005;72:339–98. doi: 10.1016/S0083-6729(05)72010-0. [DOI] [PubMed] [Google Scholar]
- 43.Lamotte O, Gould K, Lecourieux D, Sequeira-Legrand A, Lebrun-Garcia A, Durner J, et al. Analysis of nitric oxide signaling functions in tobacco cells challenged by the elicitor cryptogein. Plant Physiol. 2004;135:516–29. doi: 10.1104/pp.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wendehenne D, Durner J, Klessig DF. Nitric oxide: a new player in plant signalling and defence responses. Curr Opin Plant Biol. 2004;7:449–55. doi: 10.1016/j.pbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. On the origins of nitric oxide. Trends Plant Sci. 2011;16:160–8. doi: 10.1016/j.tplants.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Horchani F, Prévot M, Boscari A, Evangelisti E, Meilhoc E, Bruand C, et al. Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 2011;155:1023–36. doi: 10.1104/pp.110.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foyer CH, Valadier MH, Migge A, Becker TW. Drought-induced effects on nitrate reductase activity and mRNA and on the coordination of nitrogen and carbon metabolism in maize leaves. Plant Physiol. 1998;117:283–92. doi: 10.1104/pp.117.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosales EP, Iannone MF, Groppa MD, Benavides MP. Nitric oxide inhibits nitrate reductase activity in wheat leaves. Plant Physiol Biochem. 2011;49:124–30. doi: 10.1016/j.plaphy.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Graziano M, Lamattina L. Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J. 2007;52:949–60. doi: 10.1111/j.1365-313X.2007.03283.x. [DOI] [PubMed] [Google Scholar]
- 50.Du S, Zhang Y, Lin XY, Wang Y, Tang CX. Regulation of nitrate reductase by nitric oxide in Chinese cabbage pakchoi (Brassica chinensis L.) Plant Cell Environ. 2008;31:195–204. doi: 10.1111/j.1365-3040.2007.01750.x. [DOI] [PubMed] [Google Scholar]
- 51.Buchanan BB, Gruissem W, Jones RL. Biochemistry and Molecular Biology of Plants, American Society of Plant Physiologists, Rockville, MD, USA, 2000. [Google Scholar]
- 52.Cheng CL, Acedo GN, Dewdney J, Goodman HM, Conkling MA. Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 1991;96:275–9. doi: 10.1104/pp.96.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–41. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraisier V, Gojon A, Tillard P, Daniel-Vedele F. Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J. 2000;23:489–96. doi: 10.1046/j.1365-313x.2000.00813.x. [DOI] [PubMed] [Google Scholar]
- 55.Forde BG. Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta. 2000;1465:219–35. doi: 10.1016/S0005-2736(00)00140-1. [DOI] [PubMed] [Google Scholar]
- 56.Geiger M, Walch-Liu P, Engels C, Harnecker J, Schulze ED, Ludewig F, et al. Enhanced carbon dioxide leads to a modified diurnal rhythm of nitrate reductase activity in older plants, and a large stimulation of nitrate reductase activity and higher levels of amino acids in young tobacco plants. Plant Cell Environ. 1998;21:253–68. doi: 10.1046/j.1365-3040.1998.00277.x. [DOI] [Google Scholar]
- 57.Lin JF, Wu SH. Molecular events in senescing Arabidopsis leaves. Plant J. 2004;39:612–28. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- 58.Astier J, Lindermayr C. Nitric oxide-dependent posttranslational modification in plants: an update. Int J Mol Sci. 2012;13:15193–208. doi: 10.3390/ijms131115193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta KJ, Igamberdiev AU, Manjunatha G, Segu S, Moran JF, Neelawarne B, et al. The emerging roles of nitric oxide (NO) in plant mitochondria. Plant Sci. 2011;181:520–6. doi: 10.1016/j.plantsci.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 60.Bethke PC, Libourel IGL, Reinöhl V, Jones RL. Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta. 2006;223:805–12. doi: 10.1007/s00425-005-0116-9. [DOI] [PubMed] [Google Scholar]
- 61.Filippou P, Antoniou C, Fotopoulos V. The nitric oxide donor sodium nitroprusside regulates proline and polyamine biosynthesis in Medicago truncatula plants. Free Radic Biol Med. 2013;56:172–83. doi: 10.1016/j.freeradbiomed.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 62.Filippou P, Antoniou C, Yelamanchili S, Fotopoulos V. NO loading: Efficiency assessment of five commonly used application methods of sodium nitroprusside in Medicago truncatula plants. Plant Physiol Biochem. 2012;60:115–8. doi: 10.1016/j.plaphy.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 63.Richardson AD, Duigan SP, Berlyn GP. An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 2002;153:185–94. doi: 10.1046/j.0028-646X.2001.00289.x. [DOI] [Google Scholar]
- 64.Sims DA, Gamon JA. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens Environ. 2002;81:337–54. doi: 10.1016/S0034-4257(02)00010-X. [DOI] [Google Scholar]
- 65.Hodges DM, DeLong JM, Forney CF, Prange RK. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta. 1999;207:604–11. doi: 10.1007/s004250050524. [DOI] [PubMed] [Google Scholar]
- 66.Velikova V, Yordanov I, Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants protective role of exogenous polyamines. Plant Sci. 2000;151:59–66. doi: 10.1016/S0168-9452(99)00197-1. [DOI] [Google Scholar]
- 67.Zhou B, Guo Z, Xing J, Huang B. Nitric oxide is involved in abscisic acid-induced antioxidant activities in Stylosanthes guianensis. J Exp Bot. 2005;56:3223–8. doi: 10.1093/jxb/eri319. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, He J, Jiang L, Wu H, Xiao Y, Liu Y, et al. Nitric oxide production is associated with response to brown planthopper infestation in rice. J Plant Physiol. 2011;168:739–45. doi: 10.1016/j.jplph.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 69.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J. 1997;11:1187–94. doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- 70.Yamamoto Y, Kobayashi Y, Matsumoto H. Lipid peroxidation is an early symptom triggered by aluminum, but not the primary cause of elongation inhibition in pea roots. Plant Physiol. 2001;125:199–208. doi: 10.1104/pp.125.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mhadhbi H, Fotopoulos V, Mylona PV, Jebara M, Aouani ME, Polidoros AN. Differential regulation of Aox1 gene expression in roots of Medicago truncatula as a genotype-specific component of salt stress tolerance. J Plant Physiol. 2013;170:111–4. doi: 10.1016/j.jplph.2012.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.