Abstract
Rho GTPases are key regulators of cellular protrusion and are involved in many developmental events including axon guidance during nervous system development. Rho GTPase pathways display functional redundancy in developmental events, including axon guidance. Therefore, their roles can often be masked when using simple loss-of-function genetic approaches. As a complement to loss-of-function genetics, we constructed a constitutively activated CDC-42(G12V) expressed in C. elegans neurons. CDC-42(G12V) drove the formation of ectopic lamellipodial and filopodial protrusions in the PDE neurons, which resembled protrusions normally found on migrating growth cones of axons. We then used a candidate gene approach to identify molecules that mediate CDC-42(G12V)-induced ectopic protrusions by determining if loss of function of the genes could suppress CDC-42(G12V). Using this approach, we identified 3 cytoskeletal pathways previously implicated in axon guidance, the Arp2/3 complex, UNC-115/abLIM, and UNC-43/Ena. We also identified the Nck-interacting kinase MIG-15/NIK and p21-activated kinases (PAKs), also implicated in axon guidance. Finally, PI3K signaling was required, specifically the Rictor/mTORC2 branch but not the mTORC1 branch that has been implicated in other aspects of PI3K signaling including stress and aging. Our results indicate that multiple pathways can mediate CDC-42-induced neuronal protrusions that might be relevant to growth cone protrusions during axon pathfinding. Each of these pathways involves Rac GTPases, which might serve to integrate the pathways and coordinate the multiple CDC-42 pathways. These pathways might be relevant to developmental events such as axon pathfinding as well as disease states such as metastatic melanoma.
Keywords: CDC-42, axon pathfinding, AGE-1/PI3K, mTORC2, MIG-15, Arp2/3, UNC-115
Introduction
Rho GTPases are central regulators of cellular protrusions involved in cell migration and growth cone outgrowth during axon pathfinding. Many studies implicate Rho GTPases in actin cytoskeleton rearrangement and subsequent neuronal migration and axon extension.1-4 Rho family proteins are Ras-related small GTPases that regulate cytoskeletal organization and dynamics, cell adhesion, motility, trafficking, proliferation, and survival.5 These proteins function as tightly regulated molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state.6 Misregulation of Rho proteins can result in defects in cell morphology and cell migration.7
During development of the nervous system, neurons must first migrate to their final positions and then extend their axons in order to establish synaptic connections. The leading edges of migrating cells and the growth cones of axons undergo dynamic changes in their actin cytoskeletons, which mediate migration.8-11 Guidance receptors present on the leading edge of the growth cone receive extracellular cues from the environment. Ligand binding to these guidance receptors will signal either an attractive or a repulsive cue, depending on the ligand/receptor interaction and the context.12 These guidance cues are transduced by intracellular signaling pathways, resulting in alterations in the cytoskeleton in the growth cone of the migrating axon. There is evidence to suggest that both attractive and repulsive guidance cues affect protrusiveness of the filopodial and lamellipodial structures of the growth cone.13 Proper issuance and interpretation of these cues are necessary for normal axon guidance. Disruptions in the proper extension of axons in the nervous system are associated with several neurological disorders including dyslexia and autism spectrum disorders.14,15
Previous work from our lab showed that the Rac GTPases are involved in axon guidance and neuronal filopodial and lamellipodial protrusions.16-20 We identified a pathway required for lamellipodial and filopodial protrusions induced by activated CDC-42, which included the Rac GTP exchange factor TIAM-1 and the Rac GTPases MIG-2 and CED-10.16 Furthermore, we showed that MIG-2 and CED-10 likely act through the Arp2/3 complex,21 the Arp2/3 activators WSP-1/WAS and WAVE-1/WAVE19, and also UNC-115.20 Rho GTPase action in axon guidance is subject to much functional redundancy, such that loss of one GTPase causes only mild or no effects on guidance. Indeed, ced-10/Rac, mig-2/RhoG, and cdc-42 only weakly affect axon guidance alone, but have much stronger effects when double mutants are analyzed.17,22 Thus, a simple loss-of-function genetic approach might miss key players in Rho GTPase signaling in developmental events.
In this work, we take a complementary approach to identifying CDC-42 pathway members in neuronal protrusion by analyzing molecules required for ectopic neuronal protrusion induced by activated CDC-42(G12V). This epistatic approach in a sensitized background might allow us to identify new players in CDC-42-mediated protrusion. We took a candidate gene approach to identify CDC-42(G12V) suppressors. Evidence from other systems and other contexts suggested that actin modulatory proteins such as the Arp2/3 complex and the Arp2/3 activators WSP-1/WASP and WVE-1/WAVE may be involved in filopodial and lamellipodial protrusions downstream of CDC-42.19,21,23,24 Components of the WAVE activation complex, such as GEX-2/Sra-1 and GEX-3/Kette are also likely candidates to participate in this process.19 Furthermore, previous genetic studies have identified pathways that regulate the cytoskeleton, including UNC-115/abLIM and UNC-34/Enabled, which might act in parallel to the Arp2/3 complex in axon guidance.20,21,25 Additionally, other molecules that are involved in CDC-42-mediated actin assembly such as Transducer of Cdc42-dependent actin assembly (Toca-1) may also be involved in this process.26 Taking a candidate approach, we also examined several other known CDC-42 effectors including p21-activated kinase (PAK), NCK, NCK-interacting kinase (NIK/MIG-15), phospholipase C-γ (PLC-γ/PLC-3), IQGAP/PES-7, and protein kinase C (PKC/PKC-3).27-31 We also hypothesized that PI3K, a known CDC-42 effector and activator that is implicated in cell migration and axon guidance, may be involved in this process.32-34
Here, we show that multiple pathways are required for ectopic protrusions induced by CDC-42(G12V). As expected, deletion alleles of components of the Arp2/3 complex and the Arp2/3 activators wsp-1 and wve-1 suppressed ectopic protrusions mediated by CDC-42(G12V). Furthermore, mutations in regulators of the WAVE complex, such as gex-2 and gex-3, were also able to suppress this phenotype. We also show that toca-1 but not toca-2 was able to suppress CDC-42(G12V). Furthermore, alleles of p21-activated kinase (PAK), the adaptor protein NCK, and MIG-15/NIK also suppressed this phenotype.
We also show evidence that the PI3K signaling pathway acts downstream of CDC-42 in protrusion. Interestingly, components of the mTORC2 but not the mTORC1 complex appear to mediate CDC-42-induced protrusion. Taken together, our data suggest that CDC-42 sits atop of a complex signaling cascade involving a variety of molecules that regulate the actin cytoskeleton to induce ectopic lamellipodia protrusions, including PI3K/mTORC2 signaling. A common feature of these pathways is that each involves Rac GTPases, which might serve to integrate and coordinate the functions of these pathways in similar or discrete cellular and developmental events. Many of these molecules have been implicated in axon guidance, therefore the mechanisms of ectopic protrusion found here might be relevant to growth cone protrusion during axon guidance.
Results
Expression of activated CDC-42(G12V) in the PDE neuron results in ectopic protrusions
Ras superfamily GTPases, including the Rho-family GTPases, cycle between the active GTP-bound form and the inactive GDP-bound form, and the latter is driven by the intrinsic GTPase activity of the molecules. The glycine 12 to valine (G12V) substitution has been previously shown to result in inhibition of the intrinsic GTPase activity of the GTPase, favoring the active, GTP-bound state.35 The G12V mutation also makes the protein insensitive to GTPase activating proteins (GAPs, which facilitate the GDP-bound form),36 rendering the protein constitutively GTP bound and active. Previously, we showed that expression of the Rac GTPases CED-10 and MIG-2 harboring the G12V mutation in neurons in vivo resulted in the formation of ectopic lamellipodial and filopodial protrusions,20 and that CDC-42(G12V) expression caused similar lamellipodial and filopodial protrusions.22
C. elegans has 2 bilateral PDE neurons that are located in the post-deirid ganglia of the animal.37 In the wild-type N2 strain, PDE neurons extend their axons ventrally in a straight line from the cell body to the ventral nerve cord (Fig. 1A, left panel). When the axon reaches the ventral cord, it then branches and extends toward both the anterior and the posterior. The osm-6 promoter is expressed in all ciliated sensory neurons including the PDE neurons.38 Transgenic animals expressing cdc-42(G12V) driven by the osm-6 promoter (Posm-6::cdc-42(G12V)) displayed PDE neurons with ectopic lamellipodia and filopodia protrusions (Fig. 1A, right panel). Two independently generated transgenic lines, lqIs36 and lqIs37, had similar effects (45 and 48% of PDE neurons with ectopic lamellipodia protrusions, respectively) (Fig. 1B). Presumably, activated CDC-42(G12V) constitutively engages downstream effectors, leading to ectopic lamellipodia protrusions. While the initial guidance of the PDE neuron is disrupted in these lines (Fig. 1B), the axon eventually is guided correctly to the ventral nerve cord, suggesting that CDC-42(G12V) does not significantly interfere with PDE axon guidance. Rather, it leads to ectopic protrusions from the axon and cell body of the PDE neuron.
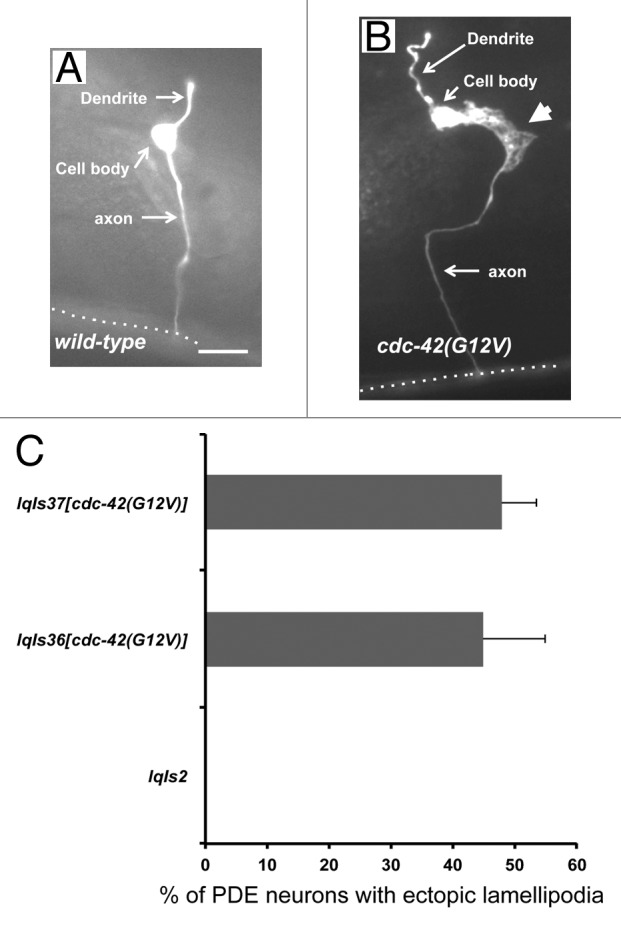
Figure 1. Expression of CDC-42(G12V) results in the formation of ectopic lamellipodia in PDE neurons. (A) A micrograph of a PDE neuron of a WT adult animal (B) A PDE neuron of an adult animal expressing CDC-42(G12V). An ectopic lamellipodial protrusion is indicated by the large arrowhead. The dotted line indicates the ventral nerve cord. The scale bar represents 5 μm. (C) Quantitation of PDE defects. lqIs2 is the osm-6::gfp control transgene and lqIs36 and lqIs37 are the Posm-6::CDC-42(G12V) activated CDC-42 transgenes. At least 100 neurons were scored for each genotype. p < 0.0001 as determined by Fisher Exact Analysis. The error bars represent 2x the standard error of the proportion (2x SEP).
Arp2/3-related signaling pathways are required for CDC-42(G12V)-induced protrusions
We used a candidate gene approach to identify pathways required for CDC-42(G12V)-induced neuronal protrusion. Previous results indicated that a pathway involving the Rac guanine nucleotide exchange factor (GEF) TIAM-1 and the Rac GTPases MIG-2 and CED-10 were required for the ectopic lamellipodia and filopodia protrusions induced by activated CDC-42(G12V).22 Mutations in tiam-1, mig-2, and ced-10 suppressed ectopic CDC-42(G12V) protrusions. We also found previously that MIG-2 and CED-10 likely act through the Arp2/3 activators WSP-1/WASP and WVE-1/WAVE.19
We tested if mutations in wsp-1, wve-1, arx-4, and arx-7 could suppress CDC-42(G12V). Two alleles of wsp-1 (wsp-1(gm324) and wsp-1(tm2299M+)) suppressed the ectopic lamellipodial protrusions of CDC-42(G12V), and wsp-1(gm324) suppressed completely (Fig. 2A). wve-1(ne350M+) showed incomplete but significant suppression, however, systemic RNAi against wve-1 did not suppress significantly (Fig. 2A). Canonically, WAVE acts with the Rac GTPases and WASP acts with CDC-42, so it was surprising that wve-1(ne350M+) suppressed CDC-42(G12V). WAVE associates with other molecules in a complex (which is activated by Rac GTPases) called the WAVE activation complex.39,40 Mutations and RNAi for 2 genes that encode components of this complex, gex-2/Sra-1 and gex-3/Kette,41 also significantly suppressed CDC-42(G12V) ectopic protrusions (Fig. 2A).
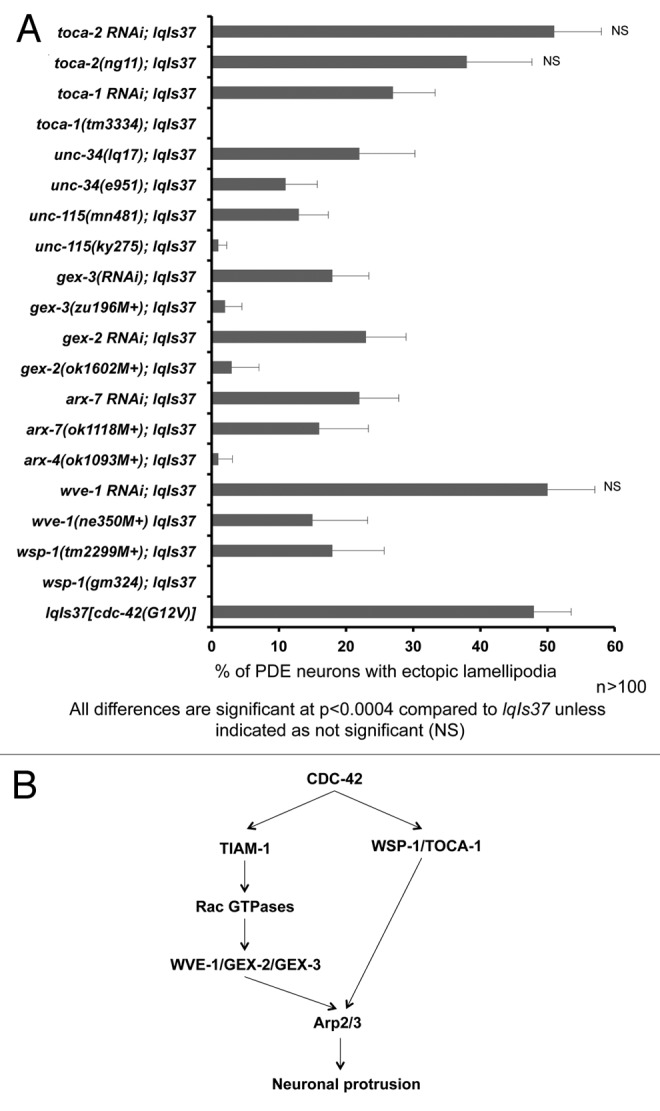
Figure 2. Multiple cytoskeletal pathways suppress activated CDC-42 in PDE neurons. (A) Quantitation of PDE defects. lqIs37 is Posm-6::CDC-42(G12V) activated CDC-42 transgene. The Y-axis denotes the genotype and the X-axis represents the percentage of ectopic lamellipodia formation. “M+” denotes that the animals had wild-type maternal contribution. The number of axons scored > 100 for each genotype. p < 0.0004 as determined by Fisher Exact Analysis. The error bars represent 2x SEP. (B) A diagram depicting CDC-42 signaling to the Arp2/3 complex based on genetic results in this study and in other work.
WAVE and WASP both activate the Arp2/3 complex. Mutations in or systemic RNAi against genes encoding Arp2/3 complex components (arx-4 and arx-7) suppressed the ectopic lamellipodia (Fig. 2A). WASP and the Arp2/3 complex are known downstream effectors of CDC-42,24,42 and wsp-1 and arx suppression of CDC-42(G12V) is consistent with this idea (Fig. 2B). WAVE is thought to act downstream of Rac signaling.19,24 This is consistent with our previous results showing that CED-10/Rac and MIG-2/RhoG act downstream of CDC-42 and TIAM-1/Rac GEF in PDE ectopic protrusions16 (Fig. 2B). Thus, CDC-42 might influence Arp2/3 via WSP-1 and WVE-1, the latter through TIAM-1 and Rac signaling (Fig. 2B).
The Transducer of Cdc-42-dependent actin assembly (Toca-1) molecule interacts with WASP and CDC-42 in actin assembly and membrane dynamics.26 C. elegans encodes 2 Toca-1-like molecules, TOCA-1 and TOCA-2/CIP-4. We found that toca-1(tm3334) and toca-1(RNAi) suppressed cdc-42(G12V) ectopic protrusions whereas toca-2(ng11) and toca-2(RNAi) did not, suggesting that TOCA-1 but not TOCA-2 acts with CDC-42 in PDE neuronal protrusion.
The cytoskeletal regulators UNC-34/Ena and UNC-115/abLIM are required for CDC-42(G12V)-induced protrusions
Genetic studies have identified cytoskeletal pathways defined by UNC-115/abLIM and UNC-34/Enabled, which might act in parallel to Arp2/3 in axon guidance.18,21,25,43 We found that unc-34(e951) and unc-34(lq17) significantly suppressed CDC-42(G12V) (Fig. 2A). Furthermore, unc-115 mutations also suppressed CDC-42(G12V) (Fig. 2A). In each case, the null alleles unc-34(e951) and unc-115(ky275) showed stronger suppression than the hypomorphic alleles unc-34(lq17) and unc-115(mn481).
In sum, these results indicate that CDC-42 regulates a cytoskeletal signaling network consisting of at least three pathways to mediate neuronal protrusion including the Arp/2/3 complex, UNC-115/abLIM, and UNC-34/Enabled. Genetic studies of axon guidance indicate cross talk between these pathways, as CED-10/Rac might act with both Arp2/3 and UNC-115,19,21 and WSP-1 might act with MIG-2/RhoG as well as CDC-42.19
MIG-15/NIK, NCK-1/Nck, and p21 activated kinase (PAK) are required for CDC-42(G12V)-induced protrusion
MIG-15, similar to Nck-Interacting Kinase (NIK), has been previously implicated in axon pathfinding, neuroblast migration, and Rac GTPase signaling.18,44 Furthermore, NIK has been shown to be regulated by a well-characterized CDC-42 effector, p21 activated kinase (PAK).45 PAK is a known effector of CDC-42 and the Rac GTPases, which regulate actin reorganization.27 Therefore, we sought to determine whether MIG-15 and PAK were involved in the ectopic lamellipodial protrusions produced by CDC-42(G12V). Five independent mutations in mig-1544 suppressed the ectopic lamellipodial protrusions (Fig. 3). Three genes in C. elegans encode PAK-like kinases: pak-1, pak-2, and max-2. Mutations in each of these significantly suppressed the ectopic lamellipodia protrusions driven by CDC-42(G12V). A deletion that removes coding exons of the nck-1/Nck gene, ok694, suppressed, but a deletion in an intron of nck-1, ok383, did not significantly suppress.
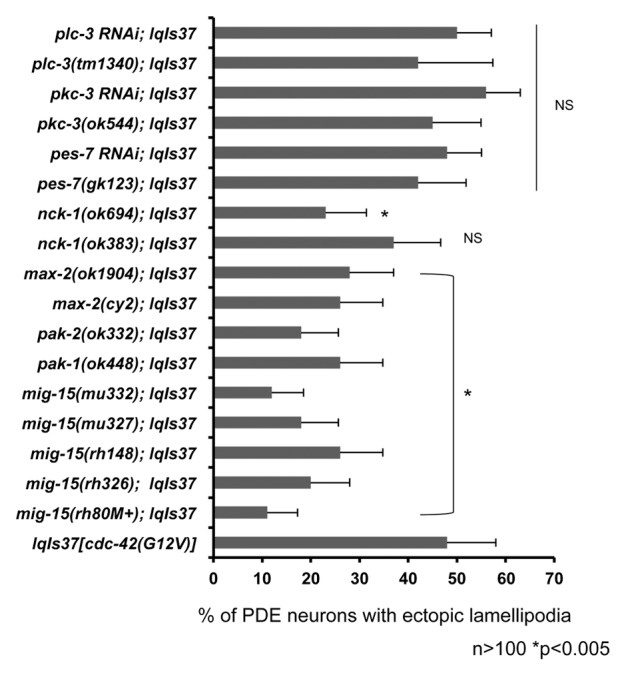
Figure 3. MIG-15(NIK) and p21 activated kinase (PAK) suppress activated CDC-42 in PDE neurons. Quantitation of PDE defects. lqIs2 is the osm-6::gfp control transgene and lqIs37 is Posm-6::CDC-42(G12V) activated CDC-42 transgene. The Y-axis denotes the genotype and the X-axis represents the percentage of ectopic lamellipodia formation. “M+” denotes that the animals had wild-type maternal contribution. The number of axons scored > 100. *p < 0.005 as determined by Fisher Exact Analysis. The error bars represent 2x SEP.
Not all genes associated with axon pathfinding and CDC-42 activity are required for CDC-42(G12V)-induced neuronal protrusion
Figures 2 and 3 show that multiple pathways are required for the ectopic neuronal protrusions induced by CDC-42(G12V). It is important to note that not all mutations tested had a suppressive effect, including some in genes previously shown to be involved in CDC-42 signaling in other contexts. For example, mutations in IQGAP (pes-7(gk123)), PKC-3 (pkc-3(ok544)), and PLC-γ (plc-3(tm1340)) did not suppress CDC-42(G12V) (Fig. 3), nor did toca-2 (Fig. 2). It is also important to note that while mutations in many of the genes required for CDC-42(G12V) protrusion alone cause axon guidance defects, none of these displayed any ectopic protrusions similar to CDC-42(G12V) (data not shown). However, pkc-3(ok544), which did not suppress CDC-42(G12V), displayed ectopic lamellipodia in 6% of PDE neurons (data not shown). Furthermore, mutation of the unc-73/Trio Rac GEF gene (rh40 and e936) did not significantly suppress CDC-42(G12V) (data not shown), despite having severe PDE axon defects alone.17 Thus, not all mutations in genes affecting axon pathfinding and CDC-42 function suppressed CDC-42(G12V)-induced neuronal protrusion, indicating that the suppression observed is specific to the role of CDC-42 in neuronal protrusion.
CDC-42(G12V) utilizes the PI3K pathway to drive ectopic protrusion
The phosphoinositide 3-kinase (PI3K) pathway is a complex pathway involved in many aspects of development and physiology including insulin signaling, cell proliferation, and aging. The PI3K pathway is well conserved in C. elegans. Previously, it has been shown that CDC-42 activates PI3K in certain contexts.33 Recent work also showed that RAC-1 and CDC-42 can bind to and activate the p110β catalytic subunit of PI3K.46 Furthermore, PI3K has been shown to regulate axon guidance, although its relationship to CDC-42 in this specific context has not been determined.32,34 We sought to determine whether the PI3K pathway was involved downstream of CDC-42(G12V). PI3K is composed of 2 subunits, the p110 catalytic subunit and the p85 adaptor subunit. Mutations in the p110 subunit (age-1[m333] and age-1[mg44]) suppressed CDC-42(G12V), as did 2 mutations in the p85 subunit, aap-1(m889) and aap-1(ok282) (Fig. 4A). atm-1 encodes a PI3K-like molecule similar to human ATM, and atm-1(gk186) also suppressed CDC-42(G12V) (Fig. 4A).
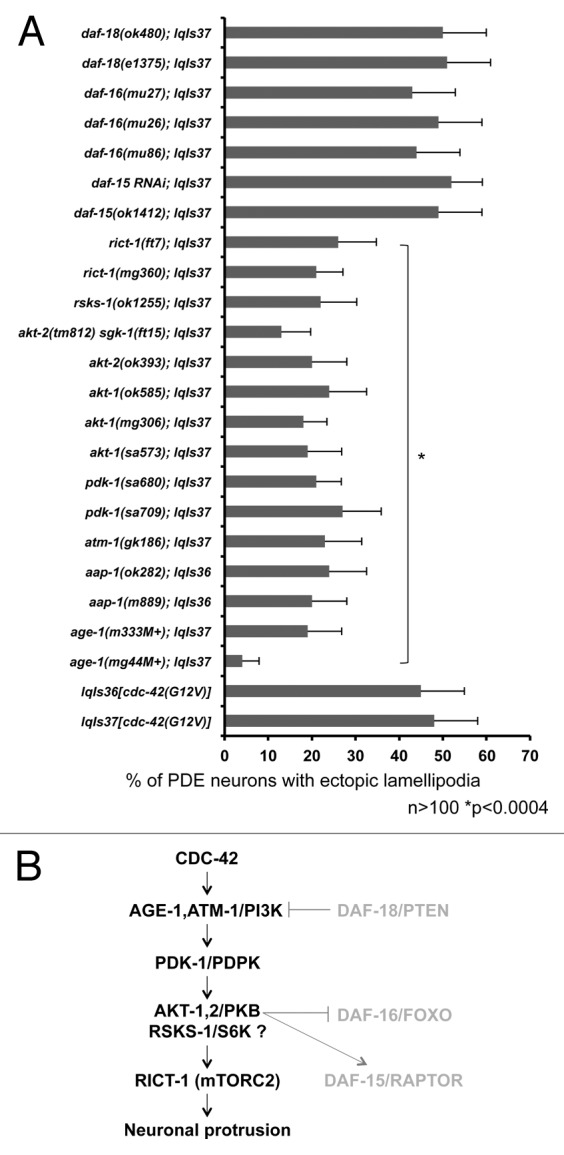
Figure 4. CDC-42 utilizes specific components of the PI3K pathway to drive ectopic lamellipodia formation in PDE neurons. (A) PI3K signaling mutants suppress CDC-42(G12V). The Y-axis denotes the genotype and the X-axis represents the percentage of ectopic lamellipodia formation. “M+” denotes that the animals had wild-type maternal contribution. The number of axons scored > 100. *p < 0.0004 as determined by Fisher Exact Analysis. The error bars represent 2x SEP. (B) A diagram of the genetic interaction with CDC-42 and the PI3K signaling pathway. Molecules in gray are those not involved in CDC-42-induced neuronal protrusion.
We next asked which known components of PI3K signaling were required for CDC-42(G12V)-induced neuronal protrusion. We tested pdk-1, which encodes for a 3-phosphoinositide-dependent kinase (PDPK-1/PDK). PDPK/PDK-1 is an upstream activator of Akt and is required for Akt activation. PDPK/PDK-1 also activates other downstream kinases, such as s6 kinase (s6K) and PKC. We found that multiple alleles of pdk-1 (pdk-1[sa680] and pdk-1[sa709]) suppressed the ectopic protrusions induced by CDC-42(G12V) (Fig. 4A). The AKT/PKB serine threonine kinase acts in PI3K signaling, and AKT-1 and AKT-2 in C. elegans genetically interact with AGE-1/PI3K in insulin signaling and other processes. Multiple mutations of akt-1 and akt-2 suppressed the ectopic protrusions induced by CDC-42(G12V) (Fig. 4A), further implicating the PI3K pathway in this biological process. PTEN is a negative regulator of PI3K signaling, and mutations in the C. elegans PTEN gene daf-18 did not suppress. In sum, these results indicate that core components of the PI3K signaling pathway, including PI3K, PDPK/PDK-1 and AKT are required for CDC-42(G12V)-induced neuronal protrusion.
Components of mTORC2, but not mTORC1, are required for neuronal protrusion
PI3K signaling impinges on the TOR (Target of Rapamycin) complexes mTORC1 and mTORC2. rict-1 encodes a component of the mTORC2 complex, and rict-1(mg360) and rict-1(ft7) suppressed ectopic lamellipodia formation (Fig. 4A). However, perturbation of a component of the mTORC1 complex (daf-15[mu86] and daf-15[RNAi]) did not suppress, nor did daf-16(mu26) and daf-16(mu27), which encodes a FoxO protein (Fig. 4A). The role of the FoxO transcription factor in PI3K signaling is complex. FoxO is inhibited by Akt and can also activate Akt. Furthermore, FoxO can also negatively regulate mTor1. In addition to its role in the mTORC1 pathway and Akt regulation, FoxO has been shown to elevate the expression of Rictor, leading to increased mTORC2 activity.47 However, FoxO, did not suppress the phenotype of CDC-42 in our hands, suggesting that FoxO is not required for this phenotype in this context. Taken together, these data suggest that CDC-42(G12V) utilizes the mTORC2 complex downstream of PI3K and not the mTORC1 complex to form ectopic protrusions in PDE neurons (Fig. 4B). These data also suggest that this assay is sensitive enough to resolve and analyze the specific contribution of complex pathways to this process. Interestingly, mutation of s6 kinase (S6K, rsks-1[ok1255]) also suppressed. While rsks-1/S6K is downstream of the mTORC1 complex, it has also been shown to be directly downstream of pdk-1.48 Furthermore, S6K has also been shown to directly phosphorylate Rictor (rict-1), a component of the mTORC2 complex. This phosphorylation mediates 14–3-3 protein binding to Rictor, altering interactions of this complex.49 These data are consistent with a role of mTORC2, but not mTORC1, in CDC-42(G12V)-induced neuronal protrusion.
An activated version of AKT-1 results in the formation of ectopic lamellipodia
If the mutations we found that suppress activated CDC-42(G12V) affect molecules that act in the CDC-42 pathway, we expect that activated versions of these molecules might also drive ectopic protrusions similar to activated CDC-42(G12V). Indeed, loss of function of the Rac GTPase genes mig-2 and ced-10 suppress CDC-42(G12V), and activated versions of both result in the formation of ectopic lamellipodia in the PDE neurons.16,20 The akt-1(mg144) mutation results in an activated AKT-1 molecule,50 and we found that akt-1(mg144) mutants displayed ectopic protrusion in the PDE neuron (Fig. 5) similar to but less severe than CDC-42(G12V) (Fig. 5). Furthermore, rict-1(mg360) and rict-1(ft7) (mTORC2) but not daf-15(mu86) nor daf-15(RNAi) (mTORC1) suppressed akt-1(mg144)-mediated protrusions (Fig. 6). These data lend further support to the idea that CDC-42(G12V) requires AGE-1/PI3K, AKT, PDPK/PDK-1, and the mTORC2 complex, but not the mTORC1 complex, to drive ectopic neuronal protrusion.
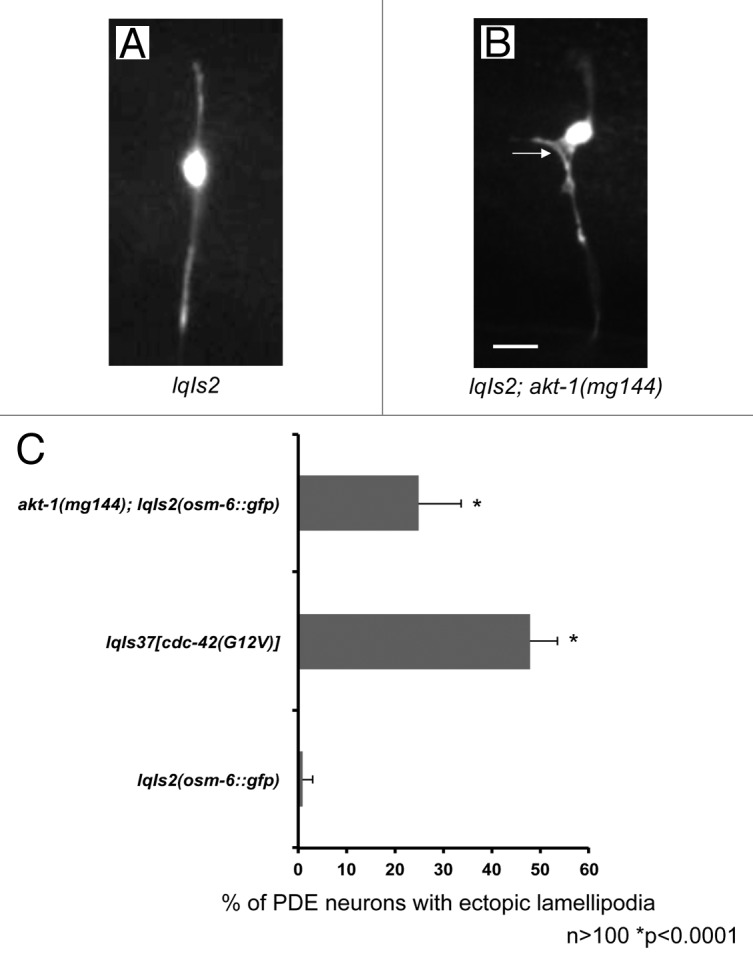
Figure 5. An activated allele of akt-1 results in the formation of ectopic lamellipodia, similar to activated CDC-42 (A) A micrograph of a PDE neuron of a WT adult animal. (B) A PDE neuron of an akt-1(gm144) adult animal. An ectopic lamellipodial protrusion is indicated by the arrow. The scale bars represents 5 μm. (C) Quantitation of PDE defects. lqIs2 is the osm-6::gfp control transgene. The Y-axis denotes the genotype and the X-axis represents the percentage of ectopic lamellipodia formation. The number of axons scored > 100. *p < 0.0001 as determined by Fisher Exact Analysis. The error bars represent 2x SEP.
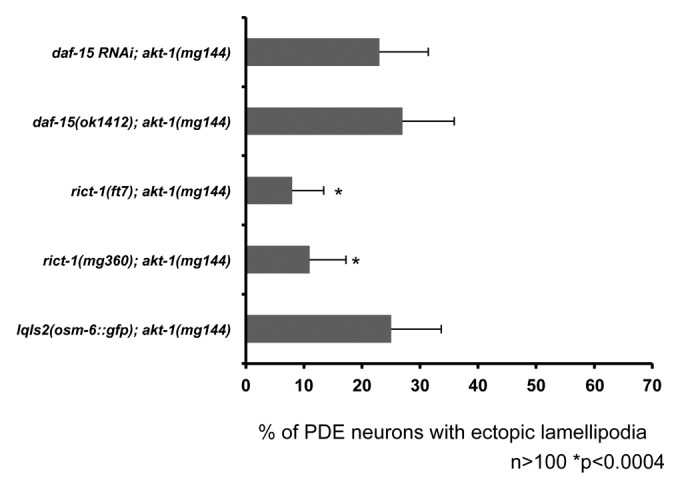
Figure 6. An activated allele of akt-1 is suppressed by components of the mTORC2 complex, but not the mTORC1 complex. Quantitation of PDE defects. lqIs2 is the Posm-6::gfp control transgene. rict-1 is in the mTORC2 complex and daf-15 is in the mTORC1 complex. The Y-axis denotes the genotype and the X-axis represents the percentage of ectopic lamellipodia formation. The number of axons scored > 100. *p < 0.0004 as determined by Fisher Exact Analysis. The error bars represent 2x SEP.
age-1 and components of the mTORC2 complex are required cell autonomously
To test whether age-1 and rict-1 (a component of the mTORC2 complex) work cell autonomously, we made cell-specific RNAi transgenes for each gene driven by the osm-6 promoter for expression in the PDE neuron. We also made a similar RNAi transgene for daf-15, a component of the mTORC1 complex. By driving RNAi expression with the osm-6 promoter we ensured expression of RNAi in only the ciliated neurons, including the PDE neuron. To limit RNAi spreading from the PDE and other neurons, we constructed these lines in a sid-1(pk3321) background. SID-1 is required for spreading of RNAi because SID-1 affects the ability of cells to accumulate extracellular dsRNA molecules.51,52 We found that cell-specific RNAi for both age-1 and rict-1 suppressed the ectopic protrusion driven by lqIs37[Posm-6::cdc-42(G12V)] (Fig. 7). daf-15(RNAi) did not suppress the ectopic protrusion (Fig. 7). These results are consistent with the model that CDC-42 utilizes the AGE-1 pathway through the mTORC2 complex to mediate protrusion in the PDE neuron in a cell autonomous manner.
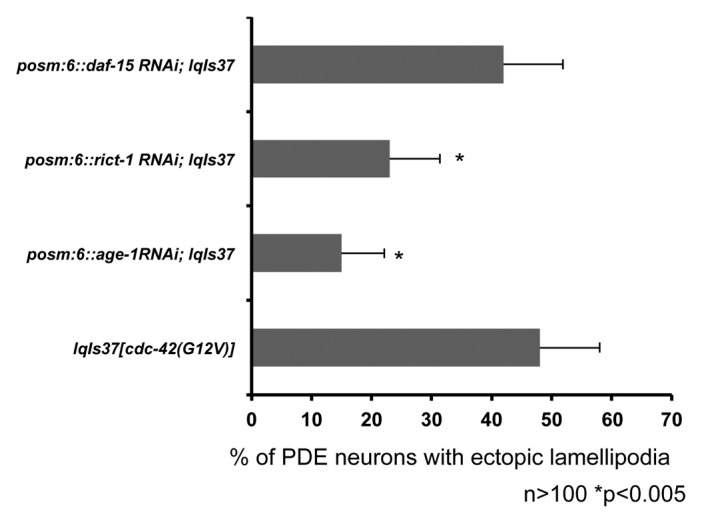
Figure 7. AGE-1 and RICT-1 act cell autonomously in the PDE neuron. Transgenic lines containing cell-specific RNAi against, age-1, rict-1 and daf-15 were constructed and crossed with lqIs37. All of the strains are in a sid-1(pk3321) background, which limits RNAi spreading. The Y-axis denotes the genotype and the X-axis represents the percentage of ectopic lamellipodia formation. The number of axons scored > 100. *p < 0.005 as determined by Fisher Exact Analysis. The error bars represent 2x SEP.
Expression of CDC-42(G12V) results in pathfinding defects in the VD/DD motor neurons
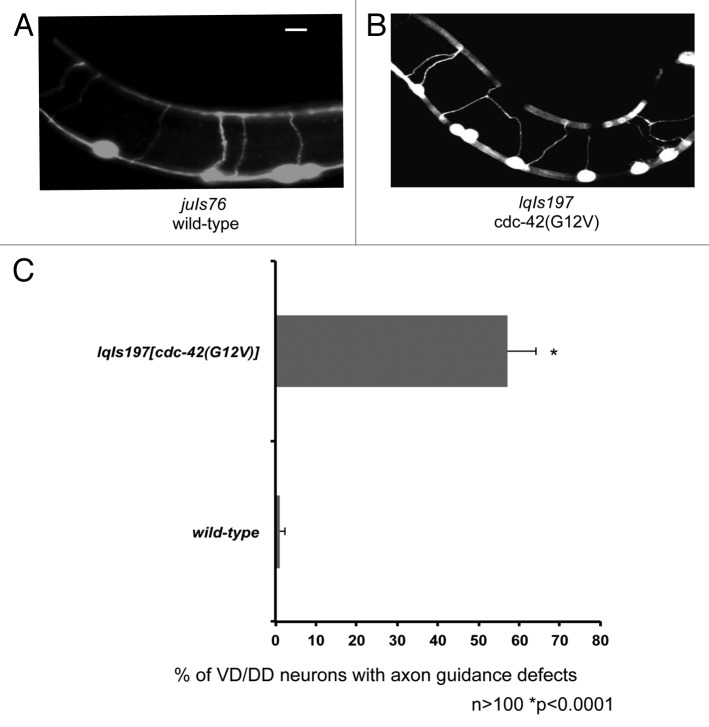
To assay the effects of CDC-42(G12V) on other neurons, we drove expression of CDC-42(G12V) using the unc-25 promoter. The unc-25 promoter is active in the VD/DD type motor neurons, which are born in the ventral nerve cord and extend ventral to dorsal commissural axons. We found that expression of CDC-42(G12V) resulted in significant axon pathfinding defects in the VD/DD motor neurons (Fig. 8). CDC-42(G12V) did not induce the formation of significant ectopic protrusions in the VD/DD neurons as it did in the PDEs. This could be because these neuron classes have distinct guidance and protrusion mechanisms reflecting distinct roles of CDC-42 in the neurons. Possibly, CDC-42(G12V) interferes with guidance mechanisms in the VD/DD neurons but does not drive ectopic protrusion as it does in the PDE neurons.
Figure 8. Expression of CDC-42(G12V) results in the axon guidance defects in the VD/DD motor neurons. (A) A micrograph of transgenic animals expressing Punc-25::gfp. (B) A transgenic animal expressing Punc-25::CDC-42(G12V) with axon guidance defects. The scale bars represent 5 μm. (B) Quantitation of VD/DD defects. juIs76 is the Punc-25::gfp control transgene. The Y-axis denotes the genotype and the X-axis represents the percentage of axon pathfinding defects. The number of axons scored > 100. *p < 0.0001 determined by Fisher Exact Analysis. The error bars represent 2x SEP.
Components of the AGE-1/PI3K pathway suppress the pathfinding defects driven by activated CDC-42 in VD/DD motor neurons
Next, we wanted to determine whether components of the AGE-1/PI3K pathway were required for CDC-42(G12V)-induced VD/DD axon guidance defects. age-1(m333), (age-1mg44), akt-1(ok585), and akt-2(ok383) suppressed (Fig. 9). Additionally, rict-1(RNAi) but not daf-15(RNAi) suppressed (Fig. 9), indicating that CDC-42/AGE-1 signaling through the mTORC2 complex may be important in multiple neuronal types.
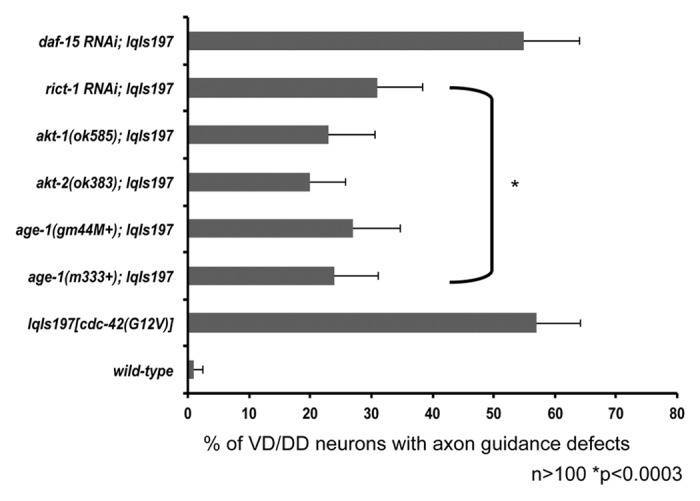
Figure 9. Components of the AGE-1 pathway and the mTORC2 complex suppress the axon guidance defects driven by activated CDC-42 in the VD/DD motor neurons. Quantitation of axon guidance defects in the VD/DD motor neurons. Null alleles several downstream CDC-42 effectors including: age-1, akt-1, and akt-2 suppress the axon pathfinding defects driven by activated CDC-42 in VD/DD motor neurons. Components of the mTORC2 pathway (rict-1) but not the mTORC2 pathway (daf-15) suppress the axon pathfinding defects driven by CDC-42(G12V). The Y-axis denotes genotype and the X-axis represents percentage of axon pathfinding defects. The number of axons scored > 100. *p < 0.0003 determined by Fisher Exact Analysis. The error bars represent 2x SEP.
Discussion
Here, we present evidence, which strongly suggests that CDC-42 acts through a complex network of downstream effectors to control neuronal protrusion. First, we show that a variety of cytoskeletal regulators are required downstream of CDC-42 including components of the Arp2/3 complex, the WAVE complex, WASP, Toca-1, UNC-34/Enabled, and UNC-115/abLIM. Second, we show that CDC-42 utilizes other signaling molecules such as MIG-15/NIK, NCK, and PAK to drive PDE protrusions. Finally, we show that CDC-42 utilizes several components of the PI3K pathway to drive neuronal protrusion. We found that CDC-42 signals through the mTOR2 complex (mTORC2) but not the mTORC1 complex in neuronal protrusion. There have been several studies focused on molecules that function in axon guidance. However, the entire picture of how these molecules function together in signaling pathways is still unclear. CDC-42, a Rho GTPase, has been previously shown to function in axon guidance.2,53,54 Axon guidance pathways exhibit much functional redundancy, and thus the roles of molecules in axon guidance can be masked when using loss of function studies.18,19 Indeed, cdc-42 mutations themselves have only very weak effects on axon guidance.22 Here, we used an activated version of CDC-42 as a sensitized background to dissect downstream mechanisms of neuronal protrusion that might be masked by genetic redundancy. The ectopic protrusion induced by CDC-42(G12V) resembles the protrusions normally found in growth cones of axons during their outgrowth.13,55 Indeed, the pathways that we have identified acting downstream of CDC-42 in neuronal protrusion also affect axon guidance, including Arp2/3, UNC-115/abLIM, UNC-34/Ena,19,21 MIG-15/NIK,18 and PI3K signaling.32 Thus, pathways identified by suppression of CDC-42-induced ectopic protrusion might normally act to regulate protrusion in the growth cone during outgrowth. Many of the protrusions in lqIs37 are near or in contact with the PDE cell body. However, roughly half are distinctly separate from the cell body and are located on the axon. This suggests that the protrusions are in fact axon-based. Therefore, the signaling pathways delineated in this study may participate in both protrusion and axon guidance. Future studies on the roles of these pathways in growth cone protrusion will delineate their normal roles in axon guidance.
Rho family members (most notably Rho, Rac and Cdc-42) have been extensively studied for their roles in cytoskeletal dynamics.5 Seminal studies in cell culture highlighted a role for Rho in stress fiber formation, Rac in lamellipodia formation, and Cdc-42 in filopodial formation.56-58 Further work in this field has uncovered that considerable crosstalk between these GTPases and their effectors occurs.59 In our system of neuronal protrusion, we found that Rac and Cdc-42 GTPases both regulate the formation of lamellipodia and filopodia. Thus, the roles of Rho GTPases in protrusion might be dependent upon biological context. Cdc-42 acts thorough a number of effectors, and can influence a number of signaling events, most notably cytoskeletal arrangement.60,61 Although Cdc-42 has many validated effectors, a complete picture of how Cdc-42 works in vivo to regulate axon guidance is not clear. Here, we used CDC-42-induced protrusion as an in-road into dissection of CDC-42 pathways that regulate neuronal protrusion and thus possibly growth cone protrusion and axon guidance.
Cytoskeletal regulators downstream of CDC-42 in axon guidance
Previous work indicated that the Rac GTPases MIG-2 and CED-10 along with the Rac GTP exchange factor TIAM-1 were required for ectopic lamellipodia formation mediated by CDC-42(G12V).16 Additionally, work from our lab has shown that MIG-2 and CED-10 likely act through the Arp2/3 activators WSP-1/WASP and WVE-1/WAVE.19,21 This study confirmed that CDC-42 does in fact signal through components of the Arp2/3 complex (ARX-4/ArpC2 and ARX-7/ArpC5) as well as WSP-1/WASP to induce ectopic lamellipodia formation in PDE neurons. Although a deletion allele of WVE-1/WAVE produced incomplete but significant suppression, RNAi knockdown of WVE-1/WAVE was unable to replicate these results. These results may be due to incomplete knockdown of the protein with RNAi. WAVE is activated by other molecules in the cell as part of the WAVE activation complex, which includes GEX-2/Sra-1, and GEX-3/kette24 This study showed two genes that encode components of this complex, gex-2/Sra-1 and gex-3/Kette, also significantly suppressed CDC-42(G12V) ectopic protrusions, further supporting a role for WAVE and the WAVE activation complex downstream of CDC-42. WASP and the Arp2/3 complex are known downstream effectors of CDC-42,24,62 and wsp-1 and arx suppression of CDC-42(G12V) is consistent with this data, providing validation of our results. Furthermore, WAVE is thought to act downstream of RAC signaling.24 This is consistent with data from this study showing that wve-1 and gex mutations suppress CDC-42(G12V) and consistent with previous results showing that mig-2 and ced-10 also suppress CDC-42(G12V).16
There have been genetic studies identifying pathways that include UNC-115/abLIM and UNC-34/Enabled, which potentially act in parallel to Arp2/3 in axon guidance. UNC-115/abLIM and UNC-34/Enabled along with Arp2/3 have been shown to regulate growth cone dynamics and axon pathfinding.21 Furthermore, work in our lab has shown that UNC-115/abLIM works downstream of the Rac GTPases in axon guidance.21 This study was able to show that both unc-34 and unc-115 mutations suppressed activated CDC-42(G12V), suggesting that these genes works downstream of CDC-42, potentially through the Rac GTPases MIG-2 and CED-10.
Toca-1 (Transducer of Cdc42-dependent actin assembly) and Toca-2/CIP4 have been shown to interact with WASP and CDC-42 in actin assembly.26 In this study, we found that TOCA-1 but not TOCA-2 works downstream of CDC-42 in PDE neuronal protrusion. TOCA-1 and TOCA-2 have been shown to be direct downstream effectors of CDC-42 via binding of their imperfect HR1/CRIB-like domain. Furthermore, TOCA-1 association with the WASP-WIP complex was shown to be essential for Arp2/3 activation downstream of CDC-42.26,63 In this study, we found that TOCA-1 but not TOCA-2 was downstream of CDC-42 in PDE axon guidance.
In the study, we were able to show that CDC-42 sits at the top of a complex cytoskeletal signaling network to mediate neuronal protrusion (Figs. 2B and 10). The cytoskeletal pathways consist of at least 3 distinct proteins including the Arp2/3 complex, UNC-115/abLIM, and UNC-34/Enabled. Furthermore, our work combined the other studies indicates that there is considerable cross talk between pathways. For example, CED-10/Rac likely acts with Arp2/3 and UNC-115, and WSP-1 might also interact with MIG-2/RhoG.18,19,21Furthermore, CDC-42 is also capable of signaling through CED-10/Rac and MIG-2/RhoG in this context.16
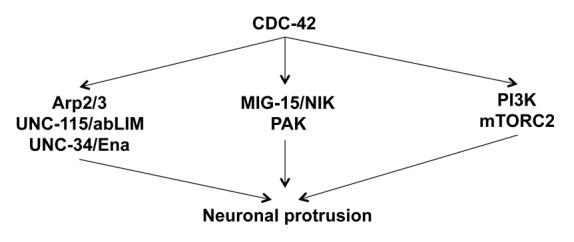
Figure 10. Multiple pathways mediate CDC-42-induced neuronal protrusion. A diagram of the multiple pathways mediating CDC-42(G12V)-induced neuronal protrusion described in this work.
MIG-15, PAK-1 and NCK are downstream of CDC-42 in axon guidance
In cell culture experiments, one of the best-characterized Cdc-42 effectors is p21-activated kinase (PAK). Cdc-42 shares this effector with Rac, further underlying the complexity of these signaling pathways.27 GTP-bound Cdc-42 and Rac regulate PAK activity through binding to its Cdc-42/Rac interactive binding (CRIB) domain. Cdc-42 binding relieves the auto-inhibition of the N-terminal domain of PAK.27 There is data in other model organisms, which suggests that PAK likely functions downstream of Cdc-42 in axon guidance. In Drosophila, overexpression of PAK results in axon guidance defects, which can be suppressed by genetically removing one copy of CDC-42.64
Evidence in cell culture and other systems indicates that the scaffolding protein Nck-1 is also involved in Cdc-42 signaling. Furthermore, there is evidence from Drosophila indicating that Nck-1 may be involved downstream of CDC-42 with Pak-1 in axon guidance.65 Furthermore, Pak and Nck function are required cell autonomously for proper axon targeting in Drosophila olfactory neurons.65,66 To date, there have been no documented interactions between CDC-42 and the NIK-interacting kinase MIG-15. However, there is evidence that MIG-15 works downstream of Rac to regulate axon pathfinding.18 Here we show evidence that CDC-42 does utilize PAK, NCK, and MIG-15 to mediate axon pathfinding.
PI3K signaling in axon guidance
There is a large body of work describing the relationship between CDC-42 and phosphatidylinositol 3-kinase. There is evidence that PI3K is both upstream and downstream of CDC-42, depending on the context.33 PI3K is well known for its role in the insulin-signaling pathway and in the aging process67 and is involved in axon pathfinding.32 However, the role of the PI3K pathway and its relationship to CDC-42 in axon pathfinding is not well understood. There is 1 study showing that age-1 is required downstream of netrin signaling for proper axon guidance.32 There are also several studies that implicate PI3K in axon specification, because pharmacologic inhibition of PI3K prevents axon formation.68,69 AKT, a protein downstream of PI3K well-known for its role in survival,67 has also been implicated in axon pathfinding.70,71 AKT is critical for neuronal polarity and has been shown to undergo selective degradation. This selective degradation leaves an active pool of AKT in a single neuronal process that is designated to become the axon.72
Mammalian target of rapamycin (mTor) is an evolutionarily conserved Ser/Thr protein kinase that plays an essential role in protein translation and cell growth. Dysregulation of mTor is involved in many disease states including cancer, diabetes, and more recently Alzheimer disease.73 mTor is also important for long-term synaptic plasticity and axon pathfinding.74
mTor signals through 2 distinct complexes downstream of PI3K, namely mTORC1 and mTORC2. mTORC1 is sensitive to rapamycin and activates S6K1 and 4EBP1. mTORC1 is regulated by DAF-15/Raptor, and is inhibited by FOXO1. mTORC2 is reported to be resistant to rapamycin and activates PKC-α, AKT, and Rho GTPases. mTORC2 is regulated by RICT-1/Rictor.75 In our study, we showed that RICT-1 is involved in neuronal protrusion downstream of CDC-42. Many previous studies on Rictor have addressed its role as an essential component of the mTORC2 complex. However, there are recent studies that suggest that Rictor can mediate migration and cell survival independent of mTORC2.76,77 Another recent study found that loss of Rictor results in induction of RhoGDI2. This induction of RhoGDI2 inhibits Rac1 and Cdc-42 function, resulting in impaired cell migration.78 Therefore, future work describing the role of Rictor in axon guidance is warranted. This study is the first to show that CDC-42 utilizes the AGE-1/PI3K pathway to guide axon pathfinding through the mTORC2 complex and not the mTORC1 complex. Specific inhibitors of mTORC1 and mTORC2 are currently in development.79 It would be of interest to determine whether pharmacologic inhibition would parallel the genetic data that we have presented here.
Multiple pathways are required for CDC-42-induced neuronal protrusion
While our genetic studies presented here lead to linear models of distinct pathways interacting downstream of CDC-42, there is likely extensive cross-talk between these pathways (Fig. 10). For example, Rac GTPases might be involved in each of the pathways and thus serve as an integrating node in coordinating CDC-42 activity in protrusion. Work from our lab has shown that the Rac GTPases MIG-2 and CED-10 work through WSP-1/WASP and WVE-1/WAVE to modulate the Arp2/3 complex.19,21 The Arp 2/3 complex in turn results in cytoskeletal changes, which mediates protrusion. TOCA-1 may also be involved in this process, because it has previously been shown to be essential for Arp2/3 activation downstream of CDC-42.26 UNC-115/abLIM and UNC-34/Enabled, which are also activated by Rac GTPases, may act in parallel in to Arp2/3 to mediate protrusion.21 PAK is an effector of both CDC-42 and Rac GTPases, and it signals through cofilin to regulate actin dynamics in the cell.27 Furthermore, NCK may act as a scaffold to facilitate CDC-42/PAK signaling.80 MIG-15/NIK may be acting directly downstream of CDC-42 or it may be activated indirectly by CDC-42 through Rac, because there is evidence that MIG-15 works downstream of Rac in axon guidance.18 CDC-42/PI3K/mTORC2 signaling is currently not well understood, and our study suggests that it mediates protrusion. There is evidence that PI3K is both upstream and downstream of CDC-42 depending on the context.33 Furthermore, PI3K can also activate Rac to mediate actin cytoskeletal rearrangement and protrusion.81 There is also evidence that the mTORC2 component Rictor can mediate cytoskeleton dynamics independent of other mTORC2 components.78 Rictor interacts with RhoGDI2, relieving inhibition of CDC-42 and RAC, which then results in actin cytoskeleton changes and altered cell migration.78 These pathways might all act generally in protrusive activity, but likely they have distinct roles in specific events. For example, subsets of the pathways might act in response to distinct signals that regulate cell movement and axon guidance. Further studies of the roles of these pathways in specific axon guidance events and growth cone dynamics will begin to delineate these relationships.
Several recent studies have identified mutations in Rho family members in various cancer types82-84, including one that identified CDC42(G12V) as a driver mutation in malignant melanoma.83 Because the signaling pathways in C. elegans are well-conserved, pathways studied in C. elegans often have conserved roles in humans. For examples, studies of the Ras oncogene in C. elegans have uncovered many aspects of its signal transduction and developmental pathways (reviewed in ref. 85), which led to a greater understanding the mechanisms of Ras-mediated tumorigenesis. Thus, studies reported here might have broader relevance beyond axon pathfinding, as these same pathways might mediate tumorigenic events such as cell movements and shape changes during metastasis.
Materials and Methods
C. elegans genetics and transgenics
C. elegans were cultured using standard techniques.86,87 All of the experiments were done at 20°C. Transgenic worms were made by gonadal micro-injection using standard techniques.88 RNAi was done using clones from the Ahringer library, as previously described.89 The following mutations and constructs were used in this work:
LGI- lqIs37 [Posm-6::CDC-42(G12V)], arx-7(ok1118), hT2 [bli-4(e937)] (I:III), pes-7(gk123), atm-1(gk186), daf-16(mu86), daf-16(mu26), daf-16(mu27), hIn1[unc-54(h1040)]
LGII- max-2(ok1904), max-2(cy2), pkc-3(ok544), plc-3(tm1340), mIn1, age-1(m333), age-1(mg44), mnC1 dpy-10(e128) unc-52(e444), aap-1(m889), aap-1(ok282), rict-1(mg360), rict-1(ft7), juIs76 (Punc-25::gfp)
LGIII- wve-1(ne350), hT2 [bli-4(e937)(I:III), arx-4(ok1093), sC1 [dpy-1(s2170)], toca-2(ng11), rsks-1(ok1255)
LGIV- wsp-1(gm324), wsp-1(tm2299), gex-2(ok1602), gex-3(zu196), dpy-9(e12), nT1 qIs51 (IV; V), daf-15(ok1412),nT1, daf-18(e1375), daf-18(ok480), lqIs3 (Posm-6::gfp)
LGV- pak-2(ok332), akt-1(sa573), akt-1(mg306), akt-1(ok585), akt-1(mg144), sid-1(pk3321)
LGX- unc-115(mn481), unc-115(ky275), toca-1(tm3334), mig-15(rh80), mig-15(rh326), szT1 (X:I), mig-15(rh148), mig-15(mu327), mig-15(mu332), pak-1(ok448), nck-1(ok383), nck-1(ok694), pdk-1(sa608), pdk-1(sa709) akt-2(ok393), akt-2(tm812) sgk-1(ft15), lqIs2 (Posm-6::gfp)
LG unassigned lqIs36 [Posm-6::CDC-42(G12V)], lqIs197 [Punc-25::CDC-42(G12V)]
The mutants were maintained as homozygous stocks, when possible. In some cases, this was not possible because the mutants were lethal or maternal-effect lethal, and thus, could not be maintained as homozygous stocks. If this was the case, the mutation was maintained in a heterozygous state over a balancer chromosome. arx-4, and arx-7 mutations were balanced by the rearrangements sC1 and hT2, respectively. hT2 harbors a transgene that drives gfp expression in the pharynx, whereas sC1 had no gfp marker. arx-7 homozygotes were identified by lack of pharyngeal green fluorescent protein (GFP), whereas arx-4 homozygotes were identified by the protruding vulva (Pvl) phenotype in sterile young adults. wve-1(ne350) was balanced with the inversion chromosome hIn1, which carries a recessive paralyzed uncoordinated (Unc) mutation. The presence of wve-1(ne350) was assessed by the presence of maternal-effect gut on the exterior (Gex), egg laying abnormal (Egl) and Pvl animals. mig-15(rh80) was balanced by szT1. mig-15 homozygotes were selected by the Unc, dumpy (Dpy) and Pvl phenotypes characteristic of mig-15 alleles. pkc-3(ok544) and plc-3(tm1340) were balanced by mIn1, which also harbors a transgene that drives gfp expression in the pharynx. pkc-3(ok544) and plc-3(tm1340) homozygotes were identified by lack of pharyngeal GFP. age-1(m333) and age-1(mg44) were balanced by mnC1. age-1 homozygotes were identified as Egl animals. daf-15(ok1412) was balanced by nT1.
Scoring of PDE and VD/DD axon defects
Neuronal protrusion defects were scored in the fourth larval stage (L4) or in young pre-gravid adult animals harboring green fluorescent protein (gfp), which was expressed in specific cell types. In the event that the animals did not survive to this stage, they were counted at an earlier stage with appropriate controls. The PDE neurons and axons were visualized in cells that had an osm-6::gfp transgene (lqIs3 IV or lqIs2 X), which is expressed in all ciliated neurons including the PDE.20,38 The neurons were considered to have ectopic protrusions if they had a lamellipodia-like structures protruding from anywhere on the cell body, axon, or dendrite.
The VD/DD motor neuron morphology was scored in animals that had the unc-25::gfp transgene (juIs76 II).90 The unc-25 promoter is expressed in all GABAergic neurons, including the VDs and DDs. In wild-type animals, the VD/DD commissures extend directly from the ventral nerve cord (VNC) toward the dorsal surface of the animals. At the dorsal surface, they form the dorsal nerve cord. VD/DD pathfinding defects were noted when the commissural axons were misguided or terminated prematurely.
In all experiments, at least 100 axons were scored, and statistical significance between genotypes was determined using the Fisher Exact Analysis. The error bars in the graphs represent 2 times the standard error of the proportion (2x SEP).
Molecular biology
Recombinant DNA, polymerase chain reaction (PCR) and other molecular biology techniques were performed using standard procedures.91 All primer sequences used in the PCR reactions are available upon request. The previously described cdc-42(G12V) transgenes were used.22
Microscopy and imaging
The animals used for the imaging analyses were mounted for microscopy in a drop of M9 buffer on an agarose pad.87 Both the buffer and the agarose pad contained 5 mM sodium azide, which was used as an anesthetic. Then, a coverslip was placed over the sample, and the slides were analyzed by epifluorescence. A Leica DMRE microscope with a Qimaging Rolera MGi EMCCD camera was used, along with Metamorph and ImageJ software.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors wish to thank C Giuliani and G Scita for the toca-1 and toca-2 strains. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was supported by NIH grants R01NS040945 and R21NS070417 to E.A.L., and NIH grant P20 RR016475 from the INBRE/IDEA Program of the National Center for Research Resources (J Hunt, PI).
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/26602
References
- 1.Lundquist EA. Rac proteins and the control of axon development. Curr Opin Neurobiol. 2003;13:384–90. doi: 10.1016/S0959-4388(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 2.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:173–80. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 3.Dickson BJ. Rho GTPases in growth cone guidance. Curr Opin Neurobiol. 2001;11:103–10. doi: 10.1016/S0959-4388(00)00180-X. [DOI] [PubMed] [Google Scholar]
- 4.Sulston J. J H. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1988. [Google Scholar]
- 5.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–35. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 7.Ridley AJ. Rho proteins and cancer. Breast Cancer Res Treat. 2004;84:13–9. doi: 10.1023/B:BREA.0000018423.47497.c6. [DOI] [PubMed] [Google Scholar]
- 8.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–33. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 9.Abd El-Rehim DM, Pinder SE, Paish CE, Bell JA, Rampaul RS, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer. 2004;91:1532–42. doi: 10.1038/sj.bjc.6602184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–64. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 11.Gallo G, Letourneau P. Axon guidance: proteins turnover in turning growth cones. Curr Biol. 2002;12:R560–2. doi: 10.1016/S0960-9822(02)01054-0. [DOI] [PubMed] [Google Scholar]
- 12.Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–63. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- 13.Norris AD, Lundquist EA. UNC-6/netrin and its receptors UNC-5 and UNC-40/DCC modulate growth cone protrusion in vivo in C. elegans. Development. 2011;138:4433–42. doi: 10.1242/dev.068841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galaburda AM. Dyslexia--a molecular disorder of neuronal migration: the 2004 Norman Geschwind Memorial Lecture. Ann Dyslexia. 2005;55:151–65. doi: 10.1007/s11881-005-0009-4. [DOI] [PubMed] [Google Scholar]
- 15.Wegiel J, Kuchna I, Nowicki K, Imaki H, Wegiel J, Marchi E, Ma SY, Chauhan A, Chauhan V, Bobrowicz TW, et al. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119:755–70. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demarco RS, Lundquist EA. RACK-1 acts with Rac GTPase signaling and UNC-115/abLIM in Caenorhabditis elegans axon pathfinding and cell migration. PLoS Genet. 2010;6:e1001215. doi: 10.1371/journal.pgen.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–88. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 18.Shakir MA, Gill JS, Lundquist EA. Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics. 2006;172:893–913. doi: 10.1534/genetics.105.046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shakir MA, Jiang K, Struckhoff EC, Demarco RS, Patel FB, Soto MC, Lundquist EA. The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in Caenorhabditis elegans axon guidance. Genetics. 2008;179:1957–71. doi: 10.1534/genetics.108.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struckhoff EC, Lundquist EA. The actin-binding protein UNC-115 is an effector of Rac signaling during axon pathfinding in C. elegans. Development. 2003;130:693–704. doi: 10.1242/dev.00300. [DOI] [PubMed] [Google Scholar]
- 21.Norris AD, Dyer JO, Lundquist EA. The Arp2/3 complex, UNC-115/abLIM, and UNC-34/Enabled regulate axon guidance and growth cone filopodia formation in Caenorhabditis elegans. Neural Dev. 2009;4:38. doi: 10.1186/1749-8104-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demarco RS, Struckhoff EC, Lundquist EA. The Rac GTP exchange factor TIAM-1 acts with CDC-42 and the guidance receptor UNC-40/DCC in neuronal protrusion and axon guidance. PLoS Genet. 2012;8:e1002665. doi: 10.1371/journal.pgen.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–74. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurisu S, Takenawa T. The WASP and WAVE family proteins. Genome Biol. 2009;10:226. doi: 10.1186/gb-2009-10-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundquist EA, Herman RK, Shaw JE, Bargmann CI. UNC-115, a conserved protein with predicted LIM and actin-binding domains, mediates axon guidance in C. elegans. Neuron. 1998;21:385–92. doi: 10.1016/S0896-6273(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 26.Ho HY, Rohatgi R, Lebensohn AM, Le Ma, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–16. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–81. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–7. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- 29.Papakonstanti EA, Stournaras C. Tumor necrosis factor-alpha promotes survival of opossum kidney cells via Cdc42-induced phospholipase C-gamma1 activation and actin filament redistribution. Mol Biol Cell. 2004;15:1273–86. doi: 10.1091/mbc.E03-07-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slater SJ, Seiz JL, Stagliano BA, Stubbs CD. Interaction of protein kinase C isozymes with Rho GTPases. Biochemistry. 2001;40:4437–45. doi: 10.1021/bi001654n. [DOI] [PubMed] [Google Scholar]
- 31.Quilliam LA, Lambert QT, Mickelson-Young LA, Westwick JK, Sparks AB, Kay BK, Jenkins NA, Gilbert DJ, Copeland NG, Der CJ. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem. 1996;271:28772–6. doi: 10.1074/jbc.271.46.28772. [DOI] [PubMed] [Google Scholar]
- 32.Chang C, Adler CE, Krause M, Clark SG, Gertler FB, Tessier-Lavigne M, Bargmann CI. MIG-10/lamellipodin and AGE-1/PI3K promote axon guidance and outgrowth in response to slit and netrin. Curr Biol. 2006;16:854–62. doi: 10.1016/j.cub.2006.03.083. [DOI] [PubMed] [Google Scholar]
- 33.Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, Meyer T, Heo WD. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell. 2012;47:281–90. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akiyama H, Kamiguchi H. Phosphatidylinositol 3-kinase facilitates microtubule-dependent membrane transport for neuronal growth cone guidance. J Biol Chem. 2010;285:41740–8. doi: 10.1074/jbc.M110.156489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittinghofer F, Krengel U, John J, Kabsch W, Pai EF. Three-dimensional structure of p21 in the active conformation and analysis of an oncogenic mutant. Environ Health Perspect. 1991;93:11–5. doi: 10.1289/ehp.919311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–41. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- 37.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 38.Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–3. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 40.Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6:319–27. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 41.Soto MC, Qadota H, Kasuya K, Inoue M, Tsuboi D, Mello CC, Kaibuchi K. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 2002;16:620–32. doi: 10.1101/gad.955702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/S0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 43.Withee J, Galligan B, Hawkins N, Garriga G. Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics. 2004;167:1165–76. doi: 10.1534/genetics.103.025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman JO, Li H, Lundquist EA. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev Biol. 2008;324:245–57. doi: 10.1016/j.ydbio.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao ZS, Manser E, Lim L. Interaction between PAK and nck: a template for Nck targets and role of PAK autophosphorylation. Mol Cell Biol. 2000;20:3906–17. doi: 10.1128/MCB.20.11.3906-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–63. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen CC, Jeon SM, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, Park Y, Hay N. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell. 2010;18:592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rintelen F, Stocker H, Thomas G, Hafen E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15020–5. doi: 10.1073/pnas.011318098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treins C, Warne PH, Magnuson MA, Pende M, Downward J. Rictor is a novel target of p70 S6 kinase-1. Oncogene. 2010;29:1003–16. doi: 10.1038/onc.2009.401. [DOI] [PubMed] [Google Scholar]
- 50.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–9. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 51.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–9. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 52.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–9. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallo G, Letourneau PC. Axon guidance: GTPases help axons reach their targets. Current biology. 1998;8:R80–2. doi: 10.1016/s0960-9822(98)70051-x. [DOI] [PubMed] [Google Scholar]
- 54.Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knobel KM, Jorgensen EM, Bastiani MJ. Growth cones stall and collapse during axon outgrowth in Caenorhabditis elegans. Development. 1999;126:4489–98. doi: 10.1242/dev.126.20.4489. [DOI] [PubMed] [Google Scholar]
- 56.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–52. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 58.Ridley AJ, Hall A. Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harb Symp Quant Biol. 1992;57:661–71. doi: 10.1101/SQB.1992.057.01.072. [DOI] [PubMed] [Google Scholar]
- 59.Yuan XB, Jin M, Xu X, Song YQ, Wu CP, Poo MM, Duan S. Signalling and crosstalk of Rho GTPases in mediating axon guidance. Nat Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- 60.Aspenström P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J. 2004;377:327–37. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995;23:456–9. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- 62.Stradal TE, Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Aspenström P, Ruusala A, Pacholsky D. Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp Cell Res. 2007;313:3673–9. doi: 10.1016/j.yexcr.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 64.Ng J, Luo L. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 2004;44:779–93. doi: 10.1016/j.neuron.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 65.Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–63. doi: 10.1016/S0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 66.Newsome TP, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson BJ. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–94. doi: 10.1016/S0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- 67.Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F, Mazzarino MC, et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging (Albany NY) 2011;3:192–222. doi: 10.18632/aging.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ménager C, Arimura N, Fukata Y, Kaibuchi K. PIP3 is involved in neuronal polarization and axon formation. J Neurochem. 2004;89:109–18. doi: 10.1046/j.1471-4159.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- 69.Yoshimura T, Arimura N, Kawano Y, Kawabata S, Wang S, Kaibuchi K. Ras regulates neuronal polarity via the PI3-kinase/Akt/GSK-3beta/CRMP-2 pathway. Biochem Biophys Res Commun. 2006;340:62–8. doi: 10.1016/j.bbrc.2005.11.147. [DOI] [PubMed] [Google Scholar]
- 70.Polleux F, Snider W. Initiating and growing an axon. Cold Spring Harb Perspect Biol. 2010;2:a001925. doi: 10.1101/cshperspect.a001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henle SJ, Wang G, Liang E, Wu M, Poo MM, Henley JR. Asymmetric PI(3,4,5)P3 and Akt signaling mediates chemotaxis of axonal growth cones. J Neurosci. 2011;31:7016–27. doi: 10.1523/JNEUROSCI.0216-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan D, Guo L, Wang Y. Requirement of dendritic Akt degradation by the ubiquitin-proteasome system for neuronal polarity. J Cell Biol. 2006;174:415–24. doi: 10.1083/jcb.200511028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma T, Hoeffer CA, Capetillo-Zarate E, Yu F, Wong H, Lin MT, Tampellini D, Klann E, Blitzer RD, Gouras GK. Dysregulation of the mTOR pathway mediates impairment of synaptic plasticity in a mouse model of Alzheimer’s disease. PLoS One. 2010;5:e12845. doi: 10.1371/journal.pone.0012845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaworski J, Sheng M. The growing role of mTOR in neuronal development and plasticity. Mol Neurobiol. 2006;34:205–19. doi: 10.1385/MN:34:3:205. [DOI] [PubMed] [Google Scholar]
- 75.Pópulo H, Soares P, Lopes JM. Insights into melanoma: targeting the mTOR pathway for therapeutics. Expert Opin Ther Targets. 2012;16:689–705. doi: 10.1517/14728222.2012.691472. [DOI] [PubMed] [Google Scholar]
- 76.Hagan GN, Lin Y, Magnuson MA, Avruch J, Czech MPA. A Rictor-Myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol Cell Biol. 2008;28:4215–26. doi: 10.1128/MCB.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDonald PC, Oloumi A, Mills J, Dobreva I, Maidan M, Gray V, Wederell ED, Bally MB, Foster LJ, Dedhar S. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res. 2008;68:1618–24. doi: 10.1158/0008-5472.CAN-07-5869. [DOI] [PubMed] [Google Scholar]
- 78.Agarwal NK, Chen CH, Cho H, Boulbès DR, Spooner E, Sarbassov DD. Rictor regulates cell migration by suppressing RhoGDI2. Oncogene. 2013;32:2521–6. doi: 10.1038/onc.2012.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhagwat SV, Crew AP. Novel inhibitors of mTORC1 and mTORC2. Curr Opin Investig Drugs. 2010;11:638–45. [PubMed] [Google Scholar]
- 80.Lu W, Katz S, Gupta R, Mayer BJ. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/S0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 81.Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, Scita G. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sternberg PW, Han M. Genetics of RAS signaling in C. elegans. Trends Genet. 1998;14:466–72. doi: 10.1016/S0168-9525(98)01592-3. [DOI] [PubMed] [Google Scholar]
- 86.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hodgkin J. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1988. [Google Scholar]
- 88.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–82. doi: 10.1016/S0091-679X(08)61399-0. [DOI] [PubMed] [Google Scholar]
- 89.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–21. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 90.Jin Y, Jorgensen E, Hartwieg E, Horvitz HR. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J Neurosci. 1999;19:539–48. doi: 10.1523/JNEUROSCI.19-02-00539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sambrook J. FEFaM. Molecular Cloning: A Laboratory Manuel. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]