Abstract
Proliferation and differentiation of epidermal keratinocytes are tightly controlled to ensure proper development and homeostasis of the epidermis. The Ras family of small GTPases has emerged as a central node in the coordination of cell proliferation in the epidermis. Recent genetic evidence from mouse models has revealed that the intensity of Ras signaling modulates the proliferative capacity of epidermal keratinocytes. Interfering with Ras signaling either by combined elimination of the 3 Ras genes from the basal layer of the epidermis or by overexpression of dominant-negative Ras isoforms caused epidermal thinning due to hypoproliferation of keratinocytes. In contrast, overexpression of oncogenic Ras mutants in different epidermal cell layers led to hyperproliferative phenotypes including the development of papillomas and squamous cell carcinomas. Here, we discuss the value of loss- and gain-of-function studies in mouse models to assess the role of Ras signaling in the control of epidermal proliferation.
Keywords: Ras, mouse models, hypoproliferation, epidermis, squamous cell carcinoma, hyperproliferation
Introduction
Tight regulation of cell proliferation is critical for tissue homeostasis in embryonic as well as in postnatal life. Unbalanced proliferation could result either in tissue atrophy in the case of insufficient proliferation, or in tumor development in case of unscheduled growth. The skin epidermis is a tissue characterized by a high rate of cellular turnover that requires strict control of cell proliferation.1 The stratified epidermis is the outermost structure of the skin and protects organisms against water loss and prevents inclusion of toxic agents as well as microorganisms. This protection is achieved by the formation of a non-permeable barrier consisting of a large, intercalated network of keratinocytes.2,3 The epidermis consists of various layers of keratinocytes characterized by differential proliferative capacities as well as varying degrees of differentiation.2-4 The basal layer (stratum basale) is highly proliferative and constantly gives rise to new basal cells as well as more differentiated suprabasal cells (stratum spinosum) through balanced symmetric and asymmetric cells divisions, respectively. Whereas basal keratinocytes are characterized by expression of keratins 5 and 14, suprabasal cells display a shift toward expression of keratins 1 and 10, thereby forming a more robust keratin network accompanied by an increase in cell-cell junctions.1 As these cells further differentiate to form the granular layer (stratum granulosum), the process of stratification continuously proceeds to generate layers of dead, enucleated cells forming the corneal envelope (stratum corneum).2,4 Cells of the granular and corneal layers gradually reduce expression of keratins 1 and 10 and start to produce other proteins such as involucrin, loricrin, or filaggrin, thereby further contributing to establish a non-permeable barrier.2-5
Although the structural and morphological traits of epidermal development have been well characterized, the molecular pathways controlling these processes are only beginning to emerge.6 Accumulating evidence obtained from mouse models suggests that the Ras family of small GTPases plays a fundamental role in these processes. In mammals, Ras proteins are encoded by three independent loci, H-Ras, N-Ras and K-Ras.7,8 Genetic studies have indicated that H-Ras and N-Ras, either individually or in combination, are dispensable for mouse development and tissue homeostasis. In contrast, K-Ras is an essential gene and mice lacking this locus die between 12 and 14 d of gestation due to anemia and liver defects.8-11 However, expression of H-Ras from the K-Ras locus rescues these defects and supports embryonic development and adult homeostasis. Thus, suggesting that Ras isoforms perform redundant functions and that their unique properties are largely due to tissue distribution and/or expression levels.12 In this article we will discuss the role of Ras signaling in epidermal biology and tumorigenesis based on evidence derived from genetic studies in mouse models.
Ras Signaling in Epidermal Development and Homeostasis
In vivo genetic analyses of the role of Ras signaling in epidermal biology has been challenging due to the high redundancy of the different Ras isoforms. Dajee et al. addressed this issue by expressing a dominant-negative H-RasS17N mutant that presumably blocks signaling from all Ras isoforms under control of the keratin 14 promoter in mice.13 This study demonstrated that interfering with Ras signaling in the highly proliferative basal layer caused epidermal thinning and hyperkeratosis. Interestingly, the authors also observed a striking increase in more differentiated suprabsal cells. Likewise, overexpression of H-RasS17N in cultured keratinocytes diminished their proliferative capacity, thus suggesting a relevant role for Ras signaling in the maintenance of keratinocyte proliferation.
Dissecting the role of individual Ras isoforms by gene knockout studies is a difficult task due to the high redundancy among Ras GTPases. Mice deficient for H-Ras and N-Ras did not show any abnormalities in the skin, suggesting that K-Ras expression is sufficient for epidermal development and to maintain tissue homeostasis.11 Moreover, ubiquitous deletion of K-Ras in adult mice did not induce significant defects in the skin (our unpublished observations).14,15 Thus, it seems reasonable to assume that any of the Ras isoforms might be able to sustain cell proliferation in the epidermis. To determine whether Ras signaling is required for epidermal development, we eliminated all three Ras isoforms from the epidermis by generating a compound strain deficient for H-Ras and N-Ras loci that harbored conditional K-Ras alleles. In these mice, specific ablation of K-Ras from the epidermis was achieved by breeding this strain to mice expressing a Cre recombinase under the control of the keratin 5 promoter.16 In this model, Cre expression was turned on during embryonic development in the basal layer of the epidermis, thus leading to complete ablation of K-Ras protein expression by midgestation. Elimination of all 3 Ras loci from the epidermis was not compatible with postnatal life, indicating that Ras proteins provide essential functions in epidermal homeostasis. Indeed, combined deficiency of H-Ras, N-Ras and K-Ras was associated with epidermal thinning and a dramatic decrease in proliferation of epidermal keratinocytes.17
Removal of all Ras isoforms from keratinocytes in vitro also caused cell cycle arrest. Interestingly, cell cycle arrest, both in vitro and in vivo, was accompanied by downregulation of c-Myc and ΔNp63, 2 well-known regulators of proliferation recognized to play vital roles in epidermal homeostasis and development.18,19 The regulation of c-Myc by Ras signaling has been studied in great detail, and therefore, it was not surprising that c-Myc was absent in cells lacking Ras molecules.20 ΔNp63 on the other hand is the most abundant isoform (> 99%) expressed from the p63 locus in keratinocytes as well as in other epithelial cell types.21 Mice lacking p63 display severe defects in epidermal morphogenesis which are partially rescued by overexpression of ΔNp63, thus indicating that ΔNp63 is critical for keratinocyte proliferation.22 Given the similarities between the phenotypes observed in keratinocytes lacking ΔNp63 and the three Ras isoforms, it seems reasonable to propose that Ras signaling might directly regulate expression of ΔNp63.
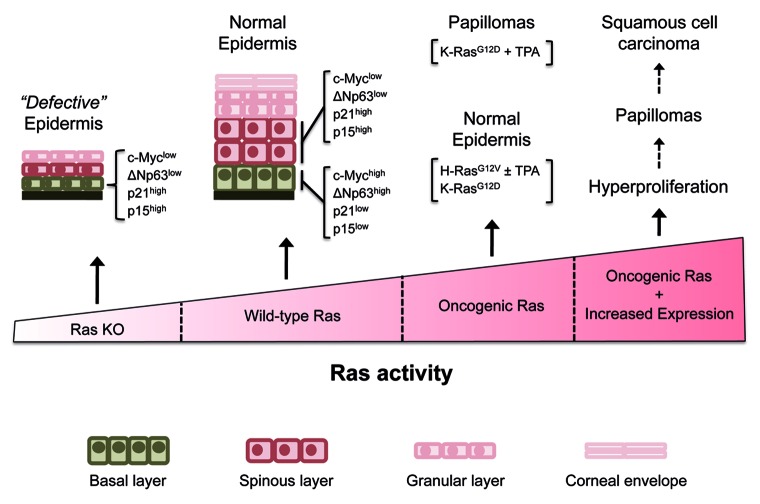
In the absence of Ras signaling, we also observed a striking increase in the expression of the cell cycle regulators p21Cip1 and p15INK4b in the basal layer of the epidermis. Similar results were obtained in cultured keratinocytes.17 Both proteins are known to act as inhibitors of cyclin-dependent kinase complexes involved in cell cycle progression. Early work has established p21Cip1 as a mediator of cell cycle arrest and induction of differentiation in keratinocytes.23,24 Accordingly, p21Cip1 levels were undetectable in the highly proliferative basal layer of the epidermis and were subsequently induced upon asymmetric cell division in the suprabasal layer.17 In contrast, we detected strong p21Cip1 expression in the basal layer of the epidermis in the absence of Ras expression. Interestingly, both c-Myc and ΔNp63 have previously been implicated as negative regulators of p21Cip1, thus suggesting that the absence of c-Myc and/or ΔNp63 may contribute to p21Cip1 induction and subsequently, to cell cycle arrest.25,26 Similarly, p15INK4b, which displayed an expression pattern similar to that of p21Cip1 in cells of the basal layer was subject to repression by c-Myc.27 These observations suggest that Ras signaling is critical for keratinocyte proliferation, possibly through positive stimulation of c-Myc and ΔNp63 expression which, in turn, prevent the accumulation of the cell cycle inhibitors p21Cip1 and p15INK4b (Fig. 1).
Figure 1. Ras activity and epidermal proliferation. Genetic elimination of H-Ras, N-Ras and K-Ras from epidermis (Ras KO) causes a defective epidermis characterized by low levels of expression of c-Myc and ΔNp63 and high expression levels of p21Cip1 and p15INK4b in the basal layer. In contrast, normal epidermis (wild-type Ras) expresses high levels of c-Myc and ΔNp63 in the basal but not in the suprabasal layer. An opposite pattern of expression is shown by p21Cip1 and p15INK4b which are highly expressed in the suprabasal layer but not in the basal layer. In most scenarios, expression of endogenous oncogenic H-Ras and K-Ras mutants in the epidermis do not affect epidermal development. However, when oncogenic Ras mutants are expressed at unphysiologically elevated levels in transgenic mice, they efficiently induce papillomas and squamous cell carcinomas. See text for details.
Previous genetic studies have indicated that Ras proteins mediate proliferative signaling through the Raf/Mek/Erk pathway.28,29 In vitro data from cultured keratinocytes lacking Ras expression also confirmed these observations.17 Ectopic expression of a constitutively active Erk2 kinase allowed keratinocytes to proliferate in the absence of Ras signaling. Furthermore, constitutive Erk signaling rescued c-Myc/ΔNp63 expression leading to inhibition of p21Cip1/p15INK4b. Likewise, genetic elimination of both Mek isoforms, which are crucial for Erk activation, caused hypoproliferation of keratinocytes in vivo, thus indicating that the linear Ras-Raf-Mek-Erk cascade plays a fundamental role in controlling keratinocyte proliferation.30
Although blocking Ras signaling by overexpression of dominant-negative Ras caused an increase in differentiated suprabasal cells, genetic elimination of the three Ras loci from the epidermis resulted in delayed appearance of differentiation markers.13,17 Disruption of both Mek isoforms displayed a phenotype similar to the one observed after Ras elimination.30 These observations suggest that interfering with Ras or its downstream target Mek in the epidermis results in hypoproliferation accompanied by defects in differentiation and possibly also barrier formation.30 The reason for the increase in differentiated suprabasal cells observed in the epidermis expressing dominant-negative Ras is currently unknown, but might involve altered signaling capacities of Ras-independent pathways, probably affecting those driven by other members of the Ras superfamily of proteins. However, elimination of the Ras loci from keratinocytes in vitro did cause induction of suprabasal marker proteins, thus indicating that arrested keratinocytes in vitro do not exactly recapitulate the behavior of keratinocytes in the basal layer of the epidermis, at least after elimination of the three Ras loci. The appearance of senescence markers in vitro, but not in vivo, further supports this hypothesis.17
Taken together, these results indicate that elimination of all Ras isoforms, or their downstream targets such as the Mek kinases, strongly interferes with the tightly regulated control of proliferation in the epidermis; thus, indicating that Ras proteins perform essential functions in epidermal development. Furthermore, these observations demonstrate that mitogenic signals cannot be effectively transmitted in their absence, thereby ultimately causing severe hypoproliferation of epidermal keratinocytes.
Ras Signaling in Epidermal Tumorigenesis: Transgenic Mice
There is ample evidence for a causative role of deregulated Ras signaling in tumor development.31 To date, mutations in one of the RAS genes have been detected in at least 16% of all human cancers, although the actual number of tumors harboring a constitutively active RAS pathway might be significantly higher, given that activating mutations were found in upstream regulators such as the EGFR, or downstream targets such as the B-RAF and MEK kinases.32 In keratinocyte-derived skin cancer, such as squamous cell carcinoma, RAS mutations occur in up to 22% of cases, with H-RAS being the most frequently mutated locus followed by K-RAS.33 Likewise, it has been observed that RAS GTPases are constitutively activated in the majority of squamous cell carcinomas in spite of carrying wild-type RAS alleles.33
To study the role of Ras signaling in skin cancer, researchers have been taking advantage of mouse models for many years. Initially, it was demonstrated that the chemical carcinogen 7,12-dimethylbenz(a)anthracene (DMBA) induced mutations in H-Ras and initiated skin tumors in combination with the inflammation promoter 12-O-tetradecanoylphorbol-13-acetate (TPA), often accompanied by amplification of the H-Ras locus.34,35 Later on, a number of transgenic strains were developed that specifically expressed H-Ras or K-Ras oncogenes in the epidermal compartment. Development of transgenic strains constitutively expressing H-Ras or K-Ras oncogenes under control of strong promoters active in the basal layer, such as the keratin 5 or keratin 14 promoters, resulted in embryonic lethality.33 To avoid this drawback, one group used a truncated keratin 5 promoter that displayed a patchy, restricted expression pattern, being mostly active only in hair follicles and a few cells of the interfollicular epidermis.36 However, despite its restricted expression pattern, expression of an H-Ras oncogene was sufficient to induce papillomas that occasionally progressed to carcinomas even in the absence of a tumor promoter.36 Other strategies such as expression of an inducible H-Ras oncogene fused with the ligand binding domain of the estrogen receptor from the keratin 14 promoter or a K-Ras oncogene under control of an inducible keratin 5 promoter also yielded hyperproliferative phenotypes ranging from benign papillomas to invasive squamous cell carcinomas even in the absence of tumor-promoting inflammatory agents.37,38 Likewise, infection of keratinocytes in mice with retroviruses expressing H-Ras oncogenes yielded benign papillomas that occasionally even progressed to carcinomas, but only when promoted with TPA.39
Targeting Ras oncogene expression to suprabasal layers by using the keratin 1 or keratin 10 promoters for transgene expression did not per se result in tumor formation, but rather required a second stimulus such as a wound or inflammation.40,41 Another transgenic strain that expressed an oncogenic H-Ras transgene under control of the mouse ζ-globin promoter developed benign papillomas as well as malignant squamous carcinomas, but only when treated with the tumor promoter phorbol-12-myristate-13-acetate (PMA).42 Other, indirect strategies to activate Ras signaling, such as expression of a constitutively active form of the guanine nucleotide exchange factor SOS under control of the keratin 5 promoter, yielded severe skin hyperplasia and papilloma development.43 Taken together, these studies indicate that overexpression of H-Ras or K-Ras oncogenes can be sufficient to initiate skin carcinogenesis.
Ras Signaling in Epidermal Tumorigenesis: Genetically Engineered Mice
A major limitation of the models described above is that papilloma or tumor development requires ectopic expression of a Ras transgene. More recently, several laboratories have generated mouse strains that harbor oncogenic mutations in their endogenous Ras loci. Expression of an endogenous K-RasG12D oncogene following activation of an inducible Cre recombinase under control of the keratin 5 promoter produced benign papillomas when promoted with TPA.44 Moreover, combining endogenous K-Ras oncogene expression with expression of a mutant p53 allele (R172H) yielded a significant increase in carcinomas after TPA promotion, thus providing evidence for a cooperation between mutated K-Ras and p53 alleles in skin carcinogenesis.44
However, expression of an endogenous H-RasG12V oncogene in the germ line of mice failed to yield any skin phenotype even in homozygosity.45 Moreover, exposure of these animals to repeated TPA treatments also failed to induce papillomas or any other type of tumor growth. However, these mutant mice developed papillomas with the same frequency as their wild-type littermates when treated with DMBA and TPA.45 Interestingly, those papillomas that developed in heterozygous H-RasG12V mice treated with DMBA and TPA harbored mutations in their wild-type allele. No mutations, other than the G12V mutation engineered in the germ line, were present in the H-Ras locus in homozygous H-RasG12V animals. Thus, indicating that DMBA must mutate other loci to initiate skin carcinogenesis in these mice. These observations also suggest that oncogenic H-Ras isoforms are not sufficient to initiate skin carcinogenesis when expressed at endogenous levels, although they may contribute to papilloma formation.45 A similar mouse model generated in another laboratory expressing endogenous H-RasG12V in heterozygosity did develop skin papillomas after a long latency even in the absence of TPA.46 Yet, all papillomas analyzed displayed elevated copy numbers of the mutated H-RasG12V allele, thus suggesting that oncogenic H-Ras needs to be expressed above a certain threshold to be able to induce skin papillomas. Although a rigorous analysis of ectopic oncogenic H-Ras expression levels in the transgenic strains mentioned earlier is lacking, it appears likely that mutated H-Ras might be expressed at higher levels than its endogenous counterpart.
The contribution of Ras signaling to skin carcinogenesis has been further supported by carcinogenesis studies using H-Ras knockout mice.47 Treatment of these mutant mice with the standard DMBA+TPA carcinogenesis protocol resulted in significantly fewer papillomas than wild-type mice. Interestingly, a high proportion of those papillomas that arose in mice lacking H-Ras alleles, carried mutated K-Ras oncogenes, suggesting that K-Ras can substitute for H-Ras in skin carcinogenesis. These observations, taken together, highlight the importance of using mouse models harboring mutations in their endogenous loci, since they might substantially differ from transgenic strains in certain settings, possibly as a result of the lack of proper transcriptional regulation of the artificial transgenes.
Conclusions
From the work generated in various laboratories over the past years, a picture is beginning to emerge where the level of Ras activity has direct consequences on the proliferation of epidermal keratinocytes (Fig. 1). Whereas lack of Ras activity causes hypoproliferation and atrophy of the epidermis, increased Ras activity might lead to hyperproliferation and eventually tumor development in the skin. Yet, the molecular mechanisms as well as the downstream targets of Ras proteins that control epidermal development, homeostasis, and tumor development remain largely unknown. A large array of molecular players and signaling pathways acting in epidermal biology have been identified over the last years with the help of mouse models.35,48 Therefore, it would not be surprising if some of these regulators eventually turned out to be connected to Ras signaling pathways. Nevertheless, genetic studies using mouse models will be an adequate way to verify these signaling networks, especially with regard to the development of new strategies for the treatment of skin cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Our work was supported by grants from the European Research Council (ERC-AG/250297-RAS AHEAD), EU-Framework Programme (LSHG-CT-2007-037665/CHEMORES, HEALTH-F2-2010-259770/LUNGTARGET and HEALTH-2010-260791/EUROCANPLATFORM), Spanish Ministry of Economy and Competitiveness (SAF2011-30173) and Autonomous Community of Madrid (S2011/BDM-2470/ONCOCYCLE) to M.B.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/26905
References
- 1.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–17. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol. 2011;12:565–80. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- 4.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E. Epidermal differentiation: the bare essentials. J Cell Biol. 1990;111:2807–14. doi: 10.1083/jcb.111.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Zhang H, Duan E. Epidermal development in mammals: key regulators, signals from beneath, and stem cells. Int J Mol Sci. 2013;14:10869–95. doi: 10.3390/ijms140610869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 8.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci U S A. 1995;92:1709–13. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–81. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteban LM, Vicario-Abejón C, Fernández-Salguero P, Fernández-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, et al. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21:1444–52. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potenza N, Vecchione C, Notte A, De Rienzo A, Rosica A, Bauer L, Affuso A, De Felice M, Russo T, Poulet R, et al. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 2005;6:432–7. doi: 10.1038/sj.embor.7400397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dajee M, Tarutani M, Deng H, Cai T, Khavari PA. Epidermal Ras blockade demonstrates spatially localized Ras promotion of proliferation and inhibition of differentiation. Oncogene. 2002;21:1527–38. doi: 10.1038/sj.onc.1205287. [DOI] [PubMed] [Google Scholar]
- 14.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–20. doi: 10.1016/S1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 15.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–26. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, Vidal M, Tusell L, Genesca A, Whitaker DA, Melton DW, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–7. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- 17.Drosten M, Lechuga CG, Barbacid M. Ras signaling is essential for skin development. Oncogene 2013; epub. [DOI] [PubMed]
- 18.Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, Metzger D, Chambon P, Hummler E, Beard P, et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev. 2006;20:2024–9. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–97. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King KE, Ha L, Camilli T, Weinberg WC. Delineating molecular mechanisms of squamous tissue homeostasis and neoplasia: focus on p63. J Skin Cancer. 2013;2013:632028. doi: 10.1155/2013/632028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, De Laurenzi V, Spagnoli LG, Catani MV, Ramadan S, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–47. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 23.Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto GP. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996;10:3065–75. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- 24.Di Cunto F, Topley G, Calautti E, Hsiao J, Ong L, Seth PK, Dotto GP. Inhibitory function of p21Cip1/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science. 1998;280:1069–72. doi: 10.1126/science.280.5366.1069. [DOI] [PubMed] [Google Scholar]
- 25.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–60. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 26.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Δ Np63 α phosphoprotein binds the p21 and 14-3-3 σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–9. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 28.Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, Castellano E, Guerra C, Santos E, Barbacid M. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 2010;29:1091–104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasco RB, Francoz S, Santamaría D, Cañamero M, Dubus P, Charron J, Baccarini M, Barbacid M. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19:652–63. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholl FA, Dumesic PA, Barragan DI, Harada K, Bissonauth V, Charron J, Khavari PA. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 2007;12:615–29. doi: 10.1016/j.devcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khavari TA, Rinn J. Ras/Erk MAPK signaling in epidermal homeostasis and neoplasia. Cell Cycle. 2007;6:2928–31. doi: 10.4161/cc.6.23.4998. [DOI] [PubMed] [Google Scholar]
- 34.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz M, Münzel PA, Braeuning A. Non-melanoma skin cancer in mouse and man. Arch Toxicol. 2013;87:783–98. doi: 10.1007/s00204-012-0998-9. [DOI] [PubMed] [Google Scholar]
- 36.Brown K, Strathdee D, Bryson S, Lambie W, Balmain A. The malignant capacity of skin tumours induced by expression of a mutant H-ras transgene depends on the cell type targeted. Curr Biol. 1998;8:516–24. doi: 10.1016/S0960-9822(98)70203-9. [DOI] [PubMed] [Google Scholar]
- 37.Tarutani M, Cai T, Dajee M, Khavari PA. Inducible activation of Ras and Raf in adult epidermis. Cancer Res. 2003;63:319–23. [PubMed] [Google Scholar]
- 38.Vitale-Cross L, Amornphimoltham P, Fisher G, Molinolo AA, Gutkind JS. Conditional expression of K-ras in an epithelial compartment that includes the stem cells is sufficient to promote squamous cell carcinogenesis. Cancer Res. 2004;64:8804–7. doi: 10.1158/0008-5472.CAN-04-2623. [DOI] [PubMed] [Google Scholar]
- 39.Brown K, Quintanilla M, Ramsden M, Kerr IB, Young S, Balmain A. v-ras genes from Harvey and BALB murine sarcoma viruses can act as initiators of two-stage mouse skin carcinogenesis. Cell. 1986;46:447–56. doi: 10.1016/0092-8674(86)90665-3. [DOI] [PubMed] [Google Scholar]
- 40.Bailleul B, Surani MA, White S, Barton SC, Brown K, Blessing M, Jorcano J, Balmain A. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell. 1990;62:697–708. doi: 10.1016/0092-8674(90)90115-U. [DOI] [PubMed] [Google Scholar]
- 41.Greenhalgh DA, Rothnagel JA, Quintanilla MI, Orengo CC, Gagne TA, Bundman DS, Longley MA, Roop DR. Induction of epidermal hyperplasia, hyperkeratosis, and papillomas in transgenic mice by a targeted v-Ha-ras oncogene. Mol Carcinog. 1993;7:99–110. doi: 10.1002/mc.2940070208. [DOI] [PubMed] [Google Scholar]
- 42.Leder A, Kuo A, Cardiff RD, Sinn E, Leder P. v-Ha-ras transgene abrogates the initiation step in mouse skin tumorigenesis: effects of phorbol esters and retinoic acid. Proc Natl Acad Sci U S A. 1990;87:9178–82. doi: 10.1073/pnas.87.23.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sibilia M, Fleischmann A, Behrens A, Stingl L, Carroll J, Watt FM, Schlessinger J, Wagner EF. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102:211–20. doi: 10.1016/S0092-8674(00)00026-X. [DOI] [PubMed] [Google Scholar]
- 44.Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest. 2007;117:1893–901. doi: 10.1172/JCI31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuhmacher AJ, Guerra C, Sauzeau V, Cañamero M, Bustelo XR, Barbacid M. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008;118:2169–79. doi: 10.1172/JCI34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Mitsutake N, LaPerle K, Akeno N, Zanzonico P, Longo VA, Mitsutake S, Kimura ET, Geiger H, Santos E, et al. Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc Natl Acad Sci U S A. 2009;106:7979–84. doi: 10.1073/pnas.0900343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ise K, Nakamura K, Nakao K, Shimizu S, Harada H, Ichise T, Miyoshi J, Gondo Y, Ishikawa T, Aiba A, et al. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene. 2000;19:2951–6. doi: 10.1038/sj.onc.1203600. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Roop DR. Genetically engineered mouse models for skin research: taking the next step. J Dermatol Sci. 2008;52:1–12. doi: 10.1016/j.jdermsci.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]