Abstract
Epithelia consisting of highly polarized columnar cells contribute to many organs during development, including the central nervous system. Epithelial organization is essential for proliferation and differentiation of progenitor cells and subsequent organ morphology and function. Small GTPases of the Rho family are important regulators of cellular morphology and polarity. We recently identified ArhGEF18 as a key regulator of RhoA-Rock2 signaling that is crucial for maintenance of polarity in the vertebrate retinal epithelium. ArhGEF18 is required to maintain apico-basal polarity, localization of tight junctions and cortical actin, thus shaping cellular morphology. Loss of ArhGEF18 activity results in increased proliferation and reduced cell cycle exit. Together, these perturbations result in a severely misshaped embryonic eye, where the stereotype arrangement of retinal cell types is randomized. Our findings reveal an important role for RhoA-Rock2 signaling to maintain apico-basal polarity in retinal progenitor cells, which is essential for subsequent cellular differentiation, morphology and eventually organ function.
Keywords: ArhGEF18, RhoA-Rock2 signaling, apico-basal polarity, vertebrate, retina, teleost, medaka
In the vertebrate retina a stereotype arrangement of 5 neuronal cell types and 1 glial cell type forms 3 nuclear layers.1,2 During embryogenesis specified retinal progenitor cells (RPCs) of the anterior neuroectoderm evaginate from the forebrain to form the optic vesicles.3 Subsequently RPCs then arrange in the pseudo-stratified single-layer neuroepithelium of the optic cup with a distinct apico-basal (a-b) polarity. Each RPC is attached to the apical and basal surface of this single-layer epithelium by cellular processes. The pseudo-stratified appearance is a result of the varied nuclear position along the apico-basal axis. During the cell cycle, nuclei move along the a-b axis in a process called interkinetic nuclear migration (IKNM). Cell fate decision depends at least in part on IKNM.4 Differentiation involves changes in the orientation of the cleavage plane5 and also morphological changes, whereby the cell soma migrates according to its fate to 1 of the 3 nuclear layers followed by dendrite and axon formation.6,7 This eventually results in the multilayered arrangement of specific cell types in the neural retina. The simple architecture and easy accessibility renders the vertebrate retina an ideal model to study neuronal development, differentiation and organ formation.
Teleost genetic model systems such as medaka (Oryzias latipes) and zebrafish (Danio rerio) are especially well suited to study vertebrate development at the organismal and cellular level. Their short generation time and high fecundity allow unbiased large-scale genetic approaches to study gene function. In addition, their embryos are completely transparent, enabling the analysis at cellular or even subcellular resolution in the living organism. It has, therefore, been possible to genetically dissect the mechanisms that underlie retina formation.2,8,9 In zebrafish a number of mutants were identified that affect epithelial a-b polarity of the retinal neuroepithelium, namely heart and soul (prkci), mosaic eyes (epb41l5: FERM domain protein), nagie oko (mpp5e, a homolog of murine Pals1), oko meduzy (Crbs2a), and parachute (cdh2).10-15 The retinal phenotypes of these mutants share several key features, namely partially pigmented and abnormally shaped eyes, perturbed a-b polarity, hyperproliferation of RPCs, and loss of neuronal lamination such that specified retinal cell types localize to abnormal positions.1,2
Small GTPases were shown to play a prominent role in the establishment and maintenance of a-b polarity.16 Cdc42 and Rac1 are required for the formation of apical adherence junctions (AJ) and tight junctions (TJ).17 Furthermore, Cdc42 and Rac1 interact with the PAR complex (Par3/Par6/aPKC), thus regulating a-b polarity.18 Using conditional inactivation it has recently been shown that RhoA plays a role in neuroepithelial development of the murine CNS. Maintenance of AJ and epithelial organization depend on RhoA activity.19,20 Furthermore region specific regulation of progenitor proliferation was observed. In the spinal cord loss of RhoA activity results in premature cell cycle exit,19 whereas in the fore- and midbrain progenitor cells exhibit accelerated proliferation and reduced cell cycle exit.20 In the cerebral cortex conditional knock out of RhoA results in increased proliferation of progenitor cells and perturbations of the actin and microtubule cytoskeleton in radial glial cells causing a defect of neuronal migration.21
Small GTPases are activated by guanine nucleotide exchange factors (GEF) that facilitate the dissociation of GDP. Subsequent binding of GTP induces conformational changes that allow the interaction of GTPases with their effector proteins.22 Dbl-related RhoGEFs contain a dbl homology (DH) domain that interacts with the switch region of the GTPase, followed by a pleckstrin homology (PH) domain with regulatory functions. Vertebrate genomes, including those of teleosts contain over 70 GEFs that regulate the activity of more than 20 GTPases.22,23 This high complexity suggests that vertebrates achieve an intricate regulation of the many functions of small GTPases in a spatio-temporal and cell-specific manner. In view of this, it is important to study the role of small GTPases in the context of the entire organism using animal models. In particular teleost model systems offer the use of unbiased mutational analysis by large-scale mutagenesis screens. For such random mutagenesis screens no previous knowledge of the gene contents and function of a given genome is required. The successful isolation of informative mutations rather relies on a suitable identification of interesting phenotypes that can be employed to a large numbers of individual animals. This allows to identify novel gene functions. We have undertaken chemical mutagenesis to identify genes that affect vertebrate development and retinal development in particular.9
The thus isolated mutant medeka (japanese: large eye) comprises several morphological hallmarks of perturbed retinal a-b polarity.24 The enlarged and rounded eyes are only partially pigmented. The stereotype lamination into 3 nuclear layers is lost and differentiated cells occupy random positions. The cellular architecture is affected such that tight junction associated proteins and components of a-b polarity complexes are mislocalized along the a-b axis. In addition, apical cortical actin localization is affected, and its colocalization with TJs is often lost. By labeling single cells to visualize their morphology, we found that in RPCs cellular morphology is initially normal at a developmental stage when TJs are already mislocalized. Thus, the morphological perturbations are a consequence of mislocalized junctional complexes and perturbed polarity.
We identified the guanine nucleotide exchange factor ArhGEF18 as the affected gene in the medeka mutant. Both alleles that were identified result in the loss of full-length ArhGEF18 protein, indicating that they are amorphic or at least strongly hypomorphic loss-of-function alleles. ArhGEF18 is ubiquitously expressed in the developing wild-type embryo. However, the CNS is differentially affected by loss of ArhGEF18 function: the mutant phenotype is most severe in the neural retina of the developing eye, whereas the brain and spinal cord are less affected with only moderate perturbations of epithelial polarity and actomyosin. Considering the vast number of GEFs in vertebrates it is possible that this region-specific susceptibility is due to partial functional redundancy. Alternatively, it may reflect a differential activation of ArhGEF18, such that the ubiquitously expressed protein is activated in a tissue- and stage-specific manner. In support of this latter hypothesis, heterotrimeric G proteins and the FERM domain containing protein Lulu have been shown to enhance the GEF activity of ArhGEF18, whereas the cytoskeletal GTPase Sept9b negatively regulates it (Fig. 2).25-27
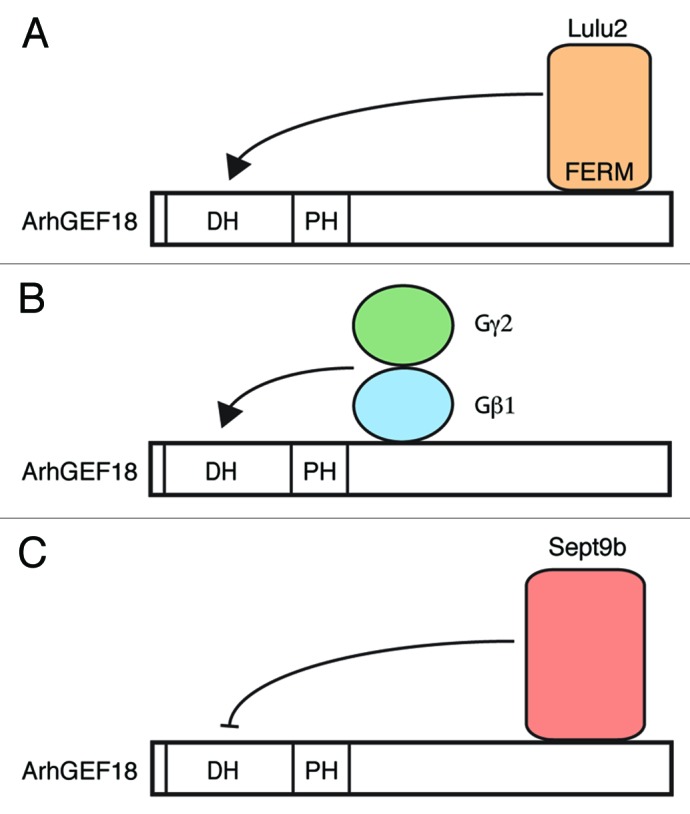
Figure 2. Regulation of ArhGEF18 activity. (A) The apical junction associated protein Lulu2 enhances the GEF activity of ArhGEF18. (B) Interaction of the Gβ1Gγ2 subunit of heterotrimeric G proteins with ArhGEF18 also enhances the GEF activity. The interaction domain of ArhGEF18 with Gβ1Gγ2 proteins has not been determined in detail. (C) The cytoskeletal GTPase Sept9b interacts with the C terminus of ArhGEF18 and negatively regulates the GEF activity.
It is interesting to note that similar to the medeka phenotype also in the zebrafish a-b polarity mutants mosaic eyes, nagie oko, and oko meduzy epithelial polarity in the retina is more severely affected than in the forebrain even though a ubiquitous expression in the CNS has been reported for the respective genes.12,13,28 The gene products of mosaic eyes, nagie oko, and oko meduzy directly interact and are part of the apical Crumbs complex.29 On the other hand, there is no evidence that ArhGEF18 is part of this complex or directly interacts with it.24-27,30-32 Thus, it is unlikely that the shared phenotype of these zebrafish and medaka mutations is due to a higher sensitivity of the retinal epithelium to perturbations of the Crbs complex. Also, it is unlikely that the functional redundancies of the zebrafish mosaic eyes, nagie oko, and oko meduzy genes and that of the medeka gene in the diverged teleost medaka vary similarly between different regions of the developing CNS. Therefore, this shared phenotype of mutations that affect different genes in different species suggests that the developing neural retina is more sensitive to genetic perturbations of a-b polarity. In contrast to the retina, the teleost neural tube develops as a mirror image arrangement of 2 pseudo-stratified epithelia separated by the midline.33 The retinal epithelium on the other hand is abutting that of the retinal pigmented epithelium consisting of thin squamous epithelial cells. It is tempting to speculate that this special architecture of the neural tube with 2 identical epithelia with mirror image arrangement could confer a higher stability with respect to a-b polarity. This could involve structural and morphological features provided by the direct contact of apical sides of progenitor cells of the opposing epithelia.
Loss of ArhGEF18 function in various cell culture systems results in abnormal cellular morphology and concomitant perturbed actomyosin localization, whereas an effect on TJ maintenance is cell type specific.26,31,32 Thus, it appears that control of actomyosin through Rho-Rock is a more general and less cell-type specific activity of ArhGEF18. It is interesting to note that ArhGEF18 depletion in MDCK cells results in abnormal cellular morphology whereas polarity per se is maintained.31 Therefore, control of epithelial polarity by ArhGEF18 also appears to be cell-type dependent. The cell-type specific susceptibility to the loss of ArhGEF18 function and the cell-type specific repertoire of its function underscores the importance of animal models to study this complex situation.
In teleosts retinal differentiation is initiated in the central neural retina and proceeds from there toward the periphery in a wave like fashion.34 As a result of this differentiation wave, there is a developmental gradient across the retina, where cells of the central retina are developmentally more advanced than peripheral cells. The peripheral most portion of the retina, the ciliary marginal zone (CMZ), contains continuously dividing stem cells which provide new progenitor cells that enable live long growth of the retina.35 We noted that a-b polarity and TJ localization are less affected in newly formed progenitor cells of the CMZ than in the more central portion where older cells reside. Furthermore, mutant embryos are initially phenotypically normal. Thus, ArhGEF18 is required for the maintenance but not initiation of polarity and TJs. The yolk-rich eggs of many teleosts, including medaka and zebrafish, contain maternally supplied RNAs and proteins. This maternal contribution may compensate for the loss of gene function during early developmental stages in homozygous mutant embryos. It is therefore possible that ArhGEF18 may have an early role, for example in the establishment of epithelial polarity that is not detectable in zygotic medeka homozygous mutant embryos. However, western blot analysis revealed only very low levels of ArhGEF18 protein at 1dpf in wild-type embryos when a-b polarity is established. Protein levels peak 30 h later, when the mutant phenotype becomes first apparent. Even though this finding does not rule out that ArhGEF18 may also function in the establishment of a-b polarity, it suggests that the main role of ArhGEF18 is maintenance of a-b polarity, which is consistent with its maintenance function in the CMZ. In some Ca-switch experiments the formation of TJs was impaired when ArhGEF18 was downregulated, indicating a requirement for TJ maturation in these in vitro conditions.31
We used the mutant ArhGEF18 loss-of-function background for a deletion analysis to test individual domains for their contribution to wild-type activity. Our analysis showed that the DH and PH domains which are responsible for activation of small GTPases are required for a phenotypic rescue of mutant embryos, indicating that with respect to polarity and TJ formation the GEF activity of ArhGEF18 is indispensable.
We and others found that ArhGEF18 activates both RhoA and Rac1.25,26,31,36 We therefore used dominant-negative mutants of RhoA and Rac1 to test their contribution to regulating polarity, cortical actin and TJ localization. Expression of dominant-negative RhoA but not dominant-negative Rac1 phenocopied all aspects of the medeka phenotype, indicating that ArhGEF18-mediated RhoA activation is crucial for normal epithelial development. Since expression of the dominant-negative version of the RhoA effector Rock2 similarly phenocopied the mutant we conclude that ArhGEF18 regulates the RhoA-Rock2 signaling pathway to control epithelial polarity, cortical actin and TJ localization. In good agreement with our findings ArhGEF18 activates Rho-Rock2 also in corneal epithelial cells to regulate the actin cytoskeleton and TJ maintenance.31 Conditional RhoA knock out analysis in the mouse showed that RhoA is required for epithelial organization and maintenance of AJ in the fore-/midbrain as well as the spinal cord.19,20 It has been suggested that RhoA functions through mDia to control maintenance of AJ and epithelial polarity of the murine fore- and midbrain.19 Thus, RhoA may signal through different effector proteins to regulate epithelial organization.
We have used GFP-tagged ArhGEF18 to visualize its subcellular localization in vivo. Since GFP-ArhGEF18 rescues the medeka mutation with efficiency comparable to the wild-type protein, we conclude that GFP does not interfere with protein function. We noted that GFP-ArhGEF18 is apically enriched with a cytoplasmic pool in neuroepithelial cells of live medaka embryos. This apically enriched localization hints at a graded activation of RhoA. A similar subcellular distribution has been reported in vitro for human ArhGEF18 in epithelial cell lines26,31 and a corresponding local activation of RhoA has been shown.31 In Drosophila epithelial invagination by apical constriction requires apical RhoGEF2 and RhoGEF64C to locally activate Rho1.37 Also in vertebrate lens invagination, apical constriction depends on apical RhoA activation by the GEF Trio, for which apical localization has been suggested.38 Thus, subcellularly-restricted activation of Rho GTPases by localized GEFs appears to be crucial for different aspects of epithelial development. A number of proteins have been identified that interact with and apically localize ArhGEF18 (Fig. 1). ArhGEF18 interacts with the apical proteins Patj, Par326, and LKB132 and has been detected in the TJ-associated complex of cingulin, Rock2, and myosin2.31 In addition to subcellular localization of ArhGEF18, also interaction with localized modulators of GEF activity lead to graded activation of RhoA. ArhGEF18 interacts with the apical FERM domain containing protein Lulu2 (epb41l4b, a mammalian homolog of zebrafish mosaic eyes) which stimulates ArhGEF18 to activate RhoA (Fig. 2).26 Thus, there are different modes with which ArhGEF18 is regulated to locally activate RhoA.
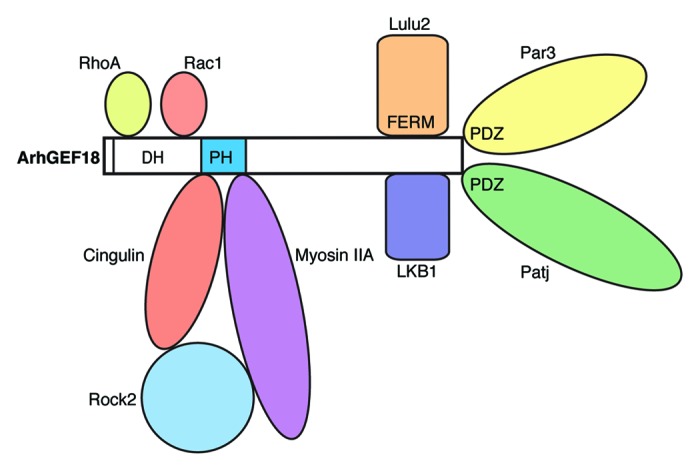
Figure 1. ArhGEF18 interacting proteins. The DH domain of ArhGEF18 binds RhoA and Rac1 and thereby catalyzes the exchange of GDP for GTP. Myosin IIA and the TJ associated protein cingulin bind to the PH domain. LKB1 and the FERM domain of Lulu2 bind to the C-terminal region. Par3 and Patj bind to the PDZ binding motif via their PDZ domains. Lulu2, LKB1, Par3, Patj, and the Cingulin/Myosin/Rock protein complex are associated with apical junctions and can thereby mediate apical localization of ArhGEF18.
Importantly, ArhGEF18 also regulates the proliferation and differentiation of epithelial cells. In the medeka mutant, progenitor cells show a dramatic increase in proliferation. Cell cycle analysis showed that the progression from S- to M-phase is normal. However the balance of proliferative to neurogenic divisions is severally perturbed. Cell cycle exit of mutant progenitor cells at the onset of neurogenesis is dramatically reduced.
As a consequence the mutant central retina, where in the wild-type neurogenesis is initiated, is occupied by proliferating progenitor cells and neurogenesis is delayed. Furthermore M-phase nuclei of dividing progenitor cells are detected at ectopic locations along the a-b axis, whereas in wild type they are apically localized. Also in the mouse embryo Rho signaling regulates proliferation of progenitor cells in the CNS. In the cerebral cortex loss of RhoA function results in transient increase of progenitor cell proliferation.21 In the fore- and mid-brain RhoA inactivation causes hyperproliferation and a decrease of cell cycle exit,20 similar to the loss of ArhGEF18 function in medaka retinal progenitor cells. Interestingly in the spinal cord RhoA inactivation has the opposite effect. There, progenitor cells exit the cell cycle prematurely and undergo precocious differentiation.19 Thus, Rho signaling plays an important role in regulating proliferation of progenitor cells in the vertebrate CNS. It has recently been shown that the Hippo pathway which controls tissue growth interacts with epithelial polarity.39 Furthermore F-actin can regulate the Hippo pathway and by that influence epithelial proliferation.40 Since loss of ArhGEF18 function severely perturbs the cortical actin cytoskeleton it is therefore possible that resulting changes in the F-actin may affect the Hippo pathway and by that cause to the observed hyperproliferation at least partially.
In summary, the analysis of the medeka mutant uncovered a key regulator of RhoA that is essential for epithelial development in the CNS. Cellular polarity, morphology and proliferation of the neural retina are perturbed by loss of ArhGEF18 function. Our analysis shows that ArhGEF18 functions as GEF to activate the Rho-Rock2 signaling pathway in this context. ArhGEF18 also regulates the cortical actin cytoskeleton and TJ in the retinal epithelium. We thus reveal a key role for ArhGEF18 regulated Rho signaling in maintaining epithelial polarity and integrity of the developing vertebrate CNS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Jakub M Swiercz and Clemens Grabher provided helpful comments to the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/27061
References
- 1.Randlett O, Norden C, Harris WA. The vertebrate retina: A model for neuronal polarization in vivo. Dev Neurobiol. 2010;71:567–83. doi: 10.1002/dneu.20841. [DOI] [PubMed] [Google Scholar]
- 2.Pujic Z, Malicki J. Retinal pattern and the genetic basis of its formation in zebrafish. Semin Cell Dev Biol. 2004;15:105–14. doi: 10.1016/j.semcdb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Rembold M, Loosli F, Adams RJ, Wittbrodt J. Individual cell migration serves as the driving force for optic vesicle evagination. Science. 2006;313:1130–4. doi: 10.1126/science.1127144. [DOI] [PubMed] [Google Scholar]
- 4.Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/S0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 6.Hinds JWJ, Hinds PLP. Early ganglion cell differentiation in the mouse retina: an electron microscopic analysis utilizing serial sections. Dev Biol. 1974;37:381–416. doi: 10.1016/0012-1606(74)90156-0. [DOI] [PubMed] [Google Scholar]
- 7.Zolessi FR, Poggi L, Wilkinson CJ, Chien C-B, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–73. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- 9.Loosli F, Del Bene F, Quiring R, Rembold M, Martinez-Morales JR, Carl M, Grabher C, Iquel C, Krone A, Wittbrodt B, et al. Mutations affecting retina development in Medaka. Mech Dev. 2004;121:703–14. doi: 10.1016/j.mod.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–502. doi: 10.1016/S0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 11.Jensen AM, Westerfield M. Zebrafish mosaic eyes is a novel FERM protein required for retinal lamination and retinal pigmented epithelial tight junction formation. Curr Biol. 2004;14:711–7. doi: 10.1016/j.cub.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–7. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- 13.Omori Y, Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol. 2006;16:945–57. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 14.Erdmann B, Kirsch F-P, Rathjen FG, Moré MI. N-cadherin is essential for retinal lamination in the zebrafish. Dev Dyn. 2003;226:570–7. doi: 10.1002/dvdy.10266. [DOI] [PubMed] [Google Scholar]
- 15.Masai I, Lele Z, Yamaguchi M, Komori A, Nakata A, Nishiwaki Y, Wada H, Tanaka H, Nojima Y, Hammerschmidt M, et al. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development. 2003;130:2479–94. doi: 10.1242/dev.00465. [DOI] [PubMed] [Google Scholar]
- 16.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–59. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 17.Samarin S, Nusrat A. Regulation of epithelial apical junctional complex by Rho family GTPases. Front Biosci (Landmark Ed) 2009;14:1129–42. doi: 10.2741/3298. [DOI] [PubMed] [Google Scholar]
- 18.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 19.Herzog D, Loetscher P, van Hengel J, Knüsel S, Brakebusch C, Taylor V, Suter U, Relvas JB. The small GTPase RhoA is required to maintain spinal cord neuroepithelium organization and the neural stem cell pool. J Neurosci. 2011;31:5120–30. doi: 10.1523/JNEUROSCI.4807-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama K, Melendez J, Baumann JM, Leslie JR, Chauhan BK, Nemkul N, Lang RA, Kuan CY, Zheng Y, Yoshida Y. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proc Natl Acad Sci U S A. 2011;108:7607–12. doi: 10.1073/pnas.1101347108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cappello S, Böhringer CRJ, Bergami M, Conzelmann KK, Ghanem A, Tomassy GS, Arlotta P, Mainardi M, Allegra M, Caleo M, et al. A radial glia-specific role of RhoA in double cortex formation. Neuron. 2012;73:911–24. doi: 10.1016/j.neuron.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 22.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 23.Salas-Vidal E, Meijer AH, Cheng X, Spaink HP. Genomic annotation and expression analysis of the zebrafish Rho small GTPase family during development and bacterial infection. Genomics. 2005;86:25–37. doi: 10.1016/j.ygeno.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Herder C, Swiercz JM, Müller C, Peravali R, Quiring R, Offermanns S, Wittbrodt J, Loosli F. ArhGEF18 regulates RhoA-Rock2 signaling to maintain neuro-epithelial apico-basal polarity and proliferation. Development. 2013;140:2787–97. doi: 10.1242/dev.096487. [DOI] [PubMed] [Google Scholar]
- 25.Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya T. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: regulation of cell shape and reactive oxygen species production. Circ Res. 2003;93:848–56. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima H, Tanoue T. Lulu2 regulates the circumferential actomyosin tensile system in epithelial cells through p114RhoGEF. J Cell Biol. 2011;195:245–61. doi: 10.1083/jcb.201104118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata K, Inagaki M. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene. 2005;24:65–76. doi: 10.1038/sj.onc.1208101. [DOI] [PubMed] [Google Scholar]
- 28.Jensen AM, Walker C, Westerfield M. mosaic eyes: a zebrafish gene required in pigmented epithelium for apical localization of retinal cell division and lamination. Development. 2001;128:95–105. doi: 10.1242/dev.128.1.95. [DOI] [PubMed] [Google Scholar]
- 29.Hsu Y-C, Willoughby JJ, Christensen AK, Jensen AM. Mosaic Eyes is a novel component of the Crumbs complex and negatively regulates photoreceptor apical size. Development. 2006;133:4849–59. doi: 10.1242/dev.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuji T, Ohta Y, Kanno Y, Hirose K, Ohashi K, Mizuno K. Involvement of p114-RhoGEF and Lfc in Wnt-3a- and dishevelled-induced RhoA activation and neurite retraction in N1E-115 mouse neuroblastoma cells. Mol Biol Cell. 2010;21:3590–600. doi: 10.1091/mbc.E10-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terry SJ, Zihni C, Elbediwy A, Vitiello E, Leefa Chong San IV, Balda MS, Matter K. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat Cell Biol. 2011;13:159–66. doi: 10.1038/ncb2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, Jin D, Durgan J, Hall A. LKB1 controls human bronchial epithelial morphogenesis through p114RhoGEF-dependent RhoA activation. Mol Cell Biol. 2013;33:2671–82. doi: 10.1128/MCB.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JD. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature. 2007;446:797–800. doi: 10.1038/nature05722. [DOI] [PubMed] [Google Scholar]
- 34.Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- 35.Centanin L, Hoeckendorf B, Wittbrodt J. Fate restriction and multipotency in retinal stem cells. Cell Stem Cell. 2011;9:553–62. doi: 10.1016/j.stem.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Blomquist A, Schwörer G, Schablowski H, Psoma A, Lehnen M, Jakobs KH, Rümenapp U. Identification and characterization of a novel Rho-specific guanine nucleotide exchange factor. Biochem J. 2000;352:319–25. doi: 10.1042/0264-6021:3520319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simões S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Hombría JC, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–67. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- 38.Plageman TF, Jr., Chauhan BK, Yang C, Jaudon F, Shang X, Zheng Y, Lou M, Debant A, Hildebrand JD, Lang RA. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development. 2011;138:5177–88. doi: 10.1242/dev.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–24. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 40.Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, Halder G. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–35. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]


