Abstract
A mono-specific antibody may recruit a second antigen binding specificity, thus converting to a dual-specific Two-in-One antibody through mutation at the light chain complementarity-determining regions (CDRs). It is, however, unknown whether mutation at the heavy chain CDRs may evolve such dual specificity. Herein, we examined the CDRs of a humanized interleukin 4 (IL4) antibody using alanine scanning and structural modeling, designed libraries of mutants in regions that tolerate mutation, and isolated dual specific antibodies harboring mutation at the heavy chain CDRs only. We then affinity improved an IL4/IL5 dual specific antibody to variants with dissociation constants in the low nanomolar range for both antigens. The results demonstrate the full capacity of antibodies to evolve dual binding specificity.
Keywords: dual specific antibody, bispecific antibody, Dual Action Fab, phage display, IL5
In the past two decades, there has been great interest in developing therapeutic antibodies that can target more than one molecule.1,2 Two-in-One antibody technology, unlike many other bispecific formats, provides an option for dual targeting antibody therapeutics that can be manufactured in regular IgG or Fab formats.3,4 The proof-of concept-study for the engineering strategy to generate a Two-in-One antibody was the conversion of a human epidermal growth factor receptor 2 (HER2)-binding antibody fragment (trastuzumab Fab) into variants that could bind vascular endothelial growth factor (VEGF) in addition to HER2.5,6 The dual specific variants of trastuzumab (Herceptin®) were isolated from a phage-displayed library of trastuzumab Fab with mutations in the light chain (LC)1 complementarity-determining regions (CDRs). The rationale behind this approach was that trastuzumab, like many natural antibodies, utilizes heavy chain (HC) CDRs as the dominant player in antigen binding and can tolerate some mutations in the LC CDRs. Structural and mutagenesis studies showed that the dual action Fab variant of trastuzumab maintained the structural epitope on HER2 of trastuzumab, while many HER2-contacting CDR residues also interacted with VEGF, which is structurally unrelated to HER2.5 Each dual specific Fab therefore interacts with one of its two antigens at a time. As an IgG with two Fab arms, a Two-in-One antibody is capable of binding both antigens simultaneously. We expect that many monospecific antibodies can be engineered to evolve dual specificity through mutations in the LC CDRs using similar engineering approaches.
The second example of engineering such a Two-in-One antibody is the development of an epidermal growth factor receptor (EGFR)/HER3 dual targeting antibody that is now in clinical trials for treating epithelial-derived cancer.7 It should be noted that the parental antibodies, trastuzumab and an anti-EGFR antibody, of both Two-in-One antibodies are identical in their framework subtypes (VH3, VLkappa1). To extend the engineering strategy, it will be useful to demonstrate the recruitment of dual specificity for an antibody of different framework subtypes. Further, we wanted to determine whether mutation in HC CDRs, instead of LC CDR, can evolve such dual specific antibodies. Some antibodies may depend heavily on LC CDRs for primary antigen binding energy;8 thus, mutation in the LC CDRs of these antibodies will likely disrupt binding to their primary antigens. We selected an IL4 antibody that was humanized to VH1/VLkappa1 frameworks and demonstrated a mutational scanning strategy to identify HC regions of the antigen-binding site that tolerate mutation to allow evolution of a second binding specificity.
We set out to recruit a second antigen binding specificity into a mouse hybridoma-derived antibody, 19C11, which binds human interleukin 4 (IL4) and blocks the interaction of IL4 to the IL4 receptor α.9 This antibody has been humanized to the frameworks of VH1 and VL kappa1 subtype (hu19C11) with high IL4 specific binding affinity (KD = 1nM).9 We cloned the variable domains of hu19C11 into a plasmid construct for Fab phage display as described previously.5,10 The upper hinge region of IgG with amino acid sequence KTHTC was included in between CH1 and M13 major coat protein p3 C-terminus domain to allow bivalent Fab display, which helps to increase the binding of displaying phage to antigens immobilized on solid surface support as previously described.10 We also verified that the Fab-displaying phage, which carried a gD expression tag fused to the C-terminus of LC, bound an anti-gD antibody and bound its primary IL4 antigen with high affinity (phage IC50 = 1nM).
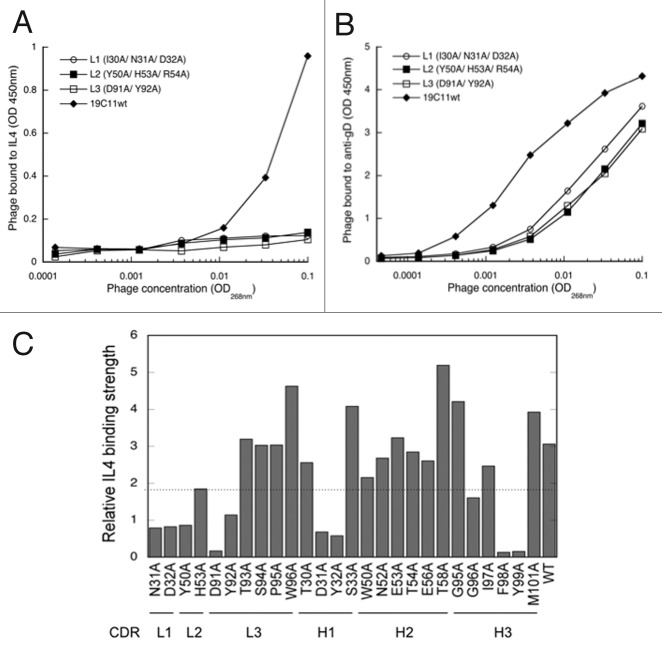
To determine the engineering strategy of recruiting a secondary binding specificity to hu19C11, we first examined the importance of its three LC CDRs for binding IL4 by mutating two or three residues of each LC CDR to alanine. These LC CDR positions were selected because they were identified previously as critical positions for combinatorial mutagenesis in the LC library approach for selecting dual specific clones from antibodies relying heavily on HC CDRs for their primary antigen binding.6 Phage displaying Fab with alanine mutations in any of three LC CDRs exhibited no detectable binding to IL4, but bound to the anti-gD antibody, indicating that these mutants were indeed displayed on phage, albeit with lower level than the wild type Fab, but their binding to IL4 was severely disrupted (Fig. 1A and B). Therefore, the previously described approach of generating randomized LC CDR libraries will not likely produce dual specific antibodies since the number of amino acid residues available for evolving secondary antigen binding is quite limited. We next mutated individual key residues of LC CDRs as well as surface accessible HC CDRs as previously described,11,12 and evaluated their role in IL4 binding by determining the relative binding affinity of these alanine mutants compared with wild type hu19C11. The results verified the importance of those LC CDR residues for IL4 binding. However, all tested positions of CDR H2 and half of the CDR H1 and H3 appeared tolerant to mutation (Fig. 1C).
Figure 1. Mutagenesis mapping of hu19C11 CDRs. To measure the relative antigen binding affinity of hu19C11 Fab variants, binding of serially diluted phage displaying anti-IL4 hu19C11 wild type (wt) or LC CDR alanine mutants to IL4 (A) or to anti-gD antibody (B) coated on ELISA wells was detected by an anti-M13 phage horseradish peroxidase (HRP) conjugate to measure antigen binding and Fab expression, respectively. Single letter codes of amino acids are used. gD is an expression peptide tag fused to the C terminus of the light chain. Anti-gD antibody was directly coated on ELISA wells, whereas IL4 was captured with a non-blocking anti-IL4 antibody coated on ELISA wells. (C) Effects of alanine mutations at individual sites of CDRs were examined by comparing the relative IL4 binding affinity of phage displaying Fab variants by performing assays as (A) and (B). Relative IL4 binding affinity was determined by first fitting the data with a linear regression model then dividing the slope of IL4 binding (vs. phage concentration) with the slope of Fab expression. Low values (below the dotted line) were deemed low IL4 binding affinity relative to hu1911wt indicating disruptive mutations.
Mapping the mutational effects on the modeled structure of hu19C11 indicated an area centered at CDR H2 that tolerated mutation and may be suitable to evolve a second specificity (Fig. 2A). This area includes residues in the CDR H1–3 and CDR L3 that were targeted for randomization. The randomization design was guided by the natural diversity in amino acid composition at these positions, as well as the variability in the length of CDR H2 and CDR L3 of human antibodies.12 The sizes of the libraries after electroporation into E. coli were in the range of 109.
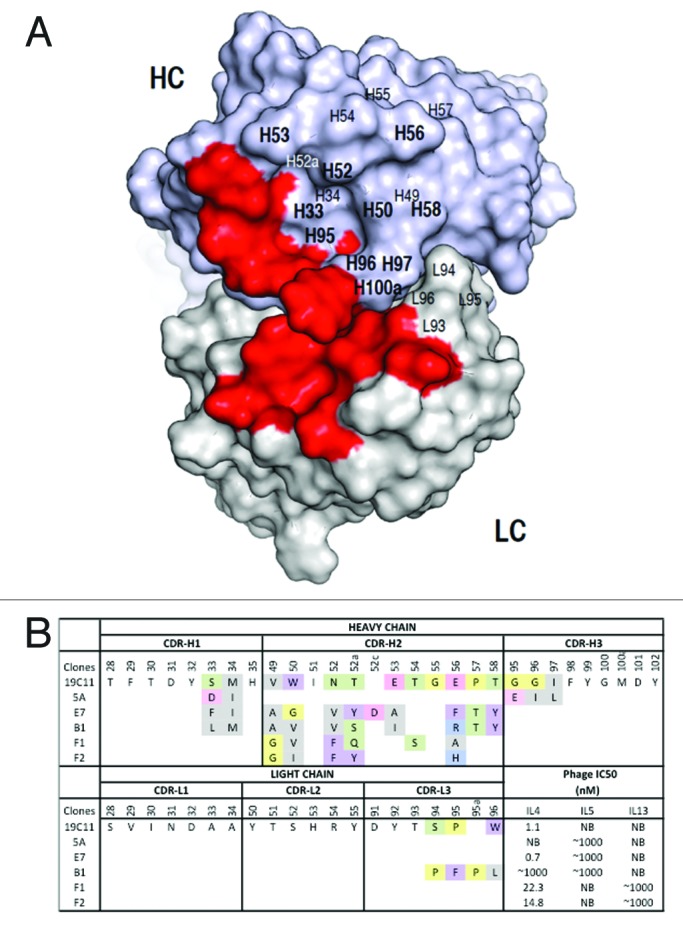
Figure 2. Dual specific clones isolated from phage libraries of h19C11 variants. (A) The structural model of hu19C11 was generated with MOE using the PDB entries 3SQO and 3BE1 as templates for the heavy and light chain, respectively. Residues important for IL4 binding, LC: 31, 32, 50, 53, 91, 92 and HC: 31, 32, 96, 98, 99 (in Kabat numbering), are colored in red on the model structure in a top down view of the antigen-binding site. Libraries of hu19C11 Fab variants displayed on phage were generated by site directed mutation. HC residues that were allowed all 20 amino acids with preference for wild type residues are labeled in bold while other HC and LC residues as labeled were allowed limited mutation to mimic natural diversity. (B) CDR sequences of selected clones including anti-IL5 specific (5A), anti-IL4/5 specific (E7, B1) and anti-IL4/13 specific (F1, F2) are shown with mutations from the parent hu19C11wt. To show the change of property upon mutation, sites included in the randomization scheme were colored according to their properties: Y, W, F in purple, hydrophobic L, I, V, A, M in gray, basic K, R, H in blue, acidic D, E in pink, polar S, T, N, Q in green, and P, G in yellow. The relative antigen binding affinity as measured by IC50 of phage competition assays are shown. Fab displaying phage was first incubated with serial dilutions of the respective antigens in solution for 2 h, unbound phage was then briefly captured by antigen coated ELISA wells and detected with anti-M13 HRP conjugate. The concentration of antigen inhibiting 50% of phage binding to antigen coated well is calculated as IC50. NB denotes no detectable binding signal of phage clone to the antigen coated wells.
We chose IL5 and IL13 as the secondary antigens. While these two interleukins, like IL4, belong to the 4-helix bundle cytokine family, they are quite divergent at the level of amino acid sequence and structural organization.13 The sequence identity between IL4 and IL13 is 12%, whereas that of IL4 and IL5 is 11%. Structurally, IL5 forms a unique intertwined homo-dimer with one α helix from one chain forming a 4-helix bundle with three α helixes from the other chain;14,15 IL4 and IL13 are both monomers. However, the three cytokines are related in their biological function, and dual specific antibodies that bind and block two of these cytokines may have potential utility as treatments for allergic diseases such as asthma.16,17
We performed several rounds of panning and enrichment for IL5 or IL13 binding clones that retained IL4 binding from the constructed phage display libraries as previously described.5 By screening approximately 100 clones each, we found 3 to 8 clones that exhibited dual binding to IL5/IL4 or IL13/IL4, respectively. From sequencing, two unique IL4/IL5 binding clones (B1, E7) and two unique IL4/IL13 (F1, F2) binding clones were identified (Fig. 2B). We also isolated clones that bound solely to IL5 (e.g., clone5A). With the exception of clone B1 which contains mutation in both HC and LC, all clones contained mutations only in HC CDRs compared with their mono-specific parent template. This demonstrated that mutation of HC CDRs of a monospecific antibody can confer dual specificity. We used phage binding competition assays to estimate clone affinity as the concentration of interleukins needed to inhibit 50% of Fab displaying phage from binding to immobilized interleukins (IC50). We found that binding to the secondary antigens was weak with IC50 in the micromolar range, while IL4 binding was maintained in the low nanomolar range.
To verify the dual binding specificity, the clones B1, E7, F2 and 5A were expressed as IgGs, and binding of the resultant IgGs to the expected antigens, but not to several other proteins, was confirmed (Fig. 3). Using flow cytometry, we further tested binding specificity by demonstrating minimal binding of IgGs to the human epithelial kidney cell line 293 cells, which do not express IL4, IL5, and IL13, as well as to baculovirus particles generated from insect cell lines by ELISA (data not shown).18 Furthermore, the IL4/IL5 dual specific antibodies B1 and E7, along with the monospecific IL5 binding antibody, were shown to block IL5 from binding the IL5 receptor α, suggesting that the binding epitopes on IL5 overlapped with that of IL5 receptor (data not shown). By surface plasmon resonance (SPR) measurements, the IL4/IL5 dual specific clone E7 as Fab had low monovalent affinity toward IL5 (KD = 905 nM), but maintained the high affinity IL4 binding (KD = 3.4 nM) of its parent antibody hu19C11.
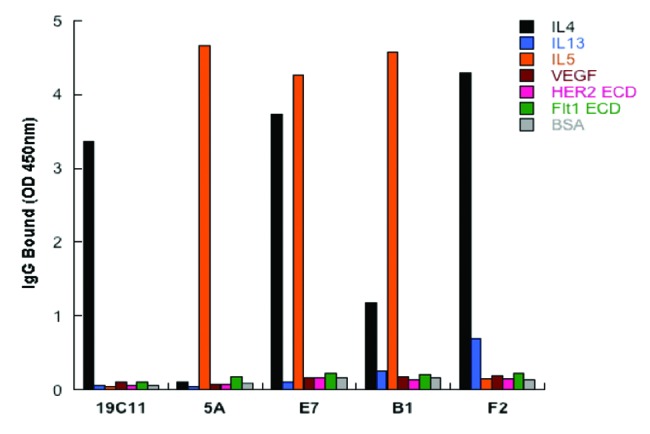
Figure 3. Dual specificity of the selected variants of hu19C11 as IgG. Antigen binding specificity of selected variants of hu19C11 was assessed as binding of these variants in human IgG1 format at 250nM to target antigen(s) or several irrelevant proteins coated on ELISA wells with detection by anti-Fc antibody HRP conjugates.
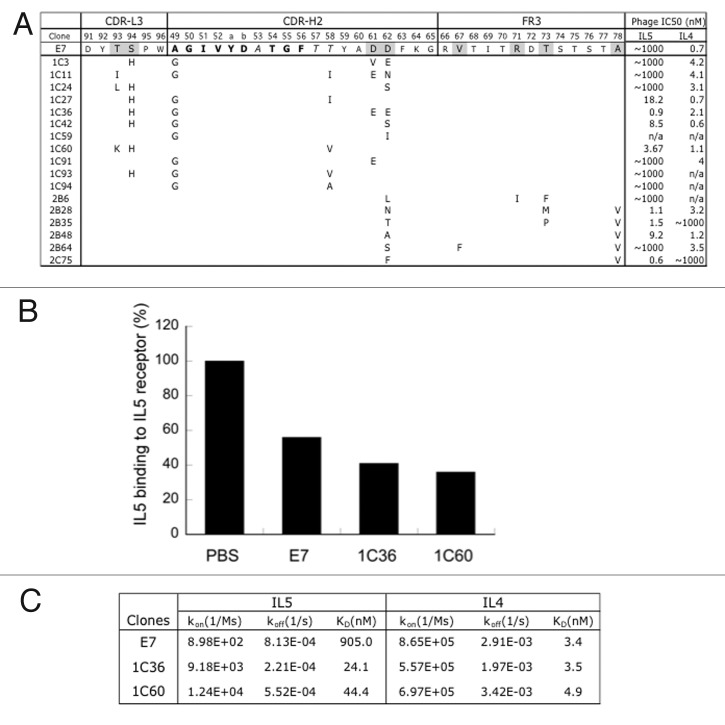
To improve the dual affinity of E7, we randomized E7 CDRs through site-directed mutagenesis and displayed the variants on phage for binding selection. Three libraries were generated targeting residues of CDR H2 and CDR L3 (H2/L3 library), CDR H1, H2 and H3 (H1/H2/H3 library), or CDR H2 plus selected sites in framework region 3 (FR3) of HC (H2/FR3 library) for randomization as described12,19,20 (Fig. 4A). We chose CDR H1–3 and L3 positions that were not critical for IL4 binding for “soft” randomization primarily, which allowed ~50% chance of wild type residues and 50% of the other amino acid types. CDR H2 containing most of the mutation in recruiting IL5 binding, hence likely quite important for IL5 binding function, is allowed gentle homologous mutation to the wild type residues in order to subtly optimize the recruited binding function toward IL5. For H2/FR3 libraries, FR positions that may be structurally proximal to CDR H2 positions were selected for randomization. These FR vernier positions have been shown to affect antibody binding affinity.21 From the H1/H2/H3 library, we found many clones with improved affinity toward IL5 but with greatly reduced affinity toward IL4, whereas clones from the H2/L3 library and the H2/FR3 library showed improved IL5 binding without loss of IL4-binding affinity (Fig. 4A). Select clones were purified as IgGs and compared with E7. Many variants from the H2/L3 library exhibited higher dual binding as IgG without increasing binding to a set of non-targeted proteins. We also confirmed the low off-target binding using baculovirus binding and 293 cell FACS as above, and that the improved clones still blocked IL5 from binding to its receptor (Fig. 4B). Some affinity-improved clones from H2/FR3 library on the other hand exhibited increased nonspecific binding. We therefore focused on two improved variants (1C36, 1C60) and determined their dual affinity by SPR measurements as Fab binding to immobilized IL4 or IL5. Both variants maintained high affinity toward IL4 (KD = 3–4 nM) and improved affinity toward IL5 by 37-fold and 20-fold, respectively (KD = 24.1nM for 1C36 and 44.4nM for 1C60 by SPR) (Fig. 4C).
Figure 4. Affinity improvements of IL4/IL5 dual specific clone E7. (A) For affinity improvement, phage libraries displaying E7 variants were constructed with selected residues mutated with the strategies of “homolog” (bold), “limited” (italic) and “soft” (gray) randomization, which allows wild type and homologous amino acids, limited diversity based on natural antibodies, or ~50% of wild type and 50% of all the other amino acids, respectively. The sequences of selected clones were aligned against E7 and mutations as shown. The relative affinity of each clone was assessed by phage IC50 as above. (B) IL5 as biotinylated protein binding to IL5 receptor coated on ELISA wells in the presence of buffer (PBS), or 50nM of E7 or two affinity improved variants (1C36, 1C60) of E7 was detected with streptavidin-HPR conjugate. (C) E7 and the improved variants (1C36 and 1C60) purified as Fabs were used as analyte in Biacore SPR measurements using a CM5 sensor chip immobilized with human IL5 (R&D Systems) or IL4 at 25 °C to determine the monovalent affinities.
In summary, we showed that mutation of HC CDRs of a monospecific antibody with human framework subtype of VH1/VLkappa1 can recruit a secondary binding specificity. Previously, we demonstrated that trastuzumab Fab and an anti-EGFR Fab (VH3/VLkappa1) can evolve dual specificity through mutations in the LC CDRs.5,7 By mutagenesis-mapping the antigen-binding site of a monospecific antibody, one can engineer Two-in-One antibodies by exploiting areas of the antigen binding site that are amenable to mutation to recruit a second binding specificity. In the case of anti-IL4 hu19C11, this area primarily encompasses the HC CDR residue, but it also includes residues in LC CDR3. In the previous case of engineering Two-in-One antibodies, we mutated residues only in LC CDRs as source libraries to isolate the initial dual specific clones. In both cases, however, the affinity improvement involves mutations in CDRs of both HC and LC. For monospecific antibodies, HC and LC of these dual specific antibodies indeed work closely together for their two binding functions, hence the requirement to adjust the residues of both chains in optimizing the binding interaction. Each Fab arm of all the Two-in-One antibodies identified so far interacts with one antigen at a time as the two antigens sterically compete with each other for the antigen-binding site. Some monospecific antibodies may utilize a large portion of the CDR residues for antigen binding, and evolving secondary specificity in these cases may prove difficult. However, typically, only a subset of residues in the antigen-binding site of an antibody or the interface of a protein-protein interaction is energetically important for binding interaction.22-24 Herein, we provided direct evidence that limited mutations in either LC or HC CDRs of antibodies can recruit a second binding specificity or change to a new binding specificity and evolve dual or new binding capabilities. Antibodies hence appear to be especially well suited to evolve binding specificity via limited mutation, a property that may play a role for the infinite capacity and function of the antibody repertoire for immune protection.
For therapeutic antibody development, Two-in-One antibodies offer the option of a dual targeting molecule that, while requiring upfront engineering efforts, can be easily manufactured as conventional IgG or Fab. It is important to note that Two-in-One molecules as IgG or in other formats with two sets of antigen-binding domains can engage either target bivalently, which may engender desired biological activity, or result in undesired biological response. With the increasing need for various bispecific antibody platforms, it is critical to continue to expand the capability of engineering Two-in-One antibodies to provide a robust option for therapeutic development.
Materials and Methods
Mutation scanning of anti-IL4
The gene segments containing VL and VH of hu19C11 were cloned into the plasmid vector pV0350–2b for displaying hu19C11 as Fab on M13 phage as described.10,12 The light chain (LC) and heavy chain (HC) are expressed bicistronically with light chain fused to a gD peptide at the C-terminus as expression tag. The second cistron comprises the heavy chain (VH and CH1) fused to the C-terminal domain of M13 g3. Alanine mutations in CDRs were introduced by oligonucleotide-directed mutagenesis.25 Phage displaying hu19C11 wild type (wt) or mutants were propagated in E. coli XL1-blue infected with M13-KO7 helper phage, purified and measured OD at 268nm as described.26 To determine the effect of mutation, serial dilutions of phage were incubated in ELISA wells coated with anti-gD antibody (Genentech) or IL4 (PeproTech) for 1 h and detected with anti-M13 phage horseradish peroxidase conjugate (GE Healthcare Life Sciences), for the measurement of relative Fab expression and antigen binding levels, respectively, as previously described.20
Phage library construction and selection
Phage vector displaying hu19C1 Fab was mutated to contain a cysteine at the C terminus of HC as Fab-C vector for bivalent display of Fab, which would increase the capturing efficiency as described.10 For library construction, stop codon was first incorporated in CDR-H2 of the hu19C1 Fab expressing phage displaying template vector. Diversity at selected sites was then incorporated by mutagenesis using mixtures of oligonucleotides coding for variations at CDR H1–3 and CDR L3 with one set of oligonucleotides for each CDR as described.26 A productive clone read through required repair of the stop within CDR H2 with the CDR H2-coding oligonucleotide only. Many clones in the library remained wild type in the other targeted CDRs (H1, H3, L3) due to less than 100% mutation efficiency. Library DNA from the mutagenesis reaction was electroporated into E. coli SS320 strain to yield ~1010 unique members as described.11,26
Selection for new binding specificity was performed by four rounds of panning on immobilized targets (IL5, IL13) followed by a final round of selection on IL4. Randomly picked clones were then assayed using ELISA for specific binding to the target and two non-target proteins. Selected clones were sequenced. The relative binding affinity was determined by competitive phage ELISA as described.12
Antibody characterization
The gene segments containing VL of selected clones was cloned into human IgG1 expressing vectors12 to expressed IgGs in 293 cells. Fabs were generated by papain treatment of IgG followed by removing the Fc and uncut IgG with Protein A column. SPR measurements with Biacore3000 were used to determine the binding affinities of Fabs as described.12 The antigens (IL4, IL5) were immobilized at low density on a Biacore CM5 chip. Serial dilutions of Fab were flowed over the immobilized antigens at 25 °C. 1:1 Langmuir binding model was used to calculate the kon, koff and KD. For E7 binding to IL5 type of weak interaction, steady-state binding analysis was utilized
For binding specificity, IgG was used to detect binding with ELISA with wells coated with cognate antigens or other irrelevant proteins, or with FACS 293 cells binding, or to baculovirus ELISA as described.18 We verified 293 cells do not express any targeted antigens on the cell surface. The bivalency of IgG should allow detection of nonspecific binding sensitively.
For IL5 binding antibodies, ELISA were performed to detect biotinylated IL5 binding to IL5 receptor α (R&D System) coated wells in the presence of antibodies to determine whether the antibody may block IL5 interaction with its receptors.
Affinity maturation library generation and selection
To generate affinity maturation library of clone E7, phage libraries of E7 Fab variants were constructed by mutagenesis using oligonucleotides for diversity in CDR H1–3, CDR L3 or framework region 3 (FR3). Three libraries with different diversity designs were generated and selected by three rounds of panning on immobilized IL5 using stringent wash condition. Randomly picked clones from each library were assayed for their relative binding affinity for IL4 or IL5 by competitive phage ELISA as described.19 High affinity clones were reformatted to IgG for specificity assay using ELISA, FACS on 293 cells and baculovirus ELISA and Fabs were generated for Biacore analysis against IL4 and IL5 as described above.
Disclosure of Potential Conflicts of Interest
All authors are paid employees of Genentech Inc., a member of the Roche Group, and are inventors for a patent application based on the work described herein.
Acknowledgments
We thank Gerald Nakamura, Nancy Chiang, Lawren Wu and Robert Kelley for providing 19C11, Mark Dennis and Yin Zhang for the humanized 19C11 clone, Mark Ultsch, Yvonne Franke, Christoph Spiess, Kurt Schroeder and Charles Eigenbrot for purification assistance, Sarah Sanowar for critical reading and editing of the manuscript and all for helpful discussion.
Glossary
Abbreviations:
- LC
light chain
- HC
heavy chain
- FR
framework region
- CDR
complementarity determining region
- ELISA
enzyme-linked immunosorbant assay
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/28483
References
- 1.Dhimolea E, Reichert JM. World Bispecific Antibody Summit, September 27-28, 2011, Boston, MA. MAbs. 2012;4:4–13. doi: 10.4161/mabs.4.1.18821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontermann R. Dual targeting strategies with bispecific antibodies. MAbs. 2012;4:182–97. doi: 10.4161/mabs.4.2.19000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eigenbrot C, Fuh G. Two-in-One antibodies with dual action Fabs. Curr Opin Chem Biol. 2013;17:400–5. doi: 10.1016/j.cbpa.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Koenig P, Fuh G. Two-in-One antibodies. In: Kontermann R, ed. Bispecific antibodies: Springer, 2011:187-98. [Google Scholar]
- 5.Bostrom J, Yu SF, Kan D, Appleton BA, Lee CV, Billeci K, Man W, Peale F, Ross S, Wiesmann C, et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–4. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- 6.Bostrom J, Haber L, Koenig P, Kelley RF, Fuh G. High affinity antigen recognition of the dual specific variants of herceptin is entropy-driven in spite of structural plasticity. PLoS One. 2011;6:e17887. doi: 10.1371/journal.pone.0017887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer G, Haber L, Crocker LM, Shia S, Shao L, Dowbenko D, Totpal K, Wong A, Lee CV, Stawicki S, et al. A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer Cell. 2011;20:472–86. doi: 10.1016/j.ccr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Colby DW, Garg P, Holden T, Chao G, Webster JM, Messer A, Ingram VM, Wittrup KD. Development of a human light chain variable domain (V(L)) intracellular antibody specific for the amino terminus of huntingtin via yeast surface display. J Mol Biol. 2004;342:901–12. doi: 10.1016/j.jmb.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 9.Spiess C, Bevers J, 3rd, Jackman J, Chiang N, Nakamura G, Dillon M, Liu H, Molina P, Elliott JM, Shatz W, et al. Development of a human IgG4 bispecific antibody for dual targeting of interleukin-4 (IL-4) and interleukin-13 (IL-13) cytokines. J Biol Chem. 2013;288:26583–93. doi: 10.1074/jbc.M113.480483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CV, Sidhu SS, Fuh G. Bivalent antibody phage display mimics natural immunoglobulin. J Immunol Methods. 2004;284:119–32. doi: 10.1016/j.jim.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Sidhu SS, Li B, Chen Y, Fellouse FA, Eigenbrot C, Fuh G. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 12.Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–93. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 13.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–72. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milburn MV, Hassell AM, Lambert MH, Jordan SR, Proudfoot AE, Graber P, Wells TN. A novel dimer configuration revealed by the crystal structure at 2.4 A resolution of human interleukin-5. Nature. 1993;363:172–6. doi: 10.1038/363172a0. [DOI] [PubMed] [Google Scholar]
- 15.Patino E, Kotzsch A, Saremba S, Nickel J, Schmitz W, Sebald W, Mueller TD. Structure analysis of the IL-5 ligand-receptor complex reveals a wrench-like architecture for IL-5Rα. Structure. 2011;19:1864–75. doi: 10.1016/j.str.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Finkelman FD, Hogan SP, Hershey GK, Rothenberg ME, Wills-Karp M. Importance of cytokines in murine allergic airway disease and human asthma. J Immunol. 2010;184:1663–74. doi: 10.4049/jimmunol.0902185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–84. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hötzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4:753–60. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bostrom J, Lee CV, Haber L, Fuh G. Improving antibody binding affinity and specificity for therapeutic development. Methods Mol Biol. 2009;525:353–76, xiii. doi: 10.1007/978-1-59745-554-1_19. [DOI] [PubMed] [Google Scholar]
- 20.Lee CV, Hymowitz SG, Wallweber HJ, Gordon NC, Billeci KL, Tsai SP, Compaan DM, Yin J, Gong Q, Kelley RF, et al. Synthetic anti-BR3 antibodies that mimic BAFF binding and target both human and murine B cells. Blood. 2006;108:3103–11. doi: 10.1182/blood-2006-03-011031. [DOI] [PubMed] [Google Scholar]
- 21.Foote J, Winter G. Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol. 1992;224:487–99. doi: 10.1016/0022-2836(92)91010-M. [DOI] [PubMed] [Google Scholar]
- 22.Kelley RF, O’Connell MP. Thermodynamic analysis of an antibody functional epitope. Biochemistry. 1993;32:6828–35. doi: 10.1021/bi00078a005. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham BC, Jhurani P, Ng P, Wells JA. Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science. 1989;243:1330–6. doi: 10.1126/science.2466339. [DOI] [PubMed] [Google Scholar]
- 24.Vajdos FF, Adams CW, Breece TN, Presta LG, de Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J Mol Biol. 2002;320:415–28. doi: 10.1016/S0022-2836(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–82. doi: 10.1016/0076-6879(87)54085-X. [DOI] [PubMed] [Google Scholar]
- 26.Koenig P, Fuh G. Selection and screening using antibody phage display libraries. Methods Mol Biol. 2014;1131:133–49. doi: 10.1007/978-1-62703-992-5_9. [DOI] [PubMed] [Google Scholar]