Abstract
To gain insight into the functional antibody repertoire of rabbits, the VH and VL repertoires of bone marrow (BM) and spleen (SP) of a naïve New Zealand White rabbit (NZW; Oryctolagus cuniculus) and that of lymphocytes collected from a NZW rabbit immunized (IM) with a 16-mer peptide were deep-sequenced. Two closely related genes, IGHV1S40 (VH1a3) and IGHV1S45 (VH4), were found to dominate (~90%) the VH repertoire of BM and SP, whereas, IGHV1S69 (VH1a1) contributed significantly (~40%) to IM. BM and SP antibodies recombined predominantly with IGHJ4. A significant proportion (~30%) of IM sequences recombined with IGHJ2. The VK repertoire was encoded by nine IGKV genes recombined with one IGKJ gene, IGKJ1. No significant bias in the VK repertoire of the BM, SP and IM samples was observed. The complementarity-determining region (CDR)-H3 and -L3 length distributions were similar in the three samples following a Gaussian curve with average length of 12.2 ± 2.4 and 11.1 ± 1.1 amino acids, respectively. The amino acid composition of the predominant CDR-H3 and -L3 loop lengths was similar to that of humans and mice, rich in Tyr, Gly, Ser and, in some specific positions, Asp. The average number of mutations along the IGHV/KV genes was similar in BM, SP and IM; close to 12 and 15 mutations for VH and VL, respectively. A monoclonal antibody specific for the peptide used as immunogen was obtained from the IM rabbit. The CDR-H3 sequence was found in 1,559 of 61,728 (2.5%) sequences, at position 10, in the rank order of the CDR-H3 frequencies. The CDR-L3 was found in 24 of 11,215 (0.2%) sequences, ranking 102. No match was found in the BM and SP samples, indicating positive selection for the hybridoma sequence. Altogether, these findings lay foundations for engineering of rabbit V regions to enhance their potential as therapeutics, i.e., design of strategies for selection of specific rabbit V regions from NGS data mining, humanization and design of libraries for affinity maturation campaigns.
Keywords: immunogobulins, therapeutic antibodies, antigen-binding site, antibody gene usage
Introduction
The success of antibody-based drugs has fueled the development and refinement of methods for discovery and optimization of more efficacious, safer and developable therapeutic antibodies.1 Rabbit antibodies are well known for their high affinity, exquisite specificity and ability to recognize unique epitopes.2 Rabbits are also evolutionarily more distant from humans than mice and rats, and thus can generate antibodies against epitopes conserved between rodent and human antigens. These advantages, compounded with the possibility of obtaining large amounts of anti-sera relative to mice and rats, made rabbit polyclonal antibodies attractive reagents for research and diagnostics for decades. In the mid-1990s, development of a myeloma rabbit B-cell fusion partner3 enabled the generation of stable rabbit hybridomas, also raising interest in rabbit V genes as substrate for development of therapeutic antibodies. Currently, humanized rabbit antibodies are undergoing validation as therapeutics,4 with some having advanced to preclinical or early clinical development by emerging companies, e.g., Apexigen.
The rabbit IGH locus has been the subject of numerous studies in the last three decades. Genomic mapping have revealed the existence of over 200 IGHV germline genes.5-7 Over 50% have been found to be “non-functional,” with about 80% to 90% of circulating antibodies derived from the IGHV1 gene and expressing the IGHVa1–3 allotypic markers. The VH chains of the remaining 10–20% of circulating antibodies are encoded by the IGHVn genes, which are localized at least 100 Kb upstream of IGHV1. The coding regions of the IGHJ genes from different haplotypes are also conserved,6 and the repertoire of VDJ rearrangements is limited by the usage of a small number of IGHD and IGHJ genes. Of six IGHJ genes, IGHJ4 has been found in 80% of the VDJ gene rearrangements and IGHJ2 in the other 20%. Of the 12 IGDH gene segments, most VDJ gene rearrangements use D2a (D9), D2b (Df), D3 or D5; D4 and D6 are rarely utilized. The limited usage of IGHV, IGHD and IGHJ genes is thought to be compensated by diversity generated at N regions. The size of this repertoire has not been estimated yet and the basis for the preferential usage of the IGHV1 and IGHJ4 gene in VDJ rearrangements remains unanswered.6
Different from other species such as humans and mice where VH plays a fundamental role in antigen recognition, VL seems to be a major contributor to the rabbit antibody diversity and thus specificity.8 Estimates9 of the number of rabbit IGKV germline genes have suggested a repertoire greater than 50 IGKV functional genes8,10 and the preferential use of one IGKJ gene. Importantly, the IGKV genes encode at least seven CDR-L3 lengths, resulting in a potential larger repertoire of CDR-L3 loop lengths than its mouse or human counterparts.9 A diverse repertoire of VK chains, together with gene conversion as a means to diversify the repertoire of antibodies,9 seems to compensate for the limited repertoire of VH chains to build the rabbit immune response.
New technologies such as next-generation sequencing (NGS) are providing the means to study whole natural and man-made repertoires in expedited ways and at relatively low cost.11 Having access to the complete information encoded in antibody repertoires before and after selection under a variety of selection pressures, or after immunization, have revealed features of the antibody repertoire of diverse species that are affecting the theories addressing the origin and evolution of the antibody repertoire.12,13 NGS has also become part of the arsenal of tools to discover and optimize antibodies,14,15 providing crucial information to design repertoires to isolate antibodies with potential therapeutic use,16 as well as complementing screening strategies17 to discover antibodies against diverse targets in expedite ways.
Here, we deep-sequenced the antibody repertoires of the bone marrow (BM) and spleen (SP) of a naïve New Zealand White rabbit (NZW; Oryctolagus cuniculus), to gain insight into the functional repertoire of rabbit antibodies. Another NZW rabbit immunized (IM) with a 16-mer peptide was also deep-sequenced. Close to one million reads were obtained and analyzed to assess the IGHV, IGKV and IGHJ/KJ gene usage, CDR-H3 and CDR-L3 length distributions and compositions, and patterns of somatic mutations. Monoclonal antibodies from the IM sample were generated and the results were compared with the sequences obtained by mining the rich information generated by NGS.
Results
Data Sets
Total RNA from BM and SP of a naïve NZW rabbit and lymphocytes isolated from another NWZ rabbit immunized with a 16-mer peptide was obtained. The VH and VL regions were PCR-amplified using the primers described by Rader,18 gel-purified and submitted for 454 Roche deep-sequencing. Close to one million reads were obtained: 864,738 for VH and 102,676 for VK, with an average length depending upon the sample of 405–411 and 375–376 base pairs for VH and VK, respectively (Table S1A). Excluding reads that differed by at least one amino acid, the number of unique reads per sample was calculated in close to 200,000 for VH and ~20,000 sequences for VK (Table S1B), which is between 65% and 85% of the total sequences per sample. Although the percentage of unique reads includes sequences containing actual mutations and sequencing errors, it gives an estimate of the diversity of the repertoire. The SP sample is slightly more diverse than BM. IM is the least diverse, perhaps due to a bias introduced by the immunization.
VH repertoire
As of July 2013, International ImMunoGeneTics Information System (IMGT) compiled and annotated a total of 68 and 11 IGHV and IGHJ rabbit genes, respectively. After removing non-“01” alleles, pseudogenes, ORFs and fragments, we had 39 IGHV functional genes and 6 IGHJ genes (Table S2). These genes were used as reference to determine the IGHV germline gene usage of the BM, SP and IM samples. Consistent with a previous study,15 two closely related IGHV genes, IGHVS140 (VH1a3) and IGHVS145 (VH4) accounted for more than 90% of the BM and SP VH sequences (Fig. 1A and Table S3). Notably, as much as 40% of the IM sequences were assigned to the IGHV1S69 gene (VH1a1). As mentioned above, the BM and SP samples were obtained from the same rabbit, whereas, the IM sample was obtained from another rabbit, which may explain the difference in the IGHV usage. Alternatively, the distinct IGHV gene usage could be due to the peptide immunization.
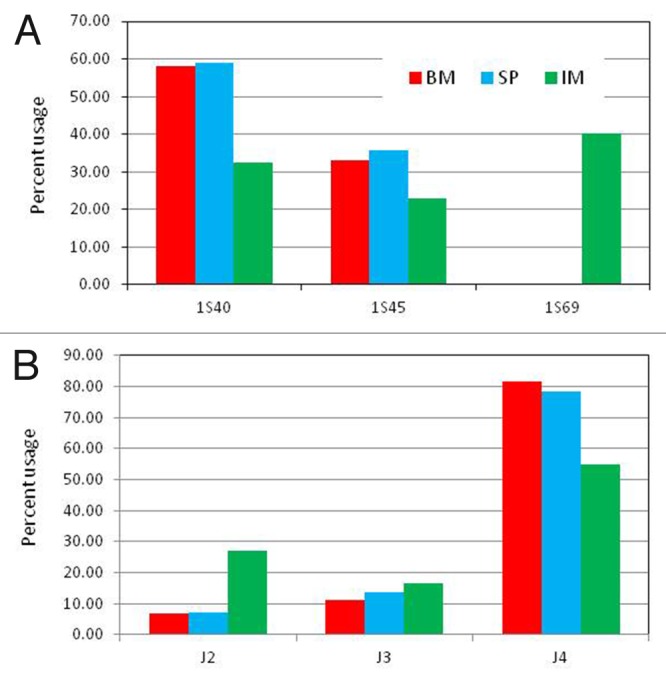
Figure 1. IGVH and IGJH usage in bone marrow (BM) and spleen (SP) of naïve and spleen lymphocytes in immunized (IM) rabbits. (A) The bars represent the % of heavy chain sequences derived from germline IGHV genes 1S40, 1S45 and 1S69. Germline genes contributing > 5% of the V-gene repertoire in at least one sample are shown. (B) The bars represent the % of heavy chain sequences containing germline IGHJ genes IGHJ2, IGHJ3 and IGHJ4. Germline genes contributing > 2% of the V-gene repertoire in at least one sample are shown. See Table S3 for a complete list of all the genes.
The IGHJ germline gene usage (Fig. 1B) of BM and SP samples was also consistent with previous studies,15 with ~80% of the sequences assigned to IGHJ4. A distinct IGHJ usage was observed in the IM sample. Only 54% of IM sequences were assigned to IGHJ4, whereas, 26% of the sequences were assigned to IGHJ2. This difference might be due to the distinct IGHV usage, i.e., IGHV1S40 or IGHV1S45 might recombine preferentially with IGHJ4, whereas, IGHV1S69 might have a preference to recombine with IGHJ2. Alternatively, the higher frequency of IGHJ2 in IM could be due to the peptide immunization.
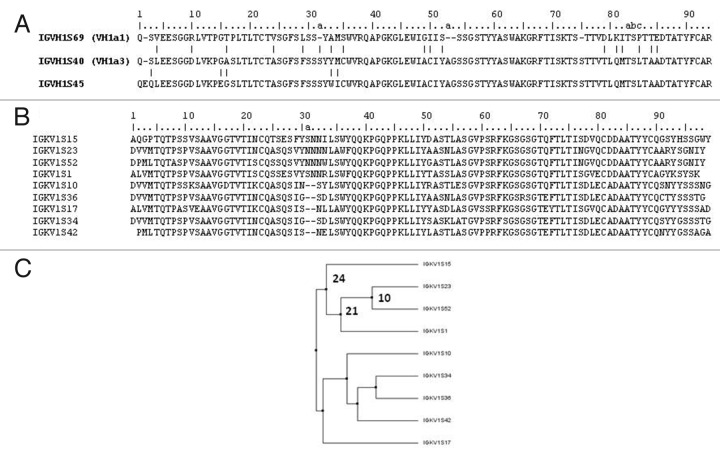
An alignment of the IGHV1S45, IGHV1S40 and IGHV1S69 amino acid sequences is shown in Figure 2A. IGHV1S40 and IGHVIS45 differ by one deletion in position 2 and five mutations, two of them in the CDR-1. In contrast, IGHV1S69 differ from IGHV1S40 by 13 mutations and three deletions. The deletions are in CDR-H1, CDR-H2 and in FR-3. Therefore, antibodies derived from IGHV1S40 and IGHVIS45 genes should generate similar antigen-binding sites, whereas antibodies encoded by IGHV1S69 should result in quite different antigen-binding sites.
Figure 2. Most prevalent IGVH and IGVK genes. Amino acid sequence alignment of the most prevalent IGVH (A) and IGVK genes (B) observed in the samples analyzed and dendogram (C) showing the diversity and relationship of the IGVK genes. Residues are numbered according to Chothia convention. The Vk amino acid sequences in the dendogram were aligned and dendogram plotted based on average distance calculated from percent identity using Jalview. The numbers close to the braches represent number of mutations
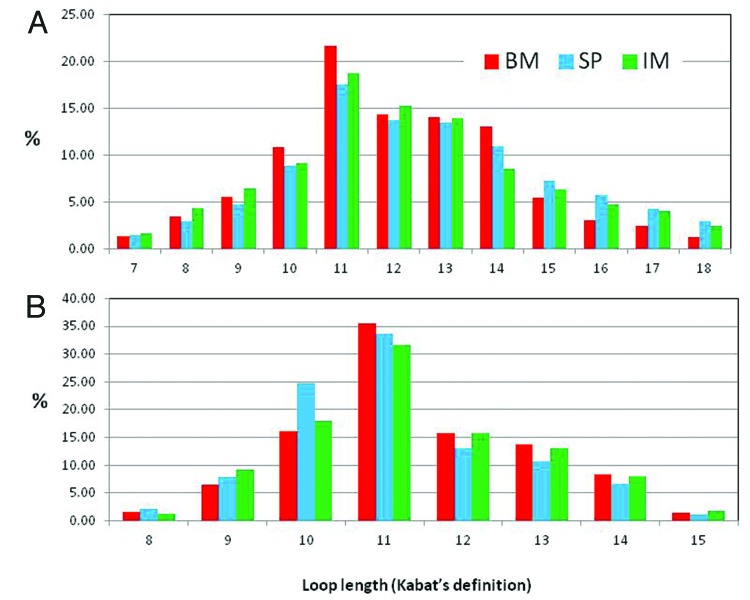
The CDR-H3 length distribution is similar in the three samples (Fig. 4A), implying that the CDR-H3 loop length distribution is independent of the bias observed in IGHV and IGHJ gene usage among the BM, SP and IM samples. Consistent with a previous report,15 the rabbit CDR-H3 lengths resemble a normal distribution with the highest frequency at 11-amino acid loops. Similar and slightly lower frequencies of 12- and 13-amino acid loop lengths generate a small shoulder in the distribution. The average lengths are 12.0 ± 1.3, 12.6 ± 2.4 and 12.3 ± 2.4 for the SP, BM and IM samples, respectively. The length of loops contributing at least 1% to the distribution in at least one of the samples ranges 7–22 amino acids. Human CDR-H3 loops tend to be slightly longer with an average of 13 amino acids21 (Kabat’s definition) with a range of 1–35. Mice CDR-H3 loops are shorter, with an average length of 9 amino acids and a range of 1–21 amino acids.21
Figure 4. CDR-H3 and L3 length distribution: Frequencies of CDR-H3 (A) and CDR-L3 (B) of different lengths expressed as percentage of the total number analyzed from naïve rabbit bone marrow (BM), spleen (SP and immunized rabbit splenocytes (IM).
In addition to the length distribution, the amino acid composition of the CDR-H3 loop is of critical importance to the antibody repertoire function and detailed characterization of the diversity encoded in this loop has aided the design of man-made libraries for antibody discovery.16,22-24 The composition of the CDR-H3 loops with frequencies above 10% (lengths 10 – 14 residues; Figure 4A) is reported in Figure 1 of Supplementary Material. The stems of the loop, i.e., Cys-92, Ala-93 and Arg-94 at the N-terminal region of the loop, and four residues at the C-terminal, before the conserved Trp-102 (YFNI/L for IGHJ4 and NAFDP for IGHJ2), reflect the amino acid composition of the IGHV and the IGHJ genes, respectively. The apical region is rich in Gly, Ser and Tyr, with frequencies that vary between 15% and 35% depending upon the position in the loop. Loop lengths of 11 and 12 amino acids also have a high content (~30%) of Asp at position 95, which is due to recombination events between the IGHV and IGHJ genes. Overall, the rabbit CDR-H3 amino acid composition is similar to the that of humans and mice,21 both species with a strong preference for the use of Tyr, Ser and Gly residues.
Vκ repertoire
IMGT compiled a total of 70 IGKV genes and 19 IGKJ genes as of July 2013. As for the VH genes, we used only “01” alleles as reference. After removing non-“01” alleles, pseudogenes, ORFs and fragments, we gather 68 IGKV genes and two IGKJ genes (Table S2). Of the 68 IGKV germline genes used as reference, 23 genes contributed with a frequency of 1% of more and seven contributed to the repertoire of rearranged sequences with a frequency of 5% or higher in at least one of the samples (Fig. 3 and Table S4). In contrast to VH, none of the genes contributed more than 20% of the sample.
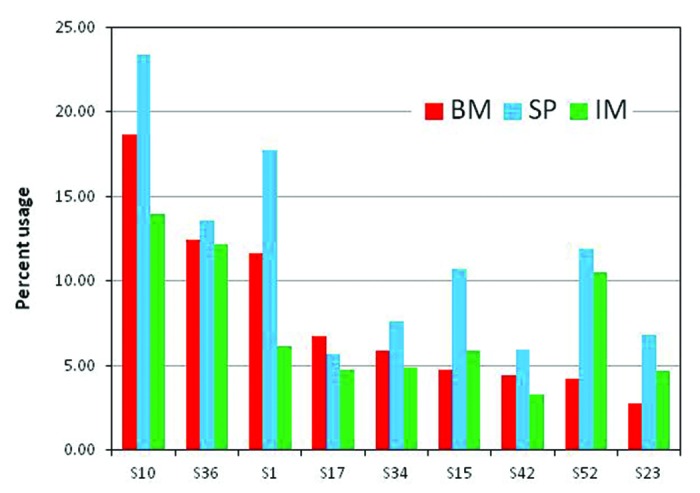
Figure 3. IGVK usage in bone marrow (BM) and spleen (SP) of naïve and spleen lymphocytes in immunized (IM) rabbits. Germline genes contributing > 5% of the V-gene repertoire in at least one sample are shown. Complete listing of all the genes is showed in Table S4 of Supplementary Material.
Differences in the IGKV gene usage were also observed between the BM, SP and IM samples. For instance, IGKVS1 is under-expressed in IM compared with BM and SP, whereas, IGKVS52 is overexpressed in SP and IM when compared with BM. However, the bias is not as apparent as in VH. An alignment of the genes contributing > 5% of BM, SP and IM samples is shown in Figure 2B. These genes are grouped into families (Fig. 2C) that differ by a deletion of two residues in the CDR-L1. This deletion should alter the conformation of the CDR-L1 loop, and together with the sequence diversity ranging 10 to more than 20 mutations should generate diverse binding sites. Nonetheless, the repertoire of loop lengths at CDR-L1 is less diverse than its humans and mice counterpart, which encode for 7 and 5 lengths respectively and encompass more than two gene families.25,26
Similar to the CDR-H3, the CDR-L3 length distribution does not differ in the BM, SP and IM samples. It is close to previous estimates14 and follows a normal distribution with average lengths of 11.1 ± 1.1, 11.4 ± 1.1 and 11.1 ± 1.1 for the SP, BM and IM samples, respectively, and a range of 8 lengths having 8 to 15 amino acids. This CDR is more diverse than its human counterpart,28 where as much as 70% of loops are seven residues in length. Few human antibodies have one deletion with respect to the 7-residue loop, or one or two insertions, yielding a range of lengths of 6–9 amino acids. Also, the conformation of rabbit CDR-L3 seems to be structurally less constrained than that of humans CDR-L3 loops. The vast majority of the human and mouse germline loops encode for Pro at position 95 (Chothia’s numbering)28 restraining the loop to a definite canonical structure, type 1,28,29 whereas such a constraint does not exist in rabbits because amino acids other than Pro are found in position 95.
The composition of the CDR-L3 loops with frequencies above 10% (lengths 10 – 13 residues; Figure 4B) is reported in Figure 1 of Supplementary Material. The frequency of amino acids per position reflects the amino acid composition of the IGKV and the IGKJ genes, with very high content (50% - 80%) of Gly, Ser, Tyr or Asp in certain positions. Approximately 10% of the loops have Cys in position 91, regardless of the length and the sample. These antibodies may present liabilities with regard to the development of therapeutic antibodies.30 Twelve-residue loops of IM sample are also rich (~65%) in Cys at position 95 and 95e, probably making of disulfide bridge. The content of Cys at those positions drops to ~20% in the BM and SP samples, suggesting a positive pressure by the peptide immunization to select for loops with a disulfide bridge between position 95 and 95e.
Somatic diversification
The immune system of rabbits has been the subject of numerous studies in part due to its peculiar use of both gene conversion and somatic hypermutation to diversify the antibody repertoire. A recent report15 described the number of amino acid substitutions in VH relative to the germline as a bimodal distribution with one peak centered at 9-amino acid substitutions and a second peak at 24- to 25-amino acids. To the best of our knowledge, the number of amino acid substitutions in VL and the distribution respect to germline genes has not been reported for a large number of sequences yet.
Figure 5 shows the distribution of VH and VK amino acid substitutions with respect to the germline of the BM, SP and IM samples except for the 8 amino acids at the N-terminal end that are encoded in the primers used for the V regions amplification. The distribution of amino acid substitution in VH (Fig. 5A) follows a Gaussian curve with an average of 12.4 ± 5.0, 10.9 ± 4.4 and 9.8 ± 4.1 mutations for BM, SP and IM, respectively. The slight difference of 1 to 3 mutations with the previous estimate15 could be due to different protocols for sample preparation, quality of the reads or procedure to assign the germline genes. The distribution of mutations in VK (Fig. 5B) also follows a Gaussian curve, with similar values in the three samples, i.e., 15.7 ± 3.0, 15.0 ± 2.8 and 14.9 ± 3.1 for MB, SP and IM, respectively. This average number of mutations in VL is slightly higher than that in VH, which may be due to the important role VL plays in the diversification of the rabbit immune response.
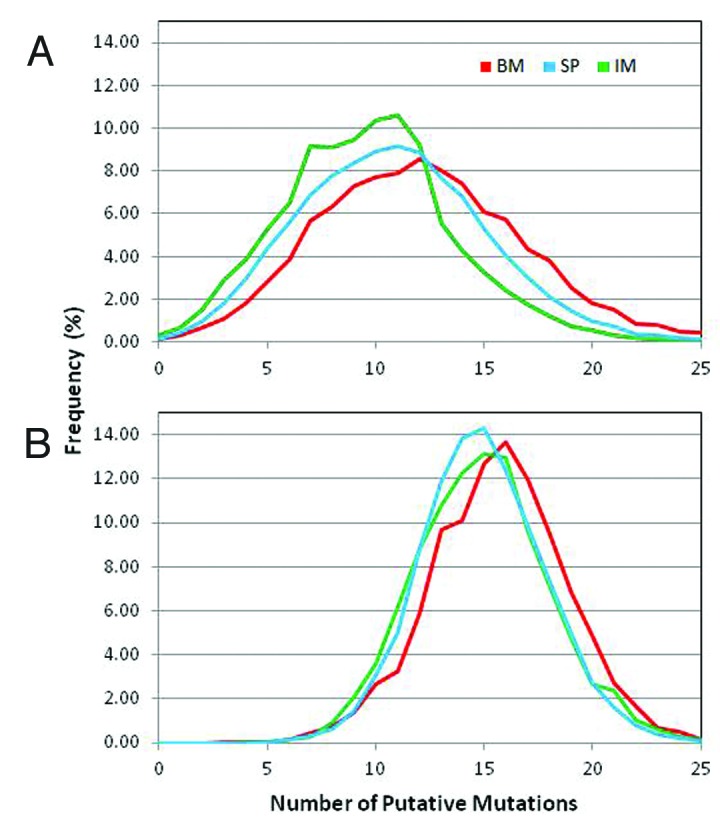
Figure 5. Distribution of total number somatic mutations per read: Total number of mutations per read observed in the three samples, bone marrow (BM), spleen (SP) from naïve rabbit and spleenocytes from immunized rabbit (IM). All reads with germline gene assignment were compared with the respective germline gene sequence and the total number of mutations, except at the first 7 amino acid residues, along the v-gene were counted and the reads were grouped based on the number of mutations. The curves represent the percent of reads with a given number of mutations for VH (A) and VL (B).
The average number of mutations observed in rabbits is significantly higher than that of other species such as humans and mice. In humans and mice, the average number of somatic mutations has been estimated31 to be close to 8 and 5 for VH and VL, respectively, which is two- to 3-fold lower than for rabbits. More mutations in rabbits compared with humans and mice might be due to the germline gene conversion mechanism and could compensate for the limited diversity of the germline genes used to build the functional repertoire of rabbit antibodies.
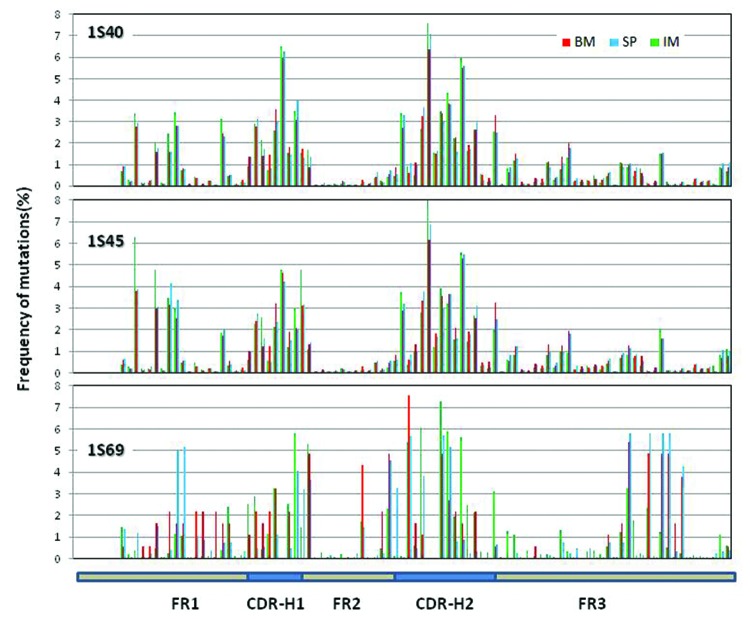
We also estimated the distribution of amino acid differences along the V regions with respect to the most used VH and VL genes. For IGHV, we estimated the mutations in sequences assigned to IGHV1S40, IGHV1S45 and IGHV1S69 (Fig. 6). For IGKV, we estimated the mutations in IGKV1S10, IGHV1S36 and IGKV1S1 (Fig. 7). IGHV1S40 and IGHV1S45 have a similar profile of mutations, centered in the CDR-H1 and CDR-H2. A significant number of mutations are also observed in FR-1, less so in FR-3. Very few mutations were found in FR-2, perhaps reflecting structural constraints at VH/VL interface. The pattern of mutations in IGHV1S69 is also centered in the CDR-H1 and CDR-H2. However, mutations in IGHV1S69 occurred at different positions than in IGHV1S40/45, probably due to different CDR conformations and thus different structural constraints. Also, IGHV1S69 had fewer mutations in FR-1 and more in FR-3 when compared with IGHV1S40/45. In contrast to the differences observed in IGHV1S69, no evident difference in the distribution of mutations in IGHV1S40/45 was observed between BM, SP and IM samples. It should be noted that very few number of sequences were assigned to IGHV1S69 in the BM and SP samples (n = 15 and n = 75, respectively), which may distort the distribution of mutations.
Figure 6. Distribution of somatic mutations along VH: Missense mutations observed in the three samples along the germline genes 1S40, 1S45 and 1S69. The number of missense mutations per position from all the reads derived from the same germline gene is plotted as percent of total number of missense mutations at all the positions for the same set of reads. Missense mutations for the first 7 positions are not calculated since these positions were encoded by the primer used in V-region amplification.
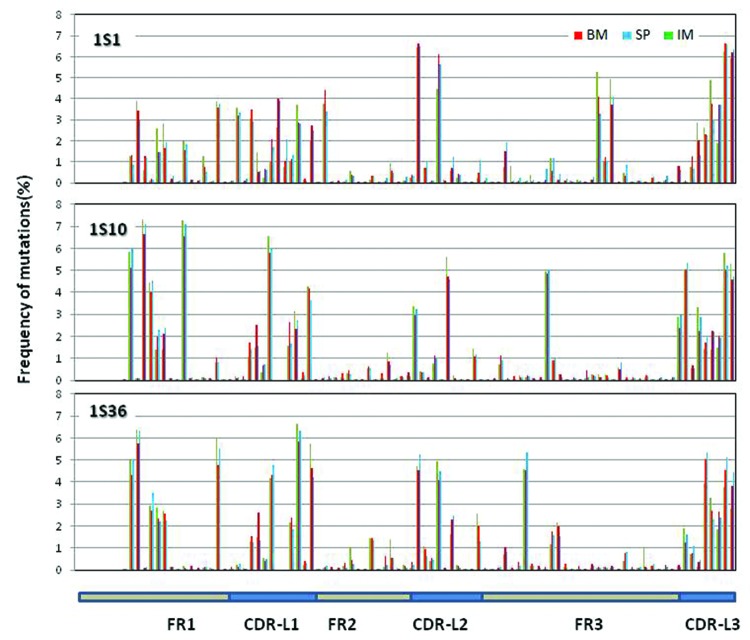
Figure 7. Distribution of somatic mutations along VL: Mutations observed in the three samples along the germline genes 1S1, 1S10 and 1S36. The number of mutations per position from all the reads derived from the same germline gene is plotted as percent of total number of mutations at all the positions for the same set of reads. Mutations for the first 7 positions are not calculated since these positions were encoded by the primer used in V-region amplification.
The distribution of mutations along IGKV (Fig. 7) is not as focused in the CDRs as in IGHV. High mutational activity is observed in FR-1 and CDR-L1, one or two positions of CDR-L2 and CDR-L3. Few mutations occur in FR-2 as in VH. The patterns of mutations differ among the genes, including the CDR-L3 and FRs, for instance around position 16 of FR-1 and position 80 at FR-3. These differences may be informative when engineering antibodies with improved affinity depending upon the V gene. Remarkably, no difference among the BM, SP and IM samples was observed.
Monoclonal antibody and comparison with NGS data mining
To assess how the peptide immunization biased the IM sample, rabbit hybridomas were generated. After sub-cloning and screening, three hybridomas were found to be positive for peptide binding. Sequencing of the V regions yielded a unique Fv, called 28–8. It was chimerized as human IgG1, transiently expressed in 293f cells and purified via Protein A chromatography. The quality and integrity of the purified antibody was assessed by SDS-PAGE and SEC. It gave the expected pattern for an IgG by gel and containing > 99% monomeric antibody by SEC. Positive binding of purified chimeric 28–8 to the 16-mer peptide used as immunogen and negative binding to a 16-mer scrambled peptide with the same amino acid composition was confirmed by AlphaScreen and ELISA.
The VH and VL sequences of 28–8 were then used in a BLASTP search against a sequence database generated from the BM, SP and IM samples. The CDR-H3 sequence of 28–8 in IGHV1S45 germline gene was found in 1,559 of 61,728 (2.5%) sequences in the IM sample, ranking 10th in frequency among the IM CDR-H3 loops (Table S5). In addition, 113 VH sequences in the IM sample were identical to the 28–8 VH sequence. Another 345 sequences matched 28–8 VH with only one amino acid difference. In sharp contrast, 28–8 VH was not found in the BM and SP samples, which comprised 127,074 and 51,705 sequences, respectively. This clearly indicates a strong selection for 28–8 VH due to the peptide immunization.
Because only one unique positive monoclonal antibody was obtained from the hybridomas, we could not determine whether or not any of the nine most frequent CDR-H3 loop sequences in IM were also specific for the peptide. The most frequent CDR-H3 that appeared 3,665 times in IM was observed only once in the BM and SP samples. However, this CDR-H3 is very short - only four amino acids - and contains two Gly residues, which are highly abundant in rabbit CDR-H3 loops (see above). Therefore, the low frequency of occurrence of this CDR-H3 in BM and SP seems to be a random event. Notably, 6 of the other 8 top ranked VH regions are combinations of CDR-H3 loops with IGHV1S69, which was over-represented in the IM sample. Hence, it is tempting to speculate that some of these sequences are the product of positive selection by the peptide or the carrier used in the immunization, and thus could lead to specific antibodies.
The CDR-L3 of 28–8 was found in 24 of 11,215 (0.2%) sequences in the IM sample. It ranked at 102 among the IM CDR-L3 loops (data not shown). Similar to the observation on CDR-H3, the CDR-L3 was not found in the BM and SP samples comprising 51,705 and 61,728 sequences, respectively, indicating positive selection. Moreover, two VL sequences from IM were identical to 28–8 VL, both derived from the IGHV1S41 germline gene. Assuming that the abundance of specific V regions in the NGS sample correlates with the abundance of circulating B cells producing the specific V regions, and that these B-cells have been selected in part by their specificity and affinity to the antigen, the difference in ranking between VH and VL suggests a stronger selection for VH than for VK. If so, it implies that, although the VH repertoire of rabbit antibodies is restricted to the use of one or two IGHV and IGHJ germline genes, VH has a leading role in the rabbit immune response as in others species such as humans and mice.
Discussion
Motivated by the attention that rabbit V regions have been gaining in the last few years as substrate for development of therapeutic antibodies, as well as the uniqueness of the rabbit adaptive immune response, we deep-sequenced the antibody repertoire of three rabbit samples: bone marrow and spleen of a naïve NZW rabbit and B cells collected from another NZW rabbit immunized with a 16-mer peptide. Close to one million reads were obtained.
Using this rich source of information, we first conducted a comparative study of the three samples in terms of IGHV/KV and IGHJ/KJ germline genes usage, CDR-H3 and CDR-L3 length distribution, and amino acid composition of the predominant CDR-H3 and CDR-L3 loop lengths. Consistent with previous reports,14,15 we found that the rabbit VH repertoire is dominated by one or two IGHV genes recombined with one or two IGHJ genes. The VL repertoire is more diverse, with nine genes contributing to the naïve repertoire and the immune response elicited in response to the peptide immunization. Also consistent with previous reports, we found that the CDR-H3 and CDR-L3 lengths followed a normal distribution with average lengths of 12 and 11 residues, respectively. The amino acid composition was found to be rich in Tyr, Gly and Ser, similar to its human counterpart.
We then analyzed in detail the number of putative somatic mutations in VH and VL. Rabbit V regions seem to accumulate two-third more mutations than humans and mice, perhaps as a mechanism to diversify the repertoire of antibodies generated by a limited number of germline genes. We also mapped the positions prone to mutations and those conserved along the V regions. In VH, the mutational activity revolved around the CDRs. In VL, the pattern is fuzzier, with high mutational activity in FR-1 and several positions of FR-3. Together with the gene usage and the CDR-H3/L3 analyses, this information provides a reference for studies aiming to understand the origin and nature of the diversity of rabbit antibodies. This knowledge has practical applications as well. For instance, it is useful in engineering of rabbit V regions to enhance their potential as therapeutics, i.e., humanization and design of libraries for affinity maturation.
Finally, we generated hybridomas from the IM sample and found three positive clones, all of which produced the same Fv sequence, 28–8. We found that as much as 2.5% of the CDR-H3 loops in IM share 28–8 CDR-H3 sequence, ranking 10th within the CDR-H3 sequences with higher frequency. 28–8 CDR-L3 ranked 102, indicating a stronger selection for VH than for VL. If this example is representative of the rabbit immune response, it implies that, although the VH repertoire of rabbit antibodies is restricted to the use of one or two IGHV/HJ germline genes, and the VL repertoire seems to be more diverse than that of other species, VH still plays an important role in the rabbit immune response.
This finding has also applications in antibody discovery via NGS data mining of rabbit antibody repertoires. To the best of our knowledge, this is the first report where NGS data mining is compared with the information generated by hybridoma technology, using the same source of V regions, i.e., same population of B cells was submitted to NGS and used as raw material for hybridoma generation. The results informed us about the VH and VL pairing and the abundance of these specific V regions in the sample obtained by NGS. Although more case studies have to be compiled before drawing a general conclusion, this first example is telling us that if one synthesizes the 20 most frequent VH sequences coming out from the NGS data mining, it is highly likely that a specific VH for the target would be identified. For VL, however, synthesis of only 20 (or 100) most frequent sequences will not be enough to identify the right pair. Thus, alternative solutions would have to be explored such as combining specific VHs with a library of VLs, as elegantly described by Wine et al.,15 or combination of VHs with the most prevalent rabbit IGKV/KJ regions in the germline gene configuration, as counterpart of the specific VH chains, as described by Rojas et al.32
Material and Methods
Peptide immunization
NWZ rabbits were immunized with KLH-conjugated 16-mer peptide (RFGAYLIQAGRMTPEG) derived from the third extracellular loop of ABCB5 isoform 2 (NP_848654.3) and subsequently given booster injections at weeks 3, 4, 6 and 8.
Titer of the polyclonal serum was estimated by standard ELISA (see below) using the same peptide, but conjugated to BSA. The specificity of the serum toward the 16-mer target peptide was assessed using a16-mer scrambled peptide with the same amino acid content of the target peptide used for immunization. Immunized animals were sacrificed on day 63 and the spleen extracted. The rabbit hybridomas were generated in collaboration with Epitomics.
Total RNA isolation from rabbit lymphocytes, spleen and bone marrow
Total RNA from lymphocytes of the immunized animal was isolated using RNeasy Plus Mini Kit (Qiagen), according to manufacturer’s instructions. Spleen and bone marrow tissue samples from naïve animal were isolated and immediately frozen in liquid nitrogen. The frozen spleen and bone marrow samples were processed in parallel. Tissues were homogenized in Trizol and total RNA was further purified using the RNeasy Plus Mini Kit.
Amplification of VH and VL chains
CDNA (cDNA) was synthesized from 1ug total RNA using the qScript cDNA Supermix (Quanta Biosciences) according to manufacturer’s instructions. PCR amplification of the VH and VL genes was performed with the PrimeSTAR HS (Premix). A 50 μl PCR reaction consisted of 25 μl of PrimeSTAR HS (Premix), 2 μl of unpurified cDNA, 0.2 μM of reverse primer, 0.2 μM of forward primer and sterilized distilled water. The reverse and forward primers used was derived from18 and listed in Table S6. Conditions were optimized for the PCR reactions based on the primer mixture used and annealing temperatures of the primer mixes. The PCR thermocycler programs were:
VK primers (Forward 21, 22 – Reverse 24, 25): 98 °C for 1 min; 30 cycles (98 °C for 15 s, 50 °C for 15 s, 72 °C for 90 s); 72 °C for 10 min; 4 °C storage.
VH primers (Forward 28 – Reverse 35, 36, 37, 38): 98 °C for 1 min; 30 cycles (98 °C for 15 s, 55 °C for 15 s, 72 °C for 90 s); 72 °C for 10 min; 4 °C storage.
VH primers (Forward 26, 27, 29, 30, 31, 32, 33, 34 – Reverse 35, 36, 37, 38): 98 °C for 1 min; 30 cycles (98 °C for 15 s, 55 °C for 15 s, 72 °C for 90 s); 72 °C for 10 min; 4 °C storage.
PCR products were gel purified and mixed at an equal mass ratio for NGS.
Antibody expression and purification
Condition media from 293f (Life Technologies) mammalian transient expressions were purified through Protein-A affinity chromatography using standard protocols. Briefly, the condition media was passed through MabSelect protein-A Sepharose resin (GE Life Sciences) previously conditioned with phosphate buffer saline at pH 7.2 (PBS: 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4). The resin was then washed with PBS, and the protein eluted with 5 column volumes of 150 mM Glycine pH 3.0. The elution fraction pH was then neutralized with 1 M Tris buffer at pH 7. The final samples were exchange back into PBS using ZebSpin buffer exchange columns (Thermo) and concentrated with 10 kDa MWCO Amicon Ultra centrifugal filters.
ELISA and AlphaScreen
Standard ELISA protocol was followed to test binding of the antibodies, at concentrations ranging from 50 μg/mL to 50 pg/mL, to the BSA-conjugated 16-mer peptide or the scrambled peptide at 10 μg/mL. Briefly, a pH 9.6 buffer containing 50 mM sodium carbonate/bicarbonate was used to dilute the peptides and coat the plate overnight at 4 °C. After 1 h of blocking reaction using 3% BSA in PBS/Tween buffer at room temperature, the primary antibody was added and incubated for 1 h. The plate was then washed and the anti-rabbit IgG HRP-conjugated detection antibody (R&D Systems HAF008) was added and incubated for 30 min. After washing, TMB One Component Substrate was used to develop the reaction and the signal was recorded using a Spectramax M5 reader in absorbance mode. Curve fitting was performed using GraphPad Prism software for a variable slope nonlinear agonist response with bottom constrained to zero.
For the AlphaScreen, equal volumes of 100 nM biotinylated peptide, either target or scrambled, and antibody at concentrations ranging from 250 nM to 0.24 nM were incubated in solution phase for 1 h at room temperature in a buffer solution containing 50 mM TRIS-HCl pH 7, 100 mM NaCl, 0.1% BSA, 0.01% Tween 20 and complete protease inhibitor in 385 microtiter gray plates. A 1:50 dilution mixture of streptavidin-coated donor beads (Perkin Elmer 6760002) and Protein-A acceptor beads (Perkin Elmer 6760137) was added to each well and the reaction was allowed to proceed overnight in the dark at room temperature. Luminometric data was captured using the Envision reader and represented graphically using GraphPad Prism software.
Next-Generation Sequencing
A 454 sequencing library was constructed by PCR amplification of the amplicon DNA using 454 fusion primers containing barcodes. After removing short fragments using Ampure beads, emulsion PCR was done following the manufacturer’s protocols.
Bioinformatics
Sequence data from Roche GS FLX+ was processed using the 454 Roche pipeline and further filtered using Mothur version 1.25.019 to remove reads with ambiguous base calls and homopolymers longer than 8 bases. The average quality score of a 50 base pair moving window was calculated and sequence trimming was done when the average quality score fell below 35. Reads of length 350 bases or more were analyzed for antibody variable domain and annotated using VDJ-Fasta.16 The sequences used as reference to determine the IGHV germline gene usage of rabbit germline genes were those annotated as “functional” by the IMGT database (http://www.imgt.org/). Reads with annotated VH chains shorter than 280 bases and VL chains shorter than 270 bases were omitted from further analysis. Similarly, reads with JH or JK segments less than 42 bases and 32 bases, respectively, were not included in the analysis of the J-segment. The number of amino acid changes from the germline V-gene sequence was calculated based on pair-wise alignments generated using MUSCLE.20 All amino acid changes except those at the first 8 positions were enumerated. Insertions and deletions were not considered as amino acid changes.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Joe Gardener and Heather Shih from External Research Sourcing (ERS), Pfizer, for coordinating the immunization and fusion of the rabbit B cells, Victoria Wong, also from ERS, for coordinating NGS externalization, Andrea Tiffany for antibody expression, Misha Shamashkin for antibody purification and characterization, and Patrice Milos, Torben Straight-Nissen and Anthony Coyle for stimulating discussions.
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/28059
References
- 1.Almagro J, Strohl W. Antibody Engineering: Humanization, Affinity Maturation and Selection Methods. In: Therapeutic Monoclonal Antibodies: From Bench to Clinic. Ed. Zhiqiang An. John Wiley & Sons, Inc. 2009; 307-327. [Google Scholar]
- 2.An Z. Rabbit Hybridoma In: Therapeutic Monoclonal Antibodies: From the Bench to the Clinic, 2009; 151-169. [Google Scholar]
- 3.Spieker-Polet H, Sethupathi P, Yam PC, Knight KL. Rabbit monoclonal antibodies: generating a fusion partner to produce rabbit-rabbit hybridomas. Proc Natl Acad Sci U S A. 1995;92:9348–52. doi: 10.1073/pnas.92.20.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Y, Lee P, Ke Y, Zhang Y, Chen J, Dai J, Li M, Zhu W, Yu GL. Development of humanized rabbit monoclonal antibodies against vascular endothelial growth factor receptor 2 with potential antitumor effects. Biochem Biophys Res Commun. 2013;436:543–50. doi: 10.1016/j.bbrc.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Currier SJ, Gallarda JL, Knight KL. Partial molecular genetic map of the rabbit VH chromosomal region. J Immunol. 1988;140:1651–9. [PubMed] [Google Scholar]
- 6.Mage RG, Lanning D, Knight KL. B cell and antibody repertoire development in rabbits: the requirement of gut-associated lymphoid tissues. Dev Comp Immunol. 2006;30:137–53. doi: 10.1016/j.dci.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Ros F, Puels J, Reichenberger N, van Schooten W, Buelow R, Platzer J. Sequence analysis of 0.5 Mb of the rabbit germline immunoglobulin heavy chain locus. Gene. 2004;330:49–59. doi: 10.1016/j.gene.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 8.Sehgal D, Johnson G, Wu TT, Mage RG. Generation of the primary antibody repertoire in rabbits: expression of a diverse set of Igk-V genes may compensate for limited combinatorial diversity at the heavy chain locus. Immunogenetics. 1999;50:31–42. doi: 10.1007/s002510050683. [DOI] [PubMed] [Google Scholar]
- 9.Ros F, Reichenberger N, Dragicevic T, van Schooten WC, Buelow R, Platzer J. Sequence analysis of 0.4 megabases of the rabbit germline immunoglobulin kappa1 light chain locus. Anim Genet. 2005;36:51–7. doi: 10.1111/j.1365-2052.2004.01221.x. [DOI] [PubMed] [Google Scholar]
- 10.Reynaud CA, Weill JC. Postrearrangement diversification processes in gut-associated lymphoid tissues. Curr Top Microbiol Immunol. 1996;212:7–15. doi: 10.1007/978-3-642-80057-3_2. [DOI] [PubMed] [Google Scholar]
- 11.Fischer N. Sequencing antibody repertoires: the next generation. MAbs. 2011;3:17–20. doi: 10.4161/mabs.3.1.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5:71ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–10. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, Wieler JS, Beaudet JG, Ramenani RK, Popova L, Comb MJ, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30:447–52. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 15.Wine Y, Boutz DR, Lavinder JJ, Miklos AE, Hughes RA, Hoi KH, Jung ST, Horton AP, Murrin EM, Ellington AD, et al. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci U S A. 2013;110:2993–8. doi: 10.1073/pnas.1213737110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–21. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, Magistrelli G, Farinelli L, Kosco-Vilbois MH, Fischer N. By-passing in vitro screening--next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 2010;38:e193. doi: 10.1093/nar/gkq789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rader C. Generation and selection of rabbit antibody libraries by phage display. Methods Mol Biol. 2009;525:101–28, xiv. doi: 10.1007/978-1-59745-554-1_5. [DOI] [PubMed] [Google Scholar]
- 19.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr., Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–49. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Schroeder HW., Jr. Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev Comp Immunol. 2006;30:119–35. doi: 10.1016/j.dci.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Schroeder HW, Jr., Hillson JL, Perlmutter RM. Early restriction of the human antibody repertoire. Science. 1987;238:791–3. doi: 10.1126/science.3118465. [DOI] [PubMed] [Google Scholar]
- 24.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–21. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almagro JC, Hernández I, Ramírez MC, Vargas-Madrazo E. Structural differences between the repertoires of mouse and human germline genes and their evolutionary implications. Immunogenetics. 1998;47:355–63. doi: 10.1007/s002510050370. [DOI] [PubMed] [Google Scholar]
- 26.Almagro JC, Hernandez I, del Carmen Ramirez M, Vargas-Madrazo E. The differences between the structural repertoires of VH germ-line gene segments of mice and humans: implication for the molecular mechanism of the immune response. Mol Immunol. 1997;34:1199–214. doi: 10.1016/S0161-5890(97)00118-1. [DOI] [PubMed] [Google Scholar]
- 27.Tomlinson IM, Cox JP, Gherardi E, Lesk AM, Chothia C. The structural repertoire of the human V kappa domain. EMBO J. 1995;14:4628–38. doi: 10.1002/j.1460-2075.1995.tb00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927–48. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda D, Shirai H, Kobori M, Nakamura H. Systematic classification of CDR-L3 in antibodies: implications of the light chain subtypes and the VL-VH interface. Proteins. 2009;75:139–46. doi: 10.1002/prot.22230. [DOI] [PubMed] [Google Scholar]
- 30.Gilliland GL, Luo J, Vafa O, Almagro JC. Leveraging SBDD in protein therapeutic development: antibody engineering. Methods Mol Biol. 2012;841:321–49. doi: 10.1007/978-1-61779-520-6_14. [DOI] [PubMed] [Google Scholar]
- 31.Finlay WJ, Almagro JC. Natural and man-made V-gene repertoires for antibody discovery. Front Immunol. 2012;3:342. doi: 10.3389/fimmu.2012.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rojas G, Almagro JC, Acevedo B, Gavilondo JV. Phage antibody fragments library combining a single human light chain variable region with immune mouse heavy chain variable regions. J Biotechnol. 2002;94:287–98. doi: 10.1016/S0168-1656(01)00432-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.