Summary
In the cell, the proteasome and lysosomes represent the most important proteolytic machineries, responsible for the protein degradation in the ubiquitin-proteasome system (UPS) and autophagy, respectively. Both the UPS and autophagy are essential to protein quality and quantity control. Alterations in cardiac proteasomal and lysosomal degradation are remarkably associated with most heart disease in humans and are implicated in the pathogenesis of congestive heart failure. Studies carried out in animal models and in cell culture have begun to establish both sufficiency and, in some cases, the necessity of proteasomal functional insufficiency or lysosomal insufficiency as a major pathogenic factor in the heart. This review article highlights some recent advances in the research into proteasome and lysosome protein degradation in relation to cardiac pathology and examines the emerging evidence for enhancing degradative capacities of the proteasome and/or lysosome as a new therapeutic strategy for heart disease.
Keywords: proteasome, lysosome, heart disease, ubiquitin, autophagy
1. Introduction
Defining how the protein complement of the heart is regulated remains fundamental to our ability to productively identify the processes and proteins that underlie normal and abnormal cardiac function. As heart disease remains the most common cause of death and significant disability worldwide, it is imperative that we understand the mechanistic bases that are responsible for controlling the normal and abnormal protein complements of the different cell types that make up the heart [1]. Protein homeostasis (also known as proteostasis) is the sum total of protein synthesis (translation), post-translational processing and transport, folding, assembly and disassembly into macromolecular complexes, stability and clearance [2]. Protein degradation by either proteasomes or lysosomes plays indispensable roles in protein quantity and quality control in the cell; therefore, proteasomal and lysosomal function is essential to proteostasis (Figure 1) [3]. Generally, the proteasome can degrade individual cellular proteins in a highly targeted fashion via the ubiquitin-proteasome system (UPS) while lysosomes degrade cytoplasmic components, including some individual proteins, protein aggregates, and defective or surplus organelles, through autophagy.
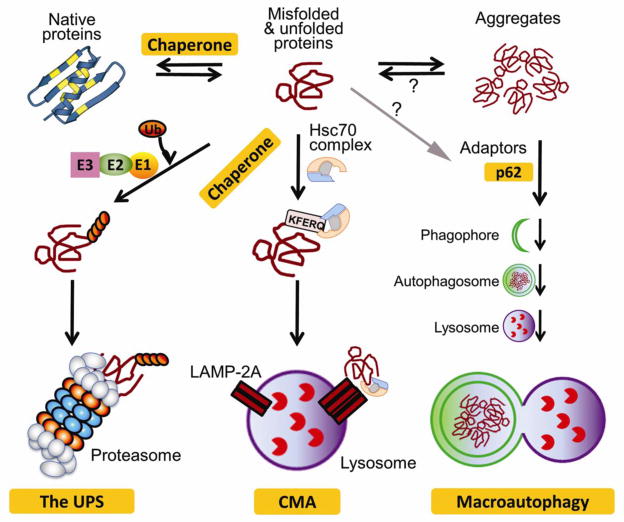
Figure 1. A schematic illustration of cellular mechanisms protecting against proteotoxicity.
Chaperones help fold nascent polypeptides, unfold misfolded proteins and refold them, and escort terminally misfolded proteins for degradation by the ubiquitin-proteasome system (UPS) or chaperone-mediated autophagy (CMA). When escaped from targeted degradation, misfolded proteins form aggregates via hydrophobic interactions. The higher order of aggregation is likely promoted by ubiquitin binding proteins and trafficking via microtubules. Aggregated proteins can be selectively targeted by macroautophagy to, and degraded by, the lysosome. (Modified from Wang et al. [29])
Both the UPS and autophagy have increasingly attracted attention from cardiovascular researchers in the past decade. Many aspects of these degradative and quality control pathways are covered elsewhere in this Special Issue. Exciting progress in elucidating the pathophysiological significance of protein degradation and protein quality control in heart disease has occurred in the past several years; this review will serve to highlight recent advances in proteasomal and lysosomal protein degradation with respect to cardiac pathogenesis, with the primary focus on proteasomes.
2. The proteasome
The UPS is responsible for the degradation of most cellular proteins, native or misfolded. As illustrated by Figure 2, UPS-mediated proteolysis is highly regulated and is capable of target degradation of individual protein molecules in an ATP-dependent manner. The targeting property is conferred by the ubiquitination step, which covalently attaches one ubiquitin (Ub) or a chain of Ub proteins to the side chain of a lysine residue of the target protein molecule. Ubiquitination is catalyzed sequentially by the Ub activating enzyme (E1), Ub conjugating enzymes (E2), and Ub ligases (E3). The E3 is substrate-specific and therefore its function determines the specificity of UPS-mediated protein degradation. Ubiquitination is countered by deubiquitination, which is catalyzed by deubiquitinases (DUBs). Poly-ubiquitinated proteins can be recognized and delivered by Ub receptors to the 26S proteasome where the substrate is de-ubiquitinated, unfolded, and degraded [4].
Figure 2. Ubiquitin-proteasome system-mediated proteolysis.
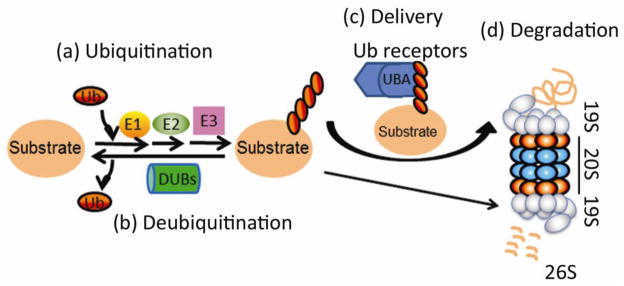
A substrate protein molecule is first covalently tagged with a chain of ubiquitin (Ub) protein molecules, a process known as ubiquitination which is performed by a cascade of enzymatic reactions catalyzed sequentially by E1 (Ub activating enzyme), E2 (Ub conjugating enzyme), and E3 (Ub ligase). The conjugated Ub can be removed from the substrate via a process known as deubiquitination, which counters ubiquitination and is performed by deubiquitinating enzymes (DUBs). Ubiquitinated substrates may be directly recognized and bound by the 19S proteasome, but often require extraproteasomal Ub receptor proteins (i.e., UBA-UBL proteins) to be delivered to, and degraded by, the 26S proteasome (26S). (Adopted from Wang and Terpstra [4])
The proteasome may be considered as a specialized organelle for targeted degradation of most cellular proteins [5, 6]. These large multi-subunit proteolytic machines are found in the cytosol, both free and attached to the endoplasmic reticulum (ER), and in the nucleus of eukaryotic cells [7]. It is generally believed that a functional proteasome in the cell is composed of two parts: a 20S proteasome and the regulatory particle (RP) that binds at one or both ends of the 20S (Figure 2). The 20S proteasome is an axial stack of four rings: two inner antiparallel β rings flanked by two outer α rings. Each β ring harbors 3 protease subunits: β1, β2, and β5, with the proteolytic activities restricted to the inner chamber formed by the two β rings. The α rings function to gate the entry of the substrates into the proteolytic chamber. Some reports show that the 20S can selectively degrade damaged/oxidized proteins [8], but this remains controversial. It is generally accepted that RP is required for proteasomal protein degradation. The RP can be the 19S proteasome, the 11S proteasome, or both [9]. The 19S associated 20S proteasome (ie, the 26S) is the most studied and is believed to mediate housekeeping protein degradation. The degradation of a polyubiquitinated protein requires the 19S, which recognizes and binds the polyubiquitinated substrates directly or via extra-proteasomal Ub receptors, deubiquitinates the substrate for Ub recycling, and unfolds the substrate and channels the unfolded polypeptide into the 20S proteasome where peptide cleavage takes place. Some polyubiquitinated proteins are recognized and bound to the intra-proteasomal Ub receptors, including Rpn10/S5a and Rpn13/ARM1. Rpn10 and Rpn13 harbor UIM (Ub interacting motif) domains and a Pru (pleckstrin-like receptor for Ub) domain, respectively; hence, they can recruit nearby polyubiquitinated proteins to the proteasome [10]. However, the recruitment of remote ubiquitinated proteins to the 26S is proposed to be performed by extra-proteasomal Ub receptors, which must be able to bind both ubiquitinated proteins and the 19S [10]. The UBL-UBA family proteins, including Ubiquilin1 and the mammalian homologs of Rad23 and Ddi1, are ideally suited to serve as shuttling receptors. They have a Ub-like (UBL) domain at N-termini and Ub associated (UBA) domain(s) at C-termini [11]. The UBA binds poly-Ub while the UBL can interact with the 19S via the UIM of RPN10/S5a [12]. Myocardial Ubiquilin1, a bona fide extra-proteasomal Ub receptor, was markedly upregulated in a mouse model of desmin-related cardiomyopathy (DRC) induced by cardiac overexpression a 7-amino-acid (R172~E178) deletion mutant desmin (D7-des) [4]; however, the pathophysiological significance of any potential Ub receptors in the heart have yet to be investigated. It will be important to fill this void because an apparent uncoupling between ubiquitination and proteasomal degradation is frequently suggested by the co-existence of increased total ubiquitinated proteins with normal or even increased proteasome peptidase activities in the heart under many pathological conditions, such as DRC, pressure overload, and myocardial infarction [13–16].
Either heteropolymerization of PA28α and PA28β or homopolymerization of PA28γ forms the 11S proteasome. The association of the 11S with the 20S was suggested to promote antigen processing during viral infection [17, 18]. However, more recent studies show that the 11S may play a wider role than just promoting antigen processing [19]. We have recently demonstrated that PA28α is essential to the degradation of a missense (R120G) mutant αB-crystallin (CryABR120G), a bona fide misfolded protein linked to human disease [20]. We have further demonstrated that forced PA28α overexpression (PA28αOE) suffices to benignly enhance proteasome-mediated removal of a surrogate as well as a bona fide misfolded protein in cultured cardiomyocytes and in transgenic mice, via up-regulating 11S proteasomes and likely increasing hybrid proteasomes [20, 21].
Proteasome peptidases are harbored in the β5, β2, and β1 subunits. Upon viral infection or treatment of interferon γ, these subunits can be replaced by their respective inducible counter parts: β5i, β2i, and β1i, forming immunoproteasomes. Immunoproteasomes, which are primarily expressed in immune cells but have also be detected in the heart [22], display higher peptidase activities and presumably enhance antigen processing during viral infection. Interestingly, interferon stimulation also increases oxidative stress to the cell, resulting in an increase in the production of polyubiquitinated proteins and protein aggregates when the immunoproteasome is impaired [23]. The immunoproteasome seemly facilitates the clearance of damaged proteins and protects against oxidative stress [23–25]. The induction of synthesis of immunoproteasomes, PA28α and PA28β, as well as 20S proteasomes by oxidative stress has been observed [25]. These new findings suggest a novel role of 11S proteasomes and immunoproteasomes in PQC under stress conditions. Notably, immunoproteasome-selective inhibitors are emerging [26], adding new tools for dissecting the immunoproteasome.
Proteasome activities are not only determined by proteasome abundance and associated partners, but are also regulated by a myriad of posttranslational modifications (PTMs) of proteasome subunits [7, 27, 28]. It will be extremely important to delineate the signaling pathways that regulate these PTMs as well as the functional consequence and (patho)physiological significance of the PTMs in cardiac proteasomes. Proteasome dysfunction is associated with common and devastating diseases; hence, normalizing proteasome function is potentially an important therapeutic strategy to treat these diseases. A better understanding of proteasome function regulation is essential to developing such a strategy.
3. Proteasomal dysfunction in cardiac disease
It has become apparent that proteasome function is highly regulated to meet the needs to maintain proteostasis in the cell [29]. Changes in proteasome abundance, posttranslational modifications, and intrinsic activities have been described in human hearts under disease conditions [30]. The pathophysiological significance of the changes has begun to be elucidated by experimental studies [31]. Emerging evidence suggests that the proteasome may be a therapeutic target for heart disease [32].
3.1 Proteasome dysfunction in human cardiomyopathies and heart failure
The literature reporting myocardial UPS changes, including proteasomal dysfunction, in human diseased hearts was comprehensively reviewed by Day [30]. Increased levels of myocardial ubiquitinated proteins are observed in human heart failure of most etiologies, including idiopathic dilated cardiomyopathy (DCM) [33–35], ischemic heart disease (IHD) [34, 36], and aortic stenosis [37]. Individual polyubiquitinated proteins are degraded by the 26S proteasome. Aggregated ubiquitinated proteins can be degraded by lysosomes via selective autophagy. Hence, there are at least five factors that can potentially increase the level of steady state ubiquitinated proteins in a cell or tissue: (i) increased ubiquitination, (ii) decreased de-ubiquitination, (iii) atypical ubiquitin linkages that do not normally target the ubiquitinated protein to the proteasome, (iv) decreased proteasomal degradation due to proteasome functional insufficiency (PFI) or inadequate delivery of ubiquitinated proteins to the proteasome, and (v) decreased lysosomal removal of the aggregated ubiquitinated proteins (see Section 4).
An increase in ubiquitination in the heart or cardiomyocytes under pathological conditions could be adaptive and maladaptive, metabolic, functional, or simply reflect structural changes in response to the demand for increased turnover of signaling proteins, enzymes, and structural proteins. Moreover, cardiac hypertrophy occurs in diseased hearts as a common response to increased stress, in which increased protein synthesis is inevitable. Approximately 30% of nascent polypeptides are never able to become mature proteins but rather are co-translationally degraded by the UPS [38]. Therefore, for quality control purposes, the demand for UPS-mediated degradation of misfolded proteins is increased by cardiac hypertrophy. Indeed, increased myocardial expression of ubiquitin E1, E2, and some E3’s (atrogin-1, MDM2) is associated with heart failure patients resulting from DCM [34–36, 39]. Hence, increased ubiquitination likely contributes to the observed increases in Ub conjugates in human failing hearts but, as discussed below, it is unlikely the sole cause. Increased polyubiquitinated proteins with atypical Ub linkages (eg, K63-, K11- linked) were observed in the brain tissue of human patients with Huntington disease and in related animal models [40]. However, the nature of the Ub linkage in the increased myocardial Ub conjugates has yet to been determined. Therefore, it is unknown whether atypical Ub linkage is a contributing factor. Approximately 100 DUBs exist in the human genome [41]. Many of them should be expressed in the heart but very few of them have been examined in human failing hearts. Increases in myocardial Ub carboxyl-terminal hydrolase (UCH) mRNA and protein, and in myocardial HAUSP proteins, and decreases in isopeptidase T proteins were described in DCM patients [34, 35, 39]. Co-immunostaining of myocardial sections from DCM human hearts showed that isopeptidase T protein expression seemed to decrease in cardiomyocytes with increased Ub staining [39]; however, myocardial DUB activities in human heart failure have not been studied. Thus, it is currently unclear whether decreased deubiquitination is a contributing factor to the increased ubiquitinated proteins.
Although the 19S proteasome in the 26S is capable of directly recognizing and binding ubiquitinated substrates that come into direct contact with the 26S proteasome, the recruitment of remote substrates to the 26S is believed to require extra-proteasomal Ub receptor proteins (eg., Ubiquilin1 and the mammalian homologs of Rad23 and Ddi1) [4]. The expression and function status of these Ub receptors in cardiomyocytes or hearts have not been studied in human heart failure. It will be important to fill this void because functional insufficiency of Ub receptors could conceivably hinder the delivery of ubiquitinated proteins to the proteasome, leading to accumulating ubiquitinated proteins in the cell. Unchanged or even increased proteasomal chymotrypsin-like activities were reported for some human hearts with DCM [35, 39]. However, a more comprehensive and more recent study shows no changes in the protein levels of 20S, 19S, and 11S proteasome subunits, but decreased proteasomal chymotrypsin-like and caspase-like activities in DCM hearts, compared with unused donor hearts [33]. The decreased 26S proteasome activities in DCM might be caused by impaired docking of the 19S to the 20S and decreased docking is associated with decreased ATPase activity of Rpt subunits and decreased phosphorylation of α7 [42]. Additional evidence for impaired proteasome activity in failing hearts comes from the longitudinal comparison of myocardial chymotrypsin-like activity in heart failure patients before and after the implantation of left ventricle assist devices (LVAD). After an average of 214 days of unloading via LVAD, the 20S proteasome chymotrypsin-like activity in DCM hearts was significantly increased [43]. Similarly, the 26S proteasome chymotrypsin-like activity was increased by ~50% in heart failure patients after an average of 31 weeks of LVAD unloading [33]. Hence, decreased proteasome peptidase activities may play an important role in accumulation of ubiquitinated proteins in human failing hearts. Another important piece of evidence indicative of cardiac PFI in human heart failure is the presence of pre-amyloid oligomers (PAO), a highly active and toxic protein conformation derived from aberrant aggregation of misfolded proteins, in the cardiomyocytes of most explanted human failing hearts resulting from DCM or hypertrophic cardiomyopathy (HCM) [44]. On one hand, the formation of PAO signifies UPS inadequacy in the cell because PAO formation would be prevented should its precursors be timely removed by the UPS; on the other hand, aberrant protein aggregation which gives rise to PAO, can impair the proteasome, causing PFI [15, 45]. Taken together, multiple lines of evidence indicate that inadequate proteasomal degradation exists in a large subset of human failing hearts and likely contributes to the rather uniformly reported increases in Ub conjugates in the failing hearts, underscoring the significance of experimental studies to delineate the underlying cause and pathophysiological importance of cardiac proteasome dysfunction.
3.2 Proteasome functional insufficiency (PFI) in animal models of heart disease
The inability of cellular proteasomes to normally degrade its substrate proteins in a cell or tissue is referred to as PFI. PFI can occur at the given intracellular location when the capacity of proteasomal degradation fails to meet the demand of timely removal of substrate proteins. Reduced amounts of functional proteasomes can cause PFI; PFI can also occur when functional proteasomes are increased but are still inadequate to meet the needs for timely degradation of target proteins. Hence, simple measurements of proteasome abundance and proteasome peptidase activities are insufficient to determine proteasome functional sufficiency. Accumulation of proteasomal substrates is the ultimate indicator of PFI. Indeed, increases in myocardial ubiquitinated proteins, which are consistent with PFI, are reported for nearly all animal models of heart disease, such as myocardial infarction, myocardial ischemia/reperfusion (I/R), pressure-overloaded hypertrophy, and DRC [46]. However, a number of factors other than PFI, as discussed above, can also cause increased Ub conjugate levels. To this end, the development of biologically inert surrogate substrates for the UPS, such as degron CL1-fused green fluorescence protein (GFPu, GFPdgn) and Ub-fused GFP (UbG76V-GFP) [47, 48], has proven to be invaluable in unraveling PFI in animal models of human heart disease. The introduction of GFPdgn into the transgenic mouse models of DRC allowed the first demonstration in intact animals that expression of a misfolded protein and resultant aberrant protein aggregation impair UPS function [15]. PFI co-exists with markedly increased proteasome peptidase activities in DRC mouse hearts, indicating that the increased proteasome function in this case is apparently compensatory but fails to adequately compensate for the increased demand on the proteasome [13]. Similarly, the use of UbG76V-GFP UPS reporter mice defined PFI in one-year-old mice with a homozygous knock-in of a human HCM-linked myosin binding protein C mutation, as well as a heterozygous mouse LmnadelK32v knock-in model of DCM [49, 50].
Ischemic heart disease is the most common cause of congestive heart failure. Although proteasome functional sufficiency in animal models of myocardial infarction has not been probed with a reporter substrate, PFI can occur in mice with acute I/R injury. Using a myocardial I/R model induced by coronary artery ligation and subsequent release in the GFPdgn mice, Tian et al showed decreases in myocardial proteasome peptidase activities and the accumulation of ubiquitinated proteins and the GFPdgn surrogate substrate proteins in all regions of the I/R hearts, demonstrating for the first time cardiac PFI in an in vivo model of myocardial I/R [51]. Pressure overload is another common primary factor for heart failure. The changes of proteasome function during the development of pressure-overloaded cardiac hypertrophy and failure remain controversial. All related reports show increased myocardial Ub conjugates in animal models of cardiac pressure overload but proteasome peptidase activities increased in some but decreased in others [14, 52]. Thus, the functional significance of proteasome functional sufficiency in pressure overloaded hearts remains ill-defined at the present. The use of a UPS surrogate substrate to probe the dynamics of UPS status will likely help solve the conflicting data.
3.3 Pathogenic role of PFI in heart disease
The proteasome plays a pivotal role in degrading soluble individual misfolded proteins. As discussed above, as a result of proteasome inhibition or PFI, misfolded proteins undergo aberrant aggregation (Figure 3). The latter appears to have two opposing impacts on cellular capacity to deal with proteotoxic stress. On one hand, aggregation ultimately sequesters highly active misfolded proteins into insoluble aggregates, including aggresomes, to reduce their toxicity; on the other hand, the intermediate soluble species of protein aggregates are believed to be harmful to the cell. It has been well demonstrated in both cardiomyocytes and non-cardiac cells that aberrant protein aggregation impairs proteasome function but the mechanism is unclear. Overexpression of human DRC-linked misfolded proteins in cardiomyocytes causes PFI in an aberrant protein aggregation-dependent fashion [15, 45]. Hence, PFI and aberrant aggregation form a vicious cycle, which likely contributes to the progressive nature of familial proteinopathies in which the synthesis of misfolded proteins resulting from genetic mutations never stops whereas the degradation of the misfolded proteins increasingly slows down. Measures to facilitate the removal of individual misfolded proteins, such as increasing the function of relevant chaperones and improved proteasome function, should break the vicious cycle and prove to be beneficial in handling proteotoxic stress. Indeed, we have recently demonstrated that enhancement of cardiac proteasome function via overexpression of PA28α significantly decreases aberrant protein aggregation, slows down the progression of DRC in a CryABR120G-based mouse model, protects against acute I/R injury [20, 21], and improves right ventricular function and animal survival in mice subject to pulmonary artery constriction [53]. Conversely, proteasome inhibition can cause or exacerbate cardiac malfunction. The first mouse model of cardiomyocyte-restricted moderate inhibition of the 20S proteasome, which was created by cardiac specific overexpression of a protease-disabled proteasome β5 subunit precursor, has been recently reported and employed to demonstrate that moderate inhibition of the 20S proteasome in cardiomyocytes exacerbates myocardial I/R injury [51]. Proteasome inhibition using bortezomib increased maladaptive cardiac remodeling and early mortality in mice subject to transverse artic constriction (TAC) [54]. Primary proteasome inhibition in pigs is sufficient to cause cardiac functional and structural abnormalities that resemble restrictive cardiomyopathy in humans [55, 56]. Clinically, the proteasome inhibitor bortezomib is FDA-approved to treat multiple myeloma but adverse cardiac effects, including severe heart failure, have been reported [57–59]. These lines of evidence support not only the sufficiency but also the necessity of PFI in cardiac pathogenesis.
Figure 3. Aberrant aggregation of misfolded proteins in cardiomyocytes.
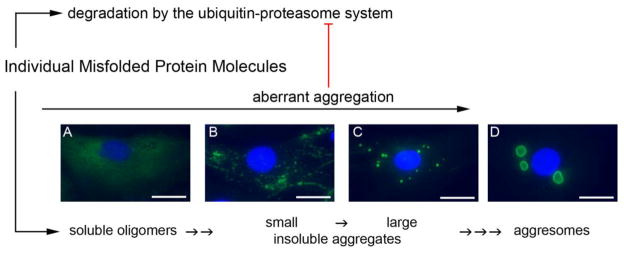
An HA-epitope tagged missense (R120G) mutant CryAB (HA-CryABR120G) was expressed in cultured neonatal rat cardiomyocytes (NRCMs) via adenoviral gene delivery. At 6 days after the viral infection, the cells were fixed with 3.8% paraformaldehyde and subject to immunofluorescence staining for the HA tag (green). The nuclei were stained blue using DAPI. The stained NRCMs were imaged with an epi-fluorescence microscope. Four main distribution patterns of CryABR120G proteins were observed and are annotated according to our perception of their representation of the different stages of the aberrant protein aggregation in the cell. Aberrant protein aggregation impairs the UPS, which in turn allows more of the misfolded proteins undergo aberrant aggregation, forming a vicious cycle. Scale bar=25 μm
3.4 Enhancing proteasome function: a potential therapeutic strategy
The ubiquitination of a substrate protein is triggered by the maturation of a degradation signal (AKA, degron) on the substrate. The maturation of a degron in a native protein molecule is coupled with its loss or absence of an essential binding partner or becoming no longer needed. The former may directly expose the degron, allowing E3 to bind. The latter situation often results from post-translational modifications, such as phosphorylation [60]. In other words, a normal or native protein would not be poly-ubiquitinated for the purpose of proteasomal degradation unless the protein is no longer needed. Generally speaking, the proteasome does not degrade normal proteins that are not ubiquitinated. Therefore, direct enhancement of proteasome activities should not increase the turnover of native proteins in the cell. In other word the regulatory degradation by the UPS should not be increased by enhancing proteasome function. Supporting this analysis, it has been shown in cultured cardiomyocytes and mice that proteasome enhancement by either PA28α overexpression or stimulating protein kinase G (PKG) does not alter the levels of endogenous native proteins that are known substrates of the UPS [20, 21, 61]. For similar reasons, we believe increased proteasomal activity should not be used to account for increased degradation of a specific native protein.
By contrast, damaged or misfolded cytosolic or nuclear proteins likely trigger ubiquitination in multiple ways, such as the exposure of a cryptic degradation signal and/or a patch of hydrophobic residues that are normally buried in the interior of the native conformation [60]; hence, misfolding per se turns on the degradation signal and the terminally misfolded proteins will be ubiquitinated as long as suitable Ub E3s are available. An E3 specific to a native substrate protein may also participate in the degradation of the misfolded form of the substrate protein; however, it would be more sensible to postulate, and indeed emerging evidence supports that there are specialized E3s for misfolded proteins. These E3s are either specialized for targeting terminally misfolded proteins at a specific site or organelle (e.g., ribosomes for protein synthesis, ER for protein folding), or specialized for recognizing a potentially specific misfolding signal shared by misfolded proteins (e.g., recognizing a patch of hydrophobic residues). Terminally misfolded ER proteins are retrotranslocated from the ER to the cytosol where they are immediately ubiquitinated and degraded primarily by the proteasome via a process known as ER-associated degradation (ERAD). Synoviolin (mammalian homolog of yeast Hrd), Parkin, and CHIP (C-terminus of Hsp70-interacting protein) are among the identified Ub E3s of ERAD in mammals [62]. CHIP, a U-box type of E3 and a co-chaperone, might also be an important player in targeting misfolded cytosolic proteins for degradation in metazoans [63]. Recently, Ub ligase San1 is identified a nuclear E3 for degradation of misfolded proteins in yeast [64]; Ltn1 is an E3 for co-translational degradation of non-stop polypeptides at the ribosome [65]; and Hul5 (HECT Ub ligase 5) is a major cytosolic Ub E3 in the degradation of cytosolic misfolded proteins in yeast [66]. It will be important to test whether the mammalian counterparts of these yeast E3s play a similar role.
Currently, it is unknown whether PQC-associated Ub E3s become insufficient in the cell under proteotoxic stress but, on the other hand, multiple lines of evidence support the hypothesis that the rate-limiting step for UPS-mediated degradation of damaged or misfolded proteins resides in the post-ubiquitination steps, including delivery to, and degradation by, the proteasome [31]. First, steady state levels of ubiquitinated proteins are always increased in cells with increased expression of misfolded proteins. Second, misfolded proteins accumulating in the cell, especially in aggregates, are often ubiquitinated. Third, under proteotoxic stress, the increase of intracellular polyubiquitinated proteins is significantly less in C. elegans with improved proteasome function than in those with normal proteasome function [67]. Finally and most importantly, enhancement of proteasome function via either overexpression of the proteasome activator 28α (PA28α) or stimulating protein kinase G, significantly reduces accumulation of stably overexpressed misfolded proteins without affecting the level of endogenous or overexpressed native proteins that are known proteasome substrates, thereby slowing down disease progression in a mouse model of cardiac proteinopathy [20, 61]. Thus, compelling data exist that measures to increase proteasome proteolytic function facilitate the degradation of misfolded proteins and thereby protect against proteotoxicity. In further support of this hypothesis, up-regulation of Rpn6 (PSMD11), a 19S proteasome subunit critical to the assembly of the 26S proteasome, increases proteasome activity, hastens the clearance of damaged proteins, and increased longevity in C. elegans [67]. Myocardial I/R injury is a common pathological process with increased production of damaged/oxidized proteins. Enhancement of proteasome function via PA28α overexpression protects against acute I/R injury in mice [20]. An important mechanism underlying the powerful cardioprotection by ischemic preconditioning is to protect the proteasome from oxidative stress damage [68]. Hence, there are compelling data for proteasome enhancement as a therapeutic strategy to treat heart disease in the context of increased proteotoxic stress.
A pressing question raised then is how to pharmacologically enhance proteasomal function in cardiomyocytes. A recent study by Ranek et al demonstrated that both genetic and pharmacological activation of PKG in cultured cardiomyocytes could stimulate the 26S proteasome chymotrypsin-like activity and improve proteasomal degradation of a surrogate (GFPu) as well as a bona fide misfolded protein (CryABR120G) that is linked to human DRC. Conversely, PKG inhibition displayed the opposite effects [61]. PKG activation in cultured cardiomyocytes also protected against proteotoxicity induced by forced expression of CryABR120G while PKG inhibition showed the opposite effects. Finally, pharmacological stimulation of PKG via systemic administration of sildenafil (an inhibitor of type 5 phosphodiesterase, PDE5) in mice increased myocardial 26S proteasome chymotrypsin-like activity and enhanced the degradation of a surrogate UPS substrate (GFPdgn), while long-term systemic administration of sildenafil reduced ubiquitinated protein accumulation and aberrant protein aggregates, conserving cardiac function in a CryABR120G-based DRC mouse model [61]. These data show that measures to activate PKG may be effective in in decreasing morbidity in heart disease with increased proteotoxic stress. Given that proteotoxic stress is increased in a large subset of heart disease as they progress to congestive heart failure, this study also suggests that improving proteasomal degradation of misfolded proteins might be a previously unappreciated mechanism underlying the efficacy of PDE5 inhibition or PKG activation in treating heart disease. Interestingly, a recent study shows excise training can prevent PFI and attenuate cardiac malfunction in an myocardial infarction-induced heart failure model in rats, suggesting improving proteasome function may contribute to the beneficial effects of exercise training on heart failure patients [69, 70].
3.5 Can proteasome inhibitors be used to treat heart disease?
The section above has argued for the notion that proteasome enhancement promotes primarily the PQC part of protein degradation and should not exert major effects on regulatory degradation. Proteasome inhibition, on the other hand, will inevitably affect the degradation of not only damaged/misfolded proteins but also native regulatory proteins. Hence, it is difficult to imagine that ubiquitous proteasome inhibitors would be useful in treating chronic cardiac conditions, such as idiopathic HCM, DCM, restrictive cardiomyopathy, and the final common pathway of virtually all cardiac disease: congestive heart failure. This is because treatment of these conditions likely requires long-term use of the drug but PFI and inadequate PQC are implicated in their pathogenesis.
Nonetheless, acute cardiac pathological conditions may be different. For example, using infract size and cardiac functional assessments as the end-point readout, several reports show that systemic administration of proteasome inhibitors ameliorates I/R injury in animal models [71, 72], perhaps through suppression of the activation of the NFκB pathway [72]. This protection may result primarily from the inhibition of proteasome activity in the non-cardiomyocyte compartment, because genetically induced cardiomyocyte-restricted moderate proteasome inhibition (40% chymotrypsin-like activity remaining) exacerbates acute myocardial I/R injury in mice [51]. The exacerbation is associated with increased prevalence of cardiomyocyte apoptosis and altered cell signaling that favors cell death, including suppression of AKT activation, increased PTEN and PKCδ protein expression, and decreased PKCε protein levels in not only the ischemic area but in the remote area of the I/R heart [51]. These findings suggest that the previously reported protective effects of proteasome inhibitors on I/R injury could be related to their action on the non-cardiomyocyte compartment, for example, anti-inflammatory effects. Thus, proteasome inhibitors may be more effective in reducing I/R injury if their effects on cardiomyocytes can be minimized.
Intriguingly, systemic administration of proteasome inhibitors suppressed pressure overload cardiac hypertrophy and benefitted long term cardiac remodeling in animal models [14, 73–75]. However, a more recent report disputes these findings. In a TAC-based pressure overload study, Tang et al showed that bortezomib administration induced cardiac hypertrophy in mice receiving the sham surgery, increased mortality in mice subject to TAC-induced pressure overload, while failing to reduce cardiac hypertrophy. In contrast to the first studies, bortezomib actually promoted maladaptive remodeling in the surviving TAC mice [54]. This study also showed that proteasome inhibition activates the calcineurin-NFAT pathway in cardiomyocytes in vitro and in vivo [54]. The cause of these conflicting data is unclear at this time but differences in drugs and dosage used, as well as the severity of pressure overload may all be contributing factors. It should be pointed out that there is a wealth of literature on the effects of proteasome inhibition on vascular function and remodeling that reduce high blood pressure [76–78], which may in turn affect cardiac afterload and cardiac remodeling in some of the hypertensive animal models.
4. Lysosomal dysfunction in heart disease
4.1 Autophagy
Based on the pathway via which a substrate is delivered to the lysosomal lumen, autophagy is classified into three forms: microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy. Microautophagy describes the direct engulfment of cytoplasmic material via the invagination of the lysosomal membrane [79]. The CMA is a specific pathway in which the heat shock cognate protein 70 (hsc70) complex recognizes the target protein and binds specifically to the target’s recognition sequence (the CMA sequence; a KFERQ motif or KFERQ-like motif), subsequently transferring the protein to the lysosomal membrane where the substrate protein is translocated into lysosomal lumen via a complex formed primarily by LAMP-2A (lysosomal membrane associated protein 2A) (Figure 1) [80]. Differing from microautophagy and CMA, formation of an autophagosome is required for macroautophagy to target bulky cytoplasmic materials including organelles to lysosomes for degradation. Self-digestion of a portion of cytoplasm provides energy and essential amino acids for the synthesis of important proteins; hence, macroautophagy is essential to cell survival during nutrient deprivation. Macroautophagy can selectively degrade defective organelles, such as depolarized mitochondria (via mitophagy), as well as aberrant protein aggregates (via aggrephagy); hence, macroautophagy is considered a major player in both organelle quality control and protein quality control in the cell.
Increases of autophagosomes in the heart are observed in many animal models of human pathological conditions and implicated in human heart failure [81]. It is important to recognize that not all increased autophagosomes result from an increased macroautophagy because both increased formation and decreased removal can lead to an elevated abundance of autophagosomes in the cell [82]. The decreased removal of autophagosomes, which can be caused by defective fusion with the lysosomes or impaired/insufficient lysosomal function, would be a decrease in macroautophagy and its consequence is obviously the opposite of increased macroautophagy. This is well illustrated in mouse hearts deficient of the subunit 8 of the COP9 signalosome (CSN8), a bona fide deneddylase known to regulate cullin-based Ub ligases [83]. Cardiac UPS-mediated protein degradation is impaired by cardiomyocyte-restricted knockout of the gene encoding CSN8 [83]. Further characterization reveals an enormous increase of autophagic vacuoles in the cardiomyocytes deficient of CSN8 [84, 85], which was initially thought to indicate a compensatory increase in macroautophagy because proteasome inhibition can activate autophagy in cardiomyocytes [86, 87]. However, additional tests of these mice reveal that autophagic flux is significantly decreased in the CSN8 deficient heart, caused by defective lysosomal removal of autophagosomes [85]. This effort unravels a previously unknown function for the COP9 signalosome in regulating intracellular proteolysis that the COP9 signalosome supports autophagosome maturation in cardiomyocytes [84, 85]. Hence, where is possible, the autophagic flux should be assessed to determine macroautophagy activity. Autophagic flux is increased in D7-des mouse hearts and suppression of macroautophagy exacerbates proteotoxicity in cardiomyocytes in cultures and intact mice [88, 89], suggesting that activation of macroautophagy protects against proteotoxicity in the heart. The increase in macroautophagy in DRC mouse hearts does not seem to be sufficient to meet the increased demand for the removal of protein aggregates because further activation of macroautophagy by pharmacological and genetic means have been demonstrated to reduce protein aggregates and cytotoxicity in cultured cardiomyocytes overexpressing DRC-linked mutant proteins [88, 90]. It will be important to verify these in vitro findings in intact animals.
4.2 Lysosomal deficiency in heart disease
Undoubtedly, the delineation of the genetic program governing the initiation and formation of autophagosomes has been instrumental in rejuvenating the field of lysosomal degradation but the successful and safe removal of autophagic substrates requires not only the formation of autophagosomes but also their fusion with, and timely degradation by, the lysosome. The heart is often affected in lysosomal storage diseases, in which the gene encoding a lysosomal component is mutated. For example, primary deficiency of LAMP-2, an essential lysosomal membrane protein, results in vacuolar cardiomyopathy and myopathy (Danon disease) in humans and mice deficient of LAMP-2 develop a similar phenotype [91]. Cathepsin D is the major protease of the lysosome. More than a decade ago, Kostin et al reported that cathepsin D protein levels were decreased by 50% in failing human hearts compared with donor controls [39]. Using double-immunofluorescence staining, they revealed further that the cathepsin D was barely detectable in cardiomyocytes with Ub-positive aggregates [39]. They also showed via electron microscopy the presence of autophagic vacuoles in cardiomyocytes with the protein aggregates and regarded these cells as undergoing “autophagic cell death”, a term that is now considered inaccurate. An alternative interpretation of the observed pathology is perhaps that lysosomal insufficiency present in cardiomyocytes of the failing heart accumulates autophagic vacuoles, which ultimately causes the cell to die. Indeed, impaired removal of autophagosomes can cause cardiomyocyte necrosis in experimental models and cardiomyopathy in humans [85, 92]. In lysosomal deoxyribonuclease (DNase) II deficient cardiomyocytes, mitochondrial DNA can escape from autophagic vacuoles and triggers cell-autonomous activation of Toll-like receptor (TLR) 9 mediated inflammatory responses in the heart [93]. Cathepsin L is also an important lysosomal protease. Cathepsin L deficiency exacerbates, while cathepsin L overexpression, attenuates pressure overloaded cardiac hypertrophy and malfunction in mice [94, 95]. Interestingly, impaired autophagosome removal was recently associated with experimental myocardial I/R injury [96, 97], disputing previous reports that had shown increased autophagy in I/R myocardium [98, 99]. This dispute remains to be resolved by future independent studies. The contribution of lysosomal insufficiency or lysosomal impairment to myocardial I/R injury or to other cardiac pathologic processes remains to be established.
There has been an influx of data that have led to exciting advances in our understanding of transcriptional regulation of lysosomal synthesis and its coupling with macroautophagy. The transcription factor EB (TFEB)-centered regulatory circuitry can couple increased lysosomal genesis with activation of macroautophagy [100, 101]. Hypoxia-inducible pro-death protein BNIP3 (BCL-2/adenovirus E1B 19-kDa interacting protein 3) is up-regulated in cardiac remodeling and failure [102]. BNIP3 overexpression provokes mitochondrial permeabilization, increases the abundance of autophagosomes, decreases lysosomes, and leads to cell death in cardiomyocyte cultures [103–105]. Enhancing lysosomal biogenesis via forced overexpression of TFEB reduced autophagosome accumulation and cell death caused by BNIP3 overexpression in cultured cardiomyocytes [105]. These recent findings suggest that increasing macroautophagy may help to reduce proteotoxicity and benefit the treatment of many heart diseases. Since autophagosome formation, fusion with lysosomes, and lysosomal degradation of the substrate are all critical parts of this pathway, it is conceivable that only the measures that are capable of correcting the weakest link of this pathway or increasing the overall capacity of macroautophagy would optimally augment macroautophagy for a therapeutic goal.
5. Concluding remarks and future directions
Although the molecular mechanisms underlying proteasomal and lysosomal dysfunction and mediating their pathogenic roles in the heart are only beginning to be unraveled, it has become apparent that altered protein degradation, especially PFI and lysosome functional insufficiency, may play an important role in the progression of a range of heart diseases to CHF. We believe that future research in this paradigm should be aimed at achieving a comprehensive understanding of: (a) the mechanisms underlying UPS and autophagic dysfunctions in various pathological conditions, (b) molecular pathways that govern the degradation of not only misfolded proteins but also key regulatory proteins or protein complexes, and (c) how different proteolytic systems including proteasomes, lysosomes, and other proteases in various cells and tissues are orchestrated to maintain proteostasis under physiological and pathological conditions. Advances in these areas will no doubt shine new light on harnessing the power of targeted protein degradation to more effectively treat heart disease.
Highlights.
The proteasome and lysosomes are the main proteolytic machineries in the cell;
Cardiac proteasome functional insufficiency is a major pathogenic factor;
Enhancement of proteasome function protects against proteotoxicity;
Lysosomal insufficiency might be involved in common cardiac pathogenesis;
Promoting proteasomal or lysosomal function may be of therapeutic benefits
Acknowledgments
This work was supported by NIH grants R01HL072166, R01HL085629, and R01HL068936 (X.W.), NIH grants P01HL69779, P01HL059408, R01HL05924, and R011062927 and a TransAtlantic Network of Excellence grant from Le Fondation Leducq (J.R.)
Footnotes
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the authors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Calamini B, Morimoto RI. Protein homeostasis as a therapeutic target for diseases of protein conformation. Curr Top Med Chem. 2012;12:2623–40. doi: 10.2174/1568026611212220014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers ET, Balch WE. Diversity in the origins of proteostasis networks--a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–48. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N Engl J Med. 2013;368:455–64. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Terpstra EJ. Ubiquitin receptors and protein quality control. J Mol Cell Cardiol. 2013;55:73–84. doi: 10.1016/j.yjmcc.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes AV, Zong C, Edmondson RD, Berhane BT, Wang GW, Le S, et al. The murine cardiac 26S proteasome: an organelle awaiting exploration. Ann N Y Acad Sci. 2005;1047:197–207. doi: 10.1196/annals.1341.018. [DOI] [PubMed] [Google Scholar]
- 6.Zong N, Li H, Li H, Lam MP, Jimenez RC, Kim CS, et al. Integration of Cardiac Proteome Biology and Medicine by a Specialized Knowledgebase. Circ Res. 2013;113:1043–53. doi: 10.1161/CIRCRESAHA.113.301151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Z, Scruggs SB, Gilda JE, Gomes AV. Regulation of cardiac proteasomes by ubiquitination, sumoylation, and beyond. J Mol Cell Cardiol. 2013 doi: 10.1016/j.yjmcc.2013.10.008. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickering AM, Davies KJ. Degradation of damaged proteins: the main function of the 20S proteasome. Prog Mol Biol Transl Sci. 2012;109:227–48. doi: 10.1016/B978-0-12-397863-9.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem. 2000;275:14336–45. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 10.Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–5. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Lee DY, Brown EJ. Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem. 2012;393:441–7. doi: 10.1515/hsz-2012-0120. [DOI] [PubMed] [Google Scholar]
- 12.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–77. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20:362–4. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 14.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–8. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–26. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 16.Adams V, Linke A, Gielen S, Erbs S, Hambrecht R, Schuler G. Modulation of Murf-1 and MAFbx expression in the myocardium by physical exercise training. Eur J Cardiovasc Prev Rehabil. 2008;15:293–9. doi: 10.1097/HJR.0b013e3282f3ec43. [DOI] [PubMed] [Google Scholar]
- 17.Kloetzel PM. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol. 2004;5:661–9. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 18.Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, et al. Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science. 1999;286:2162–5. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Zhao D, Wei H, Yao L, Dang Y, Amjad A, et al. REGgamma deficiency promotes premature aging via the casein kinase 1 pathway. Proc Natl Acad Sci U S A. 2013;110:11005–10. doi: 10.1073/pnas.1308497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. Faseb J. 2011;25:883–93. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–71. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 23.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–24. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 24.Hussong SA, Kapphahn RJ, Phillips SL, Maldonado M, Ferrington DA. Immunoproteasome deficiency alters retinal proteasome’s response to stress. J Neurochem. 2010;113:1481–90. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432:585–94. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148:727–38. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 27.Scruggs SB, Zong NC, Wang D, Stefani E, Ping P. Post-translational modification of cardiac proteasomes: functional delineation enabled by proteomics. Am J Physiol Heart Circ Physiol. 2012;303:H9–18. doi: 10.1152/ajpheart.00189.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Fang C, Zong NC, Liem DA, Cadeiras M, Scruggs SB, et al. Regulation of Acetylation Restores Proteolytic Function of Diseased Myocardium in Mouse and Human. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M113.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Pattison JS, Su H. Posttranslational modification and quality control. Circ Res. 2013;112:367–81. doi: 10.1161/CIRCRESAHA.112.268706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day SM. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am J Physiol Heart Circ Physiol. 2013;304:H1283–93. doi: 10.1152/ajpheart.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301:H2207–19. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagan J, Seto T, Pagano M, Cittadini A. Role of the ubiquitin proteasome system in the heart. Circ Res. 2013;112:1046–58. doi: 10.1161/CIRCRESAHA.112.300521. [DOI] [PubMed] [Google Scholar]
- 33.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–16. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 35.Birks EJ, Latif N, Enesa K, Folkvang T, Luong le A, Sarathchandra P, et al. Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc Res. 2008;79:472–80. doi: 10.1093/cvr/cvn083. [DOI] [PubMed] [Google Scholar]
- 36.Galasso G, De Rosa R, Piscione F, Iaccarino G, Vosa C, Sorriento D, et al. Myocardial expression of FOXO3a-Atrogin-1 pathway in human heart failure. Eur J Heart Fail. 2010;12:1290–6. doi: 10.1093/eurjhf/hfq102. [DOI] [PubMed] [Google Scholar]
- 37.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–91. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 38.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–4. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 39.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–24. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 40.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–8. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 41.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day SM, Divald A, Wang P, Davis F, Bartolone S, Jones R, et al. Impaired assembly and post-translational regulation of 26S proteasome in human end-stage heart failure. Circ Heart Fail. 2013;6:544–9. doi: 10.1161/CIRCHEARTFAILURE.112.000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassiotis C, Ballal K, Wellnitz K, Vela D, Gong M, Salazar R, et al. Markers of autophagy are downregulated in failing human heart after mechanical unloading. Circulation. 2009;120:S191–7. doi: 10.1161/CIRCULATIONAHA.108.842252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–6. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–4. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. J Mol Cell Cardiol. 2008;45:11–27. doi: 10.1016/j.yjmcc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, Liu J, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19:2051–3. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 48.Lindsten K, Menendez-Benito V, Masucci MG, Dantuma NP. A transgenic mouse model of the ubiquitin/proteasome system. Nat Biotechnol. 2003;21:897–902. doi: 10.1038/nbt851. [DOI] [PubMed] [Google Scholar]
- 49.Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, Carrier L. Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res Cardiol. 2012;107:235. doi: 10.1007/s00395-011-0235-3. [DOI] [PubMed] [Google Scholar]
- 50.Cattin ME, Bertrand AT, Schlossarek S, Le Bihan MC, Skov Jensen S, Neuber C, et al. Heterozygous LmnadelK32 mice develop dilated cardiomyopathy through a combined pathomechanism of haploinsufficiency and peptide toxicity. Hum Mol Genet. 2013;22:3152–64. doi: 10.1093/hmg/ddt172. [DOI] [PubMed] [Google Scholar]
- 51.Tian Z, Zheng H, Li J, Li YF, Su H, Wang X. Genetically Induced Moderate Inhibition of the Proteasome in Cardiomyocytes Exacerbates Myocardial Ischemia-Reperfusion Injury in Mice. Circ Res. 2012;111:532–42. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsukamoto O, Minamino T, Okada K, Shintani Y, Takashima S, Kato H, et al. Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Commun. 2006;340:1125–33. doi: 10.1016/j.bbrc.2005.12.120. [DOI] [PubMed] [Google Scholar]
- 53.Rajagopalan V, Zhao M, Reddy S, Fajardo G, Wang X, Dewey S, et al. Altered ubiquitin-proteasome signaling in right ventricular hypertrophy and failure. Am J Physiol Heart Circ Physiol. 2013;305:H551–62. doi: 10.1152/ajpheart.00771.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang M, Li J, Huang W, Su H, Liang Q, Tian Z, et al. Proteasome functional insufficiency activates the calcineurin-NFAT pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88:424–33. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrmann J, Wohlert C, Saguner AM, Flores A, Nesbitt LL, Chade A, et al. Primary proteasome inhibition results in cardiac dysfunction. Eur J Heart Fail. 2013;15:614–23. doi: 10.1093/eurjhf/hft034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X. Repeated intermittent administration of a ubiquitous proteasome inhibitor leads to restrictive cardiomyopathy. Eur J Heart Fail. 2013;15:597–8. doi: 10.1093/eurjhf/hft069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enrico O, Gabriele B, Nadia C, Sara G, Daniele V, Giulia C, et al. Unexpected cardiotoxicity in haematological bortezomib treated patients. Br J Haematol. 2007;138:396–7. doi: 10.1111/j.1365-2141.2007.06659.x. [DOI] [PubMed] [Google Scholar]
- 58.Bockorny M, Chakravarty S, Schulman P, Bockorny B, Bona R. Severe heart failure after bortezomib treatment in a patient with multiple myeloma: a case report and review of the literature. Acta Haematol. 2012;128:244–7. doi: 10.1159/000340050. [DOI] [PubMed] [Google Scholar]
- 59.Gupta A, Pandey A, Sethi S. Bortezomib-induced congestive cardiac failure in a patient with multiple myeloma. Cardiovasc Toxicol. 2012;12:184–7. doi: 10.1007/s12012-011-9146-7. [DOI] [PubMed] [Google Scholar]
- 60.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–90. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranek MJ, Terpstra EJM, Li J, Kass DA, Wang X. Protein kinase G positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128:365–76. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–75. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–6. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 64.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22:2384–95. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–3. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–52. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–8. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 68.Divald A, Kivity S, Wang P, Hochhauser E, Roberts B, Teichberg S, et al. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circ Res. 2010;106:1829–38. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 69.Campos JC, Queliconi BB, Dourado PM, Cunha TF, Zambelli VO, Bechara LR, et al. Exercise training restores cardiac protein quality control in heart failure. PLoS One. 2012;7:e52764. doi: 10.1371/journal.pone.0052764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Meer S, Zwerink M, van Brussel M, van der Valk P, Wajon E, van der Palen J. Effect of outpatient exercise training programmes in patients with chronic heart failure: a systematic review. Eur J Prev Cardiol. 2012;19:795–803. doi: 10.1177/1741826711410516. [DOI] [PubMed] [Google Scholar]
- 71.Stansfield WE, Moss NC, Willis MS, Tang R, Selzman CH. Proteasome inhibition attenuates infarct size and preserves cardiac function in a murine model of myocardial ischemia-reperfusion injury. Ann Thorac Surg. 2007;84:120–5. doi: 10.1016/j.athoracsur.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 72.Pye J, Ardeshirpour F, McCain A, Bellinger DA, Merricks E, Adams J, et al. Proteasome inhibition ablates activation of NF-kappa B in myocardial reperfusion and reduces reperfusion injury. Am J Physiol Heart Circ Physiol. 2003;284:H919–26. doi: 10.1152/ajpheart.00851.2002. [DOI] [PubMed] [Google Scholar]
- 73.Meiners S, Dreger H, Fechner M, Bieler S, Rother W, Gunther C, et al. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension. 2008;51:302–8. doi: 10.1161/HYPERTENSIONAHA.107.097816. [DOI] [PubMed] [Google Scholar]
- 74.Hedhli N, Lizano P, Hong C, Fritzky LF, Dhar SK, Liu H, et al. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am J Physiol Heart Circ Physiol. 2008;295:H1385–93. doi: 10.1152/ajpheart.00532.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stansfield WE, Tang RH, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294:H645–50. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 76.Demasi M, Laurindo FR. Physiological and pathological role of the ubiquitin-proteasome system in the vascular smooth muscle cell. Cardiovasc Res. 2012;95:183–93. doi: 10.1093/cvr/cvs128. [DOI] [PubMed] [Google Scholar]
- 77.Takaoka M, Okamoto H, Ito M, Nishioka M, Kita S, Matsumura Y. Antihypertensive effect of a proteasome inhibitor in DOCA-salt hypertensive rats. Life Sci. 1998;63:PL65–70. doi: 10.1016/s0024-3205(98)00276-8. [DOI] [PubMed] [Google Scholar]
- 78.Li S, Wang X, Li Y, Kost CK, Jr, Martin DS. Bortezomib, a proteasome inhibitor, attenuates angiotensin II-induced hypertension and aortic remodeling in rats. PLoS ONE. 2013;8:e78564. doi: 10.1371/journal.pone.0078564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–36. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–17. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lavandero S, Troncoso R, Rothermel BA, Martinet W, Sadoshima J, Hill JA. Cardiovascular autophagy: Concepts, controversies and perspectives. Autophagy. 2013;9 doi: 10.4161/auto.25969. [DOI] [PubMed] [Google Scholar]
- 82.Iwai-Kanai E, Yuan H, Huang C, Sayen MR, Perry-Garza CN, Kim L, et al. A method to measure cardiac autophagic flux in vivo. Autophagy. 2008;4:322–9. doi: 10.4161/auto.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Su H, Li J, Osinska H, Li F, Robbins J, Liu J, et al. The COP9 Signalosome Is Required for Autophagy, Proteasome-Mediated Proteolysis, and Cardiomyocyte Survival in Adult Mice. Circ Heart Fail. 2013;6:1049–57. doi: 10.1161/CIRCHEARTFAILURE.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–28. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng Q, Su H, Tian Z, Wang X. Proteasome malfunction activates macroautophagy in the heart. Am J Cardiovasc Dis. 2011;1:214–26. [PMC free article] [PubMed] [Google Scholar]
- 87.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–8. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–50. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109:151–60. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–10. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 92.Maron BJ, Roberts WC, Arad M, Haas TS, Spirito P, Wright GB, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. Jama. 2009;301:1253–9. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–5. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun M, Ouzounian M, de Couto G, Chen M, Yan R, Fukuoka M, et al. Cathepsin-L ameliorates cardiac hypertrophy through activation of the autophagy-lysosomal dependent protein processing pathways. J Am Heart Assoc. 2013;2:e000191. doi: 10.1161/JAHA.113.000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang Q, Cai J, Shen D, Bian Z, Yan L, Wang YX, et al. Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3beta signaling. J Mol Med (Berl) 2009;87:249–60. doi: 10.1007/s00109-008-0423-2. [DOI] [PubMed] [Google Scholar]
- 96.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy. 2012;8:1394–6. doi: 10.4161/auto.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, et al. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–81. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 99.Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14:2179–90. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chaanine AH, Gordon RE, Kohlbrenner E, Benard L, Jeong D, Hajjar RJ. Potential Role of BNIP3 in Cardiac Remodeling, Myocardial Stiffness, and Endoplasmic Reticulum: Mitochondrial Calcium Homeostasis in Diastolic and Systolic Heart Failure. Circ Heart Fail. 2013;6:572–83. doi: 10.1161/CIRCHEARTFAILURE.112.000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee Y, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011;301:H1924–31. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, et al. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–31. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma X, Godar RJ, Liu H, Diwan A. Enhancing lysosome biogenesis attenuates BNIP3-induced cardiomyocyte death. Autophagy. 2012;8:297–309. doi: 10.4161/auto.18658. [DOI] [PMC free article] [PubMed] [Google Scholar]