Abstract
Background
The human upper digestive tract microbial community (microbiota) is not well characterized and few studies have explored how it relates to human health. We examined the relationship between upper digestive tract microbiota and two cancer predisposing states, serum pepsinogen I/pepsinogen II ratio (PGI/II) (predictor of gastric cancer risk), and esophageal squamous dysplasia (ESD) (the precursor lesion of esophageal squamous cell carcinoma (ESCC)) in a cross-sectional design.
Methods
The Human Oral Microbe Identification Microarray was used to test for the presence of 272 bacterial species in 333 upper digestive tract samples from a Chinese cancer screening cohort. Serum PGI and PGII were determined by enzyme-linked immunosorbent assays. ESD was determined by chromoendoscopy with biopsy.
Results
Lower microbial richness (number of bacterial genera per sample) was significantly associated with lower PGI/II ratio (P=0.034) and the presence of ESD (P=0.018). We conducted principal component (PC) analysis on a β-diversity matrix (pairwise difference in microbiota), and observed significant correlations between PC1, PC3 and PGI/II (P=0.004, 0.009 respectively), and between PC1 and ESD (P=0.003).
Conclusions
lower microbial richness in upper digestive tract was independently associated with both cancer predisposing states in the esophagus and stomach (presence of ESD and lower PGI/II).
Keywords: microbiota, gastric cancer, esophageal squamous cell carcinoma, esophageal squamous dysplasia, serum pepsinogen I/pepsinogen II ratio
INTRODUCTION
The human digestive tract harbors diverse microbial organisms. These microbial organisms collectively compose a microbial community called the microbiota. Microbial organisms in the gut interact with each other and with the host immune system in ways that influence the development of disease (1). Imbalances in the composition of gut microorganisms and induced changes in their interaction with the human host have been associated with the development of obesity, diabetes, and other diseases (2–6). Helicobacter pylori is the only bacterial species that is known to thrive in the acidic environment of the normal stomach. Inflammatory responses to H. pylori, however, sometimes result in the destruction of the cells that secrete acid into the stomach, leading to a condition known as chronic atrophic gastritis. This condition results in a neutralized stomach environment that, in turn, can be colonized by many other bacterial species, and this new colonization may play an important role in gastric carcinogenesis (7). However, the relationship between the whole microbiota of the upper digestive tract, chronic atrophic gastritis and gastric cancer has not yet been established.
Pepsinogen I (PGI) and pepsinogen II (PGII) are aspartic proteinases that can hydrolyze peptides in acid environments (8). Some of the secreted molecules are released into the circulation and can be measured in serum. A low serum PGI/II ratio correlates well with the extent of chronic atrophic gastritis, and has been consistently related to gastric cancer (9). Indeed, the PGI/II ratio has been proposed as a screening test for both chronic atrophic gastritis and gastric cancer (10–12).
Esophageal squamous dysplasia (ESD) is the precursor lesion of esophageal squamous cell carcinoma (ESCC). Some previous studies suggest that chronic atrophic gastritis, defined by a low serum PGI/II ratio, is associated with risk of both ESD and ESCC (13–16). A study using the same Chinese cohort that is evaluated in the current analysis also showed that lower serum PGI/II was associated with increased risk of ESD (17), but a prospective study in the same population failed to find such an association with ESCC (9).
In the current analysis, we investigated the relationship between the upper digestive tract microbial richness and β-diversity (pairwise difference in microbiota among samples), serum PGI/II ratio and ESD in the Cytology Sampling Study 2 (CSS2), a cancer screening study in Linxian, China, a region with very high rates of ESCC and gastric cancer.
MATERIALS AND METHODS
The Study Subjects, determination of ESD status and Serological assays
The subjects were enrolled from three villages in Linxian, Henan Province, China in the spring of 2002. A description of the parent study, the Cytology Sampling Study 2 (CSS2), has been published previously (18). Subjects were volunteers, aged 40–65 years, apparently healthy and had no contraindications for endoscopy (19). In total, 720 subjects were recruited for CSS2. Within the cohort, 659 had good quality microbiota data by Human Oral Microbe Identification Microarray; 375 had previously been selected for measurement of serum PGI, PGII. The subset with PGI, PGII data were cases diagnosed with moderate or severe dysplasia in the biopsies of the squamous esophagus and the 2 sex matched controls with normal biopsies. The details of diagnosis were described in the previous publication (19). In our study, 333 subjects with both PGI, PGII and microbiota data were analyzed, including 142 with ESD and 191 without ESD. ESD status was determined by chromoendoscopy with biopsy. Serum PGI and PGII were measured by enzyme-linked immunosorbent assays (Biohit ELISA kit, Finland). The details were described previously (18–20).
Sample collection, DNA extraction
Upper digestive tract cell samples were collected by one of two devices (randomly allocated). One was an inflatable rubber balloon covered with cotton mesh attached to a 0.2 cm diameter single lumen rubber tube (manufactured in China). The second was the Cytomesh Esophageal Cytology Device (Wilson-Cook Medical, Inc, Winston-Salem, NC, USA). Each volunteer was given 2 ml of 2% lidocaine slurry by mouth prior to balloon insertion, inflation, and withdrawal. The head of the balloon was cut off and deposited in 40 ml of sterile saline. The tube with the balloon head was vortex for 30 seconds, then the balloon head was removed, and the remaining fluid was centrifuged at 1500 RPM for 5 minutes. The supernatant was discarded, and the pellet was re-suspended in 1 ml saline, frozen in liquid nitrogen and stored at −80° C. DNA was extracted by the Gentra Puregene Cell Kit (Qiagen, Valencia, CA).
DNA preparation for the array hybridization and HOMIM array
The Human Oral Microbe Identification Microarray (HOMIM), which can detect 272 known human microbial species, was conducted in the Paster laboratory and the details of the method were described previously (21). The array uses 16S rRNA-based oligonucleotide probes printed on glass slides. The extracted DNA was used as the template in PCR reactions with universal forward and reverse primers and labeled in a second nested PCR. After hybridization, the washed slides were scanned using GenePix Pro software. A normalized median intensity score was generated by subtracting the median feature intensity from the background intensity for each individual feature. The relative intensity of each probed species/strain was then estimated using feature-specific criteria to determine presence/absence of the bacterial species (21).
For quality control, five samples, selected based on the largest amount of DNA available, were measured once in each batch to assess technical replication of the array results. Our analysis showed consistent richness results between batches. Furthermore, for 10 subjects we cloned and sequenced PCR products (range 59–89) to assess whether any high abundance species were present in the samples but missing from the array. Only one species was present in multiple samples but absent from the array (Lachnospiracaeae OT107).
Statistical analysis
Since some probes could hybridize with more than one species, microbial richness was estimated as number of genera per sample. The accuracy of the bacterial richness estimation above the species level by microarrays has been validated by both quantitative PCR and 16S rRNA gene clone libraries (22, 23). Association analyses were completed using R(24). We had one outcome that was measured as a continuous response (PG I/II ratio) and one outcome measured as a dichotomous response (ESD) and there were some differences in association for PG I/II ratio when we accounted for ESD (see below). Therefore we used a variety of models to explore these associations. The associations between serum PGI/II and microbial richness were evaluated by linear regression models using richness as the dependent variable because of the heterogeneous results by ESD status (microbial richness=PGI/II + age + gender + smoking + antibiotic usage within the last 3 months + balloon sampling device). The associations between ESD status and microbial richness were evaluated by logistic regression models (ESD= microbial richness + age + gender + smoking + antibiotic usage within the last 3 months + balloon sampling device). The microbial richness was standardized by dividing it by its standard deviation.
Estimation of microbial β-diversity was measured by unweighted UniFrac distance matrix, which measures pairwise microbial difference among samples (25). UniFrac estimation was performed in FastUniFrac (25). The phylogeny used for UniFrac estimation was constructed from the Human Oral Microbiome Database (26).
UniFrac distance was used in two ways. First, the unweighted UniFrac matrix was analyzed by principal component (PC) analyses. The first three PCs were used to summarize β-diversity. As noted above, the associations between serum PGI/II and each PC were evaluated using linear regression models (PC=PGI/II + age + gender + smoking + antibiotic usage within the last 3 months + balloon sampling device). The associations between ESD status and each PC were evaluated using logistic regression models (ESD= PC + age + gender + smoking + antibiotic usage within the last 3 months + balloon sampling device). Each PC was standardized by dividing it by its standard deviation. Second, all of the samples were clustered into groups by the UniFrac distance matrix based on unsupervised clustering algorithm (the unweighted pair group method with arithmetic mean (UPGMA) (27) and then PGI/II or ESD status was regressed on these clusters and other factors.
In order to examine the association between PGI/II, ESD and presence/absence of individual strain/species targeted by each probe, we regressed PGI/II or ESD against presence/absence of individual strain/species by logistic regression model.
RESULTS
Characteristics of the study subjects
A description of the subjects evaluated in this study was provided in Table 1. Subjects with ESD had significantly lower PGI/II levels than subjects without this cancer predisposing condition (P= 0.044, data not shown).
Table 1.
Characteristics of the studied subjects from the Cytology Sampling Study 2 cancer screening study
| N(total=333) | percentage | |
|---|---|---|
| Tobacco smoker? | ||
| Yes | 42 | 12.6% |
| No | 291 | 87.4% |
| Gender | ||
| Female | 162 | 48.6% |
| Male | 171 | 51.4% |
| Balloon sampling device | ||
| Chinese Balloon | 161 | 48.3% |
| Wilson-Cook Balloon | 172 | 51.7% |
| Antibiotic usage within the last 3 months? | ||
| Yes | 42 | 12.6% |
| No | 291 | 87.4% |
| Histologically diagnosed esophageal squamous dysplasia? | ||
| Yes | 142 | 42.6% |
| No | 191 | 57.4% |
| Mean | sda | |
|---|---|---|
| Age | 55.03 | 4.54 |
| Pepsinogen I/II ratio | 9.40 | 4.12 |
| No. of genera | 22.93 | 6.11 |
| No. of families | 16.93 | 3.26 |
| No. of orders | 12.12 | 1.84 |
| No. of classes | 10.02 | 1.19 |
| No. of phyla | 5.37 | 0.72 |
standard deviation
Significant association between number of genera per sample, a measure of microbial richness, and both the PGI/II ratio and a diagnosis of ESD
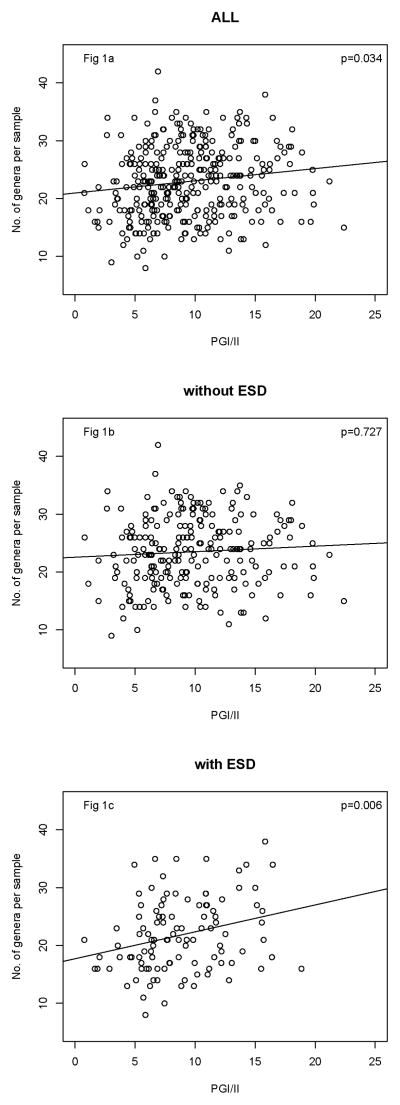
In analyses adjusted for gender, smoking, age, antibiotic usage and balloon sampling device, PGI/II was significantly positively associated with number of genera per sample (P=0.034, Fig 1a, Table 2). The magnitude of the association was little changed after further adjusted for ESD (P=0.059, Table 2). However, when we also included an interaction between PGI/II and ESD, we found that it was statistically significant (P = 0.029), a reflection of the fact that the association between richness and PGI/II was significantly stronger in subjects with ESD compared to those without ESD. Therefore, we examined the association in separate models for those with and without ESD (Fig 1b and 1c). Note the much steeper slope among subjects with ESD.
Figure 1.
Plots of serum PGI/II and number of genera per sample in all studied subjects (Fig 1a), subjects without ESD (Fig 1b), and subjects with ESD (Fig 1c). The regression lines and P value were from the adjusted linear regression models.
Table 2.
Associations between upper digestive tract microbial richness or β-diversity metrics and PGI/II ratio in the Cytology Sampling Study 2
| Adjusted Modela
|
Model Further adjusted for ESDb
|
Interaction ESD*PGI/IIb
|
|||
|---|---|---|---|---|---|
| coefficient | P value | coefficient | P value | P value | |
| Microbial richness | |||||
| No. of genera | 0.029±0.013 | 0.034 | 0.025±0.013 | 0.059 | 0.029 |
| β-diversity | |||||
| PC1 | −0.038±0.013 | 0.004 | −0.035±0.013 | 0.009 | 0.081 |
| PC2 | 0.013±0.014 | 0.346 | 0.015±0.014 | 0.287 | 0.904 |
| PC3 | 0.034±0.013 | 0.009 | 0.034±0.013 | 0.011 | 0.833 |
linear regression model adjusted for smoking, gender, age, balloon sampling device and antibiotic usage in the last three months.
linear regression model further adjusted for ESD besides smoking, gender, age, balloon sampling device and antibiotic usage in the last three months.
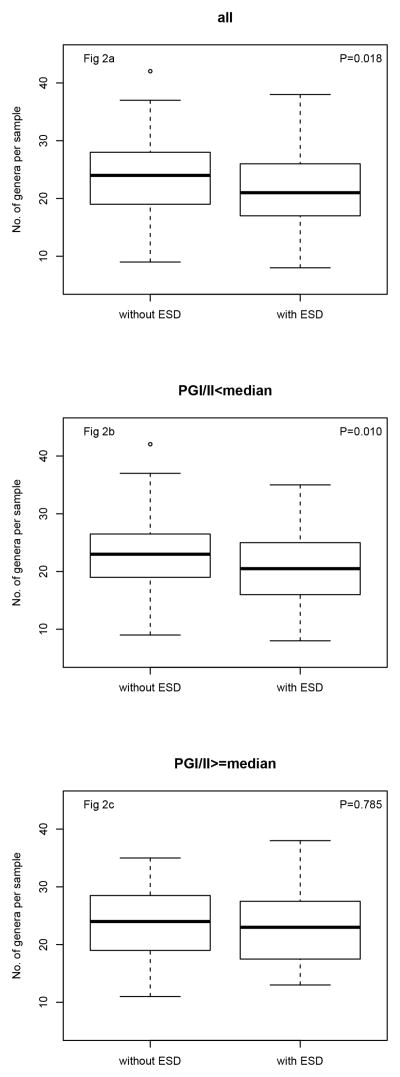
In analyses adjusted for gender, smoking, age, antibiotics usage, and balloon sampling device, the odds of ESD was decreased by a factor of 0.74 per one standard deviation increase in microbial richness (P= 0.018) (Table 3 and Figure 2a). Similar results were found with additional adjustment for PGI/II ratio (Odds ratio=0.76, P=0.032). However, the association was found in those whose PGI/II ratio is below the median, as is seen by comparing results stratified by PGI/II ratio in Table 3, Fig 2b and Fig 2c. Thus the association of low risk with increasing number of genera is mainly in those who are more likely to have chronic atrophic gastritis.
Table 3.
Odds ratio for ESD (and 95% confidence intervals) from logistic regressions on standardized upper digestive tract microbial richness or on standardized β-diversity metrics
| all (333)
|
stratified by PGI/IIa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| adjusted modela
|
Model further adjusted for PGI/IIb
|
PGI/II<median
|
PGI/II>= median
|
|||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| microbial richness | ||||||||
| No. of genera | 0.74(0.58−0.95) | 0.018 | 0.76(0.59–0.97) | 0.032 | 0.63(0.43–0.89) | 0.010 | 0.95(0.66–1.37) | 0.785 |
| β-diversity | ||||||||
| PC1 | 1.45(1.14–1.85) | 0.003 | 1.41(1.10 1.80) | 0.006 | 1.63(1.16–2.32) | 0.005 | 1.19(0.83–1.69) | 0.350 |
| PC2 | 1.13(0.90–1.44) | 0.291 | 1.15(0.91 1.45) | 0.238 | 1.27(0.91–1.78) | 0.166 | 1.06(0.74–1.52) | 0.758 |
| PC3 | 0.94(0.74–1.20) | 0.634 | 0.97(0.76–1.25) | 0.847 | 1.01(0.73–1.41) | 0.939 | 0.93(0.66–1.34) | 0.701 |
model adjusted for smoking, gender, age, balloon sampling device and antibiotic usage in the last three months
model adjusted for PGI/II besides smoking, gender, age, balloon sampling device and antibiotic usage in the last three months.
Figure 2.
Boxplots of ESD status and number of genera per sample in all studied subjects (Fig 2a), subjects with PGI/II less than median (Fig 2b), subjects with PGI/II equal or greater than median (Fig 2c). Boxes encompass the interquartile range (IQR), with central bars for medians and whiskers for 1.5-times the IQR. Outlier values are circles. P values were from the adjusted logistic regression models.
We observed no association between microbial richness and antibiotic usage although previous studies have shown the strong association between antibiotic usage and change of microbial richness in fecal samples (28, 29). This may be due to our limited number of subjects with recent antibiotic usage (antibiotic usage in the last three months, n = 42). It is also possible that upper digestive tract microbiota recover quickly from antibiotic treatment, as has been seen in fecal microbiota one week after treatment (28).
Significant association between microbial β-diversity and PGI/II, ESD status
To assess the associations between β-diversity and either PGI/II ratio or risk of ESD, we used the first three standardized principal components (PCs) of the unweighted UniFrac distance matrix. PGI/II was significantly associated with PC 1 and PC 3 (Table 2), and these associations were independent of and did not interact with ESD (PC 1, P for interaction =0.081; PC 2, P for interaction =0.833). These analyses suggest that two independent aspects of β-diversity were associated with the PGI/II ratio, because PCs are designed to be uncorrelated. In contrast, only PC 1 was associated with risk of ESD (Table 3). Of note, the magnitude of the association between PC 1, PC 3 and PGI/II were not changed by adjustment for ESD (Table 2), and the magnitude of the association between PC 1 and ESD was hardly changed by adjustment for PGI/II (Table 3).
Subjects were grouped into three clusters based on the similarities of their microbial communities, as assessed by UniFrac distances. We compared the PGI/II ratio and prevalence of ESD among subjects in the two large clusters, which are labeled B and C in Supplementary Figure 1. The odds of ESD were 1.41 fold higher in cluster C than in cluster B after adjustment for smoking, gender, age, balloon sampling device, and antibiotic usage in the last three months (95% confidence interval for odds ratio: 1.12–1.79; P=0.004), and results were little changed after further adjustment for PGI/II ratio (Odds ratio and 95% confidence interval: 1.42(1.12–1.80); P=0.004). We found no difference in PGI/II ratio between subjects in cluster B and C (P=0.766).
No significant association between PGI/II, ESD and presence/absence of specific targeted species/strain
Supplementary Tables 1 and 2 show no association between presence/absence of each targeted species/strain and PGI/II ratio or ESD status after Bonferroni correction for multiple comparisons.
DISCUSSION
In this cross-sectional study, we found a significant positive association between microbial richness and PGI/II ratio, and an inverse association between microbial richness and ESD. Because low PGI/II ratio is a marker for chronic atrophic gastritis, our results suggest that individuals with lower microbial richness were more likely to have chronic atrophic gastritis and ESD. We also observed evidence for a statistical interaction between microbial richness and these two cancer predisposing states. The association between microbial richness and PGI/II ratio was significantly stronger in subjects with ESD. Similarly, the association between microbial richness and ESD was stronger in those with a lower PGI/II ratio. While the number of genera, a measure of microbial richness, measures a property of an individual, β-diversity measures the similarity between microbiota in pairs of individuals. We also found associations between risk of ESD, PG I/II ratio, and features of β-diversity.
Decreased microbial richness has been shown to be associated with disease states such as obesity, diabetes, inflammatory bowel disease, and others (3, 4, 6). Our findings are in line with these previous results. We found that low microbial richness in the upper digestive tract was associated with cancer-predisposing conditions of the stomach and esophagus. However, as in these prior cross-sectional studies, our cross-sectional data cannot distinguish whether decreased microbial richness causes these cancer-predisposing states or is an effect of them. Our findings strongly suggest the need for prospective studies of the upper digestive tract microbiota and the development of gastric cancer and ESCC.
The normal acidic stomach is known for its paucity of bacterial genera. H. pylori is usually the only bacterial species that can grow in the stomach. Subjects with a low PGI/II ratio, however, are at high risk for chronic atrophic gastritis, a condition in which the stomach can no longer secrete normal amounts of acid. We would expect that a less acid stomach environment would allow more bacterial taxa to live, so one would predict an inverse association between gastric microbial richness and PGI/II ratio. In our study, however, we found the opposite result, a positive association between upper digestive tract microbial richness and PGI/II ratio. One possibility is that alterations in the stomach due to chronic atrophic gastritis may result in overgrowth of few taxa and extinction of others, leading to the reduction of overall microbial richness. It should be remembered, however, that our biological sample included cellular and luminal material from the esophagus and oral cavity, as well as the stomach, which complicates our interpretation. Further investigation is required to clarify our results.
Compared with subjects without ESD, subjects with ESD had a significantly lower number of genera in the upper digestive tract. This may be because ESD causes changes in the local habitat (the surface of the esophagus and the esophagus mucosa), which might lead to extinction of certain taxa in the communities associated with the normal esophagus and/or overgrowth of other taxa. Alternatively, other risk factors for ESD, such as poor oral health, may be driving changes in the upper digestive tract microbiota.
The inverse association of microbial richness with risk of ESD was stronger in subjects with low PGI/II ratios, and the positive association of microbial richness with PGI/II ratio was stronger in subjects with ESD. Lower PGI/II ratios are associated with increased prevalence of ESD (17). However, the mechanisms by which ESD might modulate the association of richness with PGI/II or by which PGI/II might modulate the association of richness with ESD are unknown.
Our other important and connected finding was the significant association between β-diversity and both PGI/II and ESD. Our results suggest that these health conditions did contribute to the differences in upper digestive tract microbial communities observed between subjects. Previous studies of the gut microbiota have shown that diet, race, and geography contribute to the taxonomic similarity between two samples (30–32). Similarly, our findings showed that the similarity of upper digestive tract microbial communities between two individuals is associated with their upper digestive tract health status.
Our study has several strengths and limitations. The strengths include the relatively large sample size, the inclusion of asymptomatic healthy subjects, and the availability of detailed clinical and demographic information on participants. The limitations include the cross-sectional design of the parent study which precludes separating cause and effect. The limitations also include the use of an array designed primarily to assess the oral microbiota, and the use a sampling device that collected cellular and luminal material from the stomach, esophagus and oral cavity. In addition, our study was conducted in a region of China with high rates of ESCC and gastric cardia adenocarcinoma. As seen previously, the risk factors in this region may differ from Western countries (33). Therefore, our result may not generalize to other populations.
Our results suggest that the microbiota in upper digestive tract may play an important role in the development of gastric and esophageal cancer. Monitoring or even modifying our upper digestive tract microbiota might provide an opportunity for predicting or even changing the risk of gastric and esophageal cancer. However, more investigation in prospective cohorts of people are needed to confirm whether and how the upper digestive tract microbiota may affect the development of gastric and esophageal cancer.
Supplementary Material
Impact.
These novel findings suggest that the upper digestive tract microbiota may play a role in the etiology of chronic atrophic gastritis and ESD, and therefore in the development of gastric and esophageal cancers.
Acknowledgments
This study was supported by the Intramural Research Program of the NIH, National Cancer Institute.
Footnotes
Conflict of interest: No
References
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS one. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. Journal of medical microbiology. 2009;58:509–16. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 8.Samloff IM, Taggart RT. Pepsinogens, pepsins, and peptic ulcer. Clinical and investigative medicine Medecine clinique et experimentale. 1987;10:215–21. [PubMed] [Google Scholar]
- 9.Ren JS, Kamangar F, Qiao YL, Taylor PR, Liang H, Dawsey SM, et al. Serum pepsinogens and risk of gastric and oesophageal cancers in the General Population Nutrition Intervention Trial cohort. Gut. 2009;58:636–42. doi: 10.1136/gut.2008.168641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda H, Saito D, Hayashi S, Hisai H, Ono H, Yoshida S, et al. Helicobacter pylori infection, serum pepsinogen level and gastric cancer: a case-control study in Japan. Japanese journal of cancer research : Gann. 1995;86:64–71. doi: 10.1111/j.1349-7006.1995.tb02989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stemmermann GN, Samloff IM, Nomura AM, Heilbrun LK. Serum pepsinogens I and II and stomach cancer. Clinica chimica acta; international journal of clinical chemistry. 1987;163:191–8. doi: 10.1016/0009-8981(87)90022-2. [DOI] [PubMed] [Google Scholar]
- 12.You WC, Blot WJ, Zhang L, Kneller RW, Li JY, Jin ML, et al. Serum pepsinogens in relation to precancerous gastric lesions in a population at high risk for gastric cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1993;2:113–7. [PubMed] [Google Scholar]
- 13.Hsing AW, Hansson LE, McLaughlin JK, Nyren O, Blot WJ, Ekbom A, et al. Pernicious anemia and subsequent cancer. A population-based cohort study. Cancer. 1993;71:745–50. doi: 10.1002/1097-0142(19930201)71:3<745::aid-cncr2820710316>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Ye W, Nyren O. Risk of cancers of the oesophagus and stomach by histology or subsite in patients hospitalised for pernicious anaemia. Gut. 2003;52:938–41. doi: 10.1136/gut.52.7.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iijima K, Koike T, Abe Y, Inomata Y, Sekine H, Imatani A, et al. Extensive gastric atrophy: an increased risk factor for superficial esophageal squamous cell carcinoma in Japan. The American journal of gastroenterology. 2007;102:1603–9. doi: 10.1111/j.1572-0241.2007.01257.x. [DOI] [PubMed] [Google Scholar]
- 16.Ye W, Held M, Lagergren J, Engstrand L, Blot WJ, McLaughlin JK, et al. Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. Journal of the National Cancer Institute. 2004;96:388–96. doi: 10.1093/jnci/djh057. [DOI] [PubMed] [Google Scholar]
- 17.Kamangar F, Diaw L, Wei WQ, Abnet CC, Wang GQ, Roth MJ, et al. Serum pepsinogens and risk of esophageal squamous dysplasia. International journal of cancer Journal international du cancer. 2009;124:456–60. doi: 10.1002/ijc.23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan QJ, Roth MJ, Guo HQ, Koclunan ML, Wang G, Henry M, et al. Cytologic detection of esophageal squamous cell carcinoma and its precursor lesions using balloon samplers and liquid-based cytology in asymptomatic adults in Linxian, China (vol 52, pg 14, 2008) Acta Cytol. 2008;52:276. doi: 10.1159/000325430. [DOI] [PubMed] [Google Scholar]
- 19.Wei WQ, Abnet CC, Lu N, Roth MJ, Wang GQ, Dye BA, et al. Risk factors for oesophageal squamous dysplasia in adult inhabitants of a high risk region of China. Gut. 2005;54:759–63. doi: 10.1136/gut.2004.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamangar F, Qiao YL, Blaser MJ, Sun XD, Katki H, Fan JH, et al. Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. British journal of cancer. 2007;96:172–6. doi: 10.1038/sj.bjc.6603517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. Journal of periodontology. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microbial ecology. 2007;53:371–83. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 23.Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 25.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. The ISME journal. 2011;5:169–72. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. Journal of bacteriology. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokal R, Michener C. University of Kansas Science Bulletin. 1958. A statistical method for evaluating systematic relationships; pp. 1409–38. [Google Scholar]
- 28.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 (Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. Plos Biol. 2008;6:2383–400. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science (New York, NY) 2013;339:548–54. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, et al. Diet Drives Convergence in Gut Microbiome Functions Across Mammalian Phylogeny and Within Humans. Science (New York, NY) 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterology clinics of North America. 2009;38(1):27–57. doi: 10.1016/j.gtc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.