Abstract
Direct counting of biomolecules within biological complexes or nanomachines is demanding. Single molecule counting using optical microscopy is challenging due to the diffraction limit. The Single Molecule Photobleaching (SMPB) technology for direct counting developed by our team (Shu et al, EMBO J, 2007, 26:527; Zhang et al, RNA, 2007, 13:1793) offers a simple and straightforward method to determine the stoichiometry of molecules or subunits within biocomplexes or nanomachines at nanometer scales. Stoichiometry is determined by real-time observation of the number of descending steps resulted from the photobleaching of individual fluorophore. This technology has now been used extensively for single molecule counting of protein, RNA, and other macromolecules in a variety of complexes or nanostructures. Here, we elucidate the SMPB technology, using the counting of RNA molecules within a bacteriophage phi29 DNA-packaging biomotor as an example. The method described here can be applied to the single molecule counting of other molecules in other systems. The construction of a concise, simple and economical single molecule total internal reflection fluorescence (TIRF) microscope combining prism-type and objective-type TIRF is described. The imaging system contains a deep-cooled sensitive EMCCD camera with single fluorophore detection sensitivity, a laser combiner for simultaneous dual-color excitation, and a Dual-View™ imager to split the multiple outcome signals to different detector channels based on their wavelengths. Methodology of the single molecule photobleaching assay used to elucidate the stoichiometry of RNA on phi29 DNA packaging motor and the mechanism of protein/RNA interaction are described. Different methods for single fluorophore labeling of RNA molecules are reviewed. The process of statistical modeling to reveal the true copy number of the biomolecules based on binomial distribution is also described.
Keywords: single molecule, photobleaching, RNA, stoichiometry, TIRF
INTRODUCTION
Fluorescence microscopy offers a way to directly visualize biomolecules carrying fluorescent labels. However, the resolution of optical microscopy is restricted by the diffraction limit, which is about 200 nm for visible light. Molecules that reside within this limit cannot be distinguished from each other by conventional optical microscopy. In recent years, many super resolution microscopy techniques have been developed, and biological samples can now be imaged with nanometer precision [1–17]. Single Molecule Photobleaching (SMPB) for direct counting, developed by our team [18,19], has been a simple and straightforward way to study stoichiometry of subunits in biocomplexes. This technology has now been used extensively for single molecule counting of proteins, RNAs and other macromolecules in a variety of biomachines and nanostructures [13,18–35]. A single fluorophore shows quantized intensity drop when photobleached (Fig. 1A). Utilizing this characteristic of single fluorophore, one can directly count the number of fluorophores within a diffraction limited fluorescent spot from the step-wise intensity drop over time. More recently, other novel approaches such as photoactivated Localization Microscopy (PALM) [36] based on localization of stochastically photoactivated molecule and Stochastic Optical Reconstruction Microscopy (STORM) [5] have also been reported for quantitative analysis of fluorescent molecules, providing information such as stoichiometry, size and arrangement of the biocomplexes [37–41]. However, some molecules may not be photoactivated during PALM and STORM imaging, making it difficult to determine the absolute number of molecules and leading to underestimation of the stoichiometry. Unlike PALM or STORM, SMPB cannot provide information on size, dimension parameter, or spatial distribution of the molecules. However, in SMPB, the choice of fluorophores is not limited to photoactivable fluorescent molecules, and mathematic algorithms are not required. The stoichiometry of packaging RNA (pRNA) in the bacteriophage phi29 DNA packaging motor has been confirmed using this method [18,19]. Further photobleaching studies on the phi29 DNA packaging motor facilitated the elucidation of a novel mechanism of protein/RNA interaction [42]. The method has also been used to determine the copy number of viral RNAs packaged in a single influenza virus particle [43]. In this paper, we focus on the studies of direct counting of RNA subunits within nanometer-sized biological nanoparticles using the SMPB approach. The single fluorophore labeling schemes for RNA molecules and the statistical analysis to conclude the true stoichiometry of the RNA subunits in the circumstance of incomplete labeling are discussed. This technology can be applied to single molecule counting of RNA, DNA, protein and other molecules in nanoparticles, macromolecule complexes, and biological complexes. Here, we describe its application focusing on RNA, while the method introduced here can be used on other nanomachines and complexes, as well.
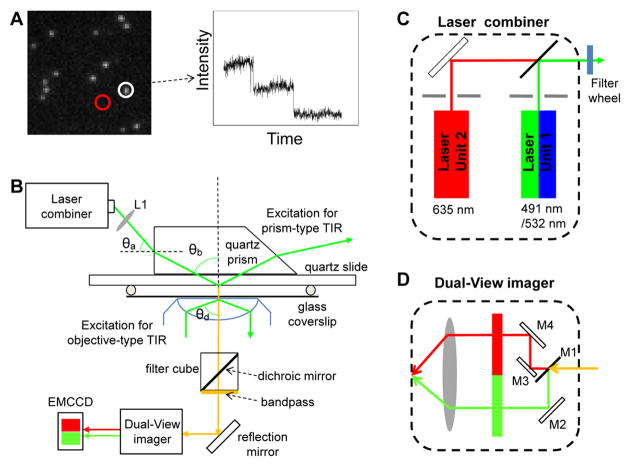
Fig. 1. Single molecule photobleaching assay for direct counting of biomolecules.
(A) Left: A typical initial fluorescence image of individual biocomplexes containing multiple copies of fluorescently labeled molecules prior to photobleaching process. Right: Photobleaching trace of average fluorescent intensity vs. time for the fluorescent spot circled in the image on the left. (B) Schematic of the single molecule TIRF microscope. Adapted from [18] © 2007 with permission from Nature Publishing Group. (C–D) Schematic designs of the optics inside the (C) laser combiner and the (D) Dual-View Imager™. Adapted from [31] © 2010 with permission from Springer.
METHODS
1. Instrumentation of single molecule total internal reflection fluorescence (TIRF) microscope: combined prism-type and objective-type
A key step to achieving single fluorophore detection in solution is to decrease the background fluorescence from the bulk solution. One approach that has been widely used in single molecule imaging is TIRF(Total Internal Reflection Fluorescence Microscope) [44–50], which is our focus here. It utilizes an evanescence excitation source generated from the total internal reflection of the excitation beam to eliminate the background fluorescence from the bulk solution. Single molecule counting using photobleaching technique is a straightforward way to determine subunit stoichiometry at the nanometer scale. It’s as easy as counting the stars. On a dark night the stars are bright and easy to see but, during the day, the bright light from the sun blocks our view of anything in space and you cannot see them. With a TIRF microscope, light from a laser is internally reflected at the water/quartz (or glass) interface, away from the objective. An evanescent field is generated, with its intensity dropping along the surface normal exponentially at a power of 6. The penetration depth of the excitation field is calculated to be around 200 nm for visible light. Only the molecules at the interface of two surfaces with different refraction indices can be excited. The molecules fluoresce brightly against a dark background towards the objective and are distinguishable due to a high contrast (Fig. 1A). There are two ways to achieve TIRF excitation, one is facilitated by a top prism that directs the excitation beam [18,19,44,47,48,50], and the other involves the beam total internally reflected through an objective with a high numerical aperture (NA) [45,48,50–52].
1.1 Prism-type TIRF
In prism-type TIRF, the sample perfusion chamber is usually made of a quartz slide on top and a glass coverslip on bottom (Fig. 1B) [2,47,53–57]. The contact between the bottom surface of the quartz prism and the top surface of the quartz slide of the chamber was mediated by glycerol, which has a refractive index close to that of quartz. The excitation laser beam was elevated through a series of optics to shine to the side of the prism, which was fixed directly above the objective’s field of view by a customized prism holder; the incident angle (θa) was adjusted so that after its refraction through the prism, the incident angle (θb) of the beam at the quartz/water interface was larger than the critical angle (θc) for total internal reflection (Fig. 1B). The critical angle (θc) for total internal reflection of a 532 nm laser beam at a quartz/water interface was 65.6°. Using a Pellin-Broca prism, as shown in Fig. 1B, θa = 33° gave an incident angle θb = 68° at the interface. The depth of penetration of the evanescent field was approximately 160 nm for the 532 nm laser beam. The size of the focused laser beam at the quartz/water was adjusted to approximately 150 μm × 50 μm by fine-tuning the position of the focusing lens (Fig. 1B, L1).
1.2 Objective-type TIRF
Objective-type TIRF [45] is achieved by using a TIRFM objective with an NA larger than 1.38, and an angular aperture allowing an incident angle larger than the critical angle (θd) for total internal reflection at the glass/water interface (Fig. 1B). The excitation beam was expanded, reflected by the dichroic mirror in the microscope filter cube and then focused onto the back focal plane of the objective through a lens next to the back aperture of the microscope. The sample was excited by the laser beam through the TIRFM objective. By adjusting the position of the lens, the beam could be aligned to the side of the objective, and thus achieve total internal reflection through the objective lens (Fig. 1B). With an NA of 1.45 for a TIRFM objective (Olympus PlanApo 100× Oil, NA 1.45), the depth of penetration into the sample chamber was about 100 nm for a 532 nm laser. Only the samples that were attached to the bottom glass surface of the perfusion chamber could be excited by the TIRF evanescent field.
1.3 Excitation source – laser combiner for multi-color excitation
When fluorophores of different spectra are used, multiple lasers are needed as excitation sources. For easy and safe beam manipulation, the lasers can be pre-aligned in a combiner and coupled to a single-mode fiber optic for further alignment with the microscope. In our experiments, lasers of three wavelengths (a Cobolt Dual Calypso™ laser of 491 nm/532 nm and a Coherent CUBE laser of 635 nm) were co-aligned inside a laser combiner (Fig. 1C) [31]. A long pass (LP) dichroic mirror with edge wavelength of 550 nm (Chroma, USA) was used inside the combiner to transmit the 635 nm beam, while reflecting the 491 nm/532 nm beam in order to co-align all three beams to the same fiber optic. The 491/532 nm laser and 635 nm laser were controlled separately by computer-controlled electronic shutters. Additional band pass filters with ranges of 488–505 nm and 519–700 nm (Semrock) were placed after the fiber optic to further separate the beam of 491 nm from 532 nm.
1.4 Detection of multi-color fluorophores
Fluorescence signals were detected with a back-illuminated electron-multiplied CCD (EMCCD) camera (Andor Technology), which was operated at −70ºC to reduce the thermal-dependent dark noise. A commercially available Dual-View™ imager (Roper Scientific) (Fig. 1D) was used to split fluorescence signals of two different wavelengths before they reached the EMCCD camera. A dichroic mirror (M1) was used to transmit the signal with a longer wavelength, while reflecting the shorter one to a side mirror (M2), which brought the signal of shorter wavelength to the detector. The signal that passed through the dichroic was reflected by two additional mirrors (M3, M4) to reach the detector. Band pass filters were used before the detector to further block unwanted signals. The optics were aligned so that the two signals reached the detector side by side, and were recorded simultaneously [19,31]. Various pairs of fluorophores can be simultaneously imaged and co-localized with different combinations of lasers in the laser combiner and filter sets in the Dual-View™ imager.
2. Single fluorophore labeling of RNA
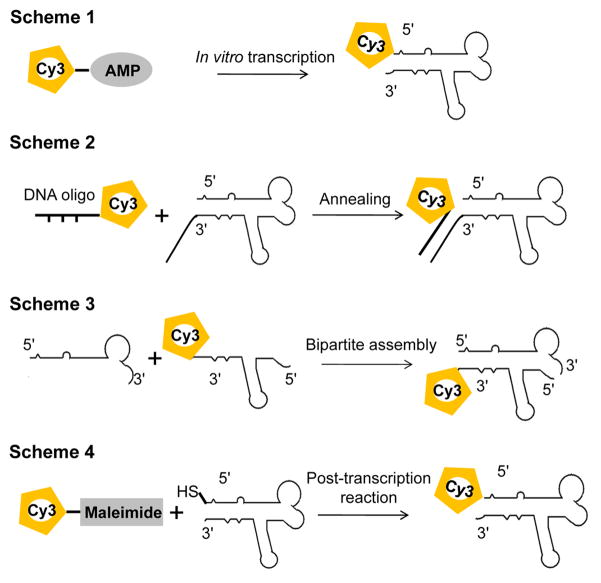
A key step in direct counting of subunits using photobleaching assay is to ensure single fluorophore labeling of each subunit. For proteins, fusion tagging with a fluorescent protein, such as GFP (green fluorescent protein) or YFP (yellow fluorescent protein) [58,59], is the most common way to fluorescently label protein with single fluorophore at high labeling efficiency [20,24,28,60]. For single labeling of RNA, chemical synthesis can produce site-specific labeled RNA strands with lengths up to 60–80 bases. For longer RNA molecules, single labeling can be achieved by in vitro transcription in a T7 RNA polymerase system with an Ø2.5 promoter [61,62] using fluorescent AMPs (adenosine monophosphates; e.g., F550/570 and F650/670 from AdeGenix) that can initiate the transcription, but cannot be incorporated for chain extension (Fig. 2, Scheme 1) [18,63,64]. Single labeling at the 3′-end can be achieved by using T4 RNA ligase [65].
Fig. 2. Different schemes of single fluorophore labeling of RNA molecules.
Alternatively, an overhang can be designed to either end of the RNA molecule and single labeling can be achieved by annealing the RNA with a short, single-fluorophore labeled DNA/RNA oligo (IDT) (Fig. 2, Scheme 2) [31]. A bipartite approach can also be used to produce singly labeled RNA molecules through assembly of two short RNA fragments. As the lengths of the RNA fragments are reduced, it is possible to make the single fluorophore labeled RNA through chemical synthesis (Fig. 2, Scheme 3) [66–68]. In addition, labeling can also be achieved by post-transcription chemical reaction of an RNA carrying terminal functional groups with a fluorescent reagent [69,70]. For example, single labeling can be achieved by reaction of a thiol end-labeled RNA with commercially available fluorescent maleimide (Fig. 2, Scheme 4) [69], or through “Click” chemistry of alkyne modified RNA with azide modified fluorophore [70–72].
The labeled RNAs were subject to further purification using gel electrophoresis. The labeling efficiency of the RNA molecules was obtained from their UV/Vis absorbance spectra. RNA concentration was determined by OD260 (1 OD260 = 40 ng/μL). The molar concentration was converted based on the molecular weight of the RNA molecule. The concentration of the fluorophore was determined from OD at its maximum absorbance wavelength, and the labeling efficiency was calculated as the molar ratio between the fluorophore and the RNA molecule.
3. Photobleaching assay and analysis of the photobleaching traces
Single molecule photobleaching causes a step-wise intensity drop for individual fluorophores and direct counting is achieved by recording the total number of steps in the intensity change over time. A fluorescent molecule is similar to a living organism concerning a variable lifespan. The counting here works in situations similar to the investigation of a cage of living mice. While active, the mice are difficult to count especially in a darkroom. Instead of counting the live mice in the cage, we seek to count the dead mice. As they die, each mouse is removed and tallyed. When the cage is empty, the total number of the mice that were in it can be recorded. The single molecule photobleaching technique enables scientists to directly count nanometer-sized biomolecules one by one without mathematical extrapolation.
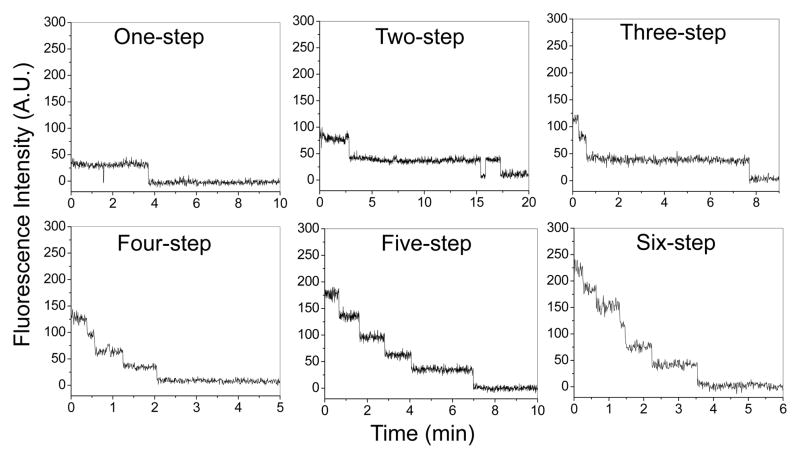
Bacteriophage phi29 DNA packaging motor is geared by a multimeric pRNA ring. The stoichiometry of the pRNA in the packaging motor has been a subject of fervent debate for a long time [18,42,73–77]. Cryo-EM studies from different laboratories have shown a hexameric [73] or a pentameric [74,77] pRNA ring on the motors. Biochemistry data has revealed that purified pRNA dimers and trimers are active in DNA packaging, implying that the number of pRNA molecules on a motor would be a common multiple of 2 and 3. Using single molecule photobleaching assay, the number of pRNA within one pRNA ring has been directly counted to be six [18].
3.1 Counting of pRNA in the phi29 DNA packaging motor
Bacteriophage phi29 DNA packaging motor is one of the strongest biological motors assembled in vitro. The motor is geared by a packaging RNA (pRNA) ring [18,42,75,78,79]. Since its discovery in 1987 [78], the 120-nucleotide pRNA has been extensively investigated, and the question of whether the pRNA is a pentamer or hexamer has been subjected to fervent debates since 1998. Such debate was ended after the application of Single Molecule Photobleaching technology to reveal that the pRNA ring contains six copy of RNA either at the initial assembly or during the active translocation of dsDNA by the motor [18]. The precise determination of the copy number of the RNA ring was further confirmed by RNA X-ray crystallography and atomic force microscopy (AFM), both revealing that the RNA ring is a hexamer with the interior angle of 120° [80].
Each pRNA molecule was singly labeled with a fluorescent dye of Cy3 or Cy5 at its 5′ end by transcription with fluorescent AMP. The biological activities of the fluorescently labeled pRNA, such as motor binding and DNA packaging, were tested before imaging to confirm that the pRNA retained its activity after labeling.
The packaging motor was first constructed by mixing the labeled pRNA with the motor in the presence of a buffer containing 100 mM NaCl, 10 mM MgCl2, and 50 mM Tris at pH 8, and were then isolated through 5–20% sucrose gradient to remove the unbound pRNA [18]. The isolated motor/pRNA complexes were tested to be active in DNA packaging and viral assembly, confirming that the complexes remained intact after purification. The complex was then immobilized to the antibody-coated surface of the chamber for TIRF imaging. The concentration of the sample was titrated so that individual complexes appeared as discrete stable fluorescent spots in the field of view with proper density. An oxygen scavenger system (0.5% β-D-glucose, 10 mM β-mercaptoethanol, and GODCAT: 0.2% Glucose Oxidase and 0.25% Catalase) was used to improve the photostability of Cy3 during the imaging by slowing down the photobleaching of Cy3 caused by photo-oxidation [81]. To avoid the possible underestimation of the step numbers due to premature photobleaching, the field of view was move to a fresh region before every data acquisition. This is especially important for counting of fluorescent proteins, as the oxygen scavenger system cannot prevent their photobleaching.
For Cy3 labeled pRNA, a 532 nm laser beam was used for excitation. The samples were excited continuously and the laser power was adjusted to 5~8 mw to ensure slow photobleaching so that multiple fluorophores were not bleached simultaneously, making it difficult to resolve the steps. Sequential images were taken at a rate of 2.3 ~ 4.3 Hz with an exposure time of 0.2 ~ 0.4 second. The exposure time could be increased to collect more photons in order to increase the signal-to-noise ratio. However, the temporal resolution would decrease in the meantime. Each fluorescent spots in the image (Fig. 1A), representing individual motor/pRNA complexes, were analyzed by Andor iQ software (Andor Technology). The average intensity of a circled area around the fluorescent spot (white circle) was measured with the nearby background fluorescence subtracted (red circle) at each time point (Fig. 1A) and plots of average fluorescence intensity versus time were produced for individual fluorescent spots (Fig. 3). To exclude the possibility of detecting signals from contaminants rather than the samples, an essential control sample without fluorescent labels was tested under the same imaging condition to ensure the signals recorded were truly from the fluorescently labeled samples.
Fig. 3. Typical photobleaching traces of single motor/Cy3-pRNA complexes showing 1 – 6 steps of intensity drop over time.
Adapted from [18] © 2007 with permission from Nature Publishing Group.
A major challenge often encountered in single molecule imaging is the requirement of subnanomolar sample concentrations to ensure the fluorescent spots are well separated from each other. The motor/pRNA complex was found to remain intact at the imaging concentrations. However, the imaging concentration may lead to the dissociation of some other biological complexes. In such cases, encapsulation of the complex within a restricted small volume can be used [82,83]. Using small unilamellar vesicles (SUVs) of 200 nm in diameter, encapsulation of one complex will result in a local concentration of a few hundred nanomolar, which may keep the complex intact.
Non-specific binding of biological samples, especially fluorescently labeled proteins, to the TIRF chamber surface represents another challenge in single molecule imaging, as it may require the use of a protein concentration much lower than its physiological concentration. Further surface coating with BSA or PEG [50,53,84] will be needed to block the non-specific protein bindings.
3.2 Analysis of the photobleaching traces
Direct counting of steps from the photobleaching traces reveals the number of fluorophores within the diffraction limited fluorescent spot. As the environment of each fluorophore differs from each other within the sample chamber, the photobleaching step size may vary. A histogram revealing the distribution in step sizes can be used to identify the one-step-drops due to bleaching of multiple fluorophores. Furthermore, due to their intrinsic properties, the intensity of fluorescent proteins fluctuates significantly during photobleaching, making it difficult to identify the steps. Mathematic filters, such as Chung-Kennedy filter [85], can be applied to the raw photobleaching traces to extract signals from noise, thus to reveal distinctive steps from the traces [20,86]. Counting programs have been developed for automatic step detection in the photobleaching traces [28,30,87].
As the number of fluorescent molecules increases in a complex, direct counting of steps become challenging. This is due to the increased possibility of synchronous photobleaching of multiple fluorophores, leading to the underestimation of the total steps. Resolving the overlapped steps will also be challenging when more molecules are involved and fluctuation in step size occurs. With the current experimental setup, we were able to directly count up to 11–12 Cy3 photobleaching steps within one complex (possibly two motors) empirically [18]. A maximum of 15 steps was however predicted to be countable without mathematical extrapolation [22]. Alternatively, number of fluorophores can be estimated by dividing the intensity of the fluorescent spot prior to photobleaching with that of a single fluorophore [20,86,88], although errors can occur due to the fluctuation of step size.
3.3 Counting of pRNA in active packaging intermediates
Similarly, the packaging motor was constructed using Cy3-labeled pRNA and Cy5-labeled genomic DNA [18]. The DNA-packaging was stalled by a non-hydrolysable ATP analog, γ-S-ATP, and isolated through sucrose gradient. The packaging intermediate was identified by the co-detection of Cy3 and Cy5 signals within the same fluorescent spots. To confirm that the overlapped signals did not come from the accidental co-localization of the two fluorophores, a control sample without packaging activity was also tested. The result confirmed that the Cy3-pRNA and Cy5-DNA would not co-exist within the same motor if DNA was not packaged. Thus, the spots that contained both Cy3 and Cy5 signals represented DNA-packaging intermediate and were then further analyzed for their Cy3 photobleaching traces. The number of fluorescently labeled pRNA in the packaging intermediates was therefore determined.
3.4 Simultaneous dual-color photobleaching of differently labeled pRNA on the phi29 DNA packaging motor
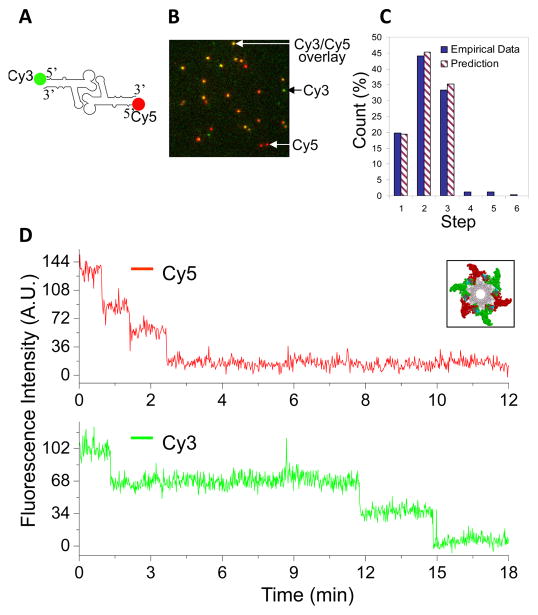
Wild-type pRNA contains two interlocking loops with complementary sequences. The interlocking loops are essential for pRNA dimerization. Two differently labeled pRNAs can be incorporated into one pRNA ring on a motor by carefully re-engineering their loop sequences. For example, with the same letter in upper and lower cases representing the complementary sequences, a pRNA-Aa’ containing two complementary interlocking loops is self-efficient to form a dimer in solution, and subsequently a closed ring on the motor. However, a pRNA-Ab’ with unmatched loop sequences can only form a dimer, and subsequently a closed ring, in the presence of a partner pRNA-Ba’. Thus, the fluorescent pRNA ring was designed to have a Cy3 labeled pRNA-Ab’ paired with a Cy5 labeled pRNA-Ba’ [18]. With the laser combiner and Dual-View™ imager, the imaging system was able to detect fluorescence signals from the two differently labeled pRNA simultaneously, using the Dual-View™ imager filter set specific for Cy3/Cy5 pair. In addition, the filter set inside the microscope turret was also carefully adjusted with the dichroic mirror to block both excitation beams of 532 nm and 635 nm, while still allowing the two fluorescence signals to transmit.
Another dual-pass filter was added to the filter cube to further block the excitation source. For simultaneously imaging of Cy3-pRNA and Cy5-pRNA on the motor, a dichroic mirror with dual-edge wavelengths of 545 nm and 650 nm (Semrock) and a dual-band bandpass filter for Cy3/Cy5 (Chroma) were used in the microscope filter cube (Fig. 1B). Sequential images were taken and each image was processed through Field-Split function to overlay the signals in the Cy3 and Cy5 channels. TetraSpeck™ fluorescent microspheres that display four different colors (Life Technology) were used to calibrate the alignment of the signals in the two channels before the images of motor/pRNA complexes were analyzed. Only the fluorescence spots that contained both Cy3 and Cy5 signals were analyzed for photobleaching to determine the number of each fluorescently labeled pRNA inside an individual motor (Fig. 4). A sample of motor containing Cy3-pRNA only was examined for the possible crosstalk of Cy3 signal to Cy5 channel, and cares were taken during the data analysis to ensure that the bleedthrough of Cy3 intensity drop was not counted for Cy5 quantification.
Fig. 4. Dual-color photobleaching of differently labeled pRNAs within the same motor/pRNA complex.
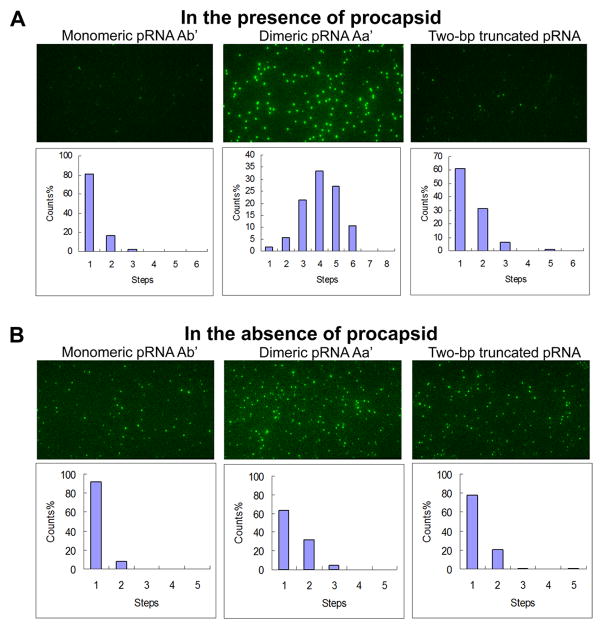
(A) Design of dual-labeled pRNA for motor binding. (B) Typical overlaid fluorescence image of the dual-color labeled motor/pRNA complexes. (Green: Cy3; Red: Cy5; Yellow: Cy3/Cy5 overlay). (C) Comparison of the experimental histogram of photobleaching steps with the theoretical histograms for Cy3-pRNA based on 70% labeling efficiency. (D) A dual-color photobleaching trace of motor/pRNA complex showing three Cy3-pRNA molecules and three Cy5-pRNA molecules on the same motor. Adapted from [18] © 2007 with permission from Nature Publishing Group.
3.5 Elucidating mechanism of pRNA ring assembly on the motor
Specificity in protein/RNA interaction is generally believed to rely on either molecular contact through surface charges, or 3D structure matching via conformational capture [89,90] or induced fit [89,91,92]. However, a single molecule photobleaching study has revealed that the specificity and affinity in the motor/pRNA interaction is dependent on a static pRNA ring formation [42]. Cy3-labeled pRNA-Aa’ has been found to have a much stronger affinity to the motor than pRNA-Ab’ that can only exist as monomers in solution, as demonstrated by the amount of fluorescent spots observed in Fig. 5A. Analysis of their photobleaching traces also showed different distributions in copy numbers, with pRNA-Aa’ having significantly more copies on the motor ; while the majority of pRNA-Ab’ showed only one copy, similar to that of free pRNA in solution (Fig. 5B). More interestingly, another mutant pRNA with the ability to form a closed ring, but with a smaller ring perimeter, was also found to be incapable of binding to the motor, showing much less binding compared to pRNA-Aa’ (Fig. 5B). The results indicate that the size of the pRNA ring is also an important factor in motor/pRNA interaction.
Fig. 5. Single molecule photobleaching comparing the oligomerization state of different pRNAs in the (A) presence and (B) absence of motor.
Adapted from [42] © 2008 with permission from Oxford University Press.
4. Statistical analysis of photobleaching histograms
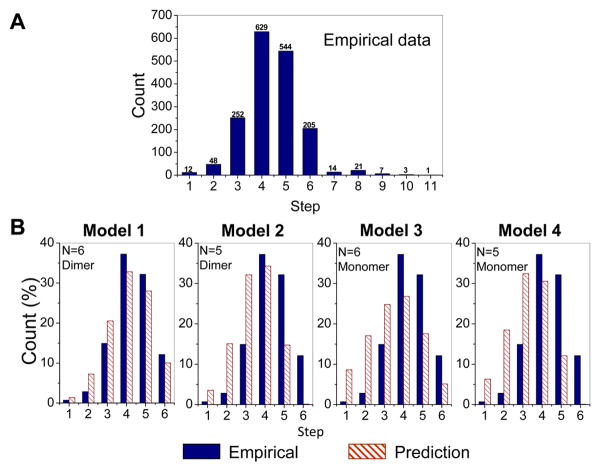
Different from proteins that are labeled through fusion technique, with which the protein and the tag are co-expressed and 100% labeling is warranted, fluorescent labeling of RNA may result in incomplete labeling. Therefore, the number of photobleaching steps revealed in the experiment data does not directly reflect how many pRNA molecules are in that biocomplex, e.g., the histogram of photobleaching steps of Cy3-pRNA in the motor/pRNA complex showing a distribution from 1 to up to 11 steps (Fig. 6A). Statistical analysis with the labeling efficiency taken into account is therefore needed to obtain the real copy number of pRNA within the complex as described below.
Fig. 6. Statistical analysis to obtain the actual copy numbers of pRNA on the motor.
(A) Experimental histogram of photobleaching steps of motor/Cy3-pRNA complexes. (B) Fitting of the experimental data with different statistic models (Models 1 – 4) that were constructed based on 70% labeling efficiency for Cy3-pRNA. Adapted from [18] © 2007 with permission from Nature Publishing Group.
4.1 Setting up models
To reveal whether the stoichiometry of pRNA is five or six on the motor, four statistical models were constructed based on different hypotheses, as shown in Fig. 6B, in shaded columns. The models also considered different assembly pathways of pRNA onto a motor. The models were built based on the following hypothesis [18]:
Model 1: pRNA assembles into a hexamer on the motor from a pRNA dimer.
Model 2: pRNA initially assembles as a hexamer on the motor from a pRNA dimer. However, one pRNA shifts away and leaves a pRNA pentamer on the motor.
Model 3: pRNA assembles as a hexamer on the motor from a pRNA monomer.
Model 4: pRNA assembles as a pentamer on the motor from a pRNA monomer.
Mathematical formulas were developed for each model to predict the fractions of motor containing i copies of Cy3-pRNA, where 1 ≤ i ≤ 6, as only the Cy3-pRNA can be counted in the experiment. In detail, using Model 1 as an example, each observable motor would contain 1, 2, or 3 pRNA dimers, and their fractions in the total motor were as defined a, b, and c, respectively. Since the binding of pRNA to motor is cooperative [79], the fractions would be a ≤ b ≤ c, where c = 1–a–b.
The possibility of i copies of Cy3 molecules to be found on each motor follows binomial distribution (k, E), where k stands for the total number of pRNA on the motor and E is the labeling efficiency of the Cy3-pRNA. The expansion of binomial (k, E) indicates that the possibility of having i Cy3-pRNA on one motor is . In the case of Model 1, k is equal to 2, 4, or 6. The fractions of motor containing i copies of Cy3-pRNA (fi) can therefore be calculated using the Eq. 1 below:
| (Eq.1) |
As the total number of observable motor s does not include the ones that have no Cy3-labeled pRNAs bound, the predicted fractions can be expressed in Eq. 2 below:
| (Eq.2) |
For the other three models, similar procedures were taken to obtain the theoretical histograms, as shown in figure 6B.
4.2 Fitting the models
Mean squared error (MSE) is defined as the average squares of discrepancy between the empirical fraction and the predicted fraction (Eq. 3).
| (Eq. 3) |
In Eq. 3, yi is the fraction empirically determined for motor containing i copies of Cy3-pRNA and can be obtained from Fig. 6A. A Matlab code was programmed to locate the pair of (a,b) that gives the best-fit to the empirical data with a smaller MSE indicating better fitting of the model.
4.3 Evaluating the models
For the stoichiometry study of pRNA bound to motor, the focus was to determine whether the pRNA ring was a hexamer or a pentamer; therefore, the focus of the evaluation of the models was on the fraction of motor containing 6 Cy3-pRNA molecules. Suppose the pair (â, b̂) gave the best fit, by inserting (â, b̂) back into the model, the z-score could be calculated using Eq. 4 below:
| (Eq. 4) |
where T is the total number of observable motor s containing at least 1 copy of Cy3-pRNA. The rationale behind the z-score is that, when the model is accurate, z-score is approximately distributed as a standard Gaussian (i.e., N(0,1)) and should be relatively small. An unusually large z-score implies that the model is not accurate. Equivalently, the p-values were determined by p=P{|N(0,1)|≥|z|}. An unusually small p-value also implies that the model is not accurate.
All four models were evaluated and the model that fit the empirical data the best was accepted. Model 1 gave the best fit with the empirical data, based on the fitting parameters of z-score and p-value [18]. It was therefore concluded that the pRNA ring on the motor was most likely to be a hexamer assembled from pRNA dimers. The other three models showed relatively large z-scores and extremely small p-values, and were thus excluded [18]. Based on the statistical analysis, it was also found that majority of the motors contained either six pRNA molecules or no pRNA at all.
FUTURE PROSPECTIVES
Single molecule imaging techniques enable the study of biomolecules one molecule at a time, and make it possible to investigate the stoichiometry of individual complexes. Single molecule photobleaching allows for the direct counting of biomolecules in a straightforward manner. Since our publication of this technique [18,19], this technology has been widely used to study protein and RNA subunits in biological complexes. More and more noncoding RNA species have been uncovered, in combination with the fluorogenic property of RNA [93–95]. It is expected that this SMPB technology will play more and more roles in the study of the biological roles of RNA. In a similar way, components of DNA, lipid, polysaccharide or metals can also be counted, by applying single fluorophore labeling technique or self-fluorescence property. Improvement of this technology will rely on the discovery of photostable, less fluctuating, less blinking fluorophores, and more sensitive optical instrument.
Acknowledgments
This technology would not have been possible without the contribution from, collaboration and communication with Peter Stockley, Wulf-Dieter Moll, David Rueda, Nils Walter, Faqing Huang, Chris Meiners, Meredith Lambert, Jiashun Jin, Taekjip Ha, Toshio Yanagida, Masasuke Yoshida, Kazuhiko Kinosita, Eckhard Jankowsky and Paul Selvin during the development of this method. The research was supported by NIH grants R01EB003730, R01EB012135 and U01CA151648 to Guo. Guo is a cofounder of Kylin Therapeutics, Inc., and Biomotor and Nucleic Acid Nanotechnology Development Corp., Ltd. Guo’s Endowed Chair of Nanobiotechnology position is supported by the William Farish Endowment Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yildiz A, Selvin PR. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc Chem Res. 2005;38:574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 2.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michelotti N, de SC, Johnson-Buck AE, Manzo AJ, Walter NG. A bird’s eye view tracking slow nanometer-scale movements of single molecular nano-assemblies. Methods Enzymol. 2010;475:121–148. doi: 10.1016/S0076-6879(10)75006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang B, Wang WQ, Bates M, Zhuang XW. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 7.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 9.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proc Natl Acad Sci USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balci H, Ha T, Sweeney HL, Selvin PR. Interhead distance measurements in myosin VI via SHRImP support a simplified hand-over-hand model. Biophys J. 2005;89:413–417. doi: 10.1529/biophysj.105.060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu XH, Wu D, Mets L, Scherer NF. Nanometer-localized multiple single-molecule fluorescence microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu Rev Phys Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jankowsky E. Biophysics: helicase snaps back. Nature. 2005;437:1245. doi: 10.1038/4371245a. [DOI] [PubMed] [Google Scholar]
- 15.Jankowsky E, Gross C, Shuman S, Pyle A. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 16.Kinosita K., Jr Real time imaging of rotating molecular machines. FASEB J Suppl. 2003;2:S201–S208. doi: 10.1096/fasebj.13.9002.s201. [DOI] [PubMed] [Google Scholar]
- 17.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 18.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Shu D, Huang F, Guo P. Instrumentation and metrology for single RNA counting in biological complexes or nanoparticles by a single molecule dual-view system. RNA. 2007;13:1793–1802. doi: 10.1261/rna.587607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 21.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das SK, Darshi M, Cheley S, Wallace MI, Bayley H. Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. Chembiochem. 2007;8:994–999. doi: 10.1002/cbic.200600474. [DOI] [PubMed] [Google Scholar]
- 23.Arumugam SR, Lee TH, Benkovic SJ. Investigation of stoichiometry of T4 bacteriophage helicase loader protein (gp59) J Biol Chem. 2009;284:29283–29289. doi: 10.1074/jbc.M109.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonson PD, Deberg HA, Ge P, Alexander JK, Jeyifous O, Green WN, Selvin PR. Counting bungarotoxin binding sites of nicotinic acetylcholine receptors in mammalian cells with high signal/noise ratios. Biophys J. 2010;99:L81–L83. doi: 10.1016/j.bpj.2010.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding H, Wong PT, Lee EL, Gafni A, Steel DG. Determination of the oligomer size of amyloidogenic protein beta-amyloid(1–40) by single-molecule spectroscopy. Biophys J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherny D, Gooding C, Eperon GE, Coelho MB, Bagshaw CR, Smith CW, Eperon IC. Stoichiometry of a regulatory splicing complex revealed by single-molecule analyses. EMBO J. 2010;29:2161–2172. doi: 10.1038/emboj.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Douglas NR, Conley NR, Miller EJ, Frydman J, Moerner WE. Sensing cooperativity in ATP hydrolysis for single multisubunit enzymes in solution. Proc Natl Acad Sci USA. 2011;108:16962–16967. doi: 10.1073/pnas.1112244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuire H, Aurousseau MR, Bowie D, Blunck R. Automating single subunit counting of membrane proteins in mammalian cells. J Biol Chem. 2012;287:35912–35921. doi: 10.1074/jbc.M112.402057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zijlstra N, Blum C, Segers-Nolten IM, Claessens MM, Subramaniam V. Molecular composition of sub-stoichiometrically labeled alpha-synuclein oligomers determined by single-molecule photobleaching. Angew Chem Int Ed Engl. 2012;51:8821–8824. doi: 10.1002/anie.201200813. [DOI] [PubMed] [Google Scholar]
- 30.Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012;14:416–423. doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Shu D, Browne M, Guo P. Construction of a laser combiner for dual fluorescent single molecule imaging of pRNA of phi29 DNA packaging motor. Biomed Microdevices. 2009;12:97–106. doi: 10.1007/s10544-009-9364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coffman VC, Wu JQ. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revyakin A, Zhang Z, Coleman RA, Li Y, Inouye C, Lucas JK, Park SR, Chu S, Tjian R. Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 2012;26:1691–1702. doi: 10.1101/gad.194936.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta P, Jovanovic G, Lenn T, Bruckbauer A, Engl C, Ying L, Buck M. Dynamics and stoichiometry of a regulated enhancer-binding protein in live Escherichia coli cells. Nat Commun. 2013;4:1997. doi: 10.1038/ncomms2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokota H, Chujo YA, Harada Y. Single-molecule imaging of the oligomer formation of the nonhexameric Escherichia coli UvrD helicase. Biophys J. 2013;104:924–933. doi: 10.1016/j.bpj.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. Bio-distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J Control Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SH, Shin JY, Lee A, Bustamante C. Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM) Proc Natl Acad Sci USA. 2012;109:17436–17441. doi: 10.1073/pnas.1215175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One. 2011;6:e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ori A, Banterle N, Iskar M, ndres-Pons A, Escher C, Khanh BH, Sparks L, Solis-Mezarino V, Rinner O, Bork P, Lemke EA, Beck M. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat Protoc. 2013;8:345–354. doi: 10.1038/nprot.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou YY, Vafabakhsh R, Doganay S, Gao Q, Ha T, Palese P. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc Natl Acad Sci USA. 2012;109:9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 1995;374:555–559. doi: 10.1038/374555a0. [DOI] [PubMed] [Google Scholar]
- 45.Tokunaga M, Kitamura K, Saito K, Iwane AH, Yanagida T. Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem Biophys Res Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
- 46.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 47.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 48.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 49.Rueda D, Walter NG. Single Molecule Fluorescence Control for Nanotechnology. J Nanosci Nanotechnol. 2005;5:1990–2000. doi: 10.1166/jnn.2005.505. [DOI] [PubMed] [Google Scholar]
- 50.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Axelrod D. Selective imaging of surface fluorescence with very high aperture microscope objectives. Journal of Biomedical Optics. 2001;6:6–13. doi: 10.1117/1.1335689. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Shu D, Wang W, Guo P. Design and application of single fluorophore dual-view imaging system containing both the objective- and prism-type TIRF. Proc SPIE. 2010;7571:757107–757108. doi: 10.1117/12.847457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi UB, Strop P, Vrljic M, Chu S, Brunger AT, Weninger KR. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat Struct Mol Biol. 2010;17:318–324. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhuang X, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating Structural Dynamics and Function in Single Ribozyme Molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 56.Jankowsky E, Fairman ME, Yang Q. RNA Helicases: Versatile ATP-Driven Nanomotors. J Nanosci Nanotechnol. 2005;5:1983–1989. doi: 10.1166/jnn.2005.508. [DOI] [PubMed] [Google Scholar]
- 57.Jankowsky E, Gross C, Shuman S, Pyle A. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 59.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 60.Lee TJ, Zhang H, Chang CL, Savran C, Guo P. Engineering of the fluorescent-energy-conversion arm of phi29 DNA packaging motor for single-molecule studies. Small. 2009;5:2453–2459. doi: 10.1002/smll.200900467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruffner DE, Dahm SC, Uhlenbeck OC. Studies on the hammerhead RNA self-cleaving domain. Gene. 1989;82:31–41. doi: 10.1016/0378-1119(89)90027-9. [DOI] [PubMed] [Google Scholar]
- 62.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acid Research. 1987;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang F, Wang G, Coleman T, Li N. Synthesis of adenosine derivatives as transcription initiators and preparation of 5′ fluorescein- and biotin-labeled RNA through one-step in vitro transcription. RNA. 2003;9:1562–1570. doi: 10.1261/rna.5106403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li N, Yu C, Huang F. Novel cyanine-AMP conjugates for efficient 5′ RNA fluorescent labeling by one-step transcription and replacement of [gamma-32P]ATP in RNA structural investigation. Nucleic Acids Res. 2005;33:e37. doi: 10.1093/nar/gni036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uhlenbeck OC, England TE. 3′-Terminal labelling of RNA with T4 RNA ligase. Nature. 1978;275:560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- 66.Fang Y, Cai Q, Qin PZ. The procapsid binding domain of phi29 packaging RNA has a modular architecture and requires 2′-hydroxyl groups in packaging RNA interaction. Biochemistry. 2005;44:9348–9358. doi: 10.1021/bi0475020. [DOI] [PubMed] [Google Scholar]
- 67.Fang Y, Shu D, Xiao F, Guo P, Qin PZ. Modular assembly of chimeric phi29 packaging RNAs that support DNA packaging. Biochemical and Biophysical Research Communications. 2008;372:589–594. doi: 10.1016/j.bbrc.2008.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shu Y, Cinier M, Fox SR, Ben-Johnathan N, Guo P. Assembly of Therapeutic pRNA-siRNA Nanoparticles Using Bipartite Approach. Molecular Therapy. 2011;19:1304–1311. doi: 10.1038/mt.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hermanson GT. Bioconjugate Techniques. Academic Press; San Diego: 1996. [Google Scholar]
- 70.Paredes E, Evans M, Das SR. RNA labeling, conjugation and ligation. Methods. 2011;54(2):251–259. doi: 10.1016/j.ymeth.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 72.Bock VD, Hiemstra H, van Maarseveen JH. Cu-I-catalyzed alkyne-azide “click” cycloadditions from a mechanistic and synthetic perspective. European Journal of Organic Chemistry. 2006;2006(1):51–68. [Google Scholar]
- 73.Ibarra B, Caston JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J Mol Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 74.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 76.Zhang F, Anderson D. In vitro selection of Bacteriophage phi29 prohead RNA aptamers for prohead binding. J Biol Chem. 1998;273:2947–2953. doi: 10.1074/jbc.273.5.2947. [DOI] [PubMed] [Google Scholar]
- 77.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson D, Rossman MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 79.Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA: A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem. 2000;275(23):17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 80.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. Crystal Structure of 3WJ Core Revealing Divalent Ion-promoted Thermostability and Assembly of the Phi29 Hexameric Motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nature Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 82.Okumus B, Wilson TJ, Lilley DM, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rhoades E, Gussakovsky E, Haran G. Watching proteins fold one molecule at a time. Proc Natl Acad Sci USA. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 85.Chung SH, Kennedy RA. Forward-backward non-linear filtering technique for extracting small biological signals from noise. J Neurosci Methods. 1991;40:71–86. doi: 10.1016/0165-0270(91)90118-j. [DOI] [PubMed] [Google Scholar]
- 86.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 88.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 90.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 91.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 92.Cruz JA, Westhof E. The Dynamic Landscapes of RNA Architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 93.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reif R, Haque F, Guo P. Fluorogenic RNA Nanoparticles for Monitoring RNA Folding and Degradation in Real Time in Living Cells. Nucleic Acid Ther. 2013;22(6):428–437. doi: 10.1089/nat.2012.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shu D, Zhang L, Khisamutdinov E, Guo P. Programmable folding of fusion RNA complex driven by the 3WJ motif of phi29 motor pRNA. Nucleic Acids Res. 2013;42(2):e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]