Abstract
Background
Data are lacking to describe gene expression-based breast cancer intrinsic subtype patterns for population-based patient groups.
Methods
We studied a diverse cohort of women with breast cancer from the Life After Cancer Epidemiology (LACE) and Pathways studies. RNA was extracted from 1 mm punches from fixed tumor tissue. Quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) was conducted for the 50 genes that comprise the PAM50 intrinsic subtype classifier.
Results
In a subcohort of 1,319 women, the overall subtype distribution based on PAM50 was 53.1% Luminal A, 20.5% Luminal B, 13.0% HER2-enriched, 9.8% Basal-like, and 3.6% Normal-like. Among low-risk endocrine positive tumors (i.e. estrogen and progesterone receptor positive by immunohistochemistry, Her2 negative, and low histologic grade), only 76.5% were categorized as Luminal A by PAM50. Continuous-scale Luminal A, Luminal B, HER2-enriched, and Normal-like scores from PAM50 were mutually positively correlated; Basal-like score was inversely correlated with other subtypes. The proportion with non-Luminal A subtype decreased with older age at diagnosis, p trend < 0.0001. Compared with non-Hispanic whites, African-American women were more likely to have Basal-like tumors, age-adjusted odds ratio (OR) 4.4 (95% CI 2.3,8.4), whereas Asian and Pacific Islander women had reduced odds of Basal-like subtype, OR 0.5 (95% CI 0.3,0.9).
Conclusions
Our data indicate that over 50% of breast cancers treated in the community have Luminal A subtype. Gene expression-based classification shifted some tumors categorized as low risk by surrogate clinicopathological criteria to higher-risk subtypes.
Impact
Subtyping in a population-based cohort revealed distinct profiles by age and race.
Keywords: breast neoplasms, cohort studies, intrinsic subtypes, PAM50
Gene expression profiling has revealed intrinsic subtypes of breast cancer that improve prognostication (1–8) and prediction of response to therapy (7, 9, 10) compared with categories defined by clinicopathological characteristics. The luminal A subtype has best prognosis and is, in most populations examined, the most frequent subtype. Defining subtypes of breast tumors for participants in breast cancer epidemiologic studies is of interest for several reasons: the distribution of subtypes by host characteristics, or associations between subtypes and risk factors, may shed light on etiologic pathways; survival differences for subtype groups should be defined in population-based studies; the influence of modifiable risk factors on recurrence and survival may vary by subtype. Much of the existing data on gene expression-based breast cancer intrinsic subtypes have been derived from clinical trial populations or other selected populations, e.g. ER positive cases only (8, 9), cases diagnosed at ages younger than 55 years (3), or patients with node-negative or low histologic grade disease (10, 11). It is not known how well the subtype distributions estimated from these studies describe the population across all ages, across a range of clinical characteristics, and across racial and ethnic groups.
Microarray gene expression assay is the gold standard for intrinsic subtyping, but because fresh-frozen tissue is required, this technology is usually not feasible for large research study populations. Instead, strategies for assigning subtypes based on clinicopathological variables, i.e. estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her2), and proliferation markers or tumor grade, have been applied in clinical and epidemiologic studies (12–22). Limitations of the clinicopathological subtyping approach are that staining and scoring of immunohistochemical (IHC) markers is subject to variability and that subtypes classified using clinicopathological variables may not align with intrinsic subtypes classified by gene expression-based assays (10, 23). Subtype classifiers based on quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) of gene products from fixed tissue are a third strategy for intrinsic subtyping. RT-qPCR classifiers are more feasible for large studies than microarray techniques and more quantitative than IHC (6, 24). Whereas the clinical utility of RT-qPCR classifiers is an active area of research (25–27), examples of research applying these classifiers in epidemiology are very limited (28).
In this study, we applied the PAM50 assay, a well-characterized RT-qPCR intrinsic subtyping classifier that measures expression of 50 genes selected as characteristic of five breast cancer intrinsic subtypes (6, 10, 11, 29), to archived primary tumor tissue from participants in the LACE and Pathways breast cancer cohorts. We describe the overall distribution of subtypes Luminal A, Luminal B, HER2-enriched, Basal-like, and Normal-like, and subtype variation in relation to clinicopathological categories and patient characteristics.
METHODS
Study Population
The study population consisted of breast cancer survivors from the LACE and Pathways cohorts. LACE participants were 18–79 years old when diagnosed with early stage breast cancer (stage I with tumor size 1 cm or greater, stage II or stage IIIA) from 1996–2000. Additional eligibility criteria at the time of LACE recruitment in 2000–2002 were that the woman was within 39 months of diagnosis (mean time from diagnosis to enrollment = 23 months, 61% between 12 and 24 months), had completed any chemotherapy or radiation therapy, was free of breast cancer recurrence, and had no other cancer diagnosis within five years. LACE study methods and baseline characteristics of participants have been described (30). For the intrinsic subtyping study, we included LACE participants recruited from the Kaiser Permanente Northern California (KPNC) Cancer Registry and the Utah Cancer Registry. The Pathways Study enrolled women diagnosed with invasive breast cancer from 2005–2013 in KPNC with no previous diagnosis of other invasive cancer, and at least 21 years of age; most women were approached for enrollment within two months of diagnosis (mean time from diagnosis to enrollment = 1.8 months, maximum 7.2 months). Details of the Pathways study methods have been described (31). For the PAM50 intrinsic subtyping study, we included Pathways women diagnosed in 2006 through 2008. Participants provided informed consent under protocols approved by the Kaiser Permanente Northern California Institutional Review Board (IRB) and the University of Utah IRB.
Self-reported race and ethnicity were obtained at study enrollment using mailed (LACE) or in-person (Pathways) questionnaires. Characteristics at the time of diagnosis, including age, disease stage (American Joint Commission on Cancer, AJCC), tumor size, node status, histologic grade, estrogen receptor (ER) status, progesterone receptor (PR) status, and Her2 overexpression or amplification in the primary tumor, were abstracted from tumor registry data and medical records review.
A stratified random sample of women meeting eligibility criteria was selected for intrinsic subtyping. This sample forms a subcohort that will be used for future case-cohort analysis of outcomes of breast cancer recurrence and death. ER, PR and Her2 status based on IHC (and/or FISH for HER2) defined the strata used for sampling, with an 18% random sample selected among cases of the common breast cancer phenotype that is positive for ER or PR expression and negative for Her2 (and has low risk of recurrence) and a 100% sample of tumors that were ER- and PR- or Her2+.
Tissue samples
For cohort members selected for the subcohort, we contacted the hospital where surgery for resection of the primary tumor was performed, or the institution's pathology storage facility, to obtain formalin-fixed, paraffin-embedded (FFPE) tissue blocks from that procedure and corresponding slides and pathology reports. Slides were reviewed by one pathologist (R.E.F.). Final eligibility for the PAM50 subtyping study was determined based on the review of slides and pathology reports. If the appearance of the primary tumor tissue in the slides indicated to the pathologist that neoadjuvant therapy had been used before resection, or if the area of invasive tumor was observed to be smaller than 0.5 cm in diameter, the case was classified as ineligible. If the pathology report indicated bilateral disease, the case was classified as ineligible. For eligible cases, the pathologist marked an area of representative tumor tissue on a slide. Tissue punches 1 mm in diameter were obtained from the area of the FFPE tissue block corresponding to the marked slide. Two punches per case, or one punch if the primary tumor was less than 0.7 cm in diameter, were placed in plastic tubes labeled with a sample identifying number.
Clinical tissue markers
For about 6% of cohort members, ER, PR, and/or Her2 clinical results were missing, precluding selection for the PAM50 assay according to the stratified random sampling scheme. We attempted to obtain tissue blocks for determination of these markers. For many of these cases with missing data, no blocks could be obtained. If blocks were obtained, sections were cut from the block and slides were made to assess expression of these markers by IHC staining. Equivocal Her2 results (IHC 2+) were subjected to fluorescence in-situ hybridization (FISH) for determination of HER2 amplification; those with a HER2/CEP17 ratio greater than 2.2 were classified as Her2 positive. For n=11 cases determined by this process to be ER and PR negative or Her2 positive, placing the case in a category with 100% sampling frequency, the case was selected for PAM50.
Gene expression assay
The tissue punch was deparaffinized and tissue was digested for RNA extraction as described previously (11). RT-qPCR was conducted for the 50 target genes that comprise the PAM50 (6). Details of RT-qPCR methods have been provided elsewhere (11, 32). Quality control included a negative control (no template) and a positive control (reference DNA template) for each gene in each plate, and RT-qPCR of five housekeeping genes from each tissue sample. Laboratory personnel were blinded to clinical information and received only a sample identification number to track the sample. Each batch of tissue punches sent to the laboratory for PAM50 assay included a mix of clinicopathological types.
Subtype classification variables
We defined two subtype classification variables for use in data analysis. The first was clinicopathological subtype, based on clinical characteristics, and the second was subtype based on the PAM50 gene expression assay. The characteristics used to define clinicopathological subtype were IHC results for ER and PR, IHC and/or FISH results for Her2, and tumor grade. Following St. Gallen conference recommendations (33) with a recent modification (34), we defined a low-risk, endocrine positive, i.e. surrogate luminal A, clinicopathological subtype category as PR+, Her−, and well or moderately differentiated. A higher-risk endocrine positive or clinicopathological surrogate luminal B category included tumors with ER+ or PR+ and any of PR−, Her2+, or tumor grade of poorly- or un-differentiated. A third clinicopathological category included Her2 positive, endocrine-negative tumors, i.e. ER−, PR−, and Her2+, and a fourth group was 'Triple negative' or ER−, PR−, and Her2−. To classify intrinsic subtypes from the gene expression data, we applied centroid-based algorithms to the calibrated log-expression ratio for the 50 genes (6). This process generates, for each sample, five continuous-scale normalized subtype scores representing degree of correlation of gene expression with that of archetypal Luminal A, Luminal B, HER2-enriched, Basal-like, and Normal-like breast tumors. Tumors were classified as the subtype with the highest normalized subtype score.
Data Analysis
All analyses incorporated the original sampling weights and the stratified sampling design for unbiased estimation of population parameters and valid estimates of standard errors (with 'svy' commands in Stata software, StataCorp, College Station, TX). Frequency distributions of intrinsic subtypes were estimated for the overall cohort and within subgroups defined by clinicopathological categories and by race and ethnicity. We calculated the kappa statistic to describe agreement between the clinicopathological subtypes and intrinsic subtypes. Tumors categorized by PAM50 as Normal-like, a subtype with no corresponding clinicopathological category, were excluded from the kappa calculation. Sensitivity and specificity were calculated for subtype classification, dichotomized as yes or no, treating clinicopathological classification from IHC as the "test" and PAM50 subtype as the gold standard.
In addition to analyses that treated each intrinsic subtype as a discrete category, we examined the distributions of the five continuous-scale normalized subtype scores from the PAM50 assay. Kernel-smoothed probability density functions were constructed, showing the distribution of the scores for each intrinsic subtype within the groups of subjects defined by subtype categories. Pairwise associations between subtype scores were assessed with Pearson correlation coefficients.
We described associations of participant and tumor characteristics with intrinsic subtypes by fitting a multinomial logistic regression model. This is similar to the case-case analysis approach widely used for dichotomous tumor characteristics (35), extended to the five subtype categories via the multinomial model. Treating the most prevalent subtype, Luminal A, as the base outcome, we estimated age-adjusted odds ratios (OR) and 95% confidence intervals (CI) for each of the non-Luminal A subtypes. Association between continuous-scale subtype scores and participant characteristics were also assessed in linear regression models.
RESULTS
The combined LACE and Pathways cohorts include women diagnosed with breast cancer at ages 25 to 91 years, with the largest proportion diagnosed between 50 and 59 years of age (Table 1). In addition to the non-Hispanic white majority of the participants, the cohort has representation of African-American, Hispanic, Asian or Pacific Islander, and American Indian or Alaska Native women. The cohort includes all stages at diagnosis, with over 90% diagnosed at AJCC stages I or II. Of 1,622 subjects selected for the intrinsic subtyping by PAM50, 103 were determined to be ineligible on pathology review due to insufficient area of invasive tumor in available blocks, 14 because of neoadjuvant therapy, and 6 due to bilateral disease. Fifty-six study participants did not consent for the tumor tissue assay, and blocks could not be obtained for another 105 participants. Laboratory assay failure prevented PAM50 subtyping for only 19 cases. After accounting for the stratified sampling scheme, the subcohort of 1,319 participants with PAM50 results was similar to the parent cohort with respect to age, race and ethnicity, and clinical characteristics (Table 1), with a mean age at diagnosis of 59 years.
Table 1.
Characteristics at time of diagnosis for LACE and Pathways breast cancer cohort participants and a subcohort subtyped by PAM50 gene expression-based classifier
| LACE Study n=2135 |
Pathways Study n=2172 |
Combined Cohort n=4307 |
Subcohorta n=1319 |
||
|---|---|---|---|---|---|
| % | % | % | raw % | weightedb % |
|
| Age at Diagnosis (years) | |||||
| <40 | 4.6 | 4.9 | 4.7 | 6.9 | 5.0 |
| 40 – 49 | 18.6 | 17.5 | 18.0 | 20.2 | 17.0 |
| 50 – 59 | 30.4 | 29.9 | 30.1 | 31.5 | 30.8 |
| 60 – 69 | 28.4 | 28.0 | 28.2 | 25.3 | 27.6 |
| 70 – 79 | 18.0 | 15.0 | 16.5 | 13.9 | 16.6 |
| 80+ | 0 | 4.7 | 2.4 | 2.2 | 3.0 |
| Race and Ethnicity | |||||
| White, non-Hispanic | 79.6 | 67.6 | 73.7 | 69.2 | 72.6 |
| Black or African-American | 5.4 | 6.9 | 6.2 | 8.7 | 6.9 |
| Hispanic | 6.5 | 11.5 | 9.0 | 9.3 | 8.8 |
| Asian or Pacific Islander | 6.5 | 10.7 | 8.6 | 9.9 | 9.2 |
| American Indian or Alaska Native | 1.0 | 2.1 | 1.6 | 1.7 | 1.6 |
| Other or Missing | 1.0 | 0.9 | 1.0 | 1.1 | 0.8 |
| AJCC Stage | |||||
| I | 46.6 | 50.7 | 48.7 | 44.3 | 49.8 |
| II | 50.6 | 33.9 | 42.2 | 47.6 | 43.6 |
| III | 2.9 | 9.3 | 6.1 | 7.2 | 6.2 |
| IV | 0.0 | 1.7 | 0.9 | 0.9 | 0.5 |
| Missing | 0 | 4.3 | 2.2 | 0 | 0 |
| Tumor Size | |||||
| <=2cm | 65.0 | 64.5 | 64.8 | 59.7 | 66.1 |
| >2cm | 35.0 | 31.5 | 33.2 | 40.3 | 33.9 |
| Tumor markers from IHC/FISHc | |||||
| ERPR+ Her2− | 67.5 | 72.7 | 70.1 | 33.0 | 74.8 |
| ERPR+ Her2+ | 11.1 | 9.3 | 10.2 | 26.0 | 9.8 |
| ERPR-2−Her22− | 10.9 | 12.5 | 11.7 | 30.7 | 11.5 |
| ERPR2− Her2+ | 3.7 | 4.5 | 4.1 | 10.3 | 3.9 |
| Missing | 6.7 | 0.9 | 3.8 | 0 | 0 |
Subjects with PAM50 subtyping results. The subcohort sample was chosen with the following selection probabilities by clinical subtype: ERPR+ Her22−, 18%; ERPR+ Her2+, 100%; Triple Negative, 100%; ERPR2− Her2+, 100%.
Estimated distribution of characteristics in the subcohort after accounting for stratified sampling.
Categories based on tumor estrogen receptor (ER) and progesterone receptor (PR) expression, as determined by clinical immunohistochemistry (IHC), and for Her2 overexpression by IHC and/or fluorescence in-situ hybridization (FISH). ERPR+ includes subjects with ER and/or PR positive; ERPR2− includes subjects with both ER and PR negative.
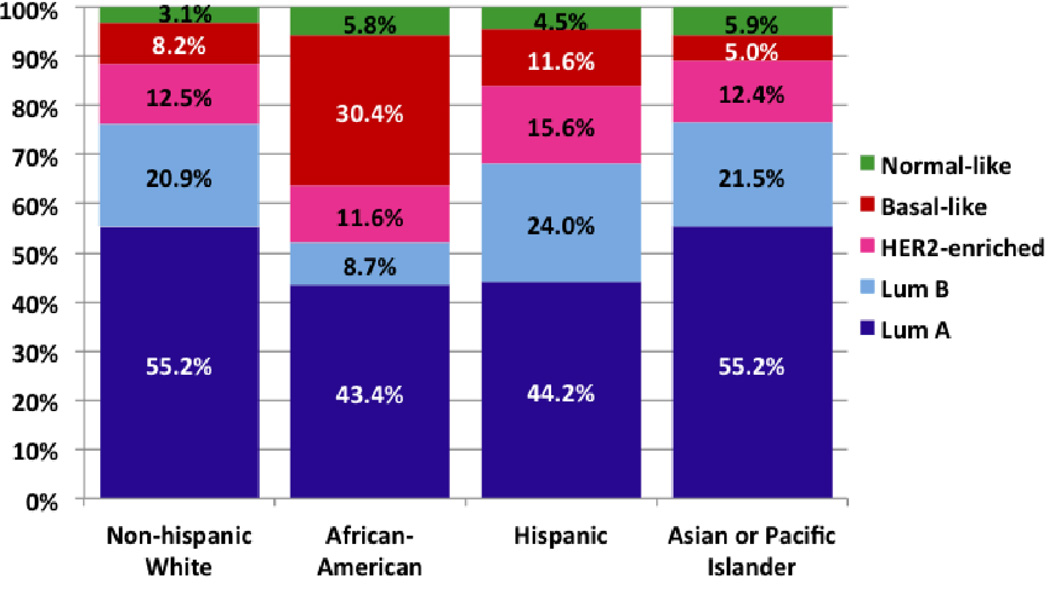
The distribution of PAM50-based intrinsic subtypes in the cohort was estimated to be 53.1% Luminal A, 20.5% Luminal B, 13.0% HER2-enriched, 9.8% Basal-like, and 3.6% Normal-like (Table 2). In an analysis restricted to the 697 cases who were enrolled in the study within one year after diagnosis, the distribution was 52.8% Luminal A, 21.6% Luminal B, 12.7% HER2-enriched, and 8.5% Basal-like and 4.3% Normal-like (p = 0.41 for comparison to those enrolled one or more years after diagnosis). Among the tumors classified as low-risk endocrine positive or surrogate luminal A by clinicopathological factors (Table 2), 76.5% were classified as Luminal A by PAM50, and a minimal percentage were classified as Basal-like, 0.3%. Within the group classified as endocrine positive, higher-risk or surrogate luminal B by clinicopathological factors, only 36.2% were classified as Luminal B by PAM50, with appreciable proportions classified as Luminal A or HER2-enriched. For the cohort members with clinicopathological Her2 positive, endocrine negative tumors, 75.7% were HER2-enriched by PAM50. Among participants with triple negative tumors, 69.8% were Basal-like by intrinsic subtype. The kappa statistic for agreement between clinicopathological and intrinsic subtype categories across four groups was 0.49. The sum of sensitivity and specificity for the clinicopathological categories was highest for triple-negative as a surrogate classifier of Basal-like subtype (Table 2). When we considered the distribution of intrinsic subtypes within racial and ethnic groups (Figure 1), the proportions with Basal-like subtype among African-American women and Asian women, 30.4% (95% CI 21.0, 41.7) and 5.0% (95% CI 3.0, 8.2), respectively, had confidence intervals that excluded the proportion classified as Basal-like among non-Hispanic whites.
Table 2.
Distribution of breast cancer intrinsic subtypes from PAM50 gene expression assay in the population-based LACE and Pathways studies, in a subcohort overall and by clinicopathological categories
| Clinicopathological Categorya | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subcohort n=1,319 |
Low-risk Endocrine +, n=298 |
High-risk Endocrine + n=480 |
Her2 +, Endocrine - n=138 |
Triple Negative n=405 |
||||||||||
| PAM50 Subtype | % | 95% CI | % | 95% CI | Sens/Specb | % | 95% CI | Sens/Specb | % | 95% CI | Sens/Specb | % | 95% CI | Sens/Specb |
| Luminal A | 53.1 | 49.8, 56.5 | 76.5 | 71.4, 81.0 | 0.74/0.74 | 40.5 | 34.5, 46.7 | 2.2 | 0.7, 6.7 | 3.0 | 1.7, 5.2 | |||
| Luminal B | 20.5 | 17.7, 23.6 | 14.8 | 11.2, 19.3 | 36.2 | 30.6, 42.3 | 0.59/0.73 | 7.4 | 4.0, 13.2 | 4.7 | 3.0, 7.2 | |||
| Her2E | 13.0 | 11.3, 15.0 | 3.7 | 2.1, 6.6 | 17.5 | 13.7, 22.0 | 75.7 | 67.8, 82.2 | 0.23/0.99 | 20.3 | 16.6, 24.5 | |||
| Basal-like | 9.8 | 8.8, 10.8 | 0.3 | 0.0, 2.0 | 2.9 | 1.3, 6.2 | 14.7 | 9.7, 21.7 | 69.9 | 65.2, 74.2 | 0.83/0.96 | |||
| Normal-like | 3.6 | 2.5, 5.4 | 4.7 | 2.8, 7.8 | 2.9 | 1.4, 5.9 | 0.0 | 2.2 | 1.2, 4.2 | |||||
Clinicopathological categories (columns), defined in methods, are based on estrogen receptor (ER) and progesterone receptor (PR) expression, as determined by immunohistochemistry (IHC), Her2 overexpression by IHC and/or fluorescence in-situ hybridization, and tumor grade determined.
Sensitivity and specificity, treating clinicopathological category as test and the most similar PAM50 subtype classification as the gold standard.
Figure 1.
Distribution of breast cancer intrinsic subtypes from PAM50 assay in a population-based cohort by race and ethnicity, LACE and Pathways studies.
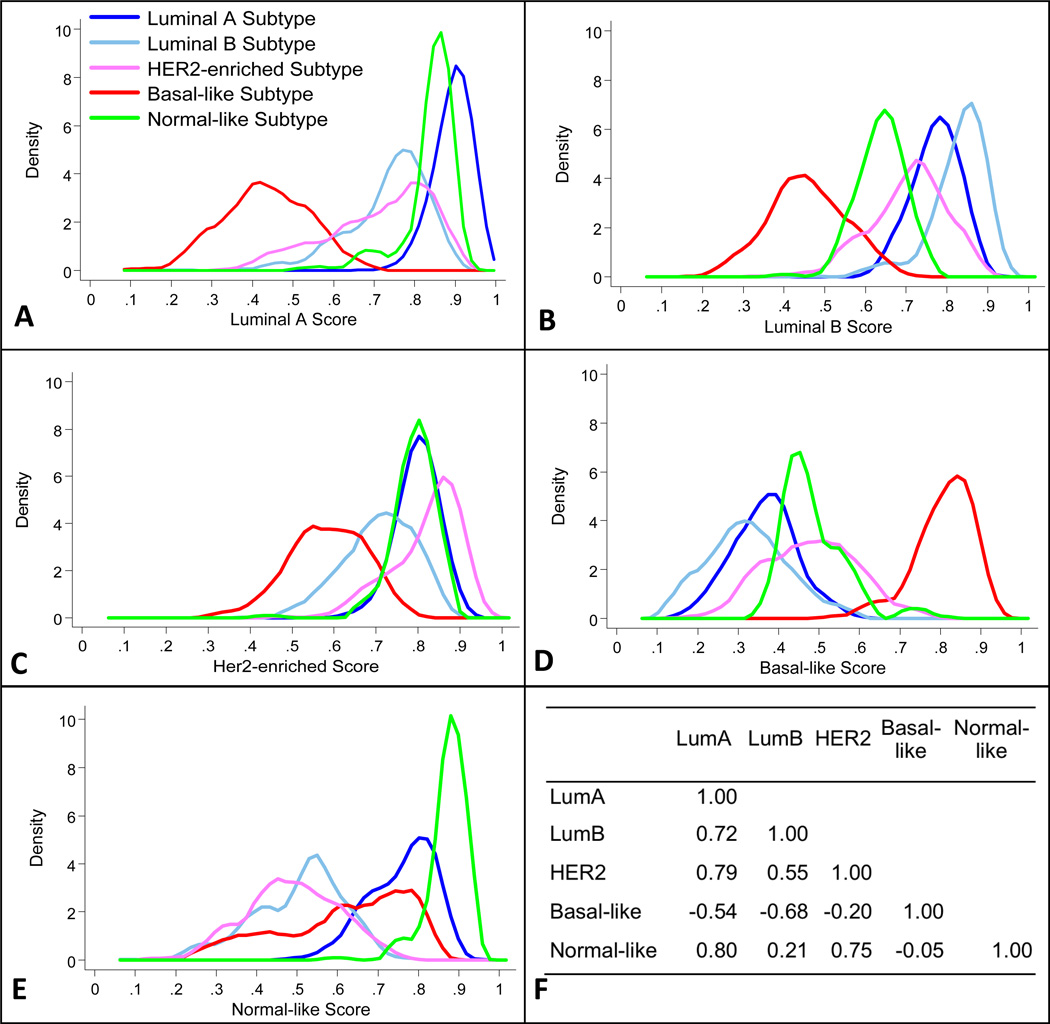
There was substantial overlap in the distribution of continuous-scale Luminal A scores between tumors classified as Luminal A and Normal-like (Figure 2A). For Luminal B and HER2-enriched scores (Figure 2B, 2C), overlap was observed between tumors classfied in different subtypes. The distribution of the Basal-like score among tumors classified by PAM50 as Basal-like was distinct, exhibiting almost no overlap with the distributions for other subtypes (Figure 2D). Tumors with subtypes other than normal-like exhibited a broad range of normal-like scores (Figure 2E). The continuous-scale PAM50 subtype scores for Luminal A, Luminal B, HER2-enriched, and Normal-like were positively correlated with each other, whereas the Basal-like score was inversely correlated with each of the other subtype scores (Figure 2F).
Figure 2.
Distributions of normalized, continuous-scale scores for breast cancer intrinsic subtypes estimated from PAM50 assay results, LACE and Pathways cohort. Panels A through E: Each panel shows, for one continuous-scale intrinsic subtype score obtained from PAM50, the kernel-smoothed probability density function of that score within PAM50 intrinsic subtype category groups of tumors, indicated by color. Panel F: Correlation coefficients for the subtype scores.
Age at diagnosis was strongly related to intrinsic subtype. Using Luminal A as the comparison group, there were significant trends of decreasing odds of having a breast tumor of Luminal B (p = 0.001), HER2-enriched (p = 0.05) or Basal-like (p < 0.0001) subtype with older age at diagnosis (Table 3). In age-adjusted models, African-American women with breast cancer had significantly higher odds of having a Basal-like tumor vs. a Luminal A tumor, OR 4.39 (95% CI 2.29, 8.29), compared with non-Hispanic whites. Hispanic women were somewhat more likely to have any of the non-Luminal A subtypes, i.e. Luminal B, HER2-enriched, or Basal-like, in univariate analysis, but the differences were attenuated after age-adjustment and did not reach statistical significance. Asian and Pacific Islander women with breast cancer had significantly reduced relative odds of having Basal-like vs. Luminal A tumors, OR 0.48 (95% CI 0.25–0.92). Results for American Indian and Alaska Native women were somewhat suggestive of higher prevalence of non-Luminal A subtypes, but confident intervals were large due to small numbers in this racial group in the cohort. In linear regression models, Luminal A subtype score was positively associated with age at diagnosis (p = 0.01) and inversely associated with African-American race (p < 0.001) among women with non-LumA subtype. Basal-like subtype score was inversely associated with age at diagnosis (p = 0.005) among women with non-Basal-like subtype.
Table 3.
Demographic and clinical characteristic associations with breast cancer intrinsic subtype from PAM50 assay, LACE and Pathways cohort: Odds ratios for non-Luminal A subtypes vs. Luminal A
| Intrinsic Subtype a from PAM50 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | LumA | LumB | LumB vs. LumA | HER2E | HER2E vs. LumA | Basal | Basal vs. LumA | |||||
| n | % a | % a | ORb | 95% CI | % a | ORb | 95% CI | %a | ORb | 95% CI | ||
| Age at diagnosis | ||||||||||||
| <40 | 91 | 2.9 | 8.1 | 2.48 | 0.98, 6.29 | 5.5 | 1.72 | 0.68, 4.34 | 10.8 | 3.05 | 1.40, 6.65 | |
| 40 – 49 | 266 | 14.2 | 20.7 | 1.27 | 0.72, 2.27 | 16.3 | 1.03 | 0.59, 1.79 | 26.5 | 1.51 | 0.93, 2.46 | |
| 50 – 59 | 415 | 28.3 | 32.4 | 1 | reference | 31.6 | 1 | reference | 35.0 | 1 | reference | |
| 60 – 69 | 334 | 31.9 | 20.8 | 0.57 | 0.33, 0.97 | 28.8 | 0.81 | 0.49, 1.32 | 17.5 | 0.44 | 0.27, 0.72 | |
| 70 – 79 | 184 | 19.1 | 15.5 | 0.71 | 0.39, 1.30 | 15.2 | 0.71 | 0.39, 1.29 | 9.5 | 0.41 | 0.22, 0.76 | |
| 80 + | 29 | 3.6 | 2.4 | 0.57 | 0.17, 1.95 | 2.6 | 0.65 | 0.19, 2.30 | 0.6 | 0.13 | 0.028, 0.61 | |
| p trend | 0.001 | 0.05 | <0.0001 | |||||||||
| Race/Ethnicity | ||||||||||||
| White | 913 | 75.5 | 74.3 | 1 | reference | 70.1 | 1 | reference | 60.9 | 1 | reference | |
| Black or African-American | 115 | 5.6 | 2.9 | 0.51 | 0.19, 1.33 | 6.1 | 1.15 | 0.55, 2.38 | 21.3 | 4.38 | 2.29, 8.39 | |
| Hispanic | 123 | 7.3 | 10.4 | 1.29 | 0.63, 2.62 | 10.6 | 1.46 | 0.71, 3.01 | 10.5 | 1.47 | 0.82, 2.63 | |
| Asian or Pacific Islander | 131 | 9.5 | 9.6 | 0.89 | 0.46, 1.73 | 9.0 | 0.93 | 0.55, 1.54 | 4.7 | 0.48 | 0.25, 0.92 | |
| American Indian or Alaska Native | 22 | 1.0 | 2.5 | 2.9 | 0.45, 10.3 | 2.9 | 2.74 | 0.61, 12.4 | 2.0 | 1.95 | 0.52, 7.33 | |
| Other | 37 | 1.0 | 0.3 | 0.30 | 0.05, 1.76 | 1.3 | 1.47 | 0.37, 5.87 | 0.6 | 0.77 | 0.13, 4.80 | |
| Grade | ||||||||||||
| Well-differentiated | 162 | 33.2 | 9.9 | 1 | reference | 8.8 | 1 | reference | 1.2 | 1 | reference | |
| Moderately differentiated | 453 | 46.9 | 47.7 | 3.19 | 1.65, 6.15 | 39.2 | 3.06 | 1.40, 6.69 | 11.7 | 6.31 | 2.11, 18.8 | |
| Poor/Undifferentiated | 630 | 9.4 | 38.6 | 12.80 | 6.14, 26.5 | 47.4 | 18.50 | 8.30, 41.3 | 82.5 | 217.40 | 74.6, 633.8 | |
| Unknown | 72 | 10.5 | 3.8 | 1.23 | 0.41, 3.72 | 4.6 | 1.68 | 0.53, 5.30 | 4.7 | 13.20 | 3.44, 50.9 | |
| p trend | <0.0001 | <0.0001 | <0.0001 | |||||||||
| Stage | ||||||||||||
| I | 562 | 57.2 | 36.1 | 1 | reference | 38.5 | 1 | reference | 44.0 | 1 | reference | |
| II | 607 | 37.8 | 52.0 | 2.04 | 1.34, 3.12 | 55.0 | 2.09 | 1.40, 3.11 | 49.6 | 1.47 | 1.01, 2.12 | |
| III | 90 | 5.0 | 10.8 | 3.05 | 1.45, 6.40 | 5.7 | 1.60 | 0.81, 3.17 | 5.2 | 1.05 | 0.51, 2.15 | |
| IV | 12 | 0.1 | 1.1 | 24.50 | 2.23, 269.6 | 0.9 | 20.70 | 2.24, 192.2 | 1.2 | 14.90 | 1.59, 139.1 | |
| p trend | <0.0001 | <0.0001 | 0.06 | |||||||||
| Tumor size | ||||||||||||
| <=2cm | 797 | 74.9 | 54.3 | 1 | reference | 55.8 | 1 | reference | 56.0 | 1 | reference | |
| >2cm | 522 | 25.1 | 45.7 | 2.36 | 1.56, 3.56 | 44.2 | 2.29 | 1.53, 3.41 | 44.0 | 2.06 | 1.43, 2.96 | |
| Positive lymph nodes | ||||||||||||
| 0 | 832 | 70.2 | 59.0 | 1 | reference | 58.0 | 1 | reference | 72.9 | 1 | reference | |
| 1–3 | 321 | 22.8 | 25.3 | 1.18 | 0.73, 1.90 | 29.9 | 1.50 | 0.97, 2.32 | 19.8 | 0.68 | 0.43, 1.07 | |
| 4+ | 139 | 6.9 | 15.6 | 2.48 | 1.30, 4.71 | 12.2 | 2.05 | 1.11, 3.76 | 7.4 | 0.87 | 0.48, 1.60 | |
| p trend | 0.01 | 0.007 | 0.19 | |||||||||
LumA: Luminal A, LumB: Luminal B, HER2E: HER2-enriched, Basal: Basal-like. The Normal-like subtype accounted for fewer than 5% of subtype results and is not shown.
Estimated distribution of characteristics within the subtype category after correction for sampling weights.
Prevalence odds ratios for cross-sectional association between subject or clinical characteristic at diagnosis and non-Luminal A subtype.
Several clinical characteristics at diagnosis were associated with intrinsic subtype (Table 3). Higher histological tumor grade was associated with increased prevalence of Luminal B, HER2-enriched, and Basal-like subtypes (p trend < 0.0001 for each). Luminal B, HER2-enriched, and Basal-like subtypes were relatively more prevalent among tumors with higher stage at diagnosis (p trend <0.0001, <0.0001, and 0.06, respectively). When we considered two of the component characteristics of stage at diagnosis, tumor size and positive lymph nodes, tumors greater than 2 cm in diameter were more likely than smaller tumors to have Luminal B, HER2-enriched, and Basal-like subtypes. Increasing number of positive nodes was associated with Luminal B (p trend = 0.01) and HER2-enriched subtypes (p trend = 0.007), but not with Basal-like subtype. The odds of having the Normal-like subtype was inversely associated with stage at diagnosis (p = 0.04) but was not significantly associated with age, race, or any other clinical characteristic (data not shown).
We conducted a sub-analysis among cohort members whose clinicopathological tumor features placed them in the low-risk endocrine positive or surrogate luminal A category, to evaluate predictors of being classified as worse-prognosis non-Luminal A (i.e. Luminal B, HER2-enriched, or Basal-like) subtypes by the PAM50 assay (data not shown). Among tumors that were low-risk, endocrine-positive from clinicopathological data, younger age at diagnosis (p trend = 0.007) showed a significant trend of being classified by the PAM50 assay as a worse-prognosis subtype.
DISCUSSION
Existence of the breast cancer intrinsic subtypes Luminal A, Luminal B, HER2-enriched, and Basal-like has been validated across multiple laboratory methods (36, 37) and multiple study populations, including confirmation that the subtypes initially characterized in European and North American populations are also present in breast cancers in Asian women (38). However, the present study is the first, to our knowledge, to describe intrinsic subtypes based on a gene expression assay in a U.S. population-based study, and in a study population that includes representation of multiple racial and ethnic groups.
Our estimated prevalence of intrinsic subtype Luminal A, 53.1% of all breast cancers, is higher than the fraction estimated by most prior studies, whereas the fraction with Normal-like subtype in our cohort, 3.6%, is lower. Others have suggested that categorization as Normal-like may be an artifact of contamination of tumor RNA with RNA from normal breast cells (11, 39, 40). The low percentage of Normal-like results in the present study suggests that the strategy of obtaining a tissue punch from the FFPE tumor block, guided by the pathologist's marked slide, was effective in minimizing normal tissue contamination. The inverse association between Normal-like subtype and tumor stage in the present study may be an indication of contamination of the tumor punch with normal tissue when smaller tumors are sampled.
Population selection likely contributes to the high prevalence of Luminal A in the present study compared with other data sets. Specifically, selection of clinical trial populations with restrictions such as lymph-node positive cases (10, 11) or young cases (3) for intrisic subtype studies, and the general tendency for clinical trial participants to be younger than the overall cancer population (41, 42), would result in skewing study populations toward more aggressive, non-Luminal A subtypes. A study population more similar to ours was based on a population-based cancer registry in Sweden with no age or stage restrictions and a mean age at diagnosis of 58 years (23). In that study, if Normal-like results were excluded, the distribution of subtypes (from microarray analysis of fresh tissue) would be 44% Luminal A, 19% Luminal B, 16% HER2-enriched, and 21% Basal-like.
Although the present study used population-based recruitment, cohort exclusions and enrollment processes may have resulted in a sample set that deviates somewhat from the true population distribution. Specifically, exclusions of tumors with less than 0.5 cm of invasive tumor or neoadjuvant chemotherapy, exclusion of late-stage cases and of cases older than 79 years from the LACE cohort, and missed enrollment for the LACE cohort of potentially eligible cases who died within about 2 years after diagnosis, introduce a limitation for describing the population distribution of subtypes. The small tumors and older women excluded would be more likely to be Luminal A subtype, whereas advanced-stage cases and those who died within a short time after diagnosis probably include more poor-prognosis, non-Luminal A subtypes, so that there is no uniform direction of bias expected from these exclusions. In analysis of women enrolled within the first year after diagnosis, the distribution of subtypes did not differ significantly, and the proportion of Luminal A remained 53%.
Whereas the literature on intrinsic subtyping initially emphasized classification and distinguishing between subtypes, and discrepancies of subtype classification for the same tumor in cross-platform validation studies are seen as problematic for clinical application (39), within-subtype molecular heterogeneity is increasingly recognized (32, 36, 43). The overlapping probability density functions for Luminal A, Luminal B, and HER2-enriched from this large, population-based study support the interpretation that, within these subtypes, a tumor of one subtype can have varying degrees of expression of genes characteristic of other subtypes (32). In a previous study, use of subtype scores resulted in better-fitting prognostic models compared with subtype category (6). Researchers should further consider applying not just subtype classification but also subtype scores, and individual gene expression data obtained from assays like PAM50, when investigating tumor characteristics in relation to prognostic and etiologic characteristics.
Results from the present study confirm the Swedish population-based study's report of a significant association of older age at diagnosis with Luminal A subtype (23). A recent report describing changes in gene expression patterns in normal breast tissue with age (44). These results taken together suggest that future research should consider the links between normal tissue context and tumor subtype.
Our observation of a significant, four-fold higher prevalence of PAM50 Basal-like subtype among African-Americans is consistent with the high proportion of clinicopathological basal-like phenotypes among African-Americans first described in a population-based study in North Carolina (12). We observed a reduced odds of having a Basal-like tumor, versus Luminal A, among Asian and Pacific Islander women. A high proportion of clinicopathological luminal A subtypes and/or low prevalence of triple negative breast cancers has been reported among Asian populations in the U.S. (45) and in Asia (22, 46).
Among participants whose clinical tumor markers placed them in the clinicopathological endocrine positive, low risk or surrogate luminal A category, about one in five were shifted to higher-risk Luminal B, HER2-enriched, or Basal-like subtypes based on subtyping by PAM50 assay. This shift in subtype is clinically significant because a St. Gallen conference (33) recommended that subtypes be used for early breast cancers to distinguish tumors for which adjuvant chemotherapy is indicated. In our analysis of cases with clinicopathological endocrine positive, low-risk surrogate luminal A tumors that shifted to other subtypes based on PAM50 results, women with younger age at diagnosis were more likely to shift to the higher-risk subtypes.
Study resources were not sufficient to support uniform IHC assessment of ER, PR, and Her2 across the full cohort. Instead, we relied on clinical determination from the time of diagnosis. Changes in scoring of ER, PR, or Her2 over time are a limitation in the data, as is our reliance on only "positive" or "negative" results, given that details on percent of cells stained or Her2 score were not consistently found in the medical records review.
Studies of RT-qPCR-based intrinsic classifiers indicate promise for prognosis, but reviewers have suggested that more evidence is needed to support application of these classifiers in clinical decision-making (25). The potential application of PCR-based classifiers for epidemiologic studies is intriguing. To our knowledge, this is the first study to apply such a tool in a large, population-based study. We were successful in obtaining PAM50 subtype results for a high proportion of cases in a population-based study using FFPE tumor blocks, many of which had been stored 10–12 years. Thus, PAM50 assay is a feasible classifier for epidemiologic studies. Subtype differences by age and by race were distinct. Epidemiologic studies have reported that the influence of breast cancer risk factors varies by clinicopathological subtype (13, 15, 16, 21, 22, 47–50); PCR-based intrinsic classifiers may prove a more sensitive tool for describing etiologic heterogeneity.
In conclusion, the PAM50 RT-qPCR assay proved feasible in an epidemiologic study. Luminal A was the intrinsic subtype for the majority of a population-based study cohort, but intrinsic subtyping shifted some tumors categorized as low risk by clinicopathological criteria to higher-risk subtypes. Gene expression-based subtyping in a population-based cohort supported the concept of molecular heterogeneity within subtypes and revealed distinct intrinsic subtype profiles by age and race.
Acknowledgments
Financial Support: Supported by U.S. National Institutes of Health awards R01CA129059 (B.J. Caan, PI) and R01CA105274 (L.H. Kushi, PI). Additional support from Bioinformatics and Biostatistics core resources of the Huntsman Cancer Institute, P30CA042014. The Utah Cancer Registry is funded by Contract No. HHSN261201000026C from the National Cancer Institute's SEER Program with additional support from the Utah State Department of Health and the University of Utah. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Potential conflict of interest: PSB has an interest in Bioclassifier LLC. The other authors declare that they have no competing interests.
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000 Aug 17;406(6797):747–752. doi: 10.1038/35021093. PubMed PMID: 10963602. eng. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10869–10874. doi: 10.1073/pnas.191367098. PubMed PMID: 11553815. Epub 2001/09/13. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002 Jan 31;415(6871):530–536. doi: 10.1038/415530a. PubMed PMID: 11823860. Epub 2002/02/02. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8418–8423. doi: 10.1073/pnas.0932692100. PubMed PMID: 12829800. Epub 2003/06/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. PubMed PMID: 16643655. Epub 2006/04/29. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009 Mar 10;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. PubMed PMID: 19204204. Epub 2009/02/11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Recommendations from the EGAPP Working Group: can tumor gene expression profiling improve outcomes in patients with breast cancer? Genet Med. 2009 Jan;11(1):66–73. doi: 10.1097/GIM.0b013e3181928f56. PubMed PMID: 19125125. Epub 2009/01/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010 Nov 1;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. PubMed PMID: 20837693. Pubmed Central PMCID: 2970720. Epub 2010/09/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbier AK, Anderson H, Ghazoui Z, Salter J, Parker JS, Perou CM, et al. Association between breast cancer subtypes and response to neoadjuvant anastrozole. Steroids. 2011 Jul;76(8):736–740. doi: 10.1016/j.steroids.2011.02.025. PubMed PMID: 21447351. Epub 2011/03/31. [DOI] [PubMed] [Google Scholar]
- 10.Chia SK, Bramwell-Wesley V, Tu D, Shepherd L, Jiang S, Vickery T, et al. A 50 Gene Intrinsic Subtype Classifier for Prognosis and Prediction of Benefit from Adjuvant Tamoxifen. Clin Cancer Res. 2012 Jun 18; doi: 10.1158/1078-0432.CCR-12-0286. PubMed PMID: 22711706. Epub 2012/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastien RR, Rodriguez-Lescure A, Ebbert MT, Prat A, Munarriz B, Rowe L, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC medical genomics. 2012;5:44. doi: 10.1186/1755-8794-5-44. PubMed PMID: 23035882. Pubmed Central PMCID: 3487945. Epub 2012/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006 Jun 7;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. PubMed PMID: 16757721. eng. [DOI] [PubMed] [Google Scholar]
- 13.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007 Mar;16(3):439–443. doi: 10.1158/1055-9965.EPI-06-0806. PubMed PMID: 17372238. Epub 2007/03/21. eng. [DOI] [PubMed] [Google Scholar]
- 14.Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007 Aug 15;110(4):876–884. doi: 10.1002/cncr.22836. PubMed PMID: 17620276. Epub 2007/07/11. eng. [DOI] [PubMed] [Google Scholar]
- 15.Phipps AI, Malone KE, Porter PL, Daling JR, Li CI. Reproductive and hormonal risk factors for postmenopausal luminal, HER-2-overexpressing, and triple-negative breast cancer. Cancer. 2008 Oct 1;113(7):1521–1526. doi: 10.1002/cncr.23786. PubMed PMID: 18726992. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwan ML, Kushi LH, Weltzien E, Maring B, Kutner SE, Fulton RS, et al. Epidemiology of breast cancer subtypes in two prospective cohort studies of breast cancer survivors. Breast Cancer Res. 2009;11(3):R31. doi: 10.1186/bcr2261. PubMed PMID: 19463150. Pubmed Central PMCID: 2716499. Epub 2009/05/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010 May;7(5):e1000279. doi: 10.1371/journal.pmed.1000279. PubMed PMID: 20520800. Pubmed Central PMCID: 2876119. Epub 2010/06/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010 Dec 15;16(24):6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. PubMed PMID: 21169259. Pubmed Central PMCID: 3029098. Epub 2010/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawood S, Hu R, Homes MD, Collins LC, Schnitt SJ, Connolly J, et al. Defining breast cancer prognosis based on molecular phenotypes: results from a large cohort study. Breast Cancer Res Treat. 2011 Feb;126(1):185–192. doi: 10.1007/s10549-010-1113-7. PubMed PMID: 20711652. Pubmed Central PMCID: 3026074. Epub 2010/08/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phipps AI, Buist DS, Malone KE, Barlow WE, Porter PL, Kerlikowske K, et al. Reproductive history and risk of three breast cancer subtypes defined by three biomarkers. Cancer Causes Control. 2011 Mar;22(3):399–405. doi: 10.1007/s10552-010-9709-0. PubMed PMID: 21184265. Pubmed Central PMCID: 3042513. Epub 2010/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamimi RM, Colditz GA, Hazra A, Baer HJ, Hankinson SE, Rosner B, et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012 Jan;131(1):159–167. doi: 10.1007/s10549-011-1702-0. PubMed PMID: 21830014. Pubmed Central PMCID: 3237947. Epub 2011/08/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sueta A, Ito H, Islam T, Hosono S, Watanabe M, Hirose K, et al. Differential impact of body mass index and its change on the risk of breast cancer by molecular subtype: A case-control study in Japanese women. SpringerPlus. 2012 Dec;1(1):39. doi: 10.1186/2193-1801-1-39. PubMed PMID: 23350064. Pubmed Central PMCID: 3550694. Epub 2013/01/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calza S, Hall P, Auer G, Bjohle J, Klaar S, Kronenwett U, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8(4):R34. doi: 10.1186/bcr1517. PubMed PMID: 16846532. Pubmed Central PMCID: 1779468. Epub 2006/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly CM, Bernard PS, Krishnamurthy S, Wang B, Ebbert MT, Bastien RR, et al. Agreement in Risk Prediction Between the 21-Gene Recurrence Score Assay (Oncotype DX(R)) and the PAM50 Breast Cancer Intrinsic Classifier™ in Early-Stage Estrogen Receptor-Positive Breast Cancer. The oncologist. 2012 Mar 14; doi: 10.1634/theoncologist.2012-0007. PubMed PMID: 22418568. Epub 2012/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azim HA, Jr, Michiels S, Zagouri F, Delaloge S, Filipits M, Namer M, et al. Utility of prognostic genomic tests in breast cancer practice: The IMPAKT 2012 Working Group Consensus Statement. Ann Oncol. 2013 Mar;24(3):647–654. doi: 10.1093/annonc/mds645. PubMed PMID: 23337633. Epub 2013/01/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007 Nov 20;25(33):5287–5312. doi: 10.1200/JCO.2007.14.2364. PubMed PMID: 17954709. Epub 2007/10/24. [DOI] [PubMed] [Google Scholar]
- 27.Colombo PE, Milanezi F, Weigelt B, Reis-Filho JS. Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res. 2011;13(3):212. doi: 10.1186/bcr2890. PubMed PMID: 21787441. Pubmed Central PMCID: 3218943. Epub 2011/07/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, Pho M, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8(3):R25. doi: 10.1186/bcr1412. PubMed PMID: 16737553. Pubmed Central PMCID: 1557737. Epub 2006/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prat A, Parker JS, Fan C, Cheang MC, Miller LD, Bergh J, et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Ann Oncol. 2012 Nov;23(11):2866–2873. doi: 10.1093/annonc/mds080. PubMed PMID: 22532584. Pubmed Central PMCID: 3477878. Epub 2012/04/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caan B, Sternfeld B, Gunderson E, Coates A, Quesenberry C, Slattery ML. Life After Cancer Epidemiology (LACE) Study: a cohort of early stage breast cancer survivors (United States) Cancer Causes Control. 2005 Jun;16(5):545–556. doi: 10.1007/s10552-004-8340-3. PubMed PMID: 15986109. [DOI] [PubMed] [Google Scholar]
- 31.Kwan ML, Ambrosone CB, Lee MM, Barlow J, Krathwohl SE, Ergas IJ, et al. The Pathways Study: a prospective study of breast cancer survivorship within Kaiser Permanente Northern California. Cancer Causes Control. 2008 Dec;19(10):1065–1076. doi: 10.1007/s10552-008-9170-5. PubMed PMID: 18478338. Pubmed Central PMCID: 2581640. Epub 2008/05/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebbert MT, Bastien RR, Boucher KM, Martin M, Carrasco E, Caballero R, et al. Characterization of uncertainty in the classification of multivariate assays: application to PAM50 centroid-based genomic predictors for breast cancer treatment plans. Journal of clinical bioinformatics. 2011;1:37. doi: 10.1186/2043-9113-1-37. PubMed PMID: 22196354. Pubmed Central PMCID: 3275466. Epub 2011/12/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011 Aug;22(8):1736–1747. doi: 10.1093/annonc/mdr304. PubMed PMID: 21709140. Pubmed Central PMCID: 3144634. Epub 2011/06/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, et al. Prognostic Significance of Progesterone Receptor-Positive Tumor Cells Within Immunohistochemically Defined Luminal A Breast Cancer. J Clin Oncol. 2012 Dec 17; doi: 10.1200/JCO.2012.43.4134. PubMed PMID: 23233704. Epub 2012/12/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begg CB, Zhang ZF. Statistical analysis of molecular epidemiology studies employing case-series. Cancer Epidemiol Biomarkers Prev. 1994 Mar;3(2):173–175. PubMed PMID: 8049640. [PubMed] [Google Scholar]
- 36.Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490(7418):61–70. doi: 10.1038/nature11412. PubMed PMID: 23000897. Pubmed Central PMCID: 3465532. Epub 2012/09/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nature reviews Clinical oncology. 2012 Jan;9(1):48–57. doi: 10.1038/nrclinonc.2011.178. PubMed PMID: 22143140. Epub 2011/12/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu K, Lee CH, Tan PH, Tan P. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin Cancer Res. 2004 Aug 15;10(16):5508–5517. doi: 10.1158/1078-0432.CCR-04-0085. PubMed PMID: 15328190. eng. [DOI] [PubMed] [Google Scholar]
- 39.Weigelt B, Mackay A, A'Hern R, Natrajan R, Tan DS, Dowsett M, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010 Apr;11(4):339–349. doi: 10.1016/S1470-2045(10)70008-5. PubMed PMID: 20181526. Epub 2010/02/26. [DOI] [PubMed] [Google Scholar]
- 40.Weigelt B, Reis-Filho JS. Molecular profiling currently offers no more than tumour morphology and basic immunohistochemistry. Breast Cancer Res. 2010;12(Suppl 4):S5. doi: 10.1186/bcr2734. PubMed PMID: 21172089. Pubmed Central PMCID: 3005725. Epub 2011/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchins LF, Unger JM, Crowley JJ, Coltman CA, Jr, Albain KS. Underrepresentation of patients 65 years of age or older in cancer-treatment trials. N Engl J Med. 1999 Dec 30;341(27):2061–2067. doi: 10.1056/NEJM199912303412706. PubMed PMID: 10615079. [DOI] [PubMed] [Google Scholar]
- 42.Scher KS, Hurria A. Under-representation of older adults in cancer registration trials: known problem, little progress. J Clin Oncol. 2012 Jun 10;30(17):2036–2038. doi: 10.1200/JCO.2012.41.6727. PubMed PMID: 22547597. Epub 2012/05/02. [DOI] [PubMed] [Google Scholar]
- 43.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology. 2011 Feb;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. PubMed PMID: 21147047. Epub 2010/12/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirone JR, D'Arcy M, Stewart DA, Hines WC, Johnson M, Gould MN, et al. Age-Associated Gene Expression in Normal Breast Tissue Mirrors Qualitative Age-at-Incidence Patterns for Breast Cancer. Cancer Epidemiol Biomarkers Prev. 2012 Oct;21(10):1735–1744. doi: 10.1158/1055-9965.EPI-12-0451. PubMed PMID: 22859400. Epub 2012/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chuang E, Paul C, Flam A, McCarville K, Forst M, Shin S, et al. Breast cancer subtypes in Asian-Americans differ according to Asian ethnic group. Journal of immigrant and minority health / Center for Minority Public Health. 2012 Oct;14(5):754–758. doi: 10.1007/s10903-012-9577-7. PubMed PMID: 22286607. Epub 2012/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurebayashi J, Moriya T, Ishida T, Hirakawa H, Kurosumi M, Akiyama F, et al. The prevalence of intrinsic subtypes and prognosis in breast cancer patients of different races. Breast. 2007 Dec;16(Suppl 2):S72–S77. doi: 10.1016/j.breast.2007.07.017. PubMed PMID: 17714947. Epub 2007/08/24. [DOI] [PubMed] [Google Scholar]
- 47.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008 May;109(1):123–139. doi: 10.1007/s10549-007-9632-6. PubMed PMID: 17578664. Epub 2007/06/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phipps AI, Malone KE, Porter PL, Daling JR, Li CI. Body size and risk of luminal, HER2-overexpressing, and triple-negative breast cancer in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2008 Aug;17(8):2078–2086. doi: 10.1158/1055-9965.EPI-08-0206. PubMed PMID: 18664548. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phipps AI, Chlebowski RT, Prentice R, McTiernan A, Wactawski-Wende J, Kuller LH, et al. Reproductive history and oral contraceptive use in relation to risk of triple-negative breast cancer. J Natl Cancer Inst. 2011 Mar 16;103(6):470–477. doi: 10.1093/jnci/djr030. PubMed PMID: 21346227. Pubmed Central PMCID: 3057984. Epub 2011/02/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phipps AI, Chlebowski RT, Prentice R, McTiernan A, Stefanick ML, Wactawski-Wende J, et al. Body size, physical activity, and risk of triple-negative and estrogen receptor-positive breast cancer. Cancer Epidemiol Biomarkers Prev. 2011 Mar;20(3):454–463. doi: 10.1158/1055-9965.EPI-10-0974. PubMed PMID: 21364029. Pubmed Central PMCID: 3064558. Epub 2011/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]