Abstract
T-helper (Th) lineages have been generated in vitro by activating CD4 cells with anti-CD3/CD28 antibodies during polarization. Physiologically, however, the generation of Th lineages is by activation with the specific antigen presented by antigen-presenting cells (APC). Here, we used T-cell receptor (TCR)-transgenic mice to compare the phenotypes of Th1, Th9 and Th17 lineages when generated by either one of the two activation modes. Lineage Th cells specific against hen egg lysozyme (HEL), were adoptively transferred into recipient mice transgenically expressing HEL in their lens. Remarkable differences were found between lineages of Th1, Th9 or Th17, generated by either one of the two modes in their capacities to migrate to and proliferate in the recipient spleen and, importantly, to induce inflammation in the recipient mouse eyes. Substantial differences were also observed between the lineage pairs in their transcript expression profiles of certain chemokines and chemokine receptors. Surprisingly, however, close similarities were observed between the transcript expression profiles of lineages of the three phenotypes, activated by the same mode. Furthermore, Th cell lineages generated by the two activation modes differed considerably in their pattern of gene expression, as monitored by microarray analysis, but exhibited commonality with lineages of other phenotypes generated by the same activation mode. This study thus shows that (i) Th lineages generated by activation with anti-CD3/CD28 antibodies differ from lineages generated by antigen/APC; and (ii) the mode of activation determines to a large extent the expression profile of major transcripts.
Keywords: Eye, Inflammation, Microarray, T-cell differentiation
Introduction
Studies in recent years revealed the heterogeneity of the T-helper (Th) cell population, with five subpopulations having been defined so far, namely, Th1, Th2, Th9, Th17 and Th22.1,2,3,4,5,6,7 Analyses of these subpopulations have been carried out mainly in vitro and accumulating data have identified the specific polarizing cytokines for each Th subpopulation. The polarization process of naïve CD4 cells requires the cells to be concurrently activated and it is assumed that in vivo, the activation is provided by interaction of the naive CD4 cells with their specific antigen (Ag), presented by Ag-presenting cells (APC). Since only minuscule proportions of CD4 cells with specificity toward tested Ags exist in preparations of CD4 cells from wild-type animals, the activation by Ag and APC has been commonly replaced by exposure of the CD4 cells to antibodies (Abs) against CD3 and CD28, two molecules that participate in the physiological process of activation.CD3 is a major component of the T-cell receptor (TCR) complex, while CD28 is a potent costimulatory molecule. Interaction of the anti-CD3 and anti-CD28 Abs with their target molecules results in vigorous activation of the CD4 cells,8,9 a process that has served as a replacement for activation with the Ag presented by APC. Activation by these Abs for generation of polarized Th cell lineages has been employed, therefore, in numerous studies that have identified and characterized the different Th subpopulations.
The availability of TCR transgenic (Tg) mice made it possible, however, to generate lines of polarized Th cells by activation of naive CD4 cells with the specific Ag presented by APC.10,11,12,13 The TCR Tg cells can also be activated by the anti-CD3/CD28 Abs, thus making it possible to compare lines of polarized cells generated by either one of the two modes of activation. In a previous study, we noticed differences in the pathogenicity between subpopulations of Th1 and Th17, generated by either one of the two modes of activation.14 The present study expanded these preliminary observations by comparing subpopulations of Th1, Th9 and Th17, specific to the same Ag (hen egg lysozyme (HEL)),but generated by activation with either HEL/APC, or anti-CD3/CD28 Abs. We compared the three lineage pairs for pathogenicity, the capacity to invade and proliferate in the recipient spleen, their major surface markers, their expression of certain chemokines and chemokine receptors transcripts, as well as their gene expression patterns by microarray analysis. The data show remarkable differences between the corresponding subpopulation pairs, generated by the two different modes. In addition, however, unexpected close similarity was found among the lineages generated by activation with either one of the modes, in their transcript expression patterns of the chemokine and chemokine receptors, as well as the gene expression as determined by the microarray analysis.
Materials and methods
Mice
All mice used in this study were (FVB/N×B10.BR) F1 hybrids, transgenically expressing either HEL in their eyes (‘HEL-Tg'), or HEL-specific TCR by their T cells (‘3A9'); see Ref. 15 for detail. The mice were housed in a pathogen-free facility and all manipulations were performed in compliance with the NIH Resolution on the Use of Animals in Research.
Reagents
IL-6, TGF-β, PE-anti-Ccr6, PE-anti-Cxcr3 and their corresponding IgG isotype controls were provided by R&D Systems. IL-1α was from PeproTech, anti-IFN-γ (clone R4-6A2) was from Harlan Bioproducts for Science, anti-IL-4 (clone 11B11) was from NCI-Frederick Repository, and IL-12 and HEL were purchased from Sigma-Aldrich. The following reagents were from BD Biosciences: IL-4, anti-CD3 antibody, anti-CD28 antibody, anti-IL-12, PE-anti-CD4, Percp-Cy5.5-anti-CD4, PE-anti-CD45RB, PE-anti-CD69, PE-anti-α4β7, PE-anti–αEβ7, PE-anti-IL-17, APC-anti-IFN-γ, IgG isotype control and 7-AAD. A clonotypicm Ab specific for the TCR of 3A9 mice, designated ‘1G12', a gift from E. Unanue (Washington University), was conjugated with FITC (Pierce).
In vitro T-cell differentiation
Naive CD4+ T cells were purified from spleen and lymph node cells of 3A9 mice, using T-cell columns (R&D Systems). CD4 cells expressing the Tg TCR were sorted by FACSAria II (BD Biosciences), using the clonotypicm Ab 1G12, and then were activated and polarized toward Th1, Th9 and Th17 lineage as follows: the CD4 cells were cultured in 12-well plates (Corning) at 25×104/ml cells in a volume of 2 ml of RPMI-1640, supplemented with 10% FCS, antibiotics and 50 µM 2-ME. Activation was induced either by HEL (1 µg/ml for Th9 cell culture, or 2 µg/ml for Th1 and Th17 cell cultures) presented by irradiated (30 Gy) syngeneic wild-type naive splenocytes serving as APCs (125×104/ml) (‘HA'), or byplate bound anti-CD3 (1 µg/ml) and anti-CD28 (10 µg/ml) Abs (‘PbAb'). Polarizing cocktails, added concurrently with the activation process included: for Th1 lineage, 10 ng/ml IL-12 and 10 µg/ml anti-IL-4; for Th9 lineage, 10 ng/ml IL-4 and 1 ng/ml TGF-β and for Th17 lineage, 3 ng/ml TGF-β, 10 ng/ml IL-6, 5 ng/ml IL-1α, 20 µg/ml anti-IFN-γ, 10 µg/ml anti-IL-4 and 10 µg/ml anti-IL-12.
In additional experiments, we titrated the capacity of the anti-CD3 Ab to activate the CD4 lymphocytes during the polarization process by testing the Ab at three different concentrations, 0.2, 1.0 and 5.0 µg/ml. All other conditions were the same as detailed above and the proportions of the generated Th1, Th9 and Th17 cells were determined by flow cytometry, as detailed below.
Adoptive transfer of polarized Th cells
Th cells (5×106) harvested after 3 or 4 days of activation were injected via the tail vein into HEL-Tg mice. Recipient mice were killed 4 or 7 days post cell injection and their eyes were collected for histological analysis by conventional hematoxylin and eosin methods.
Flow cytometric analysis
Polarized cells cultured for 3 or 4 days and lymphocytes isolated from the recipient spleen at different time points were collected for surface cytokine staining and intracellular staining according to the manufacturers' instructions (see Ref. 15 for details). Cells were acquired on a FACSCalibur (BD Bioscience). The data were analyzed by FlowJo software (Tree Star).
Quantitative (q)-PCR
Transcript levels of the tested chemokine and chemokine receptor genes of the polarized Th cells were assessed by qPCR as described elsewhere,15 using reagents and methods according to the manufacturer's instructions (Applied Biosystems).
Affymetrix microarray data collection and analysis
Total RNA was isolated from different Th cells cultured for 4 days using mirVana miRNA isolation kit (Ambion). In total, 500 ng total RNA was amplified and biotin-labelled using MessageAmp II-Biotin Enhanced Kit (Ambion). Approximately 10 µg of total labeled RNA was hybridized to GeneChip MEO430 2.0 arrays (Affymetrix) according to the manufacturer's protocols. Expression values were determined using GeneChip Operating Software v1.1.1. All data analysis was performed using GeneSpring GX 11.0 (Agilent Technologies). Expression values for each probe were normalized using Robust Multichip Average method. All probes with expression values <50 in all samples were deleted from subsequent analysis (The GEO accession number for the microarray data: GSE54938.)
Statistical analysis
Data were shown as the mean±s.e.m. The software GraphPad Prism was used to perform the statistical analyses of the data with two-tailed Student's t-test. Differences were considered significant at P<0.05 (*P<0.05; **P<0.01; ***P<0.001).
Results
Th lineages generated by activation with either anti-CD3/CD28 Abs or HEL/APC differ in their pathogenicity
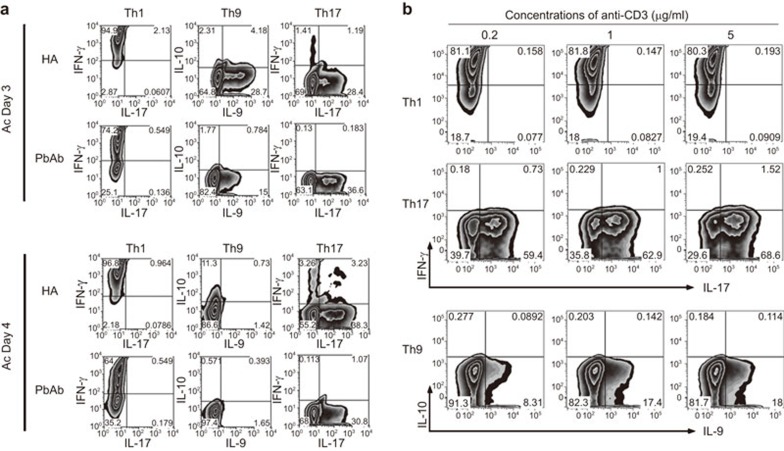
To compare between subpopulations generated by the two modes of activation, we generated lines of Th1, Th9 and Th17 cells by activating naive CD4 cells during the respective polarization process with either plate-bound anti-CD3/CD28 Abs (‘PbAb'), or the target Ag, HEL, presented by APC (‘HA'). Production of the signature cytokines of Thlineages was directly demonstrated by flow cytometry. The data of a representative experiment are recorded in Figure 1a and show that, under polarizing conditions, either activation mode drove the Th differentiation, as indicated by expression of the Th signature cytokine of each cell line.
Figure 1.
HA and PbAb Th subpopulations differ in their pathogenicity. (a) Immunostaining for intracellular expression of signature cytokines of Th cells generated by two activation modes. The cells were collected after 3 or 4 days in culture and stained with the specific mAbs. A representative experiment; similar observations were made in another experiment. (b) Titration of the anti-CD3 antibody's capacity to activate CD4 cells during polarization. The antibody was added to final concentrations of 0.2, 1.0 or 5.0 µg/ml to cultures during polarization toward Th1, Th9 or Th17 phenotypes and the antibody capacity was evaluated by flow cytometry for the expression levels of the signature cytokines, IFN-γ, IL-9 and IL-17, respectively. Ab, antibody; mAb, monoclonal antibody; Th, T-helper.
To examine the efficiency of the anti-CD3 Ab used for activation, we determined the expression levels of the three signature cytokines, IFN-γ, IL-9 and IL-17 in cultures activated by the anti-CD3 at three concentrations, 0.2, 1.0 and 5.0 µg/ml. A representative experiment is shown in Figure 1b and demonstrates that the expression of IFN-γ and IL-17 was not affected much by changes in the anti-CD3 Ab, whereas IL-9 expression declined when the Ab concentration was at 0.2 µg/ml, but was the same in cultures activated with 1.0 or 5.0 µg/ml. As mentioned above, the concentration of anti-CD3 Ab used in all experiments in this study was 1.0 µg/ml.
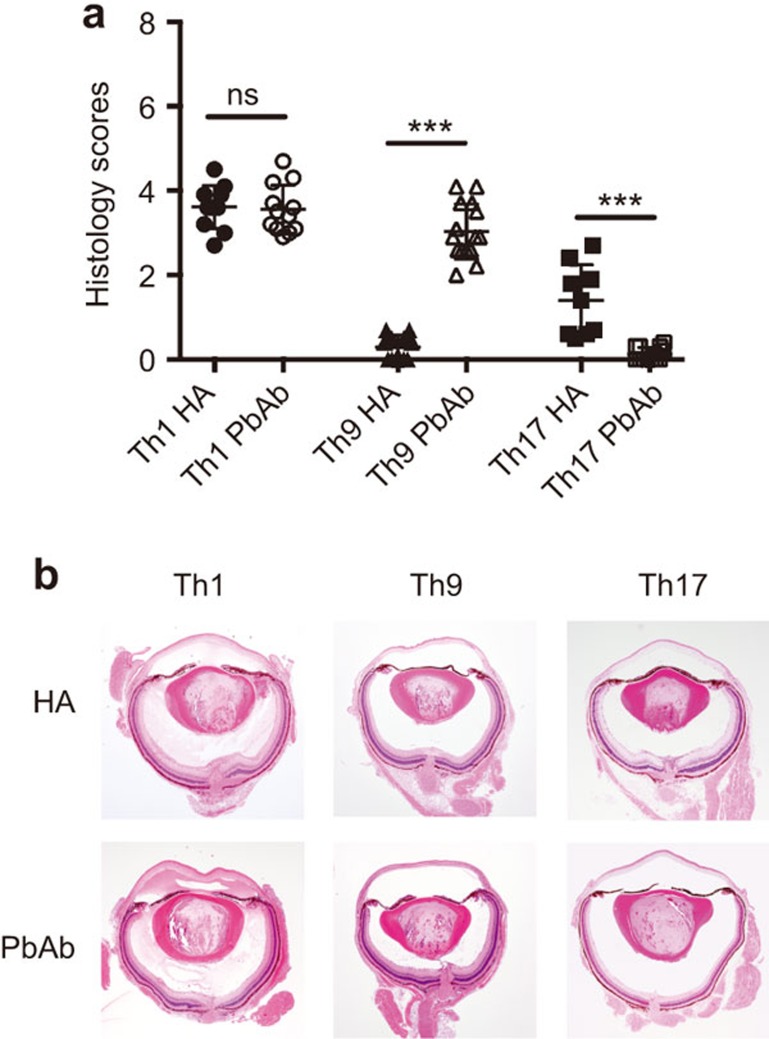
To determine the pathogenic capacity of the created line cells, we adoptively transferred them into syngeneic recipients expressing HEL in their eyes14,15,16 and examined the recipient eyes for histopathological changes. Data of three experiments are summarized in Figure 2a and eye sections representative for the typical changes for each treatment group are shown in Figure 2b. Severe inflammation was induced by Th1 cells generated by activation with either HA or PbAb. Th17 cells, however, acquired pathogenicity only following activation by HA; no disease was detected in recipients of Th17 cells activated by the PbAb. Unlike Th17, Th9 cells were consistently pathogenic only when activated by the Abs (PbAb). It is of note that moderate disease was induced by HA-activated Th9 cells, but only when collected on day 3 of incubation; no pathological changes were detected in eyes of recipients injected with cells collected on days 4 or 5 of incubation.6,16 It is also noteworthy that large portions of adoptively transferred Th17 and Th9 shift their phenotype in the recipient mice and acquire new phenotype.15,16 Additional studies are required to further discern the phenotype switching phenomenon and its effect on the pathogenicity and other biological capacities of Th cells. Interestingly, unlike with Th9 and Th17, no phenotype switching was detected with Th1 cells.
Figure 2.
(a) Variability of pathogenicity between Th lineages generated during polarization by activation with either HA or PbAb. Summaries of three independent experiments. **P<0.01, ***P<0.001. (b) Sections of representative eyes of mice recipients of Th1, Th9 or Th17, generated by HA or PbAb modes and collected on day 7 post-cell injection. Severe changes are seen in eyes of Th1 recipients, generated by either HA or PbAb. Changes are also seen in recipients of Th9 cells generated by PbAb, but not by HA; whereas Th17 cells were pathogenic only when generated by the HA mode, but not by the PbAb mode. The typical ocular pathological changes mainly include accumulation of inflammatory cells at the two ‘sites of entry', namely, the optic nerve head and limbus, and infiltration of inflammatory cells into the vitreous and anterior chamber. Also typical are the edematous changes in the retina and cornea. Scoring the histopathological changes was performed as detailed elsewhere.11 In brief, changes were scored separately in the anterior chamber, vitreous and retina, on a scale of 0–3 each, with the maximum sum score being 9.0. Ab, antibody; Th, T-helper.
Th lineages generated by HA or PbAb differ in their migration to and proliferation in the recipient spleen
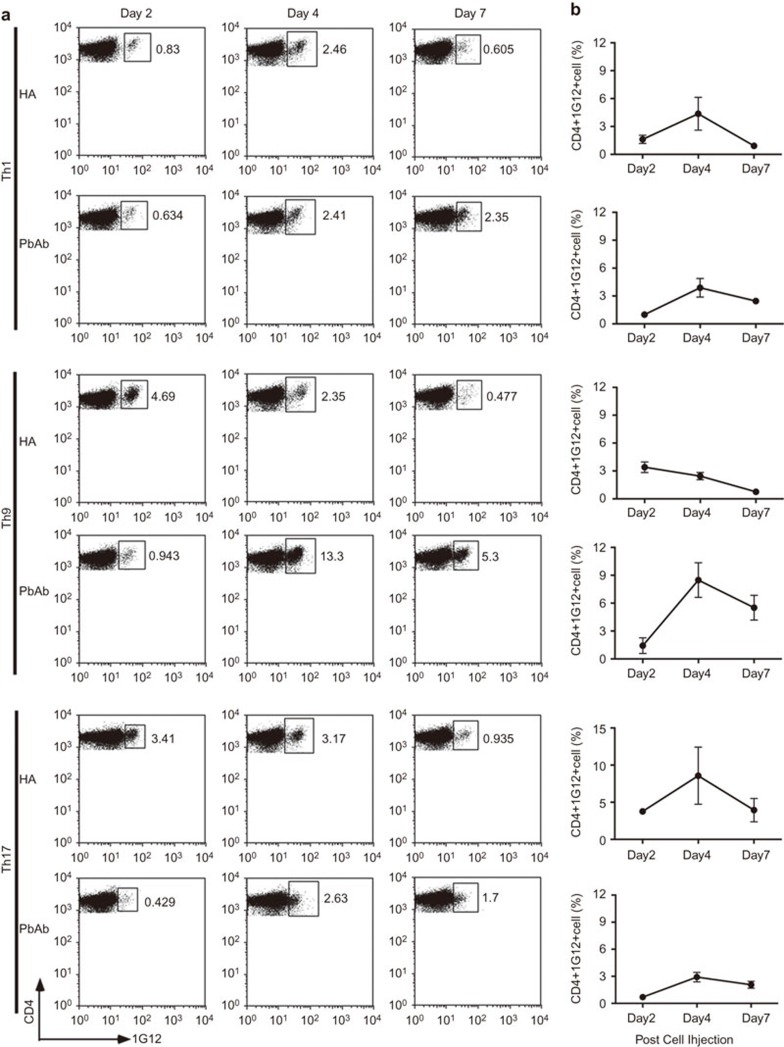
We have previously shown that major portions of adoptively transferred activated and pathogenic Th cells specific to HEL migrate to the recipient's spleen, where they proliferate,17 prior to their invasion of the eye, the only organ in which the target Ag, HEL, is expressed in this line of Tg mice.18 The Th transferred cells in our experimental system are traced in the recipient mouse by a clonotypic monoclonal Ab, ‘1G12', specific to the Tg TCR of these cells. To compare the cell lineages generated by activation with either PbAb or HA for their capacity to migrate to and proliferate in the recipient spleen, we collected the spleens 2, 4 or 7 days post-cell injection and determined by flow cytometry the percent of 1G12-positive cells among the CD4 population. Data of a representative experiment are shown in Figure 3a and are graphically summarized together with data of two other experiments in Figure 3b. Differences were noted between lineages generated by activation with HA or with PbAb and were particularly dramatic between the Th9 lineages: percentages of Th9 cells generated by activation with HA only declined gradually following their highest level on day 2, whereas the percentage of PbAb Th9 cells exhibited a steep increase between days 2 and 4. Also of note is the observation that percentages of all Th cells activated with HA were higher on day 2 than were the values found with the corresponding PbAb-activated Th lineages.
Figure 3.
Th lineages differ in their capacity to migrate to and proliferate in the recipient spleen. (a) Flow cytometric analysis carried out on recipient mouse spleen cells on days 2, 4 or 7 post-cell injection. ‘1G12' is an mAb specific to the TCR of the transferred cells. (b) Percent cell number of donor cells (CD4+/1G12+) among CD4+ cells. Summaries of three independent experiments. mAb, monoclonal antibody; TCR, T-cell receptor.
Microarray analysis reveals differences between gene expression patterns of HA and PbAb Th lineages
To further dissect the differences between Th lineages generated by activation with either HA or PbAb, we analyzed the gene expression patterns of the six lineages, using Affymetrix Mouse MOE430 2.0 GeneChips. Two independent samples for each of the six lineages were subjected to microarray analysis. In addition, to exclude the effects of residual APC RNA within HA RNA pools, we also profiled the gene expression of APCs cultured alone under Th1, Th9 and Th17 conditions. All genes that have greater expression values in APC alone cultures than the counterpart HA cultures were excluded from further analysis. In addition, we purified CD4+ T cells from HA cultures using AutoMACS (Milteyni, Auburn, CA, USA) and tested the expression of 10 major genes. Our results showed only minute differences in gene expression between the purified CD4+ T cells and the non-fractionated HA cultures, thus ruling out the possible significant contribution of APC to the results (data not shown).
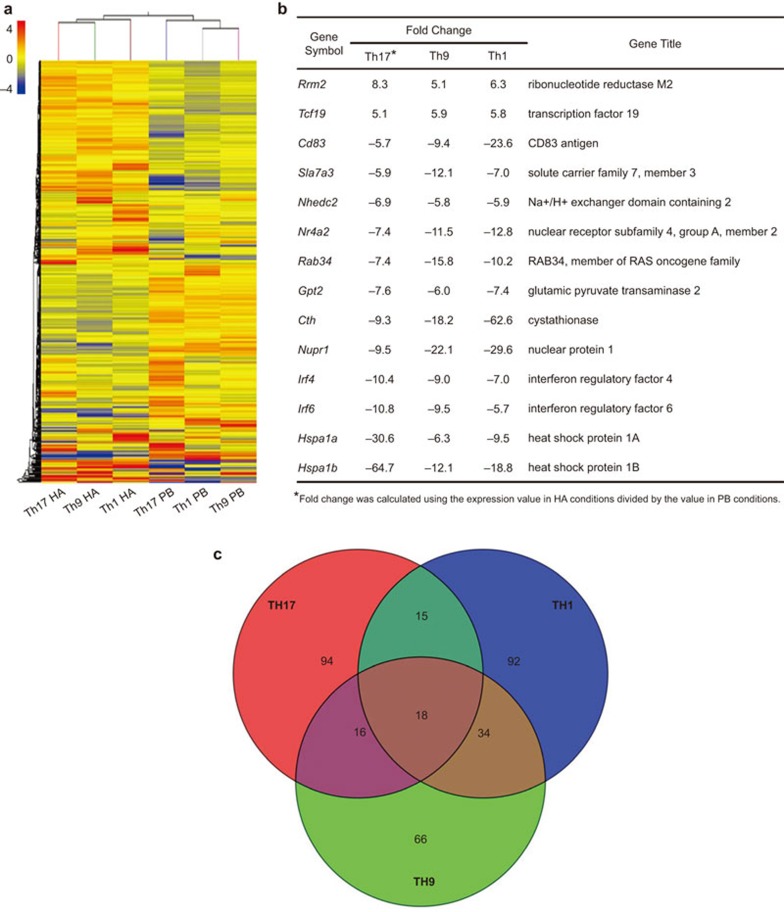
Intriguingly, a hierarchical clustering analysis of genes with expression changes of at least twofold between any two conditions revealed that in spite of being polarized under different conditions, the gene expression patterns of Th1, Th17 and Th9 activated by HA share more similarity than they do with their PbAb-activated counterparts (Figure 4a). Among 562 unique genes with significantly different expression (>twofold change; P<0.05, Student's t-test) between HA- and PbAb-activated lineages, 14 genes, including Rrm2 (ribonucleotide reductase M2)19,20 and Irf4 (interferon regulatory factor 4),21 were consistently either highly upregulated or remarkably downregulated in all three HA-activated, or three PbAb-activated lineages (Figure 4b). Therefore, our results suggest that the ways to activate CD4+ T cells may significantly affect the gene expression patterns, as well as the function of the polarized effector cells.
Figure 4.
Microarray analysis of genome-wide expression differences among the six different Th cell lineages. (a) A hierarchical clustering analysis of 20 175 probe sets with expression changes of at least twofold between any of the two conditions (‘HA' and ‘PbAb'). The color coding depicts the normalized expression value for each probe, with red representing higher expression and green representing lower expression as compared to the mean of the same probe across all samples. (b) A list of 14 unique genes (representing 18 probes) with at least fivefold increase or decrease in gene expression among all Th lineages stimulated by HA as compared to their expression in their counterpart lineages stimulated by PbAb. (b) A Venn diagram indicating the overlap and difference among Th17, Th1 and Th9 categories. Probe sets with at least fivefold increase or decrease in gene expression between HA- and PbAb-stimulated Th17, Th1 and Th9 cells are summarized in the Th17, Th1 and Th9 categories, respectively. Ab, antibody; Th, T-helper.
We next investigated whether the differences between HA- and PbAb-activated lineages were Th-specific. As shown in the Venn Diagram (Figure 4c), only 18 probe sets (encoding 14 unique genes shown in Figure 4b) shared common expression patterns among probe sets with at least fivefold difference in their expression between HA- and PbAb-activated Th1, Th17 or Th9 cells, while 94, 92 and 66 probe sets differed in their gene expression between HA- and PbAb-activated Th17, Th1, or Th9 cells in a Th lineage specific manner.
HA and PbAb Th lineages differ in their profiles of chemokine/chemokine receptor transcripts
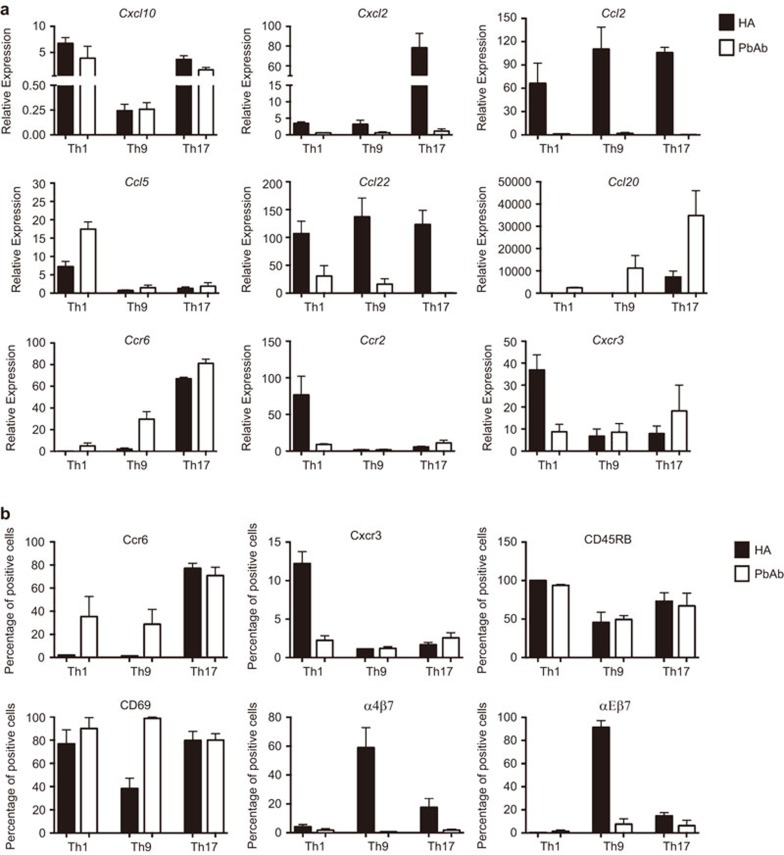
Mobility toward targets and related functions of Th cells are determined to a large extent by their profile of chemokines and chemokine receptors. To compare between the Th lineages generated by the two modes of activation, we extracted RNA samples from the cultured cells and determined by qPCR the levels of nine transcripts. These nine transcripts were found by McGeachy et al.22 to differ in their expression between the pathogenic and non-pathogenic subpopulations of Th17 cells. Data of three independent experiments are combined in Figure 5a and show in general remarkable differences between Th lineages activated by either HA or PbAb in their expression levels of the tested transcripts. Interestingly, similar patterns of difference between the pairs of transcripts were seen for five of the nine transcripts of lineages generated by activation with either HA or PbAb. Thus, the expression levels of transcripts for Cxcl2, Ccl2 and Ccl22 were higher in Th1, Th9 and Th17 cells activated by HA, whereas cells activated by PbAb expressed higher transcript levels of Ccl5 and Ccl20 than did the corresponding HA cultures. No clear consistency was seen, however, in the transcript expression levels of Ccr6, Ccr2, Cxcl10 and Cxcr3 between lineages activated by HA or PbAb.
Figure 5.
Different patterns of gene expression of immune-related molecules by the three pairs of Th lineages. The line cells were collected after 4 days in culture and examined by qPCR for expression of nine selected chemokines and chemokine receptor transcripts (a) and by flow cytometry for expression of six major cell surface Ags (b). Summaries of three independent experiments. Ag, antigen; qPCR, quantitative-PCR; Th, T-helper.
Th lineages generated by HA or PbAb differ in their profiles of surface Ag markers
Another parameter related to mobility of Th cells and their capacity to invade tissues is the profile of their surface Ags. To compare between the surface Ag profiles of cells of lineages generated by either HA or PbAb, we used flow cytometry with Abs against six major surface Ags. Data of repeated three experiments are summarized in Figure 5b. Remarkable differences were observed between the lineage pairs of the three Th phenotypes, activated by either one of the two modes, in their staining for Ccr6, Cxcr3, α4β7 and αEβ7. Only moderate or no differences were seen, however, between the three lineage pairs in their staining for CD45RB and CD69.
Discussion
Data collected in this study demonstrate that lineages of Th1, Th9 and Th17, generated by activation with plate-bound anti-CD3/CD28 Abs (‘PbAb') differ remarkably from lineages of the same named phenotypes, activated by the physiological process of interaction between the TCR on Th cells and the specific Ag, HEL, presented by APC (‘HA'). The different lineages were generated by activation by either one of the two tested modes concurrently with the polarization by the corresponding cytokines. The comparison between the two types of activation was achieved by using T-cell populations of TCR Tg mice, which were generated by activation with either PbAb or HA during polarization, thus yielding the pairs of lineages of Th1, Th9 or Th17 phenotypes we tested here.
Analysis of pathogenic capacity of the lineages, generated by activation with either HA or PbAb, revealed similar capacities by the two Th1 lineages, but sharp dissimilarities between the pairs of lineages of Th17 and Th9 (Figure 2). Th17 cells generated by activation with HA induced severe inflammation in recipient eyes, whereas no such activity could be detected in Th17 lineages activated by the PbAb. In contrast, Th9 lineages were consistently pathogenic only when activated by PbAb. The difference between the two Th17 lineages observed here is in line with our previous report.14 It is of interest that lineages of pathogenic or non-pathogenic Th17 cells were also generated in a study by McGeachy et al.,22 using a system different from ours; in their system, pre-immunized cells were stimulated by IL-23 or by IL-6/TGF-β, to generate pathogenic or non-pathogenic Th17 cells, respectively. It is possible that the differences in pathogenicity between lines generated by the different generation modes are related to variability in the cells' capacity to switch phenotypes, as shown for Th17.15
Pairs of lineages of the same phenotypes also differed in their capacity to migrate to and proliferate in spleens of the recipient mice, before moving on to invade the recipient's eyes, where the target Ag (HEL) was expressed (Figure 2). Different patterns of migration and proliferation were observed among lineages of the three Th phenotypes and, in particular, between the pairs of Th9cells generated by either one of the activation modes. The differences in the migration capacity, as well as the pathogenicity of the lineages, could be attributed, at least in part, to differences we found in expression of chemokines and their receptors (Figure 5a), as well as of surface Ags (Figure 5b). These two families of molecules determine to a large extent the cells' mobility and their capacity to invade the target tissue. Surprisingly, the pattern of differences in expression of the nine tested chemokines and chemokine receptor transcripts was found here to be similar for five transcripts among Th of the three different phenotypes (Th1, Th9 and Th17) when activated by the same mode, i.e., by either HA or PbAb.
Striking differences between lineage pairs, generated by activation with either PbAb or HA, were also found by the microarray analysis. Of particular interest is the observation we made by the hierarchical clustering analysis, namely, that the patterns of changes in gene expression were more similar among the subpopulations of Th1, Th9 and Th17, activated with either PbAb or HA, than were the similarity levels between the pairs of subpopulations of each of the three phenotypes (Th1, Th9 and Th17) (Figure 4a). Furthermore, our analysis revealed that 14 genes were highly expressed specifically by subpopulations of Th cells of the three phenotypes, activated with either HA or PbAb (Figure 4b), suggesting that the mode of activation of Th lineages critically affects the gene expression patterns during the polarization process. Moreover, this notion is in line with our other observation, mentioned above, of similarity in expression profiles of chemokine and chemokine receptortranscriptsbyTh1, Th9 and Th17 cells when activated by HA, or by PbAb. In contrast, essentially no similarity was observed between the pattern of transcript expression by Th cells of the same phenotype (Figure 5a).
A great number of published studies that have analyzed various aspects of polarized Th populations have used lineages generated by activation with the anti-CD3/CD28 Abs during the polarization process (e.g., Refs. 23–25). Our data show, however, that cell populations of the three tested phenotypes generated by activation with the Abs differ by various aspects from those generated by the ‘physiological' activation with the specific Ag presented by APC. Our findings thus underscore the limitations of biological observations made with Th lineages generated by activation of CD4 cells with the anti-CD3/CD28 Abs.
Acknowledgments
We thank Robert S Lee for tail DNA analysis, Samuel J H Hinshaw for digital microphotography, Rafael Villasmil for cell sorting and the NEI Histology Core Facility for tissue section preparations. This work was supported by the Intramural Research Program of the National Eye Institute, NIH.
References
- Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Dong C. A complex issue on CD4+ T-cell subsets. Immunol Rev. 2013;252:5–11. doi: 10.1111/imr.12041. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noelle RJ, Nowak EC. Cellular sources and immune functions of interleukin-9. Nat Rev Immunol. 2010;10:683–687. doi: 10.1038/nri2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R, Kaplan MH. A brief history of IL-9. J Immunol. 2011;186:3283–3288. doi: 10.4049/jimmunol.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Gery I. The unique features of Th9 cells and their products. Crit Rev Immunol. 2012;32:1–10. doi: 10.1615/critrevimmunol.v32.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- Lafaille JJ, Keere FV, Hsu AL, Baron JL, Hass W, Raine CS, et al. Myelin basic protein-specific T helper 2 (Th2) cells cause experimental autoimmune encephalomyelitis in immunodeficient hosts rather than protect them from the disease. J Exp Med. 1997;186:307–312. doi: 10.1084/jem.186.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell SR, Greenberg PD. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Zhang M, Vistica BP, Chan CC, Shen DF, Wawrousek EF, et al. Induction of ocular inflammation by T-helper lymphocytes type 2. Invest Ophthalmol Vis Sci. 2002;43:758–765. [PubMed] [Google Scholar]
- Iqbal N, Oliver JR, Wagner FH, Lazenby AS, Elson CO, Weaver CT. T helper 1 and T helper 2 cells are pathogenic in an antigen-specific model of colitis. J Exp Med. 2002;195:71–84. doi: 10.1084/jem.2001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trickett A, Kwan YL. T cell stimulation and expansion using anti-CD3/CD28 beads. J Immunol Methods. 2003;275:251–255. doi: 10.1016/s0022-1759(03)00010-3. [DOI] [PubMed] [Google Scholar]
- Shi G, Lovaas JD, Tan C, Vistica BP, Wawrousek EF, Aziz MK, et al. Cell–cell interaction with APC, not IL-23, is required for naive CD4 cells to acquire pathogenicity during Th17 lineage commitment. J Immunol. 2012;189:1220–1227. doi: 10.4049/jimmunol.1103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Cox CA, Vistica BP, Tan C, Wawrousek EF, Gery I. Phenotype switching by inflammation-inducing polarized Th17 cells, but not by Th1 cells. J Immunol. 2008;181:7205–7213. doi: 10.4049/jimmunol.181.10.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Aziz MK, Lovaas JD, Vistica BP, Shi G, Wawrousek EF, et al. Antigen-specific Th9 cells exhibit uniqueness in their kinetics of cytokine production and short retention at the inflammatory site. J Immunol. 2010;185:6795–6801. doi: 10.4049/jimmunol.1001676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JC, Fukushima A, Wawrousek EF, Lobanoff MC, Charukamnoetkanok P, Smith-Gill SJ, et al. Immunotolerance against a foreign antigen transgenically expressed in the lens. Invest Ophthalmol Vis Sci. 1998;39:2049–2057. [PubMed] [Google Scholar]
- Chen J, Fujimoto C, Vistica BP, He J, Wawrousek EF, Kelsall B, et al. Active participation of antigen-nonspecific lymphoid cells in immune-mediated inflammation. J Immunol. 2006;177:3362–3368. doi: 10.4049/jimmunol.177.5.3362. [DOI] [PubMed] [Google Scholar]
- Heidel JD, Liu JY, Yen Y, Zhou B, Heale BSE, Rossi JJ, et al. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res. 2007;13:2207–2215. doi: 10.1158/1078-0432.CCR-06-2218. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z, et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res. 2013;19:3416–3428. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas PS, Bhagat G, Pernis AB. IRF4 and its regulators: evolving insights into the pathogenesis of inflammatory arthritis. Immunol Rev. 2010;233:79–96. doi: 10.1111/j.0105-2896.2009.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, et al. Transforming growth factor-beta ‘reprograms' the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]