Abstract
The major cause of mortality and morbidity in human beings is bacterial infection. Bacteria have developed resistance to most of the antibiotics primarily due to large scale and “indiscriminate” usage. The need is to develop novel mechanisms to treat bacterial infections. The expression of pathogenicity during bacterial infections is mediated by a cell density dependent phenomenon known as quorum sensing (QS). A wide array of QS systems (QSS) is operative in expressing the virulent behavior of bacterial pathogens. Each QSS may be mediated largely by a few major signals along with others produced in minuscule quantities. Efforts to target signal molecules and their receptors have proved effective in alleviating the virulent behavior of such pathogenic bacteria. These QS inhibitors (QSIs) have been reported to be effective in influencing the pathogenicity without affecting bacterial growth. However, evidence is accumulating that bacteria may develop resistance to QSIs. The big question is whether QSIs will meet the same fate as antibiotics?
Keywords: Antibiotics, Biofilms, Cheaters, Quorum quenching, Quorum sensing, quorum sensing inhibitors
Introduction
Microbial infections are a major concern for health departments around the globe. The discovery of antibiotics almost a century ago brought hope and relief to many patients [1]. Antibiotics have continued to be one of the most effectual medications for treating bacterial infections since the 1950s [2, 3]. However, with the evolution of multiple drug resistant strains and evasive behavior of bacteria towards antibiotics, pharmaceutical companies are finding investments in developing novel antibiotics to be counterproductive [4]. A search for novel treatment strategies has obliged researchers to deliver a re-look on the mechanisms of bacterial infection. It has been also realized that around 80% of the infectious diseases are caused by bacteria which form biofilms [5]. Biofilms are composed of exopolysaccharides, which protect its structure and enable bacteria to resist toxic compounds and high doses of antibiotics [6]. The most important fact has been the finding that majority of the genes involved in bacterial pathogenicity are mediated through quorum sensing (QS). Bacteria are single celled organisms, which under certain conditions behave like ‘multicellular’ organisms. This generally happens only after bacterial population density crosses a threshold level. This unique bacterial behaviour has been termed as QS. QS operates through a wide range of signals such as: (1) oligopeptides, (2) acylhomoserine lactones (AHLs), (3) furanosyl borate (autinducer-2), and (4) fatty acids [7, 8]. QS in Gram negative bacteria is mediated largely by AHLs, whereas in Gram-positive bacteria, the signals are peptidic in nature [9, 10]. The quantum of QS mediated genes can constitute as much as around 10% of the total bacterial genome. Thus, pathogenic bacteria with this unique characteristic are able to evade the host’s defense while being at low cell densities [11–13].
Quorum sensing inhibition
Bacteria exist as free living forms in mixed communities or in association with plants and animals either as pathogens or as symbionts. Although interdependent, these partners are still able to keep their identities. Bioactive molecules are produced by almost all living organisms; the exact significance and the scope of their uses are yet to be elucidated [3]. Plants grow and release secondary metabolites into the rhizosphere and animals develop antibodies as defense mechanisms against bacterial invasions [14, 15]. Bacteria display different survival strategies to counter the eukaryotic attacks, including secretion of hydrolytic enzymes and antibiotics [16–24]. These protective phenomena provide clues that in nature, bacterial species must also be releasing their arsenal of QS dependent virulent factors to conform to these situations [24]. In fact, many prokaryotes and eukaryotes have been shown to produce bioactive molecules to disrupt QS process at different stages: (i) inhibiting or reducing the activity of the QS signal producing gene, (ii) disrupting the structure of the signal molecule, (iii) modulating the binding of the signal to the receptor sites, (iv) blocking the receptor site with antagonist - signal analogues [25–27]. This phenomenon of inhibiting bacterial QS especially the expression of virulence genes through the production of bioactive molecules is termed as quorum quenching [28–30].
It has been discovered that bacteria present within the QS mediated biofilm are up to 1000 times more resistant to antibiotics than the planktonic forms [31]. This change in bacterial behavior severely complicates the treatment process. The aim is thus to either prevent biofilm formation process or to disrupt it. Efforts have been made to search QS inhibitors (QSIs) for each of these stages or targets. A set of criteria has been laid down for identifying QSI molecules: (i) a low molecular mass, (ii) highly specific, (iii) stable and resistant to hydrolytic enzymes of the host (iv) no adverse effect on the host, (v) longer side chain than the native AHLs [32–34]. QSIs have been shown to be produced by plants, animals, bacteria, and other microbes [25, 26, 28, 35, 36]. The apprehension is that constant and indiscriminate usage of QSIs may put bacteria under pressure to develop means to evade this treatment procedure. The goal of this review is to provide an overview of the potential threats posed by bacteria becoming resistant to QSIs and finally what kind of QSIs should be developed to avoid this.
The Myth
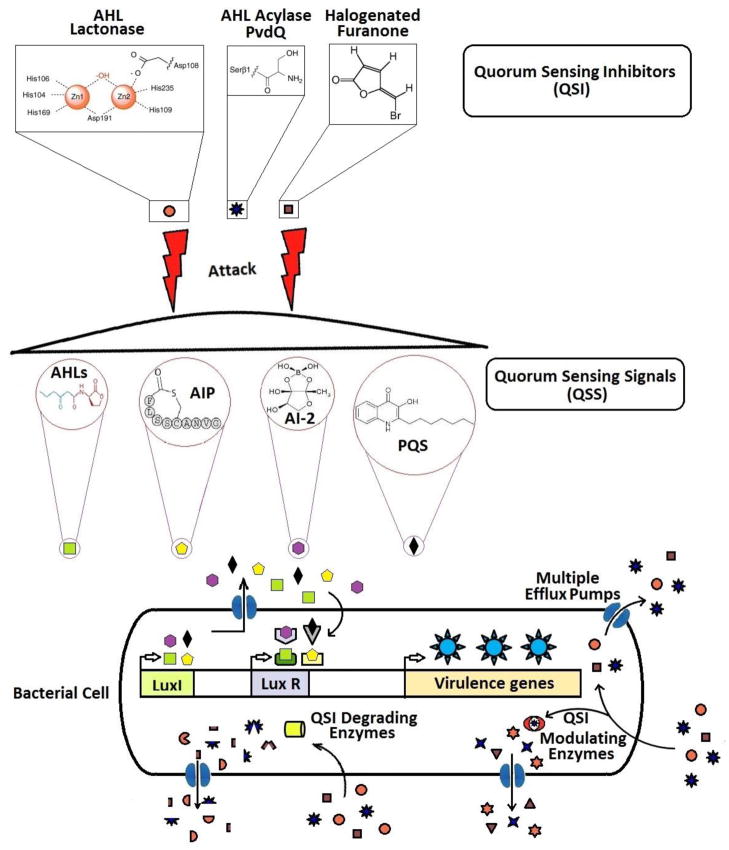
Antibiotics inhibit the growth of the microbes and may eventually kill it. The microbes under this strong selective pressure tend to develop resistance to antibiotics through natural selection or genetic mutations [37]. Bacterial pathogenicity evolves to counter environmental stress and selective pressure, especially those caused by antibacterial agents [38]. It is primarily to ensure survival that bacteria evade host defences through an attack and defence mechanism [20, 39]. Bacterial genomic plasticity and mobility of genetic material are important facets in their evolution [40]. It has been reported that in a given population there are persisters which evade the lethal effect of antibiotics [41]. These persisters thus manage to develop tolerance to antibiotics [42]. The overall mechanism of resistance to antibiotics is conferred by genes (i) responsible for the degradation of antibiotics (bla, pbp), and ii) efflux pumps (cmcT) [4, 37, 43]. It was thus thought that the role of ‘antipathogenic’ compounds will be more desirable than antibacterial compounds, since they would neither kill bacteria nor completely inhibit their growth. Since these bioactive molecules are perceived to manipulate the infection process without affecting bacterial growth, bacteria may not be compelled to develop resistance against them [7, 11, 24, 32, 44–48]. However, whether bacteria can become resistant to QSIs is now being questioned (Figure 1)[38, 49, 50]?
Figure 1.
Potential mechanisms of resistance to quorum sensing inhibitors.
The first demonstration that bacteria do indeed develop resistance to QSI was shown using growth of Pseudomonas aeruginosa on adenosine as the sole carbon source, which requires active QS [50]. When the QSI compound was added that masks the QS pathways (a brominated furanone known as C-30), growth on adenosine was impaired, and within four sequential dilutions after transposon mutagenesis, cells arose that were resistant to the QSI [50]. The gain of function mutations was in repressors of an efflux pump, and the QSI resistant strains became resistant by having greater efflux of the QSI compound, a result that had not been anticipated in regard to QSI compounds. This result was predicted using QS mimics in the absence of a QSI compound [51]. Moreover, clinical isolates from cystic fibrosis patients that had been treated with antibiotics were found to carry the same efflux-enhancing mutations and were resistant to the same QSI compound [50]; hence, QSI resistance arises even before the use of the QSI compound. Additional results identifying clinical strains resistant to the QSI C-30 were obtained using isolates from urine, blood, and catheter tips [52]. Therefore, strains in both the laboratory and in the clinic have been shown to evolve resistance to QSIs.
Multiplicity of Quorum sensing systems (QSS) and QS signals: A latent weapon to counter QSIs?
The field of QSS has made rapid progress since its discovery in Vibrio fischeri. These works have revealed that QS circuits are much more complex than envisaged initially [53]. In majority of the persistent pathogens, virulence genes are under QS control [54]. The most prevalent bacterial QSS is mediated by the signal molecules (AHLs) produced by synthase gene (luxI homologs). It binds to the receptor and activates the transcription regulator (luxR homologs). This complex leads to the transcription of a plurality of genes involved in pathogenicity [55]. In general, most bacteria possess a QSS having a single set of signal synthase gene and transcription regulator gene (I/R). A few other bacteria such as Pseudomonas, Sinorhizobium and Vibrio species have multiple (I/R) systems (Table 1)[56–66]. The complexity of these systems is reflected in the diversity of the signals produced by certain bacteria (Table 1) [62, 67–74]. The multiplicity of QSS is complicated by an overlapping regulation [75]. In multiple QSS, there are chances that transcriptional regulator from different QSS may form heterodimers [76]. The binding of these heterodimers to a promiscuous promoter might lead to different gene expression profiles, allowing bacteria to sense a wide range of environmental stresses which may include QSI [77]. The question is: Does this diversity of QSS and QS signal molecules allow bacteria to escape QSI? Is this a hidden trait, which bacteria can exploit for developing resistance to QSI? The multiplicity of QSS and their signals can prove beneficial to the bacteria to either conserve valuable resources or allow them to modulate the activity of the receptors [78]. The presence of 2–5 LuxR signal receptor homologs in Burkholderia mallei and the variability in the specificity of AHL synthases in Erwinia carotovora strains SCC3193 and SSC1 – support the likelihood of their developing resistance to QSI molecules [79, 80]. It can be implied that QSIs designed to block only the las QSS might result in the rapid appearance of the resistant strains. It may thus be necessary to block both the las and rhl QSS to efficiently reduce production of virulence factors by P. aeruginosa [75].
Table 1.
Diversity of quorum sensing systems and signal molecules
| Organism | Quorum sensing | Reference | |
|---|---|---|---|
| Systemsa | Signal moleculesb | ||
| Pseudomonas aeruginosa | LasI(Synthase)/R (Receptor) | 3OC12HSL | [56–61, 74] |
| RhlI/R | C4HSL | ||
| QscR (Quorum sensing control repressor) | 3OC12HSL (LasI regulates QscR) | ||
| PqsABCD | 2-heptyl-4-quinolone (HHQ) | [74] | |
| Pseudomonas fluorescens, and P. alcaligenes | LuxR | Diketopiperazines (DKPs): Cyclo(L-Phe-L-Pro) | [72] |
| Sinorhizobium meliloti | SinI/R | C14HSL to C18HSL | [62, 63] |
| TraM/R and Mel | C4HSL | ||
| Rhizobium leguminosorum | RhiI/R, CinI/R | OHC6HSL, 3OC6HSL, C7HSL, and 3OHC14HSL | [71, 73] |
| Vibrio harveyi | HAI-1 (Species specific) | OHC4HSL | [64] |
| CAI-1 (Intergeneric communication) | (S)-3-hydroxytridecan-4-one | [65] | |
| LuxP (Interspecies communication) | AutoInducer-2 | [64] | |
| Burkholderia cepacia | CepI/R | C8HSL | [73] |
| Burkholderia pseudomallei | BpmI2/R2 | C8HSL, C10HSL, | [73] |
| BpmI3/R3 | OHC8HSL, OHC10HSL, 3OC14HSL | ||
| PmlI1/R1 | C8HSL, C10HSL, 3OHC8HSL, 3OHC10HSL | ||
| Erwinia carotovora | ExpI/R, CarI/R | 3OC6HSL | [73] |
| Bacillus cereus | PlcR | Peptide PapR | [75] |
| Staphylococcus aureus | AgrABCD | Autoinducing peptide | [74] |
I/R system: Synthase/Receptor
- C4HSL: N-butanoyl HSL
- C7HSL: N-heptanoyl HSL
- C8HSL: N-octanoyl HSL
- C14HSL: N-tetradecanoyl-HSL
- C18HSL: N-octadecanoyl-HSL
- OHC4HSL:N-hydroxybutyrylHSL
- OHC6HSL: 3-hydroxy-N-hexanoyl-HSL
- OHC8HSL: 3-hydroxy-N-octanoyl-HSL
- OHC10HSL: 3-hydroxy-N-decanoyl-HSL
- 3OHC14HSL: 3-hydroxy-7-cis-tetradecanoyl-HSL
- 3OC6HSL: 3-oxo-N-hexanoyl-HSL
- 3OC8HSL: 3-oxo-N-octanoyl-HSL
- 3OC12HSL: 3-oxo-N-dodecanoyl-HSL
Pseudomonas, an opportunistic pathogen, expresses a wide range of genes, which help it in surviving under harsh conditions prevailing on the surface and within the host organism [20]. These are also effective in challenging the host immune system and cause infectious diseases. P. aeruginosa causes diseases such as cystic fibrosis and microbial keratitis largely through AHL dependent QSS, which activates genes responsible for biofilm formation (chronic infections) and represses genes involved in the expression of Type III secretion system (TTSS) [81]. More recent works have shown that TTSS can also be expressed in biofilms [82]. It was opined that, AHL-dependent QS partially represses TTSS expression and that other QS signals of P. aeruginosa may be instrumental in modulating the expression of TTSS within biofilms [83]. Enterohemorrhagic Escherichia coli activates the transcription of their virulence genes through three types of signals i) Aromatic autoinducer (AI-3), ii) hormones- epinephrine and nonepinephrine [89]. The QseC membrane bound sensor kinase can be triggered by any one of these signals resulting in transcription of virulence genes [90, 91].
Brominated furanone produced by Delisea pulchra could inhibit the QS regulated swarming motility of Serratia liquefaciens. Nevertheless, the inhibitory effect of QSI was reversed by higher concentrations of exogenously supplied N-butanoyl HSL (C4HSL) [92]. In a mouse model, the inhibition of LuxR controlled PluxI-gfp (ASV) fusion by QSI - Furanone C-30 was negated by increasing the dose of 3-hydroxy-N-hexanoyl-HSL from 400 to 1200 μM. Here, the effect of a single C-30 injection dose was found to last for a short period of 6 hours [11]. An interesting observation on the effect of C-30 was its ability to repress the expression of a few QS mediated virulence genes such as lasA, lasB, hcnAB, rhlAB, chic, phnAB and phzABCDEFG. It however, allowed bacteria to exploit their multiple QSS to continue with an uninterrupted expression of lasI/R and rhlI/R gene clusters [11]. Since a basal concentration is sufficient to activate QS, 3OC12HSL may not be a limiting factor [11, 54, 69]. A QSI targeting 3OC12HSL alone may not affect QS and the operation of QSS in parallel may mimic a scenario where bacteria have become resistant to QSI [93]. Thus mechanisms seem to be already in place in P. aeruginosa to evade the effect of QSI by having multiple QSS and their signals [11, 38].
Mutations in QS circuitry
Another feature which helps bacteria to withstand antimicrobial agents is their innate ability to undergo mutations or acquire genes from closely or widely related organisms, through horizontal gene transfer [4, 40, 94, 95]. Genetic mutations in raiI and raiR genes of Rhizobium elti enabled the elucidation of 7 different AIs involved in nitrogen fixation process in Phaseolus vulgaris. It revealed that expression of raiI in R. elti mutants, could be activated by one or more AIs [69]. Similarly, Agrobacterium tumefaciens, P. aeruginosa, and Rhizobium species can withstand mutations in their QS circuitry because of their abilities to get activated by multiple AHL signals, which vary from 3 to 7 [96–98].
P. aeruginosa mutant PAO-R1, defective in lasR gene is unable to produce most of the QS dependent virulence factors [99, 100]. Although it was proposed to be a potential target to control P. aeruginosa infections, however, it was envisaged that a reverse mutation may lead to restoration of lasR functioning even if las QSS is absent [75]. Mutations in the LuxR receptor lead to their failure to recognize synthetic antagonist N-(propylsulfanylacetyl)-L-HSL [101]; however, resistance to a QSI was not tested, and these authors argued that such resistance was unlikely to evolve. Unfortunately, this item has not been recognized by several authors who have claimed that resistance to QSI was seen in this report and that “QSI resistance can easily be obtained” in this manner [38] although there is no proof to date that such mutations render cells resistant to QSI compounds. Spontaneous mutations arising in the QSS of Vibrio cholerae strains make them either non-functional or constitutive in nature [102]. Serratia marcescens has a unique genetic makeup - the location of spnIR QSS on a transposon (TnTIR), which may position it to take advantage to overcome any hindrance in their QSS [103]. In fact, P. aeruginosa lasI mutant PAO1-JP1, which was not able to produce 3OC12HSL could still form strong biofilms, under hydrodynamic conditions of high shear flow [104]. This observation has been underpinned by the effect of QSI from P. aeruginosa extracts, which affected virulence factors controlled by las and rhl system to different extents. However, the extract didn’t affect the biofilm formation progress [105]. P. aeruginosa mutant PAO-R1, defective in one of the 2 QSS (LasR-PAI-1), showed up-regulation of its second QSS (RhlR-PAI-2), under carbon (C) and nitrogen (N) stress conditions [75, 93]. Enhanced growth under certain physiological conditions was also recorded in lasR deficient isolates from cystic fibrosis patients and P. aeruginosa strain PA14 [106]. It raised doubts that targeting biofilm formation process may not be the best option to moderate bacterial infections [104, 105].
The inherent ability of Pseudomonas allows it to exercise a stringent control over the infection process. QscR regulator of P. aeruginosa modulates the expression of virulence gene responsible for the production of factors such as hydrogen cyanide, pyocyanin and elastase [59, 76]. At low cell densities, QscR inhibits the expression of these genes by forming inactive heterodimers with LasR and/or RhlR. This complex consequently inhibits the QS mediated gene expression [76]. On the other hand, it may also be interpreted that suppressing QscR may lead to uninterrupted supply of QS signals, leading to ‘constitutive’ virulent behavior. Thus, while QSI will be reducing the concentration of AHLs, a mutant, of P. aeruginosa lacking qscR will counter its attack [53].
Another mechanism which retards the production of QS signal is through RsaL, the global regulator of QSS in P. aeruginosa [107]. It can repress transcription of the signal synthase gene lasI. RsaL is a QS repressor, which acts by binding to LasR and rsaL-lasI two-way promoter. It is a homeostatic mechanism which limits the production of QS signal (3O12CHSL) [108, 109]. However, a mutation in the rsaL gene, allows uninterrupted production of 3OC12HSL throughout the growth process [110]. It can help bacteria to continue with its functions of QS mediated virulence and pathogenicity. This regulator protein controls more than 130 genes in P. aeruginosa physiology and can be a potential mechanism to develop resistance to QSI by counterbalancing 3O12CHSL dependent gene activation [110].
The cheats
QS is a social system where a large population of bacteria work together and release goods (signal molecules) into the extracellular space. The system also leads to the production of enzymes necessary for their survival. These bioactive molecules are ‘freely’ available for all individuals in the population to sense and take advantage. This social co-operation is susceptible to exploitation by cheaters, who can derive benefit from it without spending their energy and are likely to destabilize the QSS [42, 79, 93, 111–114]. Studies have been conducted to elucidate the role of cheaters. QS cheats can be categorized as: (i) signal negative, the lasI mutants, which can avoid the energy needed for producing the QS signals and related activities, and (ii) signal blind, the lasR mutants, which have the ability to produce QS signals but don’t use them for production of virulent factors [115].
The growth of wild-type and lasI and lasR mutants of P. aeruginosa is influenced by the nutrients in the surrounding medium [112]. In nature, the proportion of signal blind strain is more common in mixed populations [112, 116], the social cheaters grow initially but they are not able to sustain as there are fewer co-operators available for exploitation. However, this scenario is anticipated to reverse with the evolution of QSI resistant bacterial strains [112, 115, 117]. The growth and behavior pattern of these social cheaters was gathered by allowing a small proportion of QS-deficient mutants to grow along with QS-proficient wild-types strains of P. aeruginosa [51]. In a population composed of QSI-resistant (wild-type) and QSI-sensitive strains (signal-blind lasR rhlR double mutants) of P. aeruginosa, a selective pressure was created for distinguishing the two population types by providing two different carbon sources: (i) bovine serum albumin (BSA), and (ii) adenosine. The utilization of both the C sources is QS mediated. BSA is metabolized by extracellularly produced proteases and thus benefits the entire population (public goods) and adenosine is degraded in the periplasmic space by nucleoside hydrolase (Nuh), useful only to producer cell (private goods) [51]. In a co-culturing of QSI sensitive mimics with varying proportions of wild-type P. aeruginosa (QSI-resistant mimics), growth characteristics varied with C source. A significant delay in the growth on BSA (public goods) was observed in the cases of co-culture having a larger proportion of QSI sensitive-mimic population. This growth retardation was not observed in the case of adenosine (private goods). This behavior of QSI-sensitive and – resistant populations of P. aeruginosa implies that social cheating will enhance the chances of developing resistance to QSI in mixed populations, in privately acquired nutrition [51].
In mixed populations of P. aeruginosa, wild-type strains are not able to compete with the cheats - lasR mutants, which cannot release iron-scavenging siderophores into the extracellular milieu [118]. In situations, where the growth of cheaters may prove detrimental for the survival of the population as a whole, a compensatory mutation was anticipated to take place. The protease producing ability of lasR mutants (lasR5) was restored by the rhl system, which thus showed an enhanced C4HSL production [93, 119].
The impact of anti-virulence interventions on the evolution of QSS was demonstrated in a placebo controlled clinical trial. The response of antibiotic – azithromycin, which acts as QSI [120, 121] was followed by evaluating the behaviour of using lasR mutants in intubated patients colonised by P. aeruginosa [122]. Mutation in lasR resulted in reduced expression of elastase and the mutant had a reduced growth compared to wild-type P. aeruginosa. Azithromycin treatment however, prevented the selection for lasR mutants and consequently increased the proportion of wild-type. It was therefore concluded that such anti-virulence intervention may increase the prevalence of highly virulent QS wild-type isolates [122].
Efflux pumps
Bacteria develop resistance to antibiotics and biocides by various mechanisms, the most instrumental being efflux systems [37]. Active and efficient efflux system enables bacteria to evolve resistance against toxic metals, drugs and even structurally unrelated antibiotics [123–125]. P. aeruginosa has numerous efflux pumps which regulate membrane permeability and the uptake of compounds [126]. The expression of multidrug efflux systems has been found to be a consequence of mutations in regulatory genes. Antibiotics such as azithromycin (AZM), ceftazidime and ciprofloxacin affect QS possibly by altering membrane permeability and affecting the 3OC12HSL efflux, which can contribute to the consequences of selective pressure and development of QS resistance [87, 127]. Certain antimicrobial drugs at sub inhibitory antimicrobial concentrations can induce or interfere with QSS and even promote biofilm formation [128]. Vanadium resistant mutants in ncr (non-coding region), mexI and opmD showed drastic decrease in production of the QS mediated phenazine pigment pyocyanin [129, 130]. In fact, nfxC type mutants over-expressing MexEF-OprN efflux pump had reduced transcription of the AI synthase gene rhlI and also exhibited 20 times less pyocyanin production [125, 131]. The reduced transcription of rhlI was also assigned to efflux of tryptophan, the precursor of PQS and leading to its reduced intracellular levels, which consequently affected QS functions [132, 133]. Hence, AHL homeostasis may be a potential mechanism to circumvent QSI i.e. by reducing AHL production [125, 133]. P. aeruginosa mutants for mexR and nalC show increased resistance to a well studied QSI – brominated furanone C-30. P. aeruginosa mexR mutant in the presence of C-30 could infect and cause disease in Caenorhabditis elegans [50]. These mechanisms are supported by additional genes responsible for inactivation or modification of the drug [4, 37, 134, 135].
Environmental impact
Although bacteria may produce diverse QSS and also the corresponding signals, however, their stability may be affected by certain environmental conditions. Within biofilms, pH varies from 6–11 during the diurnal cycle [136, 137]. During daylight, pH values of >9 results in hydrolysis of AHLs [138]. This physiological hydrolysis due to alkaline pH does not affect AHLs with acyl chain longer than C12. In this scenario, shorter chain AHLs might be produced during night cycle and long chain AHLs would remain largely intact during photosynthetic activity period [77]. The QS mediated exopolysaccharide matrix of the biofilm regulates the mobility of a molecule through the nanopores between polymers [139, 140]. These responses to changes in environmental conditions can help bacteria to manipulate the production and release of QS signals and withstand adverse conditions by engineering the flow of signals and ions [6, 141].
Co-evolution
Marine organism, D. pulchra has been widely studied for its QSI abilities [142, 143]. In natural marine environments, quite a few bacteria have been found to be associated with D. pulchra [144, 145]. Bacterial isolates from the distal portion of the thallus of D. pulchra had the ability to form biofilms. Crude extracts of D. pulchra strongly inhibited the attachment of the bacterial isolates from rock surfaces at significantly much lower concentrations (10 ng cm−2) than those (1 ug cm−2) needed for isolates from the surface of D. pulchra [144]. Nautella italica R11 and Phaeobacter gallaeciensis LSS9 isolated from the surface of red macro alga D. pulchra, could cause bleaching disease on the host where as other isolates could grow well and form biofilms but did not induce the disease. [146–148]. It implies that isolates from the epiphytic bacterial community closely associated with the marine algae have evolved resistance to QSI - produced by the host. As a result, it is quite probable that the algae may in turn evolve mechanisms to further avoid biofouling.
A never ending battle
In the past, our enthusiasm with the large scale utilization of antibiotics was tempered by the fact that bacteria evolve to become resistant to selection pressure. In the present scenario, QSIs are proving out to be the most promising alternatives and/or supplements to antibiotics. Most of the research in this area seems to be well aligned to meet the basic criteria of a QSI but is yet to succeed to clinical stage. Another very interesting scenario, which is quite easy to visualize is that our battle against infectious diseases may be a never ending process [149]. The experts in the area are quite apprehensive about its success, hence it may be judicious to be prudent lest QSI may meet the fate of antibiotics [15]. We may presume QSI resistance mechanism(s) to be similar to those which confer resistance to antibiotics, such as, restricted availability, inactivation or even modification of the target [51]. The big question is: Why bacteria will develop resistance against QSI? We may argue that the functioning of QS is so vital for bacteria that it may “invest” up to 10% of their total genome in it. The organisms are likely to have a natural protection mechanism in place to counter attacks which may endanger their existence. Since QSI are envisaged to involve “only” QS, without affecting bacterial growth, a constant supply of AHL signals, albeit at a slower rate can even be imaged at low cell densities. Therefore, once QSI concentration gets below the threshold level, bacteria may be free to express its virulence. Apparently, bacteria do not even need to undergo any genetic change to withstand QSI’s [77]. Bacteria can thus evade QSI, by keeping its QS under control till the concentration of QSI is higher than that of the signal molecules. It is being suggested that in future, the target should be to develop QSI with lower risk of developing resistance. In contrast to QSI causing competitive inhibition [150], it may be desirable to look for non-competitive or uncompetitive inhibitors. It is expected to exert less pressure on QS gene expression [49]. The associated drawback with the use of non-competitive biomolecules as QSIs is their narrow range of activity and may become ineffective by genetic mutations in the binding sites [49–51]. Caution should also be exercised that QSI should not affect efflux pumps [151, 152]. Taking into account the multiplicity of receptors, it is unlikely that a single molecule will be able to act as a broad range QSI [53]. The existence of natural bacterial communities is closely linked with the environment. The physiochemical conditions thus drive the bacterial activities and regulate their survival. Since, survival is the essence of life, each organism will continue to evolve its defense mechanisms. The likely hood of bacteria developing resistance to QSI is less probable than that observed with conventional antibiotics [153]. Observations on bacterial resistance to QSI oblige us to be cautious while developing drugs against QSS [154]. The composition of the bacterial community will change, if they are not able to withstand environmental stress [77]. The genetic engineering of plants with AHL degrading enzyme may also be a reason to worry as it may create selective pressure for the evolution of bacterial strains, which may evolve mechanisms (i) to inhibit the activity of these enzymes and/or (ii) to become independent of AHL based expression of virulence factors [15, 155]. We may conclude that the need is to employ innovative and novel strategies to extend the range of QSIs against multidrug resistant organisms.
Acknowledgments
The authors wish to thank the Director of CSIR-Institute of Genomics and Integrative Biology (IGIB), CSIR-INDEPTH (BSC0111), Government of India for providing the necessary funds and facilities. PK is thankful to CSIR for granting Senior Research Fellowship. TKW was supported by the NIH (R01 GM089999) and is the Biotechnology Endowed Professor at the Pennsylvania State University.
Footnotes
Conflict of Interest
The authors declare that they have no competing interests.
Author’s Contributions
PK has contributed towards a collection of material and preparation of the article. VCK has contributed towards the conceptualization and writing the article. TKW helped write some passages.
References
- 1.Demain AL, Elander R. The β-lactam antibiotics: past, present, and future. Antonie van Leeuwenhoek. 1999;75:5–19. doi: 10.1023/a:1001738823146. [DOI] [PubMed] [Google Scholar]
- 2.Thykaer J, Nielsen J. Metabolic engineering of β-lactam production. Metab Eng. 2003;5:56–69. doi: 10.1016/S1096-7176(03)00003-X. [DOI] [PubMed] [Google Scholar]
- 3.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Kalia VC, Rani A, Lal S, Cheema S, Raut CP. Combing databases reveals potential antibiotic producers. Expert Opin Drug Discov. 2007;2:211–224. doi: 10.1517/17460441.2.2.211. [DOI] [PubMed] [Google Scholar]
- 5.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 6.Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial bio lms. PLoS Biol. 2008;16:e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong YH, Zhang LH. Quorum sensing and quorum-quenching enzymes. J Microbiol. 2005;43:101–109. [PubMed] [Google Scholar]
- 8.McDougald D, Rice SA, Kjelleberg S. Bacterial quorum sensing and interference by naturally occurring biomimics. Anal Bioanal Chem. 2007;387:445–453. doi: 10.1007/s00216-006-0761-2. [DOI] [PubMed] [Google Scholar]
- 9.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 10.Amara N, Krom BP, Kaufmann GF, Meijler MM. Macromolecular inhibition of quorum sensing: Enzymes, antibodies, and beyond. Chem Rev. 2011;111:195–208. doi: 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]
- 11.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teplitski M, Robinson JB, Bauer WD. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact. 2000;13:637–648. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]
- 15.Whitehead NA, Welch M, Salmond GPC. Silencing the majority. Nat Biotechnol. 2001;19:735–736. doi: 10.1038/90780. [DOI] [PubMed] [Google Scholar]
- 16.Cos P, Vlietinck AJ, Berghe DV, Maes L. Anti-infective potential of natural products: how to develop a stronger in vitro ‘proof-of-concept’. J Ethnopharmacol. 2006;106:290–302. doi: 10.1016/j.jep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann GF, Sartorio R, Lee SH, Mee JM, Altobell LJ, III, Kujawa DP, Jeffries E, Clapham B, Meijler MM, Janda KD. Antibody interference with N-acyl homoserine lactone-mediated bacterial quorum sensing. J Am Chem Soc. 2006;128:2802–2803. doi: 10.1021/ja0578698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 19.Yim G, Wang HH, Davies J. The truth about antibiotics. Int J Med Microbiol. 2006;296:163–170. doi: 10.1016/j.ijmm.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 20.van Baarlen P, van Belkum A, Summerbell RC, Crous PW, Thomma BPHJ. Molecular mechanisms of pathogenicity: how do pathogenic microorganisms develop cross-kingdom host jumps? FEMS Microbiol Rev. 2007;31:239–277. doi: 10.1111/j.1574-6976.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- 21.Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu Rev Microbiol. 2009;63:99–118. doi: 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- 22.Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 23.Dembitsky VM, Al Quntar AAA, Srebnik M. Natural and synthetic small boron-containing molecules as potential inhibitors of bacterial and fungal quorum sensing. Chem Rev. 2011;111:209–237. doi: 10.1021/cr100093b. [DOI] [PubMed] [Google Scholar]
- 24.Nazzaro F, Fratianni F, Coppola R. Quorum sensing and phytochemicals. Int J Mol Sci. 2013;14:12607–12619. doi: 10.3390/ijms140612607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37:121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- 26.Kalia VC. Quorum sensing inhibitors: An overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 27.López D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tinh NTN, Dung NV, Trung CT, Thuy VT. In vitro characterization of a recombinant AHL-Lactonase from Bacillus cereus isolated from a striped catfish (Pangasianodon hypophthalmus) Pond. Indian J Microbiol. 2013;53:485–487. doi: 10.1007/s12088-013-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C-N, Chen C-J, Liao C-T, Lee C-Y. A probable aculeacin A acylase from the Ralstonia solanacearum GMI1000 is N-acyl-homoserine lactone acylase with quorum-quenching activity. BMC Microbiol. 2009;9:89. doi: 10.1186/1471-2180-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Annapoorani A, Jabbar AKKA, Musthafa SKS, Pandian SK, Ravi AV. Inhibition of quorum sensing mediated virulence factors production in urinary pathogen Serratia marcescens PS1 by marine sponges. Indian J Microbiol. 2012;52:160–166. doi: 10.1007/s12088-012-0272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Hentzer M, Givskov M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Invest. 2003;112:1300–1307. doi: 10.1172/JCI200320074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen TB, Givskov M. Quorum-sensing inhibitors: a bargain of effects. Microbiology. 2006;152:895–904. doi: 10.1016/j.fitote.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Vattem DA, Mihalik K, Crixell SH, McLean RJC. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia. 2007;78:302–310. doi: 10.1016/j.fitote.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Bakkiyaraj D, Sivasankar C, Pandian SK. Anti-pathogenic potential of coral associated bacteria isolated from gulf of Mannar against Pseudomonas aeruginosa. Indian J Microbiol. 2013;53:111–113. doi: 10.1007/s12088-012-0342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu W, Liu Y, Jiang Y, Zhu W, Zhuang X. Production of N-acyl homoserine lactones and virulence factors of waterborne Aeromonas hydrophila. Indian J Microbiol. 2013;53:264–268. doi: 10.1007/s12088-013-0381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 38.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 39.Pitman AR, Jackson RW, Mansfield JW, Kaitell V, Thwaites R, Arnold DL. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr Biol. 2005;15:2230–2235. doi: 10.1016/j.cub.2005.10.074. [DOI] [PubMed] [Google Scholar]
- 40.Lal S, Cheema S, Kalia VC. Phylogeny vs genome reshuffling: horizontal gene transfer. Indian J Microbiol. 2008;48:228–242. doi: 10.1007/s12088-008-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc) 2005;70:267–274. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- 42.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 43.Liras P, Rodríguez-Garcia A, Martin JF. Evolution of the clusters of genes for β-lactam antibiotics: a model for evolutive combinatorial assembly of new beta-lactams. Int Microbiol. 1998;1:271–278. [PubMed] [Google Scholar]
- 44.Molina L, Constantinescu F, Michel L, Reimmann C, Duffy B, Défago G. Degradation of pathogen quorum-sensing molecules by soil bacteria: a preventive and curative biological control mechanism. FEMS Microbiol Ecol. 2003;45:71–81. doi: 10.1016/S0168-6496(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 45.Defoirdt T, Boon N, Bossier P, Verstraete W. Disruption of bacterial quorum sensing: An unexplored strategy to fight infections in aquaculture. Aquaculture. 2004;240:69–88. doi: 10.1016/j.aquaculture.2004.06.031. [DOI] [Google Scholar]
- 46.Otto M. Quorum-sensing control in Staphylococci -- a target for antimicrobial drug therapy? FEMS Microbiol Lett. 2004;241:135–141. doi: 10.1016/j.femsle.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 47.Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi lessons learned from in vivo work. ISME J. 2008;2:19–26. doi: 10.1038/ismej.2007.92. [DOI] [PubMed] [Google Scholar]
- 48.Yeon K-M, Cheong W-S, Oh H-S, Lee W-N, Hwang B-K, Lee C-H, Beyenal H, Lewandowski Z. Quorum sensing: a new biofouling control paradigm in a membrane bioreactor for advanced waste water treatment. Environ Sci Technol. 2009;43:380–385. doi: 10.1021/es8019275. [DOI] [PubMed] [Google Scholar]
- 49.Defoirdt T, Boon N, Bossier P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010;6:e1000989. doi: 10.1371/journal.ppat.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeda T, García-Contreras R, Pu M, Sheng L, Garcia LR, Tomás M, Wood TK. Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 2012;6:493–501. doi: 10.1038/ismej.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mellbye B, Schuster M. The sociomicrobiology of antivirulence drug resistance: a proof of concept. MBio. 2011;2:e00131–11. doi: 10.1128/mBio.00131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García-Contreras R, Martinez-Vazquez M, Velazquez Guadarrama N, Villegas Paneda AG, Hashimoto T, Maeda T, Quezada H, Wood TK. Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Pathog Dis. 2013;68:8–11. doi: 10.1111/2049-632X.12039. [DOI] [PubMed] [Google Scholar]
- 53.Mattmann ME, Blackwell HE. Small molecules that modulate quorum sensing and control virulence in Pseudomonas aeruginosa. J Org Chem. 2010;75:6737–6746. doi: 10.1021/j0101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 56.Patankar AV, González JE. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev. 2009;33:739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- 57.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 61.Marketon MM, González JE. Identification of two quorum sensing systems in Sinorhizobium meliloti. J Bacteriol. 2002;184:3466–3475. doi: 10.1128/JB.184.13.3466-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marketon MM, Gronquist MR, Eberhard A, González JE. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-Acyl homoserine lactones. J Bacteriol. 2002;184:5686–5695. doi: 10.1128/JB.184.20.5686-5695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 64.Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 66.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, Bally M, Chapon V, Salmond GP, Bycroft BW. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Soc USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams P, Stewart GSAB, Cámara M, Winson MK, Chhabra SR, Salmond GPC, Bycroft BW. Signal transduction through quorum sensing in Pseudomonas aeruginosa. In: Silver S, Haas D, Nakazawa T, editors. Pseudomonas, Molecular Biology and Biotechnology. American Society for Microbiology; Washington DC, USA: 1996. pp. 195–206. [Google Scholar]
- 69.Rosemeyer V, Michiels J, Verreth C, Vanderleyden J. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J Bacteriol. 1998;180:815–821. doi: 10.1128/jb.180.4.815-821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brelles-Mariño, Graciela B, Enlogio J. Detection, purification and characterization of quorum-sensing signal molecules in plant-associated bacteria. J Biotechnol. 2001;91:197–209. doi: 10.1016/S0168-1656(01)00330-3. [DOI] [PubMed] [Google Scholar]
- 71.Holden MT, Chhabra SR, de Nys R, Stead P, Bainton NJ, Hill PJ, Manefield M, Kumar N, Labatte M, England D, Rice S, Givskov M, Salmond GP, Stewart GS, Bycroft BW, Kjelleberg S, Williams P. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol Microbiol. 1999;33:1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- 72.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 73.Antunes L, Caetano M, Ferreira RBR, Buckner MMC, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 74.Grenha R, Slamti L, Nicaise M, Refes Y, Lereclus D, Nessler S. Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR. Proc Natl Acad Sci USA. 2013;110:1047–1052. doi: 10.1073/pnas.1213770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delden C, Pesci EC, Pearson JP, Iglewski BH. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum sensing mutant. Infect Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ledgham F, Ventre I, Soscia C, Foglino M, Strugis JN, Lazdunski A. Interaction of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- 77.Decho AW, Norman RS, Visscher PT. Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 2010;18:73–80. doi: 10.1016/j.tim.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 78.Geske GD, O’Neill JC, Blackwell HE. Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria. Chem Soc Rev. 2008;37:1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brader G, Sjöblom S, Hyytiöinen H, Sims-Huopaniemi K, Palva ET. Altering substrate chain length specificity of an acylhomoserine lactone synthase in bacterial communication. J Biol Chem. 2005;280:10403–10409. doi: 10.1074/jbc.M408603200. [DOI] [PubMed] [Google Scholar]
- 80.Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- 81.Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol. 2005;187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mikkelsen H, Bond NJ, Skindersoe ME, Givskov M, Lilley KS, Welch M. Biofilms and type III secretion are not mutually exclusive in Pseudomonas aeruginosa. Microbiology. 2009;155:687–698. doi: 10.1099/mic.0.025551-0. [DOI] [PubMed] [Google Scholar]
- 83.Njoroge J, Sperandio V. Jamming bacterial communication: New approaches for the treatment of infectious diseases. EMBO Mol Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoiby N, Krogh Johansen H, Moser C, Song Z, Ciofu O, Kharazmi A. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 2001;3:23–35. doi: 10.1016/S1286-4579(00)01349-6. [DOI] [PubMed] [Google Scholar]
- 85.Zhu H, Thuruthyil SJ, Willcox MDP. Determination of quorum-sensing signal molecules and virulence factors of Pseudomonas aeruginosa isolates from contact lens-induced microbial keratitis. J Med Microbiol. 2002;51:1063–1070. doi: 10.1099/0022-1317-51-12-1063. [DOI] [PubMed] [Google Scholar]
- 86.Willcox MD, Zhu H, Conibear TC, Hume EB, Givskov M, Kjelleberg S, Rice SA. Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis. Microbiology. 2008;154:2184–2194. doi: 10.1099/mic.0.2008/019281-0. [DOI] [PubMed] [Google Scholar]
- 87.Skindersoe ME, Zeuthen LH, Brix S, Fink LN, Lazenby J, Whittall C, Williams P, Diggle SP, Froekiaer H, Cooley M, Givskov M. Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol. 2009;55:335–345. doi: 10.1111/j.1574-695X.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 88.Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol. 2008;190:4408–4415. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walters M, Sperandio V. Quorum sensing in Escherichia coli and Salmonella. Int J Med Microbiol. 2006;296:125–131. doi: 10.1073/pnas.96.4.1639. [DOI] [PubMed] [Google Scholar]
- 92.Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg PD, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hogan D, Kolter R. Why are bacteria refractory to antimicrobials? Curr Opin Microbiol. 2002;5:472–477. doi: 10.1016/S1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- 95.Normark BH, Normark S. Evolution and spread of antibiotic resistance. J Intern Med. 2002;252:91–106. doi: 10.1046/j.1365-2796.2002.01026.x. [DOI] [PubMed] [Google Scholar]
- 96.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Jr, Rinehart KL, Farrand SK. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Brussel AAN, Zaat SAJ, Wijffelman CA, Pees E, Lugtenberg BJJ. Bacteriocin small of fast-growing rhizobia is chloroform soluble and is not required for effective nodulation. J Bacteriol. 1985;162:1079–1082. doi: 10.1128/jb.162.3.1079-1082.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, Bally M, Chapon V, Salmond GPC, Bycroft BW, Lazdunski A, Stewart GSAB, Williams P. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang HB, DiMango E, Bryan R, Gambello M, Iglewski BH, Goldberg JB, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koch B, Liljefors T, Persson T, Nielsen J, Kjelleberg S, Givskov M. The LuxR receptor: the sites of interaction with quorum sensing signals and inhibitors. Microbiol. 2005;151:3589–3602. doi: 10.1099/mic.0.27954-0. [DOI] [PubMed] [Google Scholar]
- 102.Joelsson A, Liu Z, Zhu J. Genetic and phenotypic diversity of quorum–sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect Immun. 2006;74:1141–1147. doi: 10.1128/IAI.74.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wei JR, Tsai Y-H, Horng Y-T, Soo P-C, Hsieh S-C, Hsueh P-R, Horng J-T, Williams P, Lai H-C. A mobile quorum-sensing system in Serratia marcescens. J Bacteriol. 2006;188:1518–1525. doi: 10.1128/JB.188.4.1518-1525.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Purevdorj B, Costerton JW, Stoodley P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2002;68:4457–4464. doi: 10.1128/AEM.68.9.4457-4464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Y, Zhang Y, Yang Y, Wang L, Weng L. Identification of a Pseudomonas sp. that inhibits RHL system of quorum sensing. Indian J Microbiol. 2013;53:28–35. doi: 10.1007/s12088-012-0340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rampioni G, Schuster M, Greenberg EP, Zennaro E, Leoni L. Contribution of the RsaL global regulation to Pseudomonas aeruginosa virulence and biofilm formation. FEMS Microbiol Lett. 2009;301:210–217. doi: 10.1111/j.1574-6968.2009.01817.x. [DOI] [PubMed] [Google Scholar]
- 108.Liu H-B, Koh KP, Lee JH, Kim JS, Park S. Characterization of LasR protein involved in bacterial quorum sensing mechanism of Pseudomonas aeruginosa. Biotechnol Bioproc Eng. 2009;14:146–154. doi: 10.1007/s12257-008-0188-z. [DOI] [Google Scholar]
- 109.Ward JP, King JR, Koerber AJ, Croft JM, Sockett RE, Williams P. Cell-signalling repression in bacterial quorum sensing. Math Med Biol. 2004;21:169–204. doi: 10.1093/imammb/21.3.169. [DOI] [PubMed] [Google Scholar]
- 110.Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2007;66:1557–1565. doi: 10.1111/j.1365-2958.2007.06029x. [DOI] [PubMed] [Google Scholar]
- 111.Köhler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci USA. 2009;106:6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 113.Brown SP, Hochberg ME, Grenfell BT. Does multiple infection select for raised virulence? Trends Microbiol. 2002;10:401–405. doi: 10.1016/S0966-842X(02)02413-7. [DOI] [PubMed] [Google Scholar]
- 114.Buckling A, Brockhurst MA. Kin selection and the evolution of virulence. Heredity. 2008;100:484–488. doi: 10.1038/sj.hdy.6801093. [DOI] [PubMed] [Google Scholar]
- 115.Rumbaugh KP, Diggle SP, Watters CM, Gillespie AR, Griffin AS, West SA. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 116.Schaber JA, Carty NL, McDonald NA, Graham ED, Cheluvappa R, Griswold JA, Hamood ANJ. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. Med Microbiol. 2004;53:841–853. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 117.Wilder CN, Diggle SP, Schuster M. Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 2011;5:1332–1343. doi: 10.1038/ismej.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 119.Van Delden C, Pesci EC, Pearson JP, Iglewski BH. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McDermott PF, Walker RD, White DG. Antimicrobials: modes of action and mechanisms of resistance. Int J Toxicol. 2003;22:135–143. doi: 10.1080/10915810305089. [DOI] [PubMed] [Google Scholar]
- 121.Tateda K, Standiford TJ, Pechere J-C, Yamaguchi K. Regulatory effects of macrolides on bacterial virulence: potential role as quorum sensing inhibitors. Curr Pharm Des. 2004;10:3055–3065. doi: 10.2174/1381612043383377. [DOI] [PubMed] [Google Scholar]
- 122.Köhler T, Perron GG, Buckling A, van Delden C. Quorum sensing inhibitors selects for virulence and cooperation in Pseudomonas aeruginosa. Plos Pathog. 2010;6:e1000883. doi: 10.1371/journal.ppat.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rosen BP. Bacterial resistance to heavy metals and metalloids. JBIC. 1996;1:273–277. [Google Scholar]
- 124.Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 125.Köhler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2004;48:1320–1328. doi: 10.1128/AAC.48.4.1320-1328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tramper-Stranders GA, Wolfs TF, Fleer A, Kimpen JL, van der Ent CK. Maintenance azithromycin treatment in pediatric patients with cystic fibrosis: long-term outcomes related to macrolide resistance and pulmonary function. Pediatr Infect Dis J. 2007;26:8–12. doi: 10.1097/01.inf.0000247109.44249.ac. [DOI] [PubMed] [Google Scholar]
- 128.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 129.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pattery T, Mondt K, Audenaert K, Höfte M, Cornelis P. Identification of phzM, a new phenazine biosynthesis gene necessary for the production of pyocyanin by Pseudomonas aeruginosa. Abstract presented at the Pseudomonas 2001 meeting; Brussels, Belgium. 17–21 September.2001. [Google Scholar]
- 131.Köhler T, Michea-Hamzehpour M, Plesiat P, Kahr AL, Pechere JC. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Holden I, Swift I, Williams I. New signal molecules on the quorum-sensing block. Trends Microbiol. 2000;8:101–104. doi: 10.1016/S0966-842X(00)01718-2. [DOI] [PubMed] [Google Scholar]
- 133.Aendekerk S, Ghysels B, Cornelis P, Baysse C. Characterization of a new ef ux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiol. 2002;148:2371–2381. doi: 10.1099/00221287-148-8-2371. [DOI] [PubMed] [Google Scholar]
- 134.Tateda K, Comte R, Pechere J-C, Köhler T, Yamaguchi K, van Delden C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wright GD. Mechanisms of resistance to antibiotics. Curr Opin Chem Biol. 2003;7:563–569. doi: 10.1016/j.cbpa.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 136.Revsbech NP, Jorgensen BB, Blackburn TH, Cohen Y. Microelectrode studies of the photosynthesis and O2, H2S and pH profiles of a microbial mat. Limnol Oceanogr. 1983;28:1062–1074. [Google Scholar]
- 137.Visscher PT, Rudolf AP, Gemerden HV. Rates of sulfate reduction and thiosulfate consumption in a marine microbial mat. FEMS Microbiol Ecol. 1992;86:283–329. doi: 10.1111/j.1574-6941.1992.tb01763.x. [DOI] [Google Scholar]
- 138.Visscher PT, Reid RP, Bebout BM. Microscale observations of sulfate reduction: Correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology. 2000;28:919–922. doi: 10.1130/0091-7613(2000)28<919:MOOSRC>2.0.CO;2. [DOI] [Google Scholar]
- 139.Decho AW. Exopolymer microdomains as a structuring agent for heterogeneity within microbial biofilms. Microbial Sediments. 2000:9–15. doi: 10.1007/978-3-662-04036-2_2. [DOI] [Google Scholar]
- 140.Lawrence JR, Swerhone GD, Kuhlicke U, Neu TR. In situ evidence for microdomains in the polymer matrix of bacterial microcolonies. Can J Microbiol. 2007;53:450–458. doi: 10.1139/W06-146. [DOI] [PubMed] [Google Scholar]
- 141.Braissant O, Decho AW, Duprazi C, Glunk C, Przekop KM, Visscher PT. Exopolymeric substances of sulfate-reducing bacteria: Interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology. 2007;5:401–411. doi: 10.1111/j.1472-4669.2007.00117.x. [DOI] [Google Scholar]
- 142.Ren D, Sims J, Wood TK. Inhibition of bio lm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- 143.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signalling system of Escherichia coli. Biotechnol Bioeng. 2004;88:630–642. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- 144.Maximilien R, de Nys R, Holmström C, Gram L, Givskov M, Crass K, Kjelleberg S, Steinberg PD. Chemical mediation of bacterial surface colonisation by secondary metabolites from the red alga Delisea pulchra. Aquat Microb Ecol. 1998;15:233–246. [Google Scholar]
- 145.Campbell AH, Harder T, Nielsen S, Kjelleberg S, Steinberg PD. Climate change and disease: bleaching of a chemically defended seaweed. Glob Change Biol. 2011;17:2958–2970. doi: 10.1111/j.1365-2486.2011.02456.x. [DOI] [Google Scholar]
- 146.Fernandes N, Case RJ, Longford SR, Seyedsayamdost MR, Steinberg PD, Kjelleberg S, Thomas T. Genomes and virulence factors of novel bacterial pathogens causing bleaching disease in the marine red alga Delisea pulchra. PLoS ONE. 2011;6:e27387. doi: 10.1371/journal.pone.0027387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Thole S, Kalhoefer D, Voget S, Berger M, Engelhardt T, Liesegang H, Wollherr A, Kjelleberg S, Daniel R, Simon M, Thomas T, Brinkhoff T. Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 2012;6:2229–2244. doi: 10.1038/ismej.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T. The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol Rev. 2012;37:462–476. doi: 10.1111/1574-6976.12011. [DOI] [PubMed] [Google Scholar]
- 149.Bjarnsholt T, Givskov M. Quorum sensing inhibitory drugs as next generation antimicrobials: worth the effort? Curr Infect Dis Rep. 2008;10:22–28. doi: 10.1007/s11908-008-0006-y. [DOI] [PubMed] [Google Scholar]
- 150.Manefield M, De Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiol. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 151.Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. Revisiting quorum sensing: Discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci USA. 2005;102:309–314. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rossbach S, Kukuk ML, Wilson TL, Feng SF, Pearson MM, Fisher MA. Cadmium-regulated gene fusions in Pseudomonas fluorescens. Environ Microbiol. 2000;2:373–382. doi: 10.1046/j.1462-2920.2000.00117.x. [DOI] [PubMed] [Google Scholar]
- 153.Defoirdt T, Sorgeloos P, Bossier P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol. 2011;14:251–258. doi: 10.1016/j.mib.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 154.Njoroge J, Sperandio V. Jamming bacterial communication: New approaches for the treatment of infectious diseases. EMBO Mol Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Galloway WRJD, Hodgkinson JT, Bowden S, Welch M, Spring DR. Applications of small molecule activators and inhibitors of quorum sensing in gram-negative bacteria. Trends Microbiol. 2012;20:449–458. doi: 10.1016/j.tim.2012.06.003. [DOI] [PubMed] [Google Scholar]