Abstract
N-acetylglucosamine kinase (GlcNAc kinase or NAGK; EC 2.7.1.59) is highly expressed and plays a critical role in the development of dendrites in brain neurons. In this study, the authors conducted structure-function analysis to verify the previously proposed 3D model structure of GlcNAc/ATP-bound NAGK. Three point NAGK mutants with different substrate binding capacities and reaction velocities were produced. Wild-type (WT) NAGK showed strong substrate preference for GlcNAc. Conversion of Cys143, which does not make direct hydrogen bonds with GlcNAc, to Ser (i.e., C143S) had the least affect on the enzymatic activity of NAGK. Conversion of Asn36, which plays a role in domain closure by making a hydrogen bond with GlcNAc, to Ala (i.e., N36A) mildly reduced NAGK enzyme activity. Conversion of Asp107, which makes hydrogen bonds with GlcNAc and would act as a proton acceptor during nucleophilic attack on the γ-phosphate of ATP, to Ala (i.e., D107A), caused a total loss in enzyme activity. The over-expression of EGFP-tagged WT or any of the mutant NAGKs in rat hippocampal neurons (DIV 5-9) increased dendritic architectural complexity. Finally, the overexpression of the small, but not of the large, domain of NAGK resulted in dendrite degeneration. Our data show the effect of structure on the functional aspects of NAGK, and in particular, that the small domain of NAGK, and not its NAGK kinase activity, plays a critical role in the upregulation of dendritogenesis.
Keywords: dendritogenesis, GlcNAc kinase, kinetics, mutation, NAGK, neuron, transfection
INTRODUCTION
N-acetylglucosamine (GlcNAc)-6-phosphate, the main intermediate for UDP-GlcNAc production, can be synthesized through a de novo pathway by using glycolytic intermediates or by a salvage pathway using the enzyme, GlcNAc kinase (NAGK; EC 2.7.1.59) (Berger et al., 2002). The energized GlcNAc moiety of UDP-GlcNAc is used in the synthesis of O-/N-glycans, sialic acids, and O-GlcNAc. NAGK mRNA and enzyme activity is present in almost all tissues tested (Hinderlich et al., 2000). Rat liver NAGK is a homodimer of 39 kDa subunits (Hinderlich et al., 1998), whereas the cDNA sequence of human NAGK encodes a protein with a predicted molecular mass of 37.4 kDa (Hinderlich et al., 2000). The amino acid sequence indicates that NAGK is a member of the sugar-kinase/Hsp70/actin super-family protein, sharing a common ATPase domain (Berger et al., 2002). Within the cell, human NAGK is highly specific towards its natural substrate β-GlcNAc (Blume et al., 2008).
Weihofen et al. (2006) elucidated two crystal structures of homodimeric human NAGK, one complexed with GlcNAc and the other with ADP and glucose. The secondary structure of human NAGK is comprised of eleven β-strands and ten α-helices. The N-terminal small and C-terminal large domains of NAGK form a “V”-shaped structure, which acts as an active center for binding GlcNAc and ATP. With ADP and glucose, NAGK makes an ‘open’ configuration, whereas with GlcNAc NAGK forms a ‘closed’ conformation via a 26° rotation of its small domain relative to its large domain. According to amino acid sequence, the GlcNAc binding site was determined to lie on two loops of the large C-domain (aa.127–130 and 145–152) and to involve residues 36, 76–79, and 107 on the small N-terminal domain. Unfortunately, Weihofen et al. (2006). were unable to produce protein crystals of NAGK complexed with its authentic substrates GlcNAc and ATP/Mg2+. Instead, they established a model NAGK in complex with GlcNAc and ATP/Mg2+ by superimposing ADP in the ADP/glucose-bound open complex with NAGK in the GlcNAc-bound closed complex. Accordingly, the derived model needs to be verified by structure-function analysis.
Recently, we found that NAGK is highly expressed in neurons of the brain. Overexpression of small hairpin RNA (shRNA) and rescue experiments revealed a non-canonical function of NAGK, namely, that it plays a critical role in dendrite development (Lee et al., 2014). During this previous study, we noticed NAGK was not distributed homogeneously, but highly concentrated in dendrite colocalizing with microtubules. These findings prompted us to hypothesize that the non-canonical function of NAGK in dendritogenesis is subserved by some structural role of NAGK and not by its enzyme activity. To explore this hypothesis, and to verify the proposed 3D model structure of GlcNAc/ATP-bound NAGK, we carried out structure-function analysis using three NAGK mutants that retained differential kinase activities, and investigated their abilities to promote dendritogenesis in rat hippocampal cultures.
MATERIALS AND METHODS
Antibodies
The following antibodies were used at the indicated dilutions: MAb green fluorescent protein (GFP; 1:1000, Chemicon International Inc., now Millipore, USA); rabbit polyclonal red fluorescent protein (RFP or DsRed2; 1:1000, Chemicon); chicken polyclonal NAGK (NAGK; 1:1000, GenWay Biotech, Inc.; USA); Alexa Flour 488-conjugated goat anti-mouse IgG, Alexa Flour 568-conjugated goat anti-rabbit IgG, and Alexa Flour 647-conjugated goat anti-chicken IgG (1:1000, Molecular Probes, Inc.; USA).
Plasmid construction
Point mutants
The murine histidine (His)-tagged NAGK clone in the pRSET C expression vector (Life Technologies Corp., USA) was kindly donated by Dr. Stephan Hinderlich (Beuth Hochschule für Technik Berlin, Department of Life Sciences and Technology, Germany). Site-directed polymerase chain reaction (PCR) mutagenesis was carried out using a previously described overlap extension PCR method (Ho et al., 1989) using the primer sets given in Table 1. Briefly, 5′ upstream and 3′ downstream fragments containing point mutations were generated, and stitching of the two products obtained was performed using another round of PCR (AccuPower→ PCR Premix; Bioneer Co.; Korea). Full length fragments were purified using a Rapid Gel Extraction Kit (A560) (Takara; Japan) and inserted into a pRSET C vector using XhoI and HindIII. In-frame insertion and gene integrity were verified by DNA sequencing.
Table 1.
PCR primer sets
| Mutant | Primer name | Sequence |
|---|---|---|
| N36A | For N-fragment | |
| NAGK-S | 5′-AGA TCT CGA GCG GCC GCC AGT GTG ATG GAT ATC-3′ | |
| N36A-AS | 5′-CAA TCA GCC AGT GGG CTG TGC TCA GTC CAT C-3′ | |
| For C-fragment | ||
| N36A-S | 5′-GAT GGA CTG AGC ACA GCC CAC TGG CTG ATT G-3′ | |
| NAGK-AS | 5′-AAT TCG AAG CTT GGT ACC GAG CTC GGA TCC AC-3′ | |
| D107A | For N-fragment | |
| NAGK-S | 5′-AGA TCT CGA GCG GCC GCC AGT GTG ATG GAT ATC-3′ | |
| D107A-AS | 5′-TGG AAC CTG CTG CAG CCG TGG TGA TTA AGT AGT-3′ | |
| For C-fragment | ||
| D107A-S | 5′-ACT ACT TAA TCA CCA CGG CTG CAG CAG GTT CCA-3′ | |
| NAGK-AS | 5′-AAT TCG AAG CTT GGT ACC GAG CTC GGA TCC AC-3′ | |
| C143S | For N-fragment | |
| NAGK-S | 5′-AGA TCT CGA GCG GCC GCC AGT GTG ATG GAT ATC-3′ | |
| C143S-AS | 5′-CCC CAG CCT CCA CTG CCA CTC TCG GAG CCA TC-3′ | |
| For C-fragment | ||
| C143S-S | 5′-GAT GGC TCC GAG AGT GGC AGT GGA GGC TGG GG-3′ | |
| NAGK-AS | 5′-AAT TCG AAG CTT GGT ACC GAG CTC GGA TCC AC-3′ |
Deletion mutants
The DNA fragments corresponding to the small (NAGK-DS; aa. 1∼117) or large (NAGK-DL; aa.118∼343) domains were amplified by PCR using sense (5′-CCAAGCTTCGGTGTGGCTGTGGCGAT-3′)/antisense (5′-CCCTCGAGCGGCCGCCAGT-3′) for the small domain, or sense (5′-GGCTCGAGCGGACGGTGGGATTGTGCTCATCTCTGGAACAGG-3′/antisense (5′-CCAAGCTTGGTACCGAGCTCGGATCCACTAGTAACGGCCGCC-3′) primer sets for the large domain. The PCR products obtained were double-digested with XhoI and HindIII (underlined in the primer sequences), and inserted in-frame into pDsRed2-C1.
Expression and the purification of NAGK
The murine His-tag NAGK clones in pRSET vectors were introduced into E. coli BL21 (DE3)-pLysS cells (Invitrogen) and chloramphenicol/ampicillin-resistant transformants were selected. Cells were grown to an OD600 of 0.5, induced with 1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3 h, and stored at −70°C. For protein purification, cells were thawed, sonicated, and enzymes were purified using a Ni-nitrilotriacetic acid column (MagneHis™ Ni-Particles) (Promega; USA) by following the manufacturer’s instructions.
NAGK activity assay
The assay was performed as described elsewhere (Asensio and Ruiz-Amil, 1966; Uehara and Park, 2004). Briefly, the final volume of reaction mixtures was 200 μl, and they contained 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM phosphoenol pyruvate, 4.0 mM ATP, 0.2 mM NADH, 1.9 mM GlcNAc, 4.0 U of lactate dehydrogenase, 4.0 U of pyruvate kinase, and 0.38 μg of purified NAGK or its mutant proteins. Assay mixtures were incubated at 37°C, and the reaction was started by adding the enzyme. The utilization of NADH and formation of NAD+ was monitored at 340 nm. To determine the Km of GlcNAc using a Lineweaver-Burk plot, different GlcNAc concentrations were used in the presence of 4.0 mM ATP. One unit of NAGK was defined as the amount of NAGK required to phosphorylate 1.0 μmol of GlcNAc per min at 37°C. On the other hand, to determine the Km of ATP, the 2nd substrate of NAGK, different ATP concentrations were used at a GlcNAc concentration of 1.9 mM. To determine the substrate selectivity of the mutant NAGK enzymes, different sugars, that is, GlcNAc, ManNAc, N-acetylgalactosamine (GalNAc), or glucose were used at a concentration of 1.9 mM and ATP at 4.0 mM.
Primary culture and the transfection of rat hippocampal neurons
Hippocampi from Sprague-Dawley rat pups at embryonic day 18 (E18) or E19 were dissected, dissociated by trypsin treatment and mechanical trituration, and plated onto 12-mm diameter polylysine/laminin-coated glass coverslips at a density of ∼150 neurons/mm2, as previously described (Brewer et al., 1993). Cells were plated initially in Neurobasal medium supplemented with B27 (Invitrogen), 25 μM glutamate, and 500 μM glutamine, and fed 5 days after plating and weekly thereafter using the same medium (without added glutamate) containing 1/3 (v/v) astrocyte-conditioned Neurobasal media (Goslin et al., 1998; Moon et al., 2007). Transfection was carried out as described by Jiang et al. (2004), using the ClonTech CalPhos™ Mammalian Transfection Kit (BD Bioscience; USA).
Fixation and immunocytochemistry (ICC)
Cells were fixed using a sequential paraformaldehyde/methanol fixation procedure [incubation in 4% paraformaldehyde in phosphate buffered saline (20 mM sodium phosphate buffer, pH 7.4, 0.9% NaCl)] at room temperature for 10 min, followed by incubation in methanol at −20°C for 20 min (Moon et al., 2007). ICC was performed as previously described (Moon et al., 2007).
Image acquisition, densitometry, and Sholl analysis
A Leica Research Microscope DM IRE2 equipped with I3 S, N2.1 S, and Y5 filter systems (Leica Microsystems AG, Germany) was used for epifluorescence microscopy. Images (1388 × 1039 pixels) were acquired using a high-resolution CoolS-NAP™ CCD camera (Photometrics; USA) under the control of Leica FW4000 software. Digital images were processed using Adobe Systems Photoshop 7.0. Morphometric analyses and quantification were performed using Image J (version 1.45) software with the simple neurite tracer plug-in (National Institute of Health, USA) and the Sholl plug-in (http://biology.ucsd.edu/labs/ghosh/software). Degrees of dendritic tree arborization were determined as previously described (Sholl, 1953). The point where a dendrite intersected a circle centered at the centroid of a soma was defined as the dendritic intersection. Transfected neurons of representative morphology (6–10 cells) were selected for analysis. Densitometric analyses were performed using Image J (version 1.45) software (National Institute of Health, Bethesda, MD, USA). To analyze correlation between the expression levels of NAGK small or large domain and the number of dendrites, ICC color images were converted into black/white and inverted. The average ICC signal strength of soma areas (circular area of ∼5 μm in diameter) was measured (x-axis) and plotted against the number of dendrites crossing a circle at 45 μm from the soma centroids (∼30 μm from the surface of a soma (y-axis).
Statistics
Data were analyzed using one-way analysis of variance (ANOVA) with Duncan’s multiple comparison post hoc test. Statistical significance was accepted for p values < 0.05, and the analysis was conducted using SPSS version 16.0.
RESULTS
Weihofen et al. (2006) proposed a 3D model of NAGK in complex with GlcNAc and ATP/Mg2+. However, this model requires verification by structure-function analysis. Based on the proposed 3D model structure of GlcNAc/ATP-bound NAGK, we created three NAGK mutants (N36A, D107A, and C143S), to determine their effects on NAGK function.
Rationale for each point mutant
N36A
Asn36 (N36) is positioned in the loop segment, after β3 (aa. 26–32), of the small domain (Fig. 1A-a). GlcNAc binds to NAGK in its ‘closed’ configuration, and Asn36NδH (located on the small domain) hydrogen bonds to the N2-acetyl carbonyl oxygen of GlcNAc (Fig. 1A-b). Furthermore, this hydrogen bonding is thought to mediate domain closure (Weihofen et al., 2006). Therefore, conversion of Asn36 of NAGK to the neutral amino acid alanine (A) was expected to reduce enzyme activity.
Fig. 1.
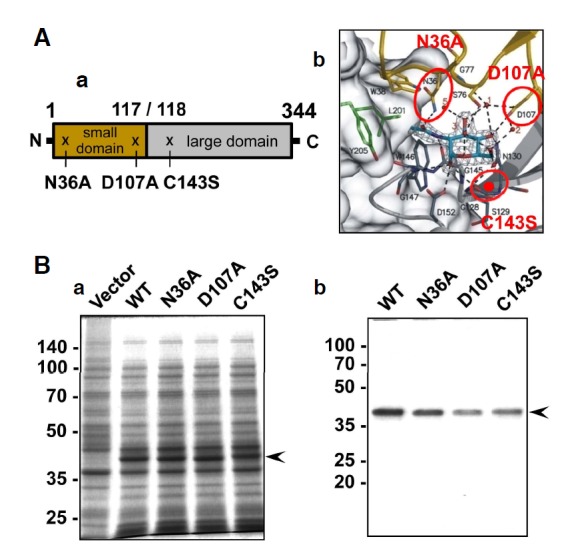
Bacterial expression and purification of NAGK. (A) Positions of the mutated amino acids. (a) Schematic representation of NAGK and the positions of the point mutations. N36A and D107A are located in the small domain (aa. 1-117), while C143 is located in the large domain (aa. 118-344). (b) The GlcNAc binding pocket of NAGK. The mutated amino acids are indicated by circles. The position of C143S is indicated by a red dot in the β8 sheet. Carbon, nitrogen, and oxygen atoms are shown in light blue, blue, and red, respectively. Modified with permission from Weihofen et al. (2006). (B) Induction and purification of NAGK. (a) SDS-PAGE result, showing induction of NAGK expression in E. coli. The expression of NAGK was induced by IPTG and same amounts of bacterial lysates were electrophoresed in 12% SDS-gels and stained with Coomassie R-250. Molecular sizes are marked on the far left in kDa. (b) SDS-PAGE of purified NAGK. Proteins were affinity-purified using Ni-nitrilotriacetic acid columns and stained with Coomassie R-250. The positions of NAGK are marked by arrowheads.
D107A
Asp107 (D107) is located in β3 (aa.107–115) of the small domain of NAGK (Fig. 1A-a). This residue plays dual important roles. First, the carboxylate group of Asp107 to GlcNAcO4 and O6 hydroxyls (Fig. 1A-b). Furthermore, these hydrogen bonds are important for producing a ‘closed’ NAGK configuration after GlcNAc binding. In addition, a model 3D structure for NAGK complexed with GlcNAc and ATP/Mg2+ indicates that D107 is directly involved in the phosphorylation of GlcNAc by acting as a proton acceptor during nucleophilic attack on the γ-phosphate of ATP (Weihofen et al., 2006). Therefore, conversion of D107 to Ala was expected to destroy the enzyme activity.
C143S
Cys143 (C143) is positioned in β8 (aa. 140–144) of the large domain of NAGK (Fig. 1A-a). This amino acid does not form any hydrogen bonds with GlcNAc or water molecules (Fig. 1A-b). However, C143 is positioned near the GlcNAc binding pocket and its mutation into serine (C143S) was thus expected to affect NAGK activity to some extent (Berger et al., 2002).
Expression of His-tag NAGK in E. coli and affinity purification
Expression of WT and mutant murine His-tag NAGK protein was induced in E. coli BL21 (DE3)-pLysS using isopropyl β-D-1-thiogalactopyranoside (IPTG) (Fig. 1B-a). The functional expression of WT NAGK and the absence of background activity in E. coli have been verified (Hinderlich et al., 2000). The expressed His-tag protein was purified to near homogeneity using the Ni-nitrilotriacetic acid column (Fig. 1B-b). The apparent molecular size of purified NAGK was 40 kDa, which included the His-tag (3 kDa).
Enzyme kinetics
Substrate utilization rate
To evaluate relative enzyme activity, we first studied substrate utilization kinetics using GlcNAc and ATP as substrates (Fig. 2A). The substrate utilization rates (reaction rate) of the mutants varied widely. For both substrates (GlcNAc and ATP), WT NAGK had the highest reaction rates, followed by the C143S and N36A mutants, indicating that the N36A mutation more severely damaged NAGK kinase activity.
Fig. 2.
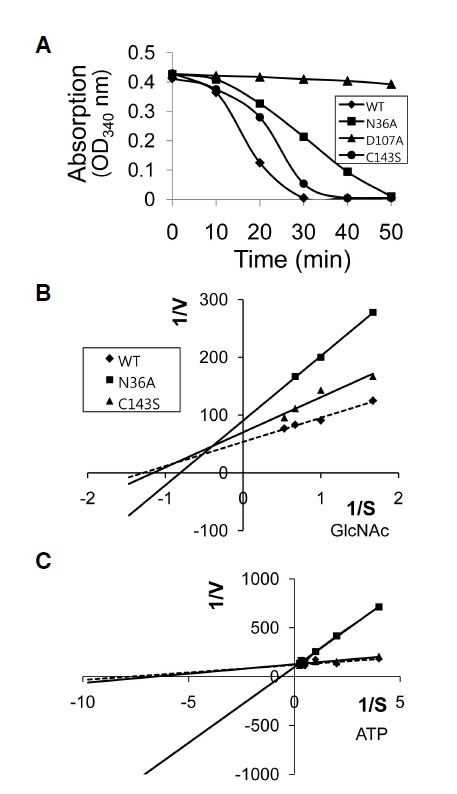
Enzyme kinetics for authentic substrates. (A) Substrate utilization kinetics. The utilization of GlcNAc (assessed by the production of GlcNAc-6-phosphate) was determined indirectly by monitoring absorption decreases at 340 nm (see text for details). The decrease in absorption at 340 nm indicates that NADH is converted to NAD+ (a surrogate of GlcNAc-6-phosphate production, or to the consumption of GlcNAc by NAGK. Note the D107A mutant had no enzyme activity. Results are the averages of three experiments. (B,C) Lineweaver-Burk plots for GlcNAc (B) and ATP (C). Reactions were carried out using GlcNAc concentrations of 0.6, 1.0, 1.5, or 1.9 mM, and ATP concentrations of 0.25, 0.5, 1.0, 2, 3, or 4.0 mM. D107A was not included because it had no enzyme activity.
Substrate affinity (Km), maximum velocity (Vmax)
Double reciprocal plots or Lineweaver-Burk plots for GlcNAc and ATP are shown in Figs. 2B and 2C, respectively.
WT and the C143S mutant
Double reciprocal plots for GlcNAc (Fig. 2B) showed that WT NAGK had the shallowest slope, indicating highest substrate affinity. From these plots we calculated substrate affinities (Km), maximum velocities (Vmax), and specific activities. In terms of specific activity, C143S retained 75% of WT activity (Table 2). The Km of GlcNAc for WT NAGK was lowest (0.78 ± 0.33 mM), followed by those of C143S and N36A (Table 2). Double-reciprocal plots for ATP (Fig. 2C) showed that these mutations also affected the ATP binding site. As was observed for the GlcNAc binding site, NAGK ATP binding affinity was less affected for C143S than N36A, as indicated by the similar slope (Fig. 2C) and Km values (0.13 mM and 0.15 mM, respectively; Table 2) of WT NAGK and C143S for ATP.
Table 2.
Kinetic data of wild-type and mutant NAGK
| GlcNAc | ATP | |||
|---|---|---|---|---|
|
|
|
|||
| Km (mM) | Vmax (mM/min) | Specific activity (U/mg protein) | Km (mM) | |
|
|
|
|||
| WT | 0.78 ± 0.33 | 0.018 ± 0.002 | 7.04 | 0.13 |
| N36A | 1.17 ± 0.13 | 0.011 ± 0.001 | 3.47 | 1.59 |
| C143S | 0.86 ± 0.18 | 0.014 ± 0.001 | 5.29 | 0.15 |
Values are the means of at least three independent experiments.
The N36A mutant
With respect to specific activity, N36A retained 49% of WT NAGK activity (Table 2). The lower specific activity of N36A was well reflected by a higher Km (1.17 mM) for GlcNAc than C143S (0.86 mM). In contrast to the mild reduction in GlcNAc binding efficiency of N36A, ATP binding was severely reduced. The Km (1.59 mM) for ATP of N36A was >10 times that of WT NAGK (0.13 mM; Table 2). These results indicate that altered GlcNAc binding affinity significantly affected ATP binding by NAGK.
The D107A mutant
The D107A mutation almost completely abrogated the kinase activity of NAGK (Fig. 2A), and thus, we did not include this mutant in double reciprocal plots.
Substrate specificity
WT NAGK
To analyze the substrate specificity of the enzymatic reaction, several isomeric substrates, that is, ManNAc, GalNAc, and glucose, were used as substrates. For this purpose, we used 1.9 mM of each substrate and 4.0 mM ATP, and monitored OD340 reductions. The rates of phosphorylation of substrates were compared with those of WT NAGK. At the initial stage of reactions (10 min), C143S and N36A showed reduced activities with respect to the phosphorylation of GlcNAc (86.6% and 67.0% of the WT, respectively). WT NAGK showed high substrate preference for GlcNAc (Fig. 3A, Table 3), and some activity on ManNAc and glucose (11.2% and 7.0% of GlcNAc, respectively) (Table 3, 40 min).
Fig. 3.
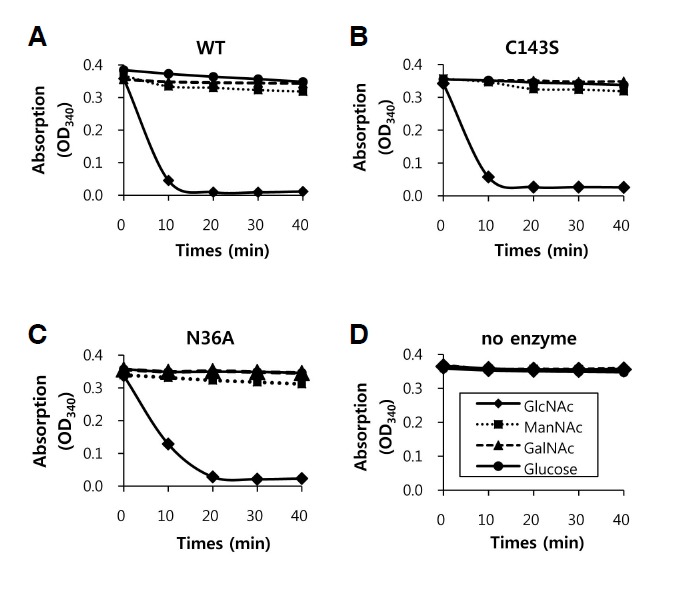
Substrate specificity. The specificities of wild-type (WT) NAGK (A), the C143S mutant (B), the N36A mutant (C), and no enzyme control (D) are shown. Reaction rates are for GlcNAc, N-acetylmannosamine (ManNAc), N-acetylgalactosamine (GalNAc), and glucose in the presence of 1.9 mM of each sugar and 4.0 mM of ATP. The plots shown are representative.
Table 3.
Substrate specificities
| Substrate | Relative activity | |||
|---|---|---|---|---|
|
| ||||
| WT | C143S | N36A | ||
|
| ||||
| 10 min | 40 min | 10 min | 10 min | |
| GlcNAc | 100 | 100 | 100 (86.6) | 100 (67.0) |
| ManNAc | 12.3 | 11.2 | 1.9 (1.6) | 0 (0) |
| GalNAc | 5.6 | 1.4 | 1.2 (1.1) | 0 (0) |
| Glucose | 5.4 | 7 | 1.2 (1.0) | 1.6 (1.0) |
Kinase activities of the affinity-purified wild-type (WT) and of the C143 and N36A mutant NAGKs were defined as the rate of formation of NAD+ at 340 nm, as described in “Materials and Methods”. Substrate concentrations were 4.0 mM for ATP and 1.9 mM for sugars. Relative activities are expressed as percentage OD340 decreases relative to when GlcNAc was used as the substrate. Parentheses indicate values relative to the reaction between WT NAGK and GlcNAc.
C143S and N36A mutants
C143S and N36A NAGK also showed strong substrate preference for GlcNAc (Figs. 3B, 3C, and Table 3), but their activities on substrates other than GlcNAc were negligible (< 2% of the WT).
Upregulation of dendritic arborization by mutant NAGKs
As a preliminary experiment, the three point mutants, i.e. N36A, D107A, or C143S, which was previously shown to retain 10% of the kinase activity of WT NAGK (Berger et al., 2002), were introduced in EGFP-tagged forms by transfection into hippocampal neurons in culture (DIV 5–9). The overexpressions of these mutant NAGKs did not cause any aberrant morphology, but increased cytoarchitectural complexity. To confirm the promotion of dendritogenesis, we carried out Scholl analysis. The typical images of live neurons transfected with control vector (pEGFP), pNAGK-WT, -N36A, or -D107A are shown in Fig. 4A. The numbers of dendrites crossing concentric circles drawn at different radii from soma centroids (Scholl analysis) were counted (Fig. 4B). Numbers of dendrites crossing at a small radius (25 μm) were similar between control (vector) and WT-or mutant-NAGK transfected neurons. However, the numbers of dendrites of neurons transfected with WT or mutant NAGK (N36A, D107A) increased significantly from those of vector-transfected neurons at radii of 35–75 μm (p < 0.05 at 35 μm, p < 0.01 at other distances). Furthermore, these mutants and the WT were no different in this respect. These results show that the promotion of dendrite development by NAGK is independent of its kinase activity.
Fig. 4.
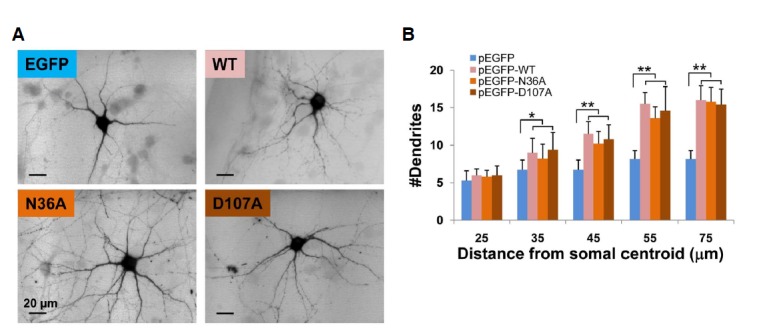
The kinase activity of NAGK is not essential for its role in the development of dendrites. (A) Epifluorescence images of typical neurons overexpressing control (EGFP) or EGFP-tagged WT, or N36A or D107A mutant NAGKs. Scale bar, 20 μm. (B) Sholl analysis. The numbers of dendrites that crossed circles of the indicated radii from stoma centroids of are indicated. *p < 0.05, **p < 0.01.
Degeneration of dendrites by the overexpression of the small domain of NAGK
To explore the structural role played by NAGK in the promotion of dendrite development, we performed dominant negative experiments. NAGK has a small N-terminal and a large C-terminal domain (Weihofen et al., 2006). To carry out dominant negative experiments, we constructed small (pDsRed2-NAGKDs) and large (pDsRed2-NAGK-DL) RFP-tagged NAGK domains and transfected them into cultured rat hippocampal neurons. Typical images of neurons expressing the large domain are shown in Fig. 5A-a. Numbers of dendrites were unaffected by the overexpression of RFP-NAGK-DL. Furthermore, when the numbers of dendrites were plotted versus NAGK-IR signal level no correlation was found (Fig. 5A-b). In contrast, neurons overexpressing RFP-NAGK-Ds exhibited dramatic dendrite degeneration (Fig. 5B-a). Furthermore, a plot of dendrite number vs. NAGK-IR signal level showed that the extent of dendritic degeneration proportional to the expression level of the small domain (Fig. 5B-b). These results indicate that the small domain of NAGK plays a structural role during dendrite development.
Fig. 5.
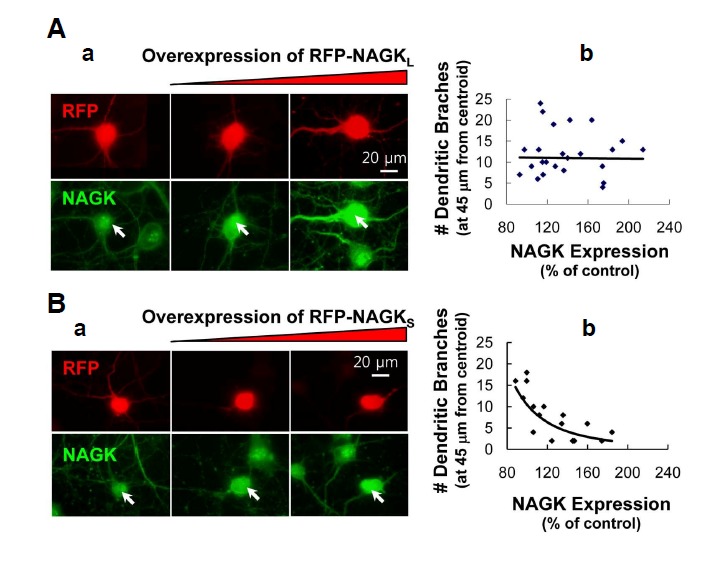
Overexpression of the small domain of NAGK resulted in dendrite degeneration. Cultured rat hippocampal neurons (DIV5-7) were transfected with the indicated plasmids and double-labeled with antibodies against RFP (red) and NAGK (green). (A) (a) Epifluorescence images of neurons expressing RFP-NAGK-DL. Transfected neurons are marked with arrows. (b) Statistics. Numbers of dendrites crossing circle of radii 45 μm from soma centroids were counted. Numbers, relative to those in the soma of untransfected control neurons, were plotted against RFP-NAGK-DL expression levels (B) Epifluorescence images of neurons expressing RFP-NAGK-Ds. Annotations are the same as in (A). Scale bar, 20 μm.
DISCUSSION
The present study has two main findings; 1) structure-function analysis using point mutants of NAGK that retained differential activities supported the previously proposed 3D model structure (Weihofen et al., 2006), and 2) dendrite upregulation by NAGK (a non-canonical function) is independent of the kinase activity of NAGK.
A previous attempt to crystallize NAGK with its substrates, GlcNAc and ATP/Mg2+ failed, and instead, a model 3D structure was proposed (Weihofen et al., 2006). To verify this model by structure-function analysis, we created three NAGK mutants (N36A, D107A, and C143S). Substrate utilization curves demonstrated the different affinities of these mutants, as was expected by 3D modeling. Conversion of Cys143 to Ser (i.e. C143S), which does not make direct hydrogen bonds with GlcNAc, had least affect on enzymatic activity (75% of the activity of wild-type NAGK).
Conversion of Asn36 (which plays a role during domain closure by hydrogen bonding with GlcNAc) to Ala (i.e. N36A) reduced enzyme activity by 50%. Conversion of Asp107 (which makes hydrogen bonds with GlcNAc and thought to act as a proton acceptor during nucleophilic attack on γ-phosphate of ATP) to Ala (i.e. D107A) caused a total loss of enzyme activity. Km and Vmax values also support the 3D model of NAGK. Changes in the GlcNAc binding affinity of NAGK were well reflected by Vmax and the specific activities. As was expected, WT NAGK, which had the lowest Km value, had the highest Vmax value, followed by C143S and N36A. These results show that WT NAGK had the highest binding affinity and GlcNAc turnover rate. Of the point mutants, C143S retained highest activity, probably because the cysteine-to-serine change retained domain polarity. Changes in the substrate binding and turnover rates of mutant NAGKs were reflected by specific enzyme activities, where WT NAGK has the highest specific activity for GlcNAc, followed by C143S and N36A. A reduction in the enzyme activity of C143S NAGK was reported in a previous study (Berger et al., 2002). C143 is located near bound GlcNAc in the substrate binding pocket. However, this residue was found not to be involved in domain closure or hydrogen bonding with GlcNAc (Weihofen et al., 2006). Thus, it appears the Cys-to-Ser change retained domain polarity and only mildly damaged NAGK functionality.
Asn36NδH hydrogen bonds to the N2-acetyl carbonyl oxygen of GlcNAc (Weihofen et al., 2006), and thus, the lack of this hydrogen bond in the N36A mutant would mildly reduce the efficiency of GlcNAc binding. It has been shown that the binding GlcNAc causes NAGK to adopt a ‘closed’ conformation, which is achieved by a 26° rotation of the small domain relative to the large domain (Weihofen et al., 2006). Thus, lack of hydrogen bonding between N36 and GlcNAc could induce a conformation change that adversely affects the binding of ATP. N36A conversion also induced non-polar characteristics within the GlcNAc binding pocket, and reduced ability to retain substrate may have reduced GlcNAc binding affinity and overall reaction rates.
According to the proposed phosphorylation reaction mechanism, D107 plays a pivotal role by acting as a proton acceptor (Weihofen et al., 2006). Furthermore, the importance of D107 is reflected by the fact that it is conserved among sugar kinases. Furthermore, the loss of enzyme activity caused by the D107A mutation well supports the proposed 3D model of NAGK (this work). Mutant enzymes with lower levels of enzyme activity would be useful in other research areas, such as, for determining non-canonical functions, protein-protein interactions, and for therapeutic studies. In this context, our results highlight the importance of the structure-function relationships of NAGK and show that more screening studies on mutational sites to better understand the enzymatic activity of NAGK.
Many proteins have several functions. The canonical function of NAGK is the phosphorylation of GlcNAc to produce GlcNAc-6-phosphate. Recently, we found that NAGK is highly expressed in neuronal dendrites and plays a critical role in their development (Lee et al., 2014), and the present study provides evidence that this non-canonical function of NAGK is independent of its kinase activity. The overexpressions of N36A and D107A mutant NAGKs, each with a point mutation in the substrate binding pocket but with retained differential kinase activity, upregulated dendrite numbers to the same extent similar as wild-type NAGK. In addition, we found that the small domain of NAGK is important in this context. In our dominant negative experiments, the overexpression of the RFP-tagged NAGK small domain (RFP-NAGK-Ds) resulted in dendritic degeneration, and the extent of dendritic degeneration was found to be proportional to the expression level of RFP-NAGK-Ds. It appears that excessive small domains may have captured, and thus, sequestered the elements necessary for assembling normal NAGK-including complexes.
During the early stage of neuronal development, neurons show dramatic re-arrangement in dendritic arbors, such as, the addition of new branches, complete retraction of branches, and extension or shortening of branches (O’Rourke, 1994). Our results indicate that NAGK excess could accelerate the processes responsible for dendrite elaboration, whereas NAGK deficiency could accelerate the processes responsible for dendrite retraction.
In conclusion, our structure-function analysis supports the 3D NAGK model previously proposed by Weihofen et al. (2006). Based on results obtained using mutant NAGKs, the present study shows that dendrite formation is not dependent on the kinase activity of NAGK. Furthermore, they show that the small domain of NAGK plays a critical structural role in this process.
Acknowledgments
We thank Dr. Stephan Hinderlich [Beuth Hochschule für Technik Berlin (University of Applied Sciences), Department of Life Sciences and Technology, Berlin, Germany] for donating the murine NAGK clone, and Eun-jung Jung in our laboratory for technical assistance. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A2006116).
REFERENCES
- Asensio C., Ruiz-Amil M. N-acetyl-D-glucosamine kinase. II. Escherichia coli. Methods Enzymol. 1966;9:421–425. [Google Scholar]
- Berger M., Chen H., Reutter W., Hinderlich S. Structure and function of N-acetylglucosamine kinase. Identification of two active site cysteines. Eur. J. Biochem. 2002;269:4212–4218. doi: 10.1046/j.1432-1033.2002.03117.x. [DOI] [PubMed] [Google Scholar]
- Blume A., Berger M., Benie A.J., Peters T., Hinderlich S. Characterization of ligand binding to N-acetylglucosamine kinase studied by STD NMR. Biochem. 2008;47:13138–13146. doi: 10.1021/bi8016894. [DOI] [PubMed] [Google Scholar]
- Brewer G.J., Torricelli J.R., Evege E.K., Price P.J. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Goslin K., Assmussen H., Banker G. Rat hippocampal neurons in low density culture. In: Banker G., Goslin K., editors. In culturing nerve cells. 2nd eds. Cambridge: MIT Press; 1998. pp. 339–370. [Google Scholar]
- Hinderlich S., Nöhring S., Weise C., Franke P., Stäsche R., Reutter W. Purification and characterization of N-acetylglucosamine kinase from rat liver: comparison with UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 1998;252:133–139. doi: 10.1046/j.1432-1327.1998.2520133.x. [DOI] [PubMed] [Google Scholar]
- Hinderlich S., Berger M., Schwarzkopf M., Effertz K., Reutter W. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 2000;267:3301–3308. doi: 10.1046/j.1432-1327.2000.01360.x. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Jiang M., Deng L., Chen G. High Ca2+-phosphate transfection efficiency enables single neuron gene analysis. Gene Ther. 2004;11:1303–1311. doi: 10.1038/sj.gt.3302305. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Cho S.-J., Moon I.S. The non-canonical effect of N-acetyl-D-glucosamine kinase on the formation of neuronal dendrites. Mol. Cells. 2014;37:248–256. doi: 10.14348/molcells.2014.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon I.S., Cho S.J., Jin I., Walikonis R. A simple method for combined fluorescence in situ hybridization and immunocytochemistry. Mol. Cells. 2007;24:76–82. [PubMed] [Google Scholar]
- O’Rourke N.A., Cline H.T., Fraser S.E. Rapid remodeling of retinal arbors in the tectum with and without blockade of synaptic transmission. Neuron. 1994;4:921–934. doi: 10.1016/0896-6273(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Sholl D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Uehara T., Park J.T. The N-acetyl-D-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 2004;186:7273–7279. doi: 10.1128/JB.186.21.7273-7279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen W.A., Berger M., Chen H., Saenger W., Hinderlich S. Structures of human N-acetylglucosamine kinase in two complexes with N-acetylglucosamine and with ADP/glucose: insights into substrate specificity and regulation. J. Mol. Biol. 2006;364:388–399. doi: 10.1016/j.jmb.2006.08.085. [DOI] [PubMed] [Google Scholar]


