Abstract
X-box binding protein 1 (XBP1) is a central regulator of the endoplasmic reticulum (ER) stress response. It is induced via activation of the IRE1 stress sensor as part of the unfolded protein response (UPR) and has been implicated in several diseases processes. XBP1 can also be activated in direct response to Toll-like receptor (TLR) ligation independently of the UPR but the pathogenic significance of this mode of XBP1 activation is not well understood. Here we show that TLR-dependent XBP1 activation is operative in the synovial fibroblasts (SF) of patients with active rheumatoid arthritis (RA). We investigated the expression of ER stress response genes in patients with active RA and also in patients in remission. The active (spliced) form of (s)XBP1 was significantly overexpressed in the active RA group compared to healthy controls and patients in remission. Paradoxically, expression of nine other ER stress response genes was reduced in active RA compared to patients in remission, suggestive of a UPR-independent process. However, sXBP1 was induced in SF by TLR4 and TLR2 stimulation, resulting in sXBP1-dependent interleukin-6 and tumour necrosis factor (TNF) production.
We also show that TNF itself induces sXBP1 in SF, thus generating a potential feedback loop for sustained SF activation. These data confirm the first link between TLR-dependent XBP1 activation and human inflammatory disease. sXBP1 appears to play a central role in this process by providing a convergence point for two different stimuli to maintain activation of SF.
Keywords: Rheumatoid arthritis (RA), Unfolded protein response (UPR), sXBP1, Toll-like receptor (TLR)
Highlights
-
•
sXBP1 is upregulated in PBMC from patients with active RA.
-
•
TLR2 and TLR4 mediated sXBP1 activation in synovial fibroblasts.
-
•
SNAPIN-induced cytokine production is dependent on sXBP1.
-
•
Proinflammatory cytokines cause XBP1 activation in synovial fibroblasts.
1. Introduction
The pathogenesis of rheumatoid arthritis (RA) is complex, and although not fully elucidated, it is widely accepted that joint inflammation and damage result from the interplay and activation of the resident synovial fibroblasts (SF) by self-reactive immune cells [1]. Activated SF demonstrate an enhanced survival and destructive phenotype, thought to result in part from dysregulation of the unfolded protein response (UPR), with accompanying ER associated degradation (ERAD) [2–4]. The activation of X-box binding protein 1 (XBP1) by inositol-requiring 1α(IRE1) is the most evolutionarily conserved component of the UPR. XBP1 is a transcription factor (TF) that regulates numerous genes involved in adaptation to ER stress. XBP1 activation depends on the IRE1-dependent removal of 23 nucleotides from the XBP1 mRNA with the resultant frameshift encoding a more stable TF with enhanced transactivation potential. More recently, XBP1 splicing has been demonstrated downstream of Toll-like receptor (TLR) 2 and TLR 4 ligation [5]. This process requires phosphorylation of IRE1, which cleaves XBP1 mRNA thereby generating the spliced, active form (s)XBP1; this occurs in the absence of ER stress or wider UPR activation and leads to synthesis of the pro-inflammatory cytokines, TNF and IL-6 [5]. Recently, it has been demonstrated that myeloid specific deletion of IRE1 protects mice from inflammatory arthritis [6]. Furthermore, the same effect was observed in mice treated with an IRE1-specific inhibitor. In this system IRE1 was required for optimal production of inflammatory cytokines resulting from TLR stimulation.
This pathway linking TLR2- and TLR4-dependent XBP1 activation to TNF and IL-6 secretion may have particular relevance to RA, since both TLR2 and TLR4 are highly expressed in the RA joints [7] and biologic therapies that target TNF and IL-6 are effective in a large number of patients. Although TLRs are primarily involved in pathogen sensing, they also recognise several endogenous ligands and it is these that may be particularly relevant in the setting of RA [8–11]. The endogenous TLR2 ligand, SNARE associated protein (SNAPIN), was recently identified as being highly expressed in synovial tissue macrophages and consequently SNAPIN has been suggested to contribute to the pathogenesis of RA [12].
We set out to investigate the relevance of interactions between UPR and TLR signalling pathways in the serum and SF of patients with RA, using osteoarthritis (OA) samples as controls. Our results reveal that, in both serum and SF of RA patients, generation of sXBP1 is uncoupled from the canonical UPR-response, while TLR2 and TLR4 stimulation by endogenous ligands, such as SNAPIN, facilitate the production of pro-inflammatory cytokines.
2. Materials and methods
2.1. Patients and cells
The active RA group included patients with established disease who had failed at least 2 traditional disease-modifying anti-rheumatic drugs (DMARDs), and were about to commence either infliximab or adalimumab. The RA remission group included patients who were treated with anti-TNF agents, infliximab or adalimumab, and who later achieved complete clinical (DAS <1.2), biochemical (CRP <10) and radiological remission (no evidence of synovitis on ultrasound scan), and who were subsequently maintained off all therapy. Ethical committee approval was granted by the UK Central Office of Research Ethics Committee (RA cohort: 01/023, HC: 04/Q1206/107).
PBMC were obtained from patients and control groups by density gradient centrifugation (Lymphoprep-Sigma, Aberdeen, UK). SF were obtained by collagenase (StemCell Technologies, Grenoble, France) digestion of synovial biopsies from RA and OA patients who had undergone consented arthroscopy of a swollen knee joint. SF were selected as the adherent cells following culture in Dulbecco's modified Eagle's medium (DMEM) containing foetal bovine serum (FBS) (10% v/v) and penicillin-streptomycin (1% v/v). After reaching confluence, the cells were passaged and replated at a ratio of 1:3. The cells were used at passage 3 (P3).
2.2. qRT-PCR and sXBP1 detection
Total RNA was isolated using TRIzol (Invitrogen, Paisley, UK) from PBMC and cultured SF and first-strand cDNA synthesized using Superscript II Reverse transcriptase (Invitrogen).
XBP1 mRNA splicing was detected using the following set of primers: F – 5′-CTGAAGAGGAGGCGGAAGC-3′ and R – 5′-AATACCGCCAGAATCCATGG-3′, which recognise both the shorter spliced (s) and longer unspliced (u) forms of XBP1 mRNA. sXBP1 was then identified on a 3% agarose gel as the faster migrating species of the two closely spaced bands.
Primers for quantitative real-time PCR (qPCR); unspliced XBP1 (uXBP1) forward 5′-TCCGCAGCACTCAGACTACG-3′, sXBP1 forward 5′-CTGAGTCCGCAGCAGGTG-3′ and reverse primer for u/sXBP1 5′-AGTTGTCCAGAATGCCCAACA-3′, final concentration 50 fmol/μl. Forward uXBP1 primer spans the splice site to avoid amplifying sXBP1 and primer pairs span different exons. Relative gene expression (2(–ΔCT)) was analysed using SDS 2.3 (Applied Biosystems), normalising to hypoxanthine phospho- ribosyltransferase 1 (HPRT):
Forward 5′-GGAAAGAATGTCTTGATTGTGGAAG-3′ and reverse 5′-GGATTATACTGCCTGACCAAGGAA-3′, final concentration 500 fmol/μl. All reactions were carried out in duplicate using SYBR green on the ABI 7900 (Applied Biosystems), using standard cycling parameters. Standard SYBR green PCR conditions were used, with an annealing temperature of 59 °C and 40 cycles. Optimisation steps were carried out to ensure specificity of primer pairs to amplify individual u/sXBP1 cDNA products.
A custom TaqMan gene array based on a 384-well micro fluidic card was designed using the Applied Biosystems website (www.appliedbiosystems.com), to determine the expression of a number of UPR genes of interest. These included: Synoviolin (SYVN1), thioredoxin domain-containing protein 4 (TXNDC4), Inositol-requiring protein 1 (IRE1), homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain family member 2 (HERPUD2), Heat shock 70 kDa protein 5 (HSPA5), Growth arrest and DNA damage-inducible protein (GADD34), C/EBP-homologous protein (CHOP), Protein disulfide isomerase family A, member 4 (PDIA4)DnaJ (Hsp40) homolog, subfamily B, member 9 (DNAJB9). The analysis was done using DataAssist™ software from Applied Biosystems (Foster City, CA, USA). The relative gene expression was calculated with reference to the housekeeping gene, GAPDH. Difference between the cycle thresholds for GAPDH and the selected genes (ΔCt) was determined and relative gene expression calculated as 2−ΔCt Results were plotted as relative expression units (REU).
UPR genes expression from synovium 100 micron sections of OCT embedded synovial tissue were cut and the OCT manually removed. RNA was extracted using Norgen ANimal Tissue RNA kit. Reverse transcription was performed using High capacity reverse transcription (Applied Biosystems). Taqman assays were performed using following primer sets: Hs99999905_m1 GAPDH, Hs00381211_m1 SYVN1, Hs00231936_m1 XBP1, Hs99999174_m1 HSPA5, Hs01052402_m1 DNAJB9.
2.3. Flow cytometry
Intracellular staining for sXBP1 was performed fixation/permeabilization buffer (Bioscience, Hatfield, UK). The primary antibody was mouse monoclonal anti-human sXBP1 [gift from Dr. G. Doody and Dr. R. Tooze [13]]. Mouse polyclonal IgG2 (AbD Serotec, Oxford, UK) was used as an isotype control, while polyclonal goat anti-mouse FITC (fluorescein isothiocyanate) conjugate and FITC labelled polyclonal (Southern Biotech, Birmingham, AL, USA) was used as the secondary antibody. Samples were analysed using a 1-colour protocol on the BD LSRII flow cytometer (BD Biosciences, Cowley, UK). Median of fluorescence intensity (MFI) was used to measure the expression levels of the intracellular protein.
2.4. Immunofluorescence
SF for IF were grown on coverslips. Adherent cells were fixed using ice-cold 100% methanol for 5 min at −20 °C. Appropriate dilutions of the primary anti-human sXBP1 antibody (as above) were added. The primary antibodies were visualized using Alexa Fluor 680 goat anti-mouse secondary Ab or FITC. DAPI (1:1000) was used to visualise nuclei (blue). Images were analysed using Andor IQ software.
2.5. Cell stimulation
RA and OA SF were treated with 10 ng/ml of LPS (Sigma) for 6 h, with or without prior incubation with the specific TLR4 inhibitor CLI-095 (Invivogen) or a TLR2/4 inhibitor, OxPAP (Invivogen), for 30 min. Stimulation of SF with 5 µg/ml of SNAPIN (Reagent Proteins) for 24 h. Treatment with Tg (Sigma) was used as a control for induction of ER stress by treating the SF with 300 nM for 6 h.
2.6. Quantification of cytokines
Cytokine levels were quantified from media supernatants using ELISA kits for IL-6 (BD Biosciences) and TNF (PeproTech), according to the manufacturers' instructions.
2.7. siRNA knock down
Human XBP1 was targeted for knock down by RNA interference using siRNA (ON-TARGET plus SMART pool, Thermo Scientific) in RA SF for 48 h. Non-targeting control (ON-TARGET plus Non-targeting siRNA, Thermo Scientific) was also used according to manufacturers' instructions.
2.8. Statistical analyses
The Kruskal–Wallis and Dunn's Multiple Comparison Test (pot-hoc analysis) were used for one-way analysis of variance between multiple groups of patients. The Mann–Whitney test was used to analyse differences between sXBP1 protein expression in RA-ACT and HC PBMC. Spearman's rank correlation coefficient was used to determine the degree of association between sXBP1 mRNA expression and joint inflammation.
3. Results
3.1. sXBP1 is upregulated in PBMC from patients with active RA
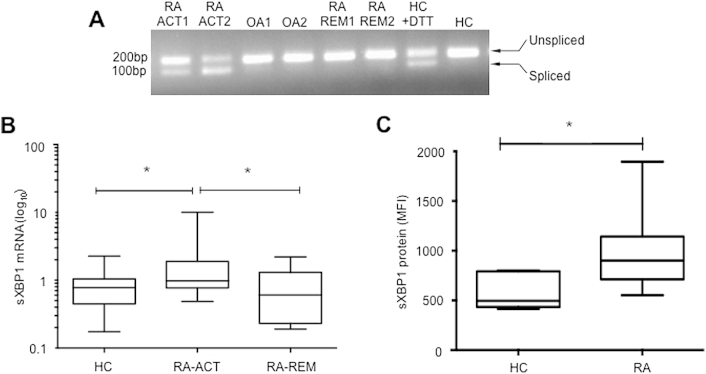
We reasoned that if XBP1 activation is involved in RA pathogenesis then the activation of XBP1, detectable by splicing of its mRNA, should be associated with disease activity. We therefore compared the expression of XBP1 and sXBP1 mRNA, using reverse transcription (RT)-PCR, in patients at different stages of disease. We used mRNA from peripheral blood mononuclear cells (PBMC) obtained from RA patients with active disease (RA-ACT), or remission (RA-REM), and compared to either healthy individuals (HC) or those with OA as controls. The sXBP1 species was detected in PBMC of patients with active RA, but not those in remission, and neither was it detected in OA patients or healthy controls (Fig. 1A). We then used quantitative (q)RT-PCR to confirm these results in a wider group of patients [active RA (n = 47), and remission (n = 12)] (demographics Table 1) as well as HC (n = 24). This confirmed that the highest expression levels of sXBP1 were observed in PBMC of RA patients with active disease (Fig. 1B).
Fig. 1.
sXBP1 expression in PBMC. (A) XBP1 splicing was determined by PCR, using primers that amplify both unspliced and spliced mRNA species. RA-ACT1 and 2 (RA patients with active disease); OA1- and OA2-osteoarthritis patients; RA-REM1 and 2 (RA patients in remission); HC, healthy control. (B) sXBP1 mRNA levels were determined by qRT-PCR and expressed as relative to levels of GAPDH. Data are shown as median with interquartile range. Statistical analysis of variance between different of groups was performed using the Kruskal–Wallis test and Dunn's Multiple Comparison Test was used to compare all pairs of data. (* = p < 0.05). (C) sXBP1 protein expression in PBMC was analysed by flow cytometry. Expression levels are shown as median fluorescence intensity (MFI) on the y-axis. Statistical significance was determined using non-parametric Mann–Whitney U test.
Table 1.
Demographics of patients with RA and HC used in UPR gene expression studies.
| Groups | HC | RA-ACT | RA-REM |
|---|---|---|---|
| Number | 24 | 47 | 12 |
| Av Age (Range) | 45.4 (24–68) | 56.9 (23–83) | 57.0 (47–64) |
| F:M (%) | 15F:9M (62.5–37.5) | 40F:7M (85:15) | 10F:2M (83:17) |
| % ACPA pos | 0 | 83 | 80 |
ACPA- anti citrullinated protein antibody.
We also confirmed sXBP1 protein expression in PBMC using flow cytometry; 10 RA-ACT patients had significantly higher mean sXBP1 expression levels compared to 7 sex- and age-matched HC (p = 0.01, Fig. 1C). Collectively, these results reveal a significant association between sXBP1 expression and the active RA phenotype (Fig. 1).
3.2. UPR responsive genes are downregulated in PBMC of RA patients
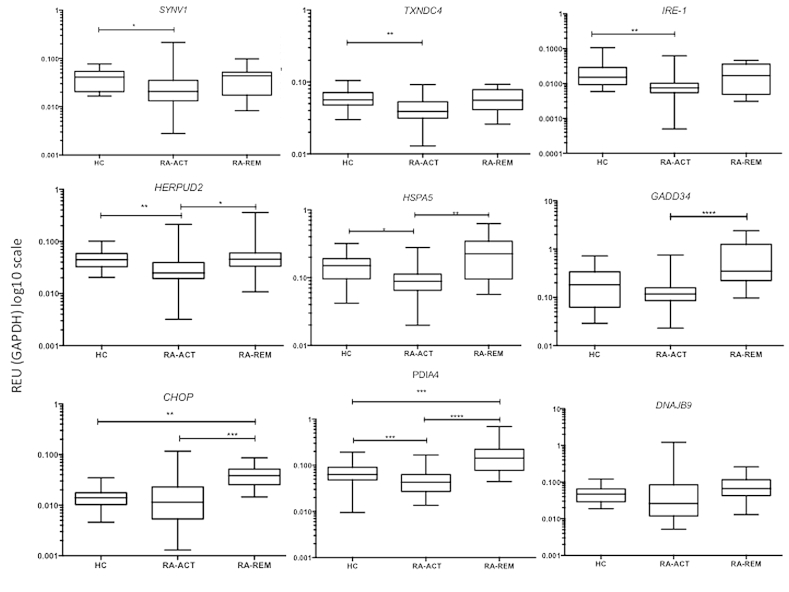
The association of sXBP1 expression with RA pathogenesis suggested a potential role for the UPR in this disease. The UPR is characterized by three primary arms leading to activation of XBP1 and the activating transcription factors (ATF) 4 and ATF6. Several target genes have been defined for these TFs, providing a signature of the canonical UPR. In order to assess whether the observed XBP1 activation in active RA was associated with a canonical UPR we measured expression of 9 genes from this UPR signature (Table 2). Surprisingly, the expression of eight of nine signature genes was downregulated in RA patients with active disease when compared to the RA remission group and the HC group (Fig. 2). For example, the expression levels of SYNV1, TXNDC4, HERPUD2, IRE-1, HSPA5 and PDIA4 were all significantly reduced in active RA patients compared to HC (all p < 0.05). In the case of HERPUD2, HSPA5 and PDIA4, their expression in the RA group with active disease was also significantly lower compared to the group of patients in remission (p < 0.05, p < 0.01 and p < 0.001 respectively). The RA-remission group also had significantly higher expression of GADD34, and CHOP compared to patients with active RA (p < 0.001 and p < 0.005) and, in the case of CHOP and PDIA4, the expression levels in remission patients were significantly higher in comparison to HC (p < 0.01 and p < 0.005). Only in the case of DNAJB9 was expression unaffected by disease activity. Overall, patients with active RA had the lowest expression levels of UPR target genes compared to other groups, despite increased expression of the sXBP1 species in this group. These results suggested that XBP1 pathway was uncoupled from other elements of a canonical UPR.
Table 2.
List of UPR genes alternative names and function.
| UPR gene | Full and alternative names | Transcription control | Function |
|---|---|---|---|
| SYVN1 | Synovial apoptosis inhibitor 1 | sXBP1, ?ATF6 |
ERAD, E3ligase |
| TXNDC4 | ERP44 (ER protein 44) | sXBP1 | Chaperone Disulfide bond formation Cell redox homeostasis |
| PDIA4 (ERP70/72) | Protein disulfide isomerase family A, member 4 | sXBP1 | Cell redox homeostasis |
| DNAJB9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | sXBP1 | Heat shock protein Protein folding |
| HERPUD2 | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain family member 2 | ATF4 ATF6 |
Co-chaperone |
| BIP (HSPA5) | Immunoglobulin heavy chain-binding protein (Heat shock 70 kDa protein 5) |
ATF4 ATF6 |
Chaperone |
| GADD34 (PPP1R15A) | Growth arrest and DNA damage-inducible protein (Protein phosphatase 1, regulatory (inhibitor) subunit 15A) |
ATF4 ATF6 |
Apoptosis Cell cycle arrest Regulation of translation Response to DNA damage |
| CHOP (DDIT3) | C/EBP-homologous protein (DNA-damage-inducible transcript 3) | ATF4 ATF6 |
Transcription regulation Growth arrest Apoptosis |
| IRE1 (ERN1) | Inositol-requiring protein 1 (Endoplasmic reticulum to nucleus signalling 1) |
? | sXBP1 activation Apoptosis UPR |
Fig. 2.
Expression of UPR genes in PBMC of RA patients and healthy controls. Levels of UPR genes (as indicated) were determined by qRT-PCR and expressed relative to levels of GAPDH. Samples were healthy controls (HC; n = 24), RA with active disease (RA-ACT; n = 47) and RA in remission (RA-REM; n = 12). Values on the Y-axis are presented on a logarithmic scale (Log 10). Statistical analysis of variance between different of groups was performed using the Kruskal–Wallis test whilst Dunn's Multiple Comparison Test was used to compare all pairs of data. Data are shown as median with interquartile range. (* = p < 0.05, ** = p < 0.01, *** = p < 0.005 and **** = p < 0.001).
3.3. TLR2 and TLR4 mediated sXBP1 activation in synovial fibroblasts (SF)
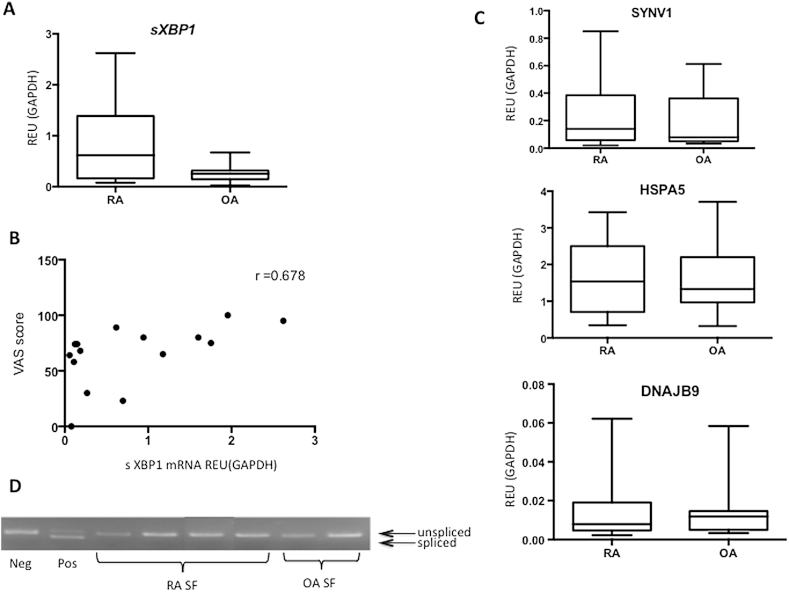
The association of sXBP1 with RA pathogenesis was further supported by the finding that there was a trend for higher expression of sXBP1 in RA (p = 0.131) compared to OA synovial biopsies (20 RA and 12 OA) (Fig. 3A). There was also a direct relationship with levels of inflammation in the joint (RA and OA combined), as measured using arthroscopic inspection (Spearman r = 0.678, p = 0.068) (Fig. 3B). Expression of the HSPA5, DNAJB9 and SYVN1 was not different between patients with RA and OA and was unrelated to the degree of inflammation (Fig. 3C).
Fig. 3.
Expression of sXBP1 and UPR genes in synovium. (A) sXBP1 mRNA levels in RA and OA-SF determined by qRT-PCR. Statistical significance determined by Mann–Whitney test. (B) Correlation between sXBP1 mRNA levels and synovial inflammation, which was determined using visual analogue scale following arthroscopic inspection of the joint. Statistical significance determined using Spearman rank correlation coefficient (C) SYNV1, HSPA5 and DNAJB9 mRNA levels in RA and OA-SF determined by qRT-PCR. Statistical significance determined by Mann–Whitney test. (D) XBP1 splicing determined by RT-PCR, using primers that amplify both unspliced and spliced mRNA species. Neg, cells at rest; Pos, cells treated with DTT (to induce a UPR). RA SF were obtained from a rheumatoid joint (knee) and cultured to P3, OA SF obtained from an OA patient (knee joint) and cultured to P3.
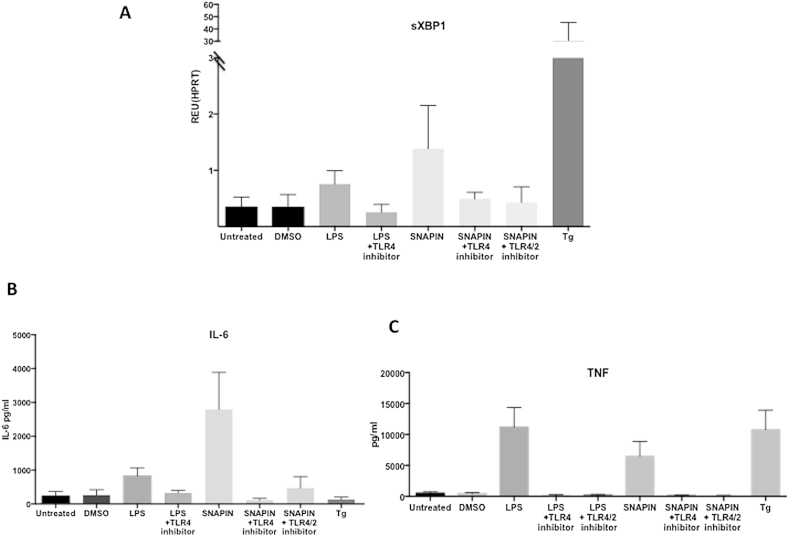
However, sXBP1 mRNA was not detectable in cultured SF obtained from either the active RA or OA synovial biopsies (Fig. 3D). The lack of detectable sXBP1 in these cultured RA synovial fibroblasts suggested the removal of an in vivo stimulus. Martinon et al. have previously shown that XBP1 splicing can occur in response to TLR2 or TLR4 ligation, via IRE1 phosphorylation, and that this occurs in the absence of a more generalized UPR [5]. We hypothesised that a similar mechanism might be responsible for the UPR-independent sXBP1 induction observed in RA patients. Stimulation of SF with LPS (the prototype TLR4 ligand) induced sXBP1 expression, and this effect was antagonized by a TLR4 pathway inhibitor, CLI-095 (Fig. 4A). However, as the inflammatory response in RA is sterile, we tested the role of the endogenous TLR ligand, SNAPIN, in sXBP1 induction. SNAPIN, a component of SNARE complexes, is a TLR2/4 ligand that has been implicated in the pathogenesis of RA [12]; SNAPIN induced sXBP1 to similar levels as that seen with LPS, and this effect was abrogated by using a selective TLR4 inhibitor or as a result of combined TLR4/2 inhibition by OxPAPC (Fig. 4A).
Fig. 4.
sXBP1, IL-6 and TNF expression in SF following stimulation with LPS and SNAPIN. 4. (A–C) sXBP1 mRNA levels in RA SF measured by qRT-PCR after treatment with LPS (10 ng/ml) for 6 h, TLR4 inhibitor (1 μg/ml) for 30 min followed by LPS (10 ng/ml) for 6 h, SNAPIN (5 μg/ml) for 24 h, with or without prior TLR4 (1 μg/ml) and TLR2/4 (30 μg/ml) inhibition for 30 min, and Tg (300 nm/ml) for 6 h with or without TLR2/4 (30 μg/ml) treatment, with DMSO as vehicle control. Results are expressed relative to HPRT. Error bars represent the standard deviation of 3 independent experiments. (B–C) Supernatants were harvested from RA SF and IL-6 and TNF levels were determined by ELISA, the X-axis represents the different treatments described above. Error bars represent the standard deviation of 3 independent experiments.
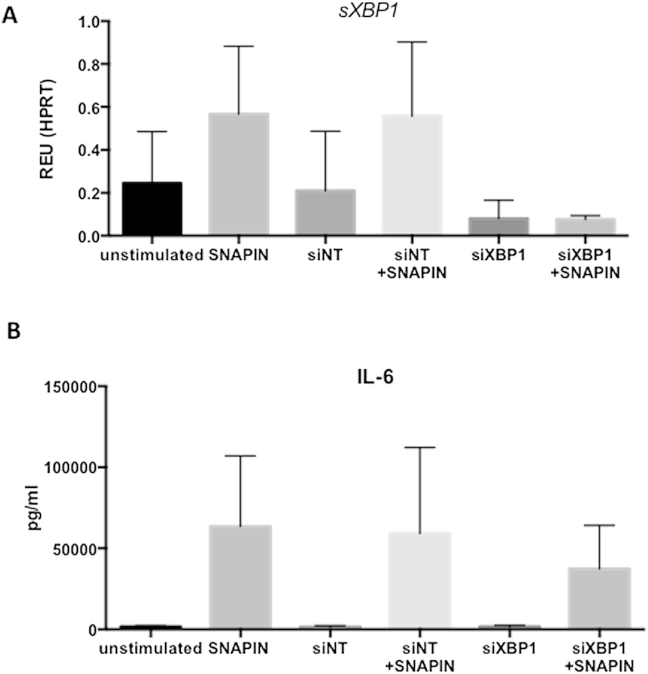
3.4. SNAPIN-induced cytokine production is dependent on sXBP1
Martinon et al. have demonstrated that TLR activation of XBP1 is required for the production of inflammatory mediators, such as IL-6 and TNF, by macrophages. Both LPS and SNAPIN induced IL-6 and TNF secretion by RA SF (Fig. 4B and C). This correlation of cytokine secretion and sXBP1 expression lead us to investigate if SNAPIN-induced cytokine production in SF is dependent on sXBP1 expression. We used siRNA to selectively inhibit expression of sXBP1 prior to stimulation with SNAPIN (Fig. 5A). Furthermore, SNAPIN-induced IL-6 production was inhibited in the absence of sXBP1 (Fig. 5B). That this was a non-canonical response was further revealed by the fact that thapsigargin (Tg), a potent activator of the UPR, failed to induce IL-6 despite generating significant amounts of sXBP1 (Fig. 4B). However, this situation was different in the case of TNF, which was produced by RA SF following stimulation with Tg (Fig. 4C).
Fig. 5.
sXBP1 knock down and IL-6 expression following stimulation with SNAPIN. (A) qRT-PCR determining sXBP1 levels in RA SF after siRNA transfection, targeting either XBP1 (siXBP1) or a non-targeting control (siNT) for 48 h in total. Cells were treated with SNAPIN (5 μg/ml) for 24 h. (B) IL-6 levels in culture supernatants measured by ELISA after XBP1 knock down and SNAPIN treatment.
3.5. Activation of RA SF is sustained by an autocrine loop downstream of XBP1 activation by pro-inflammatory cytokines
Pro-inflammatory conditions may themselves contribute to XBP1 activation. To replicate the pro-inflammatory conditions of the RA joint and to determine their effect on XBP1 mRNA splicing, we incubated RA synoviocytes with a combination of different pro-inflammatory cytokines known to have a role in the pathogenesis of RA. sXBP1 mRNA was generated following incubation of the SF with 10 ng/ml TNF for 24 h, but not after similar stimulation with IL-6 or IL-1β. However, the combination of TNF, IL-6 and IL-1β induced stronger activation of sXBP1 then TNF alone (Fig. 6A). Furthermore, immunofluorescence microscopy of TNF stimulated SF demonstrated the localisation of sXBP1 to the nucleus, similar to the localisation observed after Tg treatment (Fig. 6C). Thus RA SF can respond to TNF secretion by inducing sXBP1, and sXBP1, in turn, is linked to inducible cytokine secretion.
Fig. 6.
sXBP1 expression in response to pro-inflammatory cytokines (A) sXBP1 mRNA levels in RA-SF determined by qRT-PCR following treatment with LPS (10 ng/ml) for 6 h, TNF, IL-1β, IL-6 (all used at concentration of 10 ng/ml) for 24 h (B) sXBP1 expression and subcellular localisation was determined by IF after SF were treated with Tg (1 μM for 3 h) as a positive control and TNF (10 ng/ml for 48 h). Blue (DAPI) = nucleus; green (FITC) = SYVN1; red (TRITC) = sXBP1. Images taken at ×640 magnification.
4. Discussion
In this study we have demonstrated that XBP1 activation is implicated in RA pathogenesis and that this is uncoupled from a canonical UPR signature in this disease. Furthermore, XBP1 splicing is induced in response to TLR activation and is involved in TLR-dependent cytokine secretion. Finally, the same cytokines induced in response to TLR2 and TLR4 activation, can themselves induce splicing of XBP1 in synoviocytes. As a result of these interactions there is a potential autocrine loop, linking specific TLRs to sXBP1 formation, cytokine secretion and sustained synthesis of sXBP1 mRNA. During preparation of this manuscript Qui et al. published findings, which also show a critical role for IRE1 in the pathogenesis of a murine serum transfer model of RA [6]. Similar to our findings they also demonstrated overexpression of sXBP1 in RA patients compared to OA, but the majority of their results, which showed a critical role for TLR-dependent IRE1 activation in the production of variety of inflammatory mediators, were based on an animal model. Our study not only further supports this proinflammatory role for IRE1-sXBP1 signalling axis, but it also demonstrates the direct relevance of this biological process in patients with RA, and that this pathway can be altered by therapies such as anti-TNF biologics.
Dysregulation of the UPR has previously been implicated in the pathogenesis of RA. This notion was first proposed after SYNV1 was found to be highly expressed in RA synovium and was associated with increased survival of RA SF [2]. More recently, glucose-regulated protein-78 (GRP78), (also known as Bip and HSPA5), has been identified as a marker of ER stress and UPR dysregulation in RA [14]. The expression of GRP78 was induced by TNF and its expression was associated with SF proliferation and angiogenesis, two key pathological features of RA. However, GRP78 has also been implicated in resolution of inflammation and has been proposed to have an immunoregulatory role [15]. For this reason it has been included in a group of endogenous proteins collectively named the resolution-associated molecular pattern (RAMP) family of molecules. Clinical trials delivering GRP78 to RA patients are currently in progress and preliminary data from phase I and II studies have shown some efficacy [15].
The full extent of UPR involvement in RA pathogenesis remains to be established. Indeed it is likely that proteins such as XBP1 might have more then one role in this process. Traditionally initiation of the UPR was thought to involve activation of three pathways: protein kinase-like endoplasmic reticulum kinase (PERK), ATF6 and IRE1α, which leads to splicing of XBP1. However under certain circumstances, IRE1 activation and generation of sXBP1 can also occur independently of the other two pathways. For example, LPS-mediated engagement of TLR4leads to inhibition of CHOP expression and translational activation of ATF4 (regulated by PERK) [16]. Subsequently it was shown that that XBP1 splicing can occur in response to TLR2 or TLR4 ligation via IRE1 phosphorylation, and this occurs in the absence of PERK or ATF6 activation [5]. More recently we demonstrated involvement of (s)XBP1 and reactive oxygen species (ROS) in the pathogenesis of tumour necrosis factor receptor-associated periodic syndrome, a rare inherited autoinflammatory syndrome [17]. This condition is also characterized by hyper-responsiveness to the LPS, suggesting a role for sXBP1/TLR signalling axis.
This might explain why we detected a discrepancy between the expression of sXBP1 and other UPR genes. The downregulation of classical UPR associated genes in the context of sXBP1 expression, shown here in PBMCs from active RA, supports the concept of selective pathway and target gene activation under inflammatory conditions. Considering the disease context this could reflect either the effects of TNF or other pro-inflammatory cytokines, or, an as yet to be identified danger associated molecular pattern (DAMP) molecule(s) released as a result of tissue damage that selectively engage(s) the IRE1 pathway.
We decided to investigate this latter possibility, since TLR activation has been reported to play an important role in the pathogenesis of RA. We have demonstrated TLR4 and TLR2 activation of sXBP1 in RA SF that resulted in production of pro-inflammatory cytokines, IL-6 and TNF, which are relevant to the pathogenesis of RA. We then demonstrated that the TLR2/4 ligand, SNAPIN, can induce IL-6 and TNF secretion by SF that is dependent on sXBP1 expression. Lastly we showed that TNF itself causes splicing of XBP1 in SF. The splicing of XBP1 in response to TNF stimulation was also recently demonstrated by another group which investigated the role of TNF in protein degradation pathways in RA SF [18]. Although we have demonstrated a role for SNAPIN in TLR signalling in SF, it remains unclear if this same ligand, under physiological conditions, is likely to cause TLR activation and therefore XBP1 splicing in PBMC. The most probable targets here would be macrophages and a possible source of SNAPIN could be platelet-derived microparticles, which have been demonstrated in both the synovial fluid and systemic circulation of patients with RA [19].
Although our study showed a discrepancy between expression of sXBP1 and of other UPR genes, this does not necessarily contradict the findings of previous studies published on this theme. Abnormalities of UPR were still suggested in our experiments by the fact that almost all the genes were downregulated in the active RA group and their expression recovered in remission patients. This pattern of UPR gene expression may result from metabolic adaptations occurring in cells following a period of chronic ER stress, which ultimately might be damaging to the cell due to the associated global reduction in protein synthesis, and the risk of activating the PERK axis and CHOP-mediated death pathways. To eliminate this possibility, cells have evolved negative-feedback mechanisms to control the magnitude of the UPR response. PERK, for example, is negatively regulated by p58IPF, which is a transcriptional target of both sXBP1 and ATF6 [20,21]. In addition, the negative effects of PERK activation on global protein synthesis are reduced by GADD34, which dephosphorylates eIF2α to re-initiate protein synthesis [22].
5. Conclusions
sXBP1 plays a central role in two different cellular processes which may first appear unconnected. However the coupling of TLR signalling with XBP1 activation may have evolved to protect the inflammatory TLR-expressing cells, which, as a result of host engagement against invading pathogens, may be exposed to prolonged levels of ER stress [23]. Physiologically, the linking of pro-inflammatory pathways with activation of IRE1 provides these cells with a timely, coordinated and protective response. XBP1 activation may therefore be a suitable target in the treatment of RA, since it forms a cornerstone of two different molecular processes implicated in the pathology of RA, namely hyper-ERAD and TLR-mediated inflammatory response.
Conflict of interest disclosure
The authors declare that they have no conflicts of interest.
Acknowledgements
The work was funded by grant from Arthritis Research UK (grant number 19269), the Leeds Teaching Hospitals Special Trustees (9/R01/2002), the European Union who funded Masterswitch, an FP7-integrated project (grant number 223404), and the NIHR-Leeds Musculoskeletal Biomedical Research Unit.
Footnotes
This is an open access article under the CC BY-NC-SA license (http://creativecommons.org/licenses/by-nc-sa/3.0/).
References
- 1.El-Gabalawy H. The preclinical stages of RA: lessons from human studies and animal models. Best Pract Res Clin Rheumatol. 2009;23:49–58. doi: 10.1016/j.berh.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 2003;17:2436–2449. doi: 10.1101/gad.1096603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamasaki S., Yagishita N., Tsuchimochi K., Nishioka K., Nakajima T. Rheumatoid arthritis as a hyper-endoplasmic-reticulum-associated degradation disease. Arthritis Res Ther. 2005;7:181–186. doi: 10.1186/ar1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yagishita N., Yamasaki S., Nishioka K., Nakajima T. Synoviolin, protein folding and the maintenance of joint homeostasis. Nat Clin Pract Rheumatol. 2008;4:91–97. doi: 10.1038/ncprheum0699. [DOI] [PubMed] [Google Scholar]
- 5.Martinon F., Chen X., Lee A.H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu Q., Zheng Z., Chang L., Zhao Y.S., Tan C., Dandekar A. Toll-like receptor-mediated IRE1alpha activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32:2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radstake T.R., Roelofs M.F., Jenniskens Y.M., Oppers-Walgreen B., van Riel P.L., Barrera P. Expression of toll-like receptors 2 and 4 in rheumatoid synovial tissue and regulation by proinflammatory cytokines interleukin-12 and interleukin-18 via interferon-gamma. Arthritis Rheum. 2004;50:3856–3865. doi: 10.1002/art.20678. [DOI] [PubMed] [Google Scholar]
- 8.Saber T., Veale D.J., Balogh E., McCormick J., NicAnUltaigh S., Connolly M. Toll-like receptor 2 induced angiogenesis and invasion is mediated through the Tie2 signalling pathway in rheumatoid arthritis. PloS One. 2011;6:e23540. doi: 10.1371/journal.pone.0023540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor K.R., Trowbridge J.M., Rudisill J.A., Termeer C.C., Simon J.C., Gallo R.L. Hyaluronan fragments stimulate endothelial recognition of injury through TLR4. J Biol Chem. 2004;279:17079–17084. doi: 10.1074/jbc.M310859200. [DOI] [PubMed] [Google Scholar]
- 10.Sokolove J., Zhao X., Chandra P.E., Robinson W.H. Immune complexes containing citrullinated fibrinogen costimulate macrophages via Toll-like receptor 4 and Fc gamma receptor. Arthritis Rheum. 2011;63:53–62. doi: 10.1002/art.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavel C., Nogueira L., Laurent L., Iobagiu C., Vincent C., Sebbag M. Induction of macrophage secretion of tumor necrosis factor alpha through Fc gamma receptor IIa engagement by rheumatoid arthritis-specific autoantibodies to citrullinated proteins complexed with fibrinogen. Arthritis Rheum. 2008;58:678–688. doi: 10.1002/art.23284. [DOI] [PubMed] [Google Scholar]
- 12.Shi B., Huang Q., Tak P.P., Vervoordeldonk M.J., Huang C.C., Dorfleutner A. SNAPIN: an endogenous toll-like receptor ligand in rheumatoid arthritis. Ann Rheum Dis. 2012;71:1411–1417. doi: 10.1136/annrheumdis-2011-200899. [DOI] [PubMed] [Google Scholar]
- 13.Maestre L., Tooze R., Canamero M., Montes-Moreno S., Ramos R., Doody G. Expression pattern of XBP1(S) in human B-cell lymphomas. Haematologica. 2009;94:419–422. doi: 10.3324/haematol.2008.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo S.A., You S., Yoon H.J., Kim D.H., Kim H.S., Lee K. A novel pathogenic role of the ER chaperone GRP78/BiP in rheumatoid arthritis. J Exp Med. 2012;209:871–886. doi: 10.1084/jem.20111783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shields A.M., Thompson S.J., Panayi G.S., Corrigall V.M. Pro-resolution immunological networks: binding immunoglobulin protein and other resolution-associated molecular patterns. Rheumatology (Oxford) 2012;51:780–788. doi: 10.1093/rheumatology/ker412. [DOI] [PubMed] [Google Scholar]
- 16.Woo C.W., Cui D., Arellano J., Dorweiler B., Harding H., Fitzgerald K.A. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol. 2009;11:1473–1480. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickie L.J., Aziz A.M., Savic S., Lucherini O.M., Cantarini L., Geiler J. Involvement of X-box binding protein 1 and reactive oxygen species pathways in the pathogenesis of tumour necrosis factor receptor-associated periodic syndrome. Ann Rheum Dis. 2012;71:2035–2043. doi: 10.1136/annrheumdis-2011-201197. [DOI] [PubMed] [Google Scholar]
- 18.Connor A.M., Mahomed N., Gandhi R., Keystone E.C., Berger S.A. TNFalpha modulates protein degradation pathways in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2012;14:R62. doi: 10.1186/ar3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boilard E., Nigrovic P.A., Larabee K., Watts G.F., Coblyn J.S., Weinblatt M.E. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science. 2010;327:580–583. doi: 10.1126/science.1181928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan W., Frank C.L., Korth M.J., Sopher B.L., Novoa I., Ron D. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Huizen R., Martindale J.L., Gorospe M., Holbrook N.J. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J Biol Chem. 2003;278:15558–15564. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- 22.Novoa I., Zeng H., Harding H.P., Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinon F., Glimcher L.H. Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr Opin Immunol. 2011;23:35–40. doi: 10.1016/j.coi.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]