Abstract
A correct three-dimensional structure is crucial for the physiological functions of a protein, yet the folding of proteins to acquire native conformation is a fundamentally error-prone process. Eukaryotic organisms have evolved a highly conserved endoplasmic reticulum-mediated protein quality control (ERQC) mechanism to monitor folding processes of secretory and membrane proteins, allowing export of only correctly folded proteins to their physiological destinations, retaining incompletely/mis-folded ones in the ER for additional folding attempts, marking and removing terminally misfolded ones via a unique multiple-step degradation process known as ER-associated degradation (ERAD). Most of our current knowledge on ERQC and ERAD came from genetic and biochemical investigations in yeast and mammalian cells. Recent studies in the reference plant Arabidopsis thaliana uncovered homologous components and similar mechanisms in plants for monitoring protein folding and for retaining, repairing, and removing misfolded proteins. These studies also revealed critical roles of the plant ERQC/ERAD systems in regulating important biochemical/physiological processes, such as abiotic stress tolerance and plant defense. In this review, we discuss our current understanding about the molecular components and biochemical mechanisms of the plant ERQC/ERAD system in comparison to yeast and mammalian systems.
Keywords: endoplsamic reticulum-associated degradation, Arabidopsis, endoplasmic reticulum-mediated quality control, misfolded glycoproteins, receptor-like kinases
INTRODUCTION
It is well known that the proper function of a protein strictly depends on its native conformation, but protein folding is a fundamentally error-prone process. The endoplasmic reticulum (ER) is the cellular port of entry for secretory and membrane proteins to enter the secretory pathway and is a folding compartment for proteins to attain their native conformations through interactions with molecular chaperones, sugar-binding lectins, and folding enzymes (Gidalevitz et al., 2013). Misfolded proteins not only lead to functional deficiency but also induce dominant-negative and cellular toxicity effects, and it is thus essential that the ER should possess several highly stringent protein quality control mechanisms to closely monitor the folding process, allowing export of only correctly folded proteins to their final destinations but retaining incompletely/mis-folded proteins for additional rounds of chaperone-assisted folding. A high-efficient ER-mediated protein quality control (ERQC) system can also differentiate terminally misfolded proteins from folding intermediates and/or reparable misfolded proteins, stopping the futile folding cycles of the former proteins and eliminating them via a multistep degradation process widely known as ER-associated degradation (ERAD) that involves ubiquitination, retrotranslocation, and cytosolic proteasome (Smith et al., 2011). Our current understanding of the eukaryotic ERQC/ERAD system derived largely from studies in yeast and mammalian cells. However, recent genetic, biochemical, and cell biological studies in the reference plant Arabidopsis thaliana and other model plant species not only identified homologous ERQC/ERAD components but also revealed evolutionarily conserved features as well as unique aspects of the plant ERQC/ERAD mechanisms (Hong and Li, 2012; Huttner and Strasser, 2012; Howell, 2013), especially their connections with the stress tolerance and plant defense pathways.
N-GLYCAN-BASED ER RETENTION MECHANISM
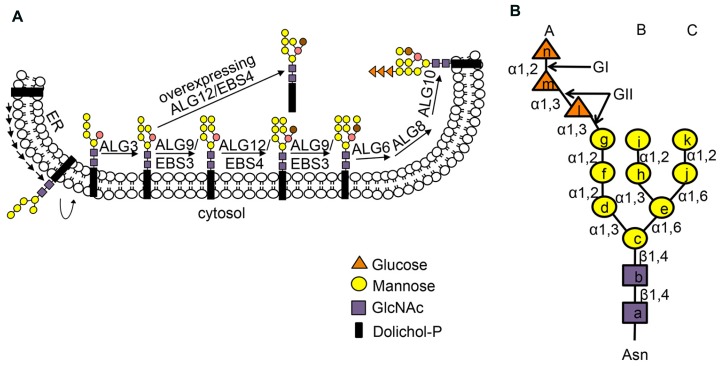
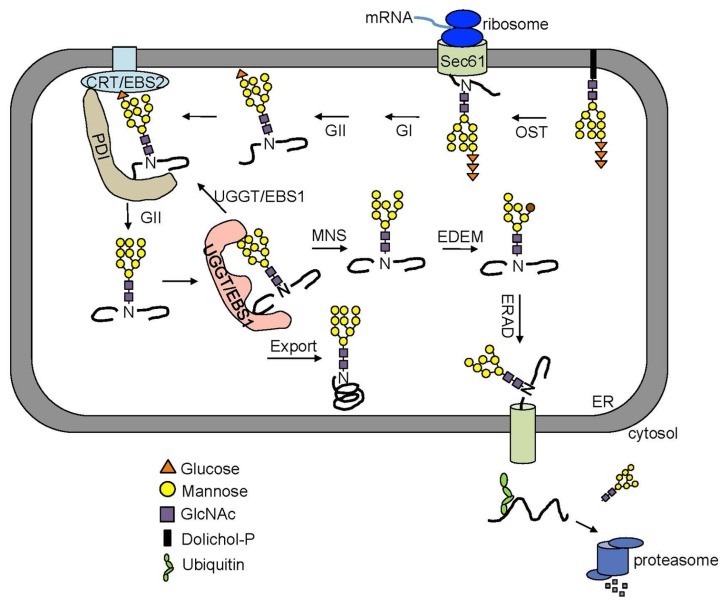
Many secretory and membrane proteins are co-translationally glycosylated when entering the ER (Aebi, 2013). The so-called N-linked glycosylation occurs on the asparagine (Asn or N) residues within the Asn-X-Ser/Thr sequons (X indicating any amino acid except proline while Ser/Thr denoting serine/threonine residue) of a nascent polypeptide. This reaction is catalyzed by the enzyme oligosaccharyltransferase (OST), an integral membrane protein complex that transfers a preassembled oligosaccharide precursor Glc3Man9GlcNac2 (Glc, Man and GlcNac denoting glucose, mannose and N-acetylglucosamine, respectively) from a membrane-anchored dolichylpyrophosphate (DolPP) carrier to the Asn residue (Figure 1; Mohorko et al., 2011). The assembly of Glc3Man9GlcNac2 involves a series of highly specific asparagine-linked glycosylation (ALG) proteins that sequentially add a monosaccharide onto the DolPP linker or a DolPP-linked oligosaccharide precursor (Aebi, 2013; Figure 1A). The structure of a N-linked glycan plays an important role in the protein folding and quality control (Aebi et al., 2010). Immediately after transferring of Glc3Man9GlcNac2 to an Asn residue, the terminal and middle Glc residues are removed sequentially by glucosidase I (GI) and glucosidase II (GII), producing a monoglucosylated N-glycan, GlcMan9GlcNac2, which is recognized by the ER chaperone-like lectins, a membrane-anchored calnexin (CNX) and its ER luminal homolog calreticulin (CRT; Caramelo and Parodi, 2008; Figure 2). The high-specificity high-affinity binding between GlcMan9GlcNac2 and CNX/CRT is crucial for folding a nascent polypeptide as CNX/CRT can recruit other ER-chaperones and folding enzymes, including protein disulfide isomerases (PDIs) essential for generating inter/intra-molecular disulfide bonds. The removal of the remaining Glc residue by GII releases the nascent glycoprotein from CNX/CRT, thus terminating its folding process (Caramelo and Parodi, 2008). If the protein folds correctly, it will be transported out of the ER to reach its final destination. However, if the protein fails to attain its native conformation, it will be recognized by UDP-glucose:glycoprotein glucosyltransferase (UGGT), an ER-resident folding sensor consisting of a large non-conserved N-terminal domain presumably involved in recognizing non-native conformations and a smaller highly conserved C-terminal catalytic domain capable of catalyzing a glucosyltransferase reaction using uridyl diphosphate-glucose (UDP-Glc) as a substrate (D’Alessio et al., 2010). As a result, a single Glc is added back to deglucosylated N-glycans of the incompletely/mis-folded protein, permitting its reassociation with CNX/CRT and their associated proteins for another round of assisted folding. The alternate reactions of GII and UGGT drive many cycles of dissociation and reassociation of CNX/CRT with an incompletely/mis-folded glycoprotein [widely known as the CNX/CRT cycle (Hammond et al., 1994)], till the protein attains its native conformation (Figure 2). It is worthy to mention that the budding yeast (Saccharomyces cerevisiae), which is widely used for studying the ERAD process, lacks the CNX/CRT-UGGT system due to the presence of a catalytically inactive UGGT homolog (Meaden et al., 1990).
FIGURE 1.
(A) Stepwise assembly of N-glycan precursor on the ER membrane. The assembly of the N-glycan precursor starts at the cytosolic face of the ER membrane by adding two GlcNAc and five Man residues to the membrane-anchored Dol-PP linker. The resulting Dol-PP-Man5GlcNAc2 flips over into the ER lumen. Four Man residues are sequentially added to the flipped Dol-PP-Man5GlcNAc2 by three mannosyltransferases, ALG3, ALG9 (known as EBS3 in Arabidopsis) and ALG12 (known as EBS4 in Arabidopsis) with the ALG9 catalyzing two reactions of adding the terminal α1,2 Man residues on the middle and right branches. Three Glc residues are subsequently added to the right branch, generating the 14-sugar precursor, Dol-PP-Glc3Man9GlcNAc2. Two α1,6 Man residues are marked by brown and salmon color. The Dol-PP linker and different sugar residues are indicated. (B) The structure of N-linked Glc3Man9GlcNac2 glycan with three dimannose branches (branch A, B and C). Lower case letters inside sugar residues represent the order of sugar addition. The sugar linkage bonds and enzymes (GI, GII) that remove the three Glc residues are indicated. Figure adapted from Hong et al. (2009).
FIGURE 2.
An overview of the ERQC/ERAD system. Two Glc residues on the N-glycan of a nascent polypeptide are rapidly trimmed by GI and GII right after being transferred from the Dol-PP-linker. The resulting monoglucosylated N-glycans bind the two ER lectins CNX and CRT chaperone-assisted folding. The removal of this last Glc by GII releases a mature polypeptide from CNX/CRT. A correctly folded protein can leave the ER while an incompletely/mis-folded glycoprotein is recognized by UGGT (known as EBS1 in Arabidopsis) that adds back a Glc residue to the A branch, permitting its reassociation with CNX/CRT. A glycoprotein that fails to gain its native structure within a certain time window is removed from the folding cycle via sequential trimming of the two terminal α1,2 Man residues of the B and C branch by MNS1 (an ER-localized α1,2-Mannosidase, known as MNS3 in Arabidopsis) and Htm1/EDEM. A terminally misfolded glycoprotein with α1,6 Man-exposed glycan is selected to enter the ERAD pathway.
The Arabidopsis genome encodes only one UGGT homolog, and its physiological function was inadvertently found in a study for identifying additional signaling proteins of the plant steroid hormones, brassinosteroids (BR; Jin et al., 2007). A genetic screening for extragenic suppressors of an Arabidopsis dwarf mutant brassinosteroid-insensitive 1-9 (bri1-9) led to the discovery of Arabidopsis UGGT (also known as EBS1 for EMS mutagenized bri1 suppressor 1; Jin et al., 2007). BRI1 is a cell surface-localized leucine-rich-repeat receptor-like-kinase that function as a BR receptor and contains a single transmembrane domain and 14 putative N-glycosylation sites in its N-terminal extracellular domain (Li and Chory, 1997). The mutant bri1-9, carrying a Ser662-Phe mutation in the BR-binding domain (Noguchi et al., 1999), was found to be retained in the ER by an EBS1/AtUGGT-dependent mechanism and subsequently degraded by a plant ERAD process (Jin et al., 2007; Hong et al., 2009). Loss-of-function mutations in EBS1/AtUGGT compromise such an ER-retention mechanism and allow some bri1-9 proteins to escape from the ER to reach the plasma membrane, resulting in phenotypic suppression of the dwarfism of the bri1-9 mutant. The same genetic screen also identified CRT3 (Jin et al., 2009), a unique member of the Arabidopsis CNX/CRT family consisting of two CNXs and three CRTs, which actually retains bri1-9 via the CRT3-GlcMan9GlcNac2 binding. Both UGGT and CRT3 were also identified from two other independent genetic screens aiming to identify key regulators of the plant innate immune response to a bacterial translational elongation factor EF-Tu (Li et al., 2009; Saijo et al., 2009). Interestingly, while loss-of-function mutations in AtUGGT/CRT3 led to regaining partial sensitivity to BRs, atuggt/crt3 mutants were insensitive to elf18, a biologically active epitope of EF-Tu. Further studies showed that both UGGT and CRT3 are absolutely required for the correct folding of EFR (EF-Tu Receptor; Saijo, 2010), a BRI1-like receptor-like kinase that binds elf18/EF-Tu to initiate a plant defense process (Zipfel et al., 2006). The importance of N-glycan-mediated folding control was further supported by discoveries that loss-of-function mutations in STT3A, a OST subunit, and GII resulted in significant reduction of the EFR protein abundance, presumably caused by incomplete folding and subsequent degradation (Lu et al., 2009; Haweker et al., 2010; von Numers et al., 2010).
In addition to the glycan-dependent ER retention system, the ER is equipped with additional retention systems to prevent export of misfolded proteins, especially those non-glycosylated ones. One system uses the family of ER-localized HSP70 proteins (known as BiPs), which have a N-terminal ATP-binding domain and a C-terminal substrate-binding domain that recognizes and binds to exposed hydrophobic patches of incompletely/mis-folded proteins in an ATP-dependent manner (Buck et al., 2007). The Arabidopsis has three BiP homologs, AtBiP1, AtBiP2 and AtBiP3, all of which were known to exhibit higher levels of gene expression under ER stresses (Sung et al., 2001). In Arabidopsis, BiPs were shown to bind both bri1-9 and bri1-5, another mutant variant of BRI1 carrying a Cys69Tyr mutation that destroys a disulfide bridge crucial for the structural integrity of the BR receptor, and were thought to contribute for the ER retention of the two mutant BR receptors (Jin et al., 2007; Hong et al., 2008). BiPs and their associated factors ERdj3B (an Arabidopsis ER-localized DNAJ homolog) and SDF2 (the Arabidopsis homolog of the murine stromal cell-derived factor 2) are also involved in the biogenesis/folding control of EFR (Nekrasov et al., 2009). BiPs were also known to interact with the orphan heavy chain of a murine IgG1 antibody or an assembly defective form of the trimeric vacuolar storage protein phaseolin in transgenic tobacco plants (Pedrazzini et al., 1997; Nuttall et al., 2002). Another glycan-independent ER retention mechanism relies on mixed disulfide bridges between incompletely/mis-folded proteins with PDIs and related ER-localized oxidoreductases (Reddy et al., 1996; Anelli et al., 2003, 2007). The Arabidopsis genome encodes 13 PDI-like proteins (Houston et al., 2005), none of which has been implicated in retaining misfolded proteins. However, a recent study on bri1-5 carrying an orphan cysteine residue (Cys62) suggested involvement of a thiol-mediated retention system in keeping the mutant BR receptor in the ER (Hong et al., 2008). Further biochemical studies are needed to verify this prediction and to identify one or more PDIs that form the predicted mixed disulfide bridge with the orphan Cys62 residue.
MARKING OF A TERMINALLY MISFOLDED GLYCOPROTEIN FOR ERAD
A protein that fails to attain its native conformation within a given time window is eliminated by ERAD (Vembar and Brodsky, 2008). One of the key events in ERQC is to terminate a futile folding cycle and to deliver an irreparable misfolded protein into the ERAD pathway. Although little is known about the marking mechanism for irreparable non-glycosylated ERAD clients, recent studies indicated that removal of the terminal α1,2-Man residue from the C-branch of N-glycan (Figure 2), catalyzed by homologous to mannosidase 1 (Htm1) in yeast and ER-degradation enhancing α-mannosidase-like proteins (EDEMs) in mammals, is required for generating the ERAD signal, an exposed α1,6 Man residue on an N-linked glycan (Quan et al., 2008; Clerc et al., 2009; Figure 1B). The Arabidopsis genome encodes at least two homologs of the yeast Htm1/mammalian EDEMs [known as MNS4 and MNS5 (Liebminger et al., 2009)], but their involvement in a plant ERAD process awaits functional investigation. Nevertheless, recent genetic screening for Arabidopsis mutants defective in ERAD of bri1-5/bri1-9 and subsequent molecular cloning and biochemical studies indicated that the glycan ERAD signal is well conserved in plants (Hong et al., 2009, 2012). Loss-of-function mutations in either EBS3 and EBS4 (homologs of the yeast/mammal ALG9 and ALG12, respectively) prevent complete assembly of the N-glycan precursor (Figure 1), resulting in glycosylation of the two ER-retained mutant BR receptor with truncated N-glycan lacking the α1,6-Man residue that would function as the ERAD signal and consequential inhibition of ERAD of bri1-5/bri1-9. In contrast, forcing the addition of the missing α1,6 Man residue to Dol-PP-Man6GlcNAc2 by overexpression of EBS4/ALG12 in an Arabidopsis ebs3/alg9 bri1-9 mutant promoted the ERAD of bri1-9 (Figure 1A; Hong et al., 2012). Similarly, the ERAD of bri1-9 was presumably accelerated when its N-linked glycans carried a different exposed α1,6 Man residue (the inner α1,6 Man; Hong et al., 2012) caused by a loss-of-function mutation in ALG3 that adds an α1,3 Man to the inner α1,6-Man (Henquet et al., 2008; Kajiura et al., 2010; Figure 1A). The exposed inner α1,6 Man residue was shown to function as an alternative ERAD signal in both yeast and mammalian cells (Clerc et al., 2009; Hosokawa et al., 2009).
RECRUITMENT OF ERAD SUBSTRATES
The N-glycan ERAD signal is decoded by one or two ER luminal lectins, osteosarcoma 9 (OS9, also known as Yos9 in yeast) and XTP3-B (Yoshida and Tanaka, 2010; Figure 3). Yos9 and its mammalian homologs contain the mannose-6-phosphate receptor homology (MRH) domain that specifically recognizes and binds N-glycans with an exposed α1,6 Man residue (Hosokawa et al., 2010). In addition to OS-9/Yos9, selection of an ERAD client requires another ER resident protein, Hrd3 (HMG-CoA reductase degradation 3) in yeast and Sel1L (Suppressor of lin-12-Like) in mammals (Hirsch et al., 2009), a type I transmembrane protein with a large ER luminal domain consisting of multiple copies of the tetratricopeptide repeat motif. It was believed that Hrd3/Sel1L, exhibiting high affinity binding to exposed hydrophobic amino acid residues on misfolded proteins, makes the initial selection of a potential ERAD client, which is subsequently inspected by OS-9/Yos9 for the presence of an N-glycan ERAD signal (Denic et al., 2006; Gauss et al., 2006; Figure 3). Such a bipartite ERAD signal of a misfolded domain plus an α1,6-Man-exposed N-glycan ensures degradation of only terminally misfolded glycoproteins but not folding intermediates carrying N-glycans with no exposed α1,6 Man residue.
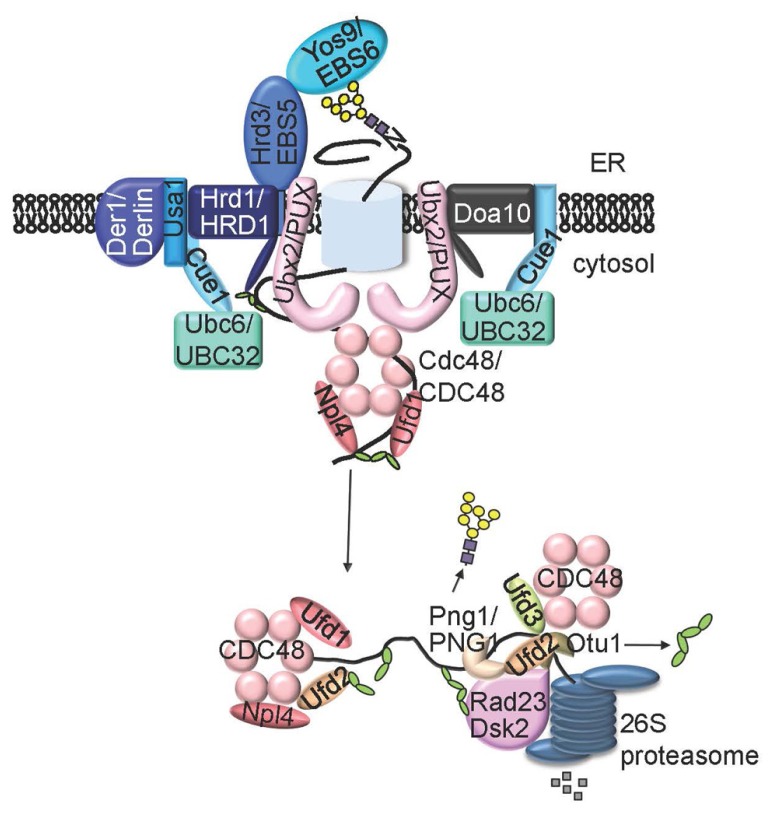
FIGURE 3.
A model of ERAD system. An ERAD client that has lesion in membrane-embedded segment (ERADM) or in ER lumen region (ERADL) is recruit by Hrd3 (EBS5 in Arabidopsis) and Yos9 (EBS6/OS9 in Arabidopsis) to the membrane-anchored E3 ligase Hrd1 (AtHrd1A and AtHrd1B in Arabidopsis) complex that also contains Cue1, Ubc6 (UBC32 in Arabidopsis), Usa1 and Der1. An ERAD substrate with a folding lesion in the cytosolic domain (ERADC is recruited to a different ER membrane-anchored E3 ligase complex that contains Doa10 and Ubc6 (or Cue1/Ubc7). The two E3 ligase complexes share similar components on the cytosolic side of the ER membrane, including the substrate-extracting Cdc48-Ufd1-Npl4 trimeric complex and its membrane recruitment factor Ubx2 (one or more AtPUX proteins in Arabidopsis). An extracted polyubiquitinated ERAD substrate is processed by Ufd2, Ufd3, Otu1, and/or Png1 (AtPNG1 in Arabidopsis), delivered to the cytosolic proteasome with help of Dsk2 and Rad23, and eventually proteolyzed by the 26S proteasome.
The Arabidopsis genome has two Hrd3/Sel1L homologous genes, AtSel1A (also known as EBS5 or HRD3A or) and AtSel1B (also known as HRD3B, an apparent pseudogene) and just one OS9/Yos9 homolog, AtOS9 (also known as EBS6; Liu et al., 2011; Su et al., 2011, 2012; Huttner et al., 2012). AtSel1A/EBS5 complemented the ERAD-defect of the yeast Δhrd3 mutant assayed by ERAD of a mutant variant of carboxypeptidase Y (CPY*; Su et al., 2011), a commonly used ERAD substrate for many ERAD studies in yeast. By contrast, AtOS9 failed to rescue the defective ERAD of CPY* when expressed in a Δ yos9 yeast strain (Huttner et al., 2012). Interestingly, a chimeric AtOS9-Yos9 protein consisting of the full-length AtOS9 and the Yos9’s C-terminal region (amino acids of 277–542) promoted CPY* degradation in Δ yos9 yeast cells (Huttner et al., 2012), suggesting that the MRH domain is interchangeable but the Yos9’s C-terminal domain might be crucial for interacting with other components of the yeast ERAD machinery. Loss-of-function mutations in either AtSel1A/EBS5 or AtOS9/EBS6 inhibit ERAD of bri1-5, bri1-9, misfolded EFR (in an ebs1/uggt mutant background), and/or the transgenically expressed MLO-1 (Liu et al., 2011; Su et al., 2011, 2012; Huttner et al., 2012), a mutant variant of barley powdery resistance o (MLO) that carries a single amino acid change in the cytoplasmic region and was previously shown to be an ERAD substrate (Muller et al., 2005). As expected, AtSel1A/EBS5 and AtOS9/EBS6 physically interacted with bri1-9 or bri1-5 in a tobacco transient expression system or an in vitro pull-down assay (Huttner et al., 2012; Su et al., 2012). Consistent with what was known in yeast and mammalian cells, AtSel1A/EBS5 binds AtOS9/EBS and seems to be required for maintaining the stability of AtOS9/EBS6 (Huttner et al., 2012; Su et al., 2012). These results strongly suggested that the selection mechanism for a terminally misfolded glycoprotein for ERAD is conserved in Arabidopsis. It is important to point out that Arabidopsis mutants of AtSel1A/EBS6 or AtOS9/EBS6 are hypersensitive to NaCl-induced salt stress, suggesting a relationship between a cellular stress response and an environmental stress pathway (Liu et al., 2011; Huttner et al., 2012). It is quite possible that environmental stresses lead to decreased folding efficiency and increased accumulation of misfolded proteins in the ER, which require a highly efficient ERAD system for their removal to maintain ER homeostasis.
UBIQUITINATION OF CHOSEN ERAD CLIENTS
Hrd3/Sel1L and Yos9/OS9 not only select irreparable misfolded glycoproteins but also bring the chosen ERAD substrates to the membrane-anchored ERAD complexes responsible for ubiquitination and retrotranslocation. The central component of these ERAD complexes is a polytopic membrane protein with a RING finger-type ubiquitin ligase (E3) activity exposed to the cytosolic surface of the ER membrane, which not only ubiquitinates ERAD substrates but also connects to various ER luminal/cytosolic adapters (Hirsch et al., 2009). Yeast contains at least two distinct E3 ligases, 6 transmembrane-spanning Hrd1 (HMG-CoA reductase degradation) and 14-transmembrane-spanning Doa10 (Degradation of alpha2), that ubiquitinate three different types of ERAD substrates differing in the location of folding lesions: ERADL (lesion in the ER luminal area), ERADM (lesion in the transmembrane segment), and ERADC (lesion in the cytosolic domain; Vashist and Ng, 2004; Carvalho et al., 2006). The Hrd1 complex ubiquitinates ERADL/M substrates while the Doa10 complex deals with ERADC clients. Mammals have at least 9 membrane-bound ERAD E3 ligases (Olzmann et al., 2013), including two Hrd1 homologs (HRD1 and gp78), one Doa10 homolog (TEB4), and several other RING-type E3 ligases such as RING membrane-anchor 1 (RMA1; Younger et al., 2006), whose founding member was initially discovered in Arabidopsis (Matsuda and Nakano, 1998).
The Arabidopsis genome encodes two Hrd1 homologs (AtHrd1A and AtHrd1B; Su et al., 2011; Huttner and Strasser, 2012), at least two Doa10 homologs (Doa10A/At4g34100 and Doa10B/At4g32670; Liu et al., 2011), and three homologs of RMA1, AtRMA1-AtRMA3 that were shown to be localized to the ER and exhibit in vitro E3 ubiquitin ligase activity (Son et al., 2009; Table 1), but it remains unclear if plants use distinct E3 ligases to removal different classes of ERAD substrates. Loss-of-function mutations in AtHrd1A or AtHrd1B had no detectable effect on bri1-5/bri1-9 degradation, but simultaneous elimination of the two Hrd1 homologs inhibited degradation of the two mutant BR receptors, indicating that AtHrd1A and AtHrd1B function redundantly in a plant ERAD pathway (Su et al., 2011). By contrast, the role of the two Doa10 homologs in the plant ERAD pathway remains unknown. Two recent genetic studies revealed important regulator roles of Doa10A (also known as SUD1 for SUPPRESSOR OF DRY2 DEFECTS1 or CER9 for ECERIFERUM9) in the cuticle lipid biosynthesis and in controlling the activity but not the protein level of an Arabidopsis HMG-CoA reductase (Lu et al., 2012; Doblas et al., 2013). Further studies are needed to determine if the Arabidopsis Doa10A is indeed involved in an ERAD pathway that regulates the protein abundance of key regulatory factors or metabolic enzymes involved in the cuticle lipid biosynthesis. Unlike yeast but similar to mammals, plants have additional membrane-anchored RING-type E3 ligases for ERAD. For example, the three Arabidopsis RMA1 homologs (Rma1H1) and a hot pepper (Capsicum annuum) Rma1H1 are involved in the degradation of a cell surface water channel to regulate its plasma membrane level (Lee et al., 2009). A recent study also suggested that a legume (Medicago truncatula) homolog of RMA1 seems to play a role in the regulation of biosynthesis of plant defense compounds, triterpene saponins that share the same biosynthetic precursors with sterols, through regulated degradation of HMG-CoA reductase (Pollier et al., 2013).
Table 1.
A list of known/predicted components of the Arabidopsis ERAD system.
| Yeast/human gene name | Arabidopsis name | Accession number | Reference |
|---|---|---|---|
| Hrd1/HRD1 | HRD1A | At3g16090 | Su et al. (2011) |
| HRD1B | At1g65040 | ||
| Hrd3/SEL1L | EBS5/HRD3A | At1g18260 | Su et al. (2011) |
| HRD3B | At1g73570 | ||
| Yos9/OS-9 | EBS6/OS9 | At5g35080 | Huttner et al. (2012), Su et al. (2012) |
| Der1/DERLIN | DER1 | At4g29330 | Kirst et al. (2005) |
| DER2.1 | At4g04860 | Wang et al. (2008), Kamauchi et al. (2005) | |
| DER2.2 | At4g21810 | ||
| Ubc6/UBE2J1 | UBC32 | At3g17000 | Kraft et al. (2005), Cui et al. (2012) |
| UBC33 | At5g50430 | ||
| UBC34 | At1g17280 | ||
| Htm1/EDEM | MNS4 | At5g43710 | Liebminger et al. (2009) |
| MNS5 | At1g27520 | ||
| Doa10/TEB4 | SUD1/CER9/DOA10A | At4g34100 | Liu et al. (2011), Doblas et al. (2013), Lu et al. (2012) |
| DOA10B | At4g32670 | ||
| RMA1 | RMA1 | At4G03510 | Matsuda and Nakano (1998), Son et al. (2009) |
| RMA2 | At4g28270 | ||
| RMA3 | At4g27470 | ||
| Ubx2/ERASIN | PUX1 | At3g27310 | Park et al. (2007), Rancour et al. (2004) |
| PUX2 | At2g01650 | Suzuki et al. (2001), Ueda et al. (2000) | |
| PUX3 | At4g22150 | ||
| PUX4 | At4g04210 | ||
| PUX5 | At4g15410 | ||
| PUX6 | At3g21660 | ||
| PUX7 | At1g14570 | ||
| PUX8/SAY1 | At4g11740 | ||
| PUX9 | At4g00752 | ||
| PUX10 | At4g10790 | ||
| PUX11 | At2g43210 | ||
| PUX12 | At3g23605 | ||
| PUX13 | At4g23040 | ||
| PUX14 | At4g14250 | ||
| PUX15 | At1g59550 | ||
| Cdc48/p97 | CDC48A | At3g09840 | Rancour et al. (2002) |
| CDC48B | At2g03670 | ||
| CDC48C | At3g01610 | ||
| Npl4/NPL4 | At2g47970 | ||
| At3g63000 | |||
| Ufd1/UFD1 | UFD1 | At2g21270 | Galvao et al. (2008) |
| At4g38930 | |||
| At2g29070 | |||
| At4g15420 | |||
| Ufd2/UFD2 | At5g15400 | Bachmair et al. (2001) | |
| Png1/PNG1 | PNG1 | At5g49570 | Diepold et al. (2007) |
| Rad23/RAD23 | RAD23A | At1g16190 | Farmer et al. (2010) |
| RAD23B | At1g79650 | ||
| RAD23C | At3g02540 | ||
| RAD23D | At5g38470 |
In a typical ubiquitination reaction, ubiquitin is attached to a substrate through a three-step process consisting of activation, conjugation, and ligation catalyzed by an ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and E3 (Pickart, 2004). In yeast, the Hrd1/Doa10 E3 ligases work together with a membrane-anchored E2 (Ubc6) and two cytosolic E2s (Ubc1 and Ubc7) that are recruited to the ER membrane by an ER anchor protein Cue1 (Hirsch et al., 2009), which also activates both E2 and E3 (Bagola et al., 2013; Metzger et al., 2013). Arabidopsis has a total of 37 E2 enzymes (Kraft et al., 2005), including one potential Ubc1 homolog (UBC27), three putative homologs of Ubc7 known to be the cognate E2 for Hrd1 (UBC7 UBC13, and UBC14), and three likely homologs of Ubc6 associated mainly with Doa10 (UBC32, UBC33, and UBC34 each having a predicted transmembrane domain at their C-termini; Liu et al., 2011), but our understanding of the roles of these potential ERAD-participating E2s in the plant ERAD process is extremely limited. One of the Arabidopsis Ubc6-like E2 gene, UBC32, was recently found to be induced by salt, drought, and ER stress (Cui et al., 2012). Interestingly, the Arabidopsis ubc32 mutant seedlings are more tolerant whereas UBC32-overexpressing transgenic Arabidopsis lines are more sensitive to salt and ER stress (Cui et al., 2012). The observed salt tolerance of the ubc32 mutant is in contrast to the reduced salt tolerance of other known Arabidopsis ERAD mutants, including ebs5/hrd3a, ebs6/os9, and hrd1ahrd1b (Liu et al., 2011; Huttner and Strasser, 2012; Huttner et al., 2012). This discrepancy might be explained by different ERAD substrates being degraded by different E3 ligase complexes: ERADL/M substrates by AtHrd1A/AtHrd1B in association with cytosolic E2s and ERADC substrates by Doa10 using membrane-anchored E2s. Indeed, UBC32 was found to interact with Arabidopsis Doa10B and to stimulate the ubiquitination and degradation of a known ERAD substrate MLO-12, another variant of MLO carrying a single amino acid change in its cytosolic domain (Muller et al., 2005), in a tobacco leaf transient expression experiment (Cui et al., 2012). However, the tobacco result was quite different from the results obtained with the yeast MLO experiment showing that the ERAD of MLO-12 plus two other mutant MLOs (all carrying a cytosolic mutation) used the Ubc7-Hrd1 pathway but was unaffected by either ubc6 or doa10 deletion in yeast (Muller et al., 2005). Such inconsistency might be simply caused by heterologous expression of ERADC substrates in two different eukaryotic systems. Nevertheless, UBC32 was implicated in the Hrd1-mediated degradation of bri1-9 (a presumed ERADL substrate) as the ubc32 mutation partially inhibited the degradation of the mutant BR receptor and weakly suppressed the corresponding dwarf phenotype (Cui et al., 2012). The partial inhibition could be attributed to a redundant role of UBC32 with its two close homologs or the potential Arabidopsis homologs of Ubc1/Ubc7. However, blast searches failed to find a single homolog of the yeast Cue1 gene from published sequences of plant genomes and expressed sequence tags (our unpublished results), suggesting that plant ERAD processes might exclusively rely on ER-anchored membrane E2s. Alternatively, plants could recruit cytosolic E2s to the membrane-anchored E3 complexes via yet unknown recruiting factors that share no sequence homology but are functionally similar to Cue1.
The ubiquitination of ERAD substrates, especially those lacking N-glycan degradation signals, by the Hrd1 complex requires two additional adapters: U1-Snp1 associating-1 (Usa1; HERP in mammals), an ER membrane protein containing a ubiquitin-like (UBL) motif near it N-terminus and two predicted transmembrane domains in the middle, and Der1 (degradation in the ER; Derlins for Der1-like proteins in mammals), another integral ER membrane protein with four transmembrane segments (Kostova et al., 2007). Usa1 is thought to regulate the stability and/or oligomerization of Hrd1 and to recruit Der1 to the Hrd1 complex (Carvalho et al., 2006, 2010; Horn et al., 2009; Carroll and Hampton, 2010), while Der1 is believed to function either as a receptor for soluble non-glycosylated ERAD substrates or a potential retrotranslocation channel (Lilley and Ploegh, 2004; Ye et al., 2004; Kanehara et al., 2010). The Arabidopsis contains no homolog of Usa1/HERP1 but its genome encodes three Der1 homologs whose functional involvement in a plant ERAD pathway awaits detailed genetic and biochemical investigations (Kirst et al., 2005). An earlier study showed that at least two maize Der1 homologs could complement the yeast Δ der1 mutant, suggesting a potential role for a plant Der1 homolog in an ERAD pathway; however, there is no genetic evidence for proving the hypothesis (Kirst et al., 2005).
RETROTRANSLOCATION OF ERAD SUBSTRATES
Because the catalytic domains of the ERAD-participating E2s and E3s are on the cytosolic surface of the ER membrane, ERAD substrates need to undergo retrotranslocation for ubiquitination and to access the cytosolic proteasome system for their degradation. However, the molecular nature of this retrotranslocon remains controversial (Hampton and Sommer, 2012). It was previously thought that the Sec61 translocon, which imports nascent polypeptides into the ER lumen during protein biosynthesis, is responsible for retrotranslocation ERAD substrates through the ER membrane (Pilon et al., 1997; Plemper et al., 1997). Other studies suggested that the yeast Der1 and its mammalian orthologs Derlins are the suspected retrotranslocon (Lilley and Ploegh, 2004; Ye et al., 2004). A recent study, however, showed that the E3 ligase Hrd1 itself could serve as the retrotranslocation channel for ERADL substrates (Carvalho et al., 2010). It is quite possible that all three proteins are capable of retrotranslocation different ERAD substrates involving different adapter proteins.
Compared to the knowledge gained from the yeast and mammalian studies, we know almost nothing about the retrotranslocation step of a plant ERAD pathway. Several earlier studies did suggest the existence of a retrotranslocon in plant cells to move ERAD substrates into the cytosol. A confocal microscopic analysis of subcellular localization of a fusion protein between green fluorescent protein (GFP) with the P-domain of a maize CRT in tobacco leaf protoplasts suggested a retrotransport route from the ER to the cytosol (Brandizzi et al., 2003). In addition, a series of studies revealed that the A chain (known as RTA) of a ribosome-inactivating toxin, ricin that is normally produced as a dimeric protein of RTA covalently linked to a galactose-binding B chain via a single intramolecular disulfide bond and stored in the central vacuole of the endosperm cells of castor bean (Ricinus communis), was detected to be deglycosylated and eventual degraded in the cytosol when expressed alone in tobacco leaf protoplasts (Di Cola et al., 2001, 2005; Marshall et al., 2008). It is important to mention that ricin and a few other plant toxins were known to exploit the ERAD pathway to reach their cytosolic targets after being internalized by mammalian cells and retrograde-transported from the cell surface to the ER (Lord et al., 2003). In both yeast and mammalian systems, retrotranslocation of ERAD substrates was driven by ubiquitination (Bagola et al., 2011); however, a recent RTA study using plant protoplasts showed that retrotranslocation is independent of ubiquitination as the lysine-lacking (hence non-ubiquitinated) variant of RTA could still be retrotranslocated from the ER into the cytosol (Di Cola et al., 2005), suggesting that the ubiquitination-retrotranslocation coupling might be substrate-dependent.
SUBSTRATE EXTRACTION, PROCESSING, AND DELIVERY TO THE PROTEASOME
Without regard to the identity of the actual retrotranslocons, ubiquitinated ERAD clients are extracted from the ER lumen (ERADL substrates) or ER membrane (ERADM/C substrates) by a trimeric complex consisting of a homohexameric Cdc48 (p97 or valosin-containing protein in mammals), an AAA-type ATPase and its two substrate-recruiting factors Ufd1 and Npl4 (each having a ubiquitin-binding domain; Wolf and Stolz, 2012). The (CDC48)6-Ufd-Npl4 complex itself is recruited to the Hrd1/Doa10 E3 complexes by Ubx2 (VIMP for p97/VCP-interacting membrane protein in mammals), one of the 7 ubiquitin regulatory X (UBX) domain-containing proteins in yeast (13 UBX proteins in mammals; Neuber et al., 2005; Schuberth and Buchberger, 2005, 2008). The current working model posits that extracted ERAD substrates are further processed through antagonistic interactions between an U-box-containing E4 multiubiquitination enzyme Ufd2 and a WD40 repeat-containing protein Ufd3 with unknown enzyme activity plus a deubiquitylating enzyme Otu1, and/or through deglycosylation by the cytoplasmic peptide:N-glycanase (PNGase) Png1 (Raasi and Wolf, 2007). The processed ERAD substrates were subsequently delivered to the cytosolic proteasome by Cdc48 in association with two ubiquitin receptors Rad23 and Dsk2, each containing a UBL domain that interacts directly with the cytosolic proteasome and a polyubiquitin-interacting ubiquitin-associated (UBA) domain (Raasi and Wolf, 2007).
The Arabidopsis genome encodes three Cdc48 homologs, AtCDC48A, AtCDC48B, and AtCDC48C (Rancour et al., 2002). AtCDC48A was able to complement a yeast cdc48 mutant (Feiler et al., 1995) and was shown to play a role in the ERAD of a mutant form of MLO and a mutant variant of the Arabidopsis vacuolar carboxypeptidase carrying the same Gly-Arg mutation as the yeast CPY* and in the retrotranslocation of RTA and the orphan subunit (RCA A) of another castor bean toxin agglutinin in plant cells (Muller et al., 2005; Marshall et al., 2008; Yamamoto et al., 2010). AtCDC48A is likely to be recruited to the ER membrane by UBX-containing proteins as the Arabidopsis genome encodes a total of 15 UBX-containing proteins (known as AtPUXs; Table 1), some of which were shown to interact with AtCDC48A (Rancour et al., 2004; Park et al., 2007). It remains to be determined which of the 15 AtPUX proteins are actually involved in recruiting an AtCDC48 to the ER membrane-anchored E3 ligase complexes and play a role in degrading known plant ERAD substrates. Our BLAST searches using the known ERAD components of yeast and mammals as query identified multiple homologs of the Ufd1, Ufd2, Ufd3, Npl4, Rad23, Dsk2 but only a single PNGase homolog in Arabidopsis (Table 1). The functional involvement of these potential ERAD components in an Arabidopsis ERAD process remains unknown except AtPNG1, which was recently shown to contain the suspected PNGase activity and could stimulate the degradation of two mutant variants of RTA in an N-glycan-dependent manner in yeast cells (Diepold et al., 2007; Masahara-Negishi et al., 2012).
CONCLUSION AND CHALLENGES
Despite rapid progress in recent years for identifying molecular components of plant ERQC/ERAD systems and studying their biochemical functions, our understanding of the plant ERQC/ERAD processes remains rather limited, especially about the later stages of the ERAD pathway, such as retrotranslocation, processing of polyubiquitin chains, and delivery (to cytosolic proteasome) of the known plant ERAD substrates. While forward genetic screens in Arabidopsis identified the GII-UGGT-mediated CNX/CRT cycle in retaining incomplete/mis-folded glycoproteins and ERAD components that function inside the ER lumen to promote the degradation of the two mutant BR receptors, reverse genetic approaches using T-DNA insertional mutants or RNAi-mediated knockdown of candidate ERAD genes listed in Table 1 will certainly provide additional knowledge on the plant ERAD mechanisms. Transgenic Arabidopsis lines expressing carefully engineered substrates of glycosylated/non-glycosylated ERADC/ERADM coupled with forward genetic screens and reverse genetic studies will reveal if Arabidopsis has several distinct ERAD subpathways using different E3 ligases and adapter proteins that recruits distinct ERAD clients. Similarly, genetic screens for enhancers/suppressors of the Arabidopsis wax mutant cer9 [defective in Doa10A (Lu et al., 2012)] or drought hypersensitive 2 mutant [that led to independent discovery of Doa10A (Doblas et al., 2013)] could uncover additional ERAD components, reveal unique features of the plant ERAD processes, and a better understanding of the regulatory function of the plant ERAD system in biosynthetic processes. Proteomic studies with the existing Arabidopsis mutants of the ERAD E3 ligases could lead to the discovery of additional biochemical pathways and/or physiological processes regulated by the plant ERAD machinery. However, the biggest challenges for the plant ERQC/ERAD research is whether the forward genetic approach in Arabidopsis could identify novel ERQC/ERAD components that haven’t been discovered in other eukaryotic systems and if the combination of the Arabidopsis genetics with cutting-edge biochemical studies in Arabidopsis and transient expression systems could reveal novel biochemical functions of known or predicted ERAD components and provide satisfactory answers to some of the outstanding questions of the general ERQC/ERAD research field.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Aebi M. (2013). N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833 2430–2437 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Aebi M., Bernasconi R., Clerc S., Molinari M. (2010). N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35 74–82 10.1016/j.tibs.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Anelli T., Alessio M., Bachi A., Bergamelli L., Bertoli G., Camerini S., et al. (2003). Thiol-mediated protein retention in the endoplasmic reticulum: the role of ERp44. EMBO J. 22 5015–5022 10.1093/emboj/cdg491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T., Ceppi S., Bergamelli L., Cortini M., Masciarelli S., Valetti C., et al. (2007). Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 26 4177–4188 10.1038/sj.emboj.7601844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Novatchkova M., Potuschak T., Eisenhaber F. (2001). Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci. 6 463–470 10.1016/S1360-1385(01)02080-5 [DOI] [PubMed] [Google Scholar]
- Bagola K., Mehnert M., Jarosch E., Sommer T. (2011). Protein dislocation from the ER. Biochim. Biophys. Acta 1808 925–936 10.1016/j.bbamem.2010.06.025 [DOI] [PubMed] [Google Scholar]
- Bagola K., von Delbruck M., Dittmar G., Scheffner M., Ziv I., Glickman M. H., et al. (2013). Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol. Cell 50 528–539 10.1016/j.molcel.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Hanton S., DaSilva L. L., Boevink P., Evans D., Oparka K., et al. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34 269–281 10.1046/j.1365-313X.2003.01728.x [DOI] [PubMed] [Google Scholar]
- Buck T. M., Wright C. M., Brodsky J. L. (2007). The activities and function of molecular chaperones in the endoplasmic reticulum. Semin. Cell Dev. Biol. 18 751–761 10.1016/j.semcdb.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo J. J., Parodi A. J. (2008). Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283 10221–10225 10.1074/jbc.R700048200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. M., Hampton R. Y. (2010). Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J. Biol. Chem. 285 5146–5156 10.1074/jbc.M109.067876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. (2006). Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126 361–373 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Stanley A. M., Rapoport T. A. (2010). Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143 579–591 10.1016/j.cell.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., et al. (2009). Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184 159–172 10.1083/jcb.200809198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Liu L., Zhao Q., Zhang Z., Li Q., Lin B., et al. (2012). Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24 233–244 10.1105/tpc.111.093062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio C., Caramelo J. J., Parodi A. J. (2010). UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin. Cell Dev. Biol. 21 491–499 10.1016/j.semcdb.2009.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. (2006). A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126 349–359 10.1016/j.cell.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Di Cola A., Frigerio L., Lord J. M., Ceriotti A., Roberts L. M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. U.S.A. 98 14726–14731 10.1073/pnas.251386098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola A., Frigerio L., Lord J. M., Roberts L. M., Ceriotti A. (2005). Endoplasmic reticulum-associated degradation of ricin A chain has unique and plant-specific features. Plant Physiol. 137 287–296 10.1104/pp.104.055434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Li G., Lennarz W. J., Nurnberger T., Brunner F. (2007). The Arabidopsis AtPNG1 gene encodes a peptide: N-glycanase. Plant J. 52 94–104 10.1111/j.1365-313X.2007.03215.x [DOI] [PubMed] [Google Scholar]
- Doblas V. G., Amorim-Silva V., Pose D., Rosado A., Esteban A., Arro M., et al. (2013). The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme a reductase activity in Arabidopsis. Plant Cell 25 728–743 10.1105/tpc.112.108696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer L. M., Book A. J., Lee K. H., Lin Y. L., Fu H., Vierstra R. D. (2010). The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22 124–142 10.1105/tpc.109.072660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiler H. S., Desprez T., Santoni V., Kronenberger J., Caboche M., Traas J. (1995). The higher plant Arabidopsis thaliana encodes a functional CDC48 homologue which is highly expressed in dividing and expanding cells. EMBO J. 14 5626–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois J. L., Drouaud J., Lecureuil A., Guyon-Debast A., Bonhomme S., Guerche P. (2013). Functional characterization of the plant ubiquitin regulatory X (UBX) domain-containing protein AtPUX7 in Arabidopsis thaliana. Gene 526 299–308 10.1016/j.gene.2013.05.056 [DOI] [PubMed] [Google Scholar]
- Galvao R. M., Kota U., Soderblom E. J., Goshe M. B., Boss W. F. (2008). Characterization of a new family of protein kinases from Arabidopsis containing phosphoinositide 3/4-kinase and ubiquitin-like domains. Biochem. J. 409 117–127 10.1042/BJ20070959 [DOI] [PubMed] [Google Scholar]
- Gauss R., Sommer T., Jarosch E. (2006). The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25 1827–1835 10.1038/sj.emboj.7601088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Stevens F., Argon Y. (2013). Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 1833 2410–2424 10.1016/j.bbamcr.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. (1994). Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. U.S.A. 91 913–917 10.1073/pnas.91.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R. Y., Sommer T. (2012). Finding the will and the way of ERAD substrate retrotranslocation. Curr. Opin. Cell Biol. 24 460–466 10.1016/j.ceb.2012.05.010 [DOI] [PubMed] [Google Scholar]
- Haweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., et al. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285 4629–4636 10.1074/jbc.M109.063073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henquet M., Lehle L., Schreuder M., Rouwendal G., Molthoff J., Helsper J., et al. (2008). Identification of the gene encoding the α1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream N-glycan processing. Plant Cell 20 1652–1664 10.1105/tpc.108.060731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009). The ubiquitylation machinery of the endoplasmic reticulum. Nature 458 453–460 10.1038/nature07962 [DOI] [PubMed] [Google Scholar]
- Hong Z., Li J. (2012). The Protein Quality Control of Plant Receptor-Like Kinases in the Endoplasmic Reticulum. Berlin: Springer-Verlag [Google Scholar]
- Hong Z., Jin H., Fitchette A. C., Xia Y., Monk A. M., Faye L., et al. (2009). Mutations of an α1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell 21 3792–3802 10.1105/tpc.109.070284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jin H., Tzfira T., Li J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20 3418–3429 10.1105/tpc.108.061879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Kajiura H., Su W., Jin H., Kimura A., Fujiyama K., et al. (2012). Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 11437–11442 10.1073/pnas.1119173109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S. C., Hanna J., Hirsch C., Volkwein C., Schutz A., Heinemann U., et al. (2009). Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell 36 782–793 10.1016/j.molcel.2009.10.015 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. (2009). Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 284 17061–17068 10.1074/jbc.M809725200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Kamiya Y., Kato K. (2010). The role of MRH domain-containing lectins in ERAD. Glycobiology 20 651–660 10.1093/glycob/cwq013 [DOI] [PubMed] [Google Scholar]
- Houston N. L., Fan C., Xiang J. Q., Schulze J. M., Jung R., Boston R. S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137 762–778 10.1104/pp.104.056507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64 477–499 10.1146/annurev-arplant-050312-120053 [DOI] [PubMed] [Google Scholar]
- Huttner S., Strasser R. (2012). Endoplasmic reticulum-associated degradation of glycoproteins in plants. Front. Plant Sci. 3:67 10.3389/fpls.2012.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner S., Veit C., Schoberer J., Grass J., Strasser R. (2012). Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant Mol. Biol. 79 21–33 10.1007/s11103-012-9891-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Hong Z., Su W., Li J. (2009). A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 106 13612–13617 10.1073/pnas.0906144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yan Z., Nam K. H., Li J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26 821–830 10.1016/j.molcel.2007.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiura H., Seki T., Fujiyama K. (2010). Arabidopsis thaliana ALG3 mutant synthesizes immature oligosaccharides in the ER and accumulates unique N-glycans. Glycobiology 20 736–751 10.1093/glycob/cwq028 [DOI] [PubMed] [Google Scholar]
- Kamauchi S., Nakatani H., Nakano C., Urade R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272 3461–3476 10.1111/j.1742-4658.2005.04770.x [DOI] [PubMed] [Google Scholar]
- Kanehara K., Xie W., Ng D. T. (2010). Modularity of the Hrd1 ERAD complex underlies its diverse client range. J. Cell Biol. 188 707–716 10.1083/jcb.200907055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M. E., Meyer D. J., Gibbon B. C., Jung R., Boston R. S. (2005). Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol. 138 218–231 10.1104/pp.105.060087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z., Tsai Y. C., Weissman A. M. (2007). Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell Dev. Biol. 18 770–779 10.1016/j.semcdb.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E., Stone S. L., Ma L., Su N., Gao Y., Lau O. S., et al. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139 1597–1611 10.1104/pp.105.067983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K., Cho S. K., Son O., Xu Z., Hwang I., Kim W. T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21 622–641 10.1105/tpc.108.061994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., et al. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U.S.A. 106 15973–15978 10.1073/pnas.0905532106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E., Huttner S., Vavra U., Fischl R., Schoberer J., Grass J., et al. (2009). Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21 3850–3867 10.1105/tpc.109.072363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. (2004). A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429 834–840 10.1038/nature02592 [DOI] [PubMed] [Google Scholar]
- Liu L., Cui F., Li Q., Yin B., Zhang H., Lin B., et al. (2011). The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21 957–969 10.1038/cr.2010.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J. M., Deeks E., Marsden C. J., Moore K., Pateman C., Smith D. C., et al. (2003). Retrograde transport of toxins across the endoplasmic reticulum membrane. Biochem. Soc. Trans. 31 1260–1262 10.1042/BST0311260 [DOI] [PubMed] [Google Scholar]
- Lu S., Zhao H., Des Marais D. L., Parsons E. P., Wen X., Xu X., et al. (2012). Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 159 930–944 10.1104/pp.112.198697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Tintor N., Mentzel T., Kombrink E., Boller T., Robatzek S., et al. (2009). Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc. Natl. Acad. Sci. U.S.A. 106 22522–22527 10.1073/pnas.0907711106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R. S., Jolliffe N. A., Ceriotti A., Snowden C. J., Lord J. M., Frigerio L., et al. (2008). The role of CDC48 in the retro-translocation of non-ubiquitinated toxin substrates in plant cells. J. Biol. Chem. 283 15869–15877 10.1074/jbc.M709316200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahara-Negishi Y., Hosomi A., Della Mea M., Serafini-Fracassini D., Suzuki T. (2012). A plant peptide: N-glycanase orthologue facilitates glycoprotein ER-associated degradation in yeast. Biochim. Biophys. Acta 1820 1457–1462 10.1016/j.bbagen.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Nakano A. (1998). RMA1, an Arabidopsis thaliana gene whose cDNA suppresses the yeast sec15 mutation, encodes a novel protein with a RING finger motif and a membrane anchor. Plant Cell Physiol. 39 545–554 10.1093/oxfordjournals.pcp.a029403 [DOI] [PubMed] [Google Scholar]
- Meaden P., Hill K., Wagner J., Slipetz D., Sommer S. S., Bussey H. (1990). The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1–6)-beta-D-glucan synthesis and normal cell growth. Mol. Cell. Biol. 10 3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M. B., Liang Y. H., Das R., Mariano J., Li S., Li J., et al. (2013). A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol. Cell 50 516–527 10.1016/j.molcel.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohorko E., Glockshuber R., Aebi M. (2011). Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 34 869–878 10.1007/s10545-011-9337-1 [DOI] [PubMed] [Google Scholar]
- Muller J., Piffanelli P., Devoto A., Miklis M., Elliott C., Ortmann B., et al. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17 149–163 10.1105/tpc.104.026625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Li J., Batoux M., Roux M., Chu Z. H., Lacombe S., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28 3428–3438 10.1038/emboj.2009.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005). Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 7 993–998 10.1038/ncb1298 [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., et al. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121 743–752 10.1104/pp.121.3.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall J., Vine N., Hadlington J. L., Drake P., Frigerio L., Ma J. K. (2002). ER-resident chaperone interactions with recombinant antibodies in transgenic plants. Eur. J. Biochem. 269 6042–6051 10.1046/j.1432-1033.2002.03302.x [DOI] [PubMed] [Google Scholar]
- Olzmann J. A., Kopito R. R., Christianson J. C. (2013). The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5 a013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Rancour D. M., Bednarek S. Y. (2007). Protein domain-domain interactions and requirements for the negative regulation of Arabidopsis CDC48/p97 by the plant ubiquitin regulatory X (UBX) domain-containing protein, PUX1. J. Biol. Chem. 282 5217–5224 10.1074/jbc.M609042200 [DOI] [PubMed] [Google Scholar]
- Pedrazzini E., Giovinazzo G., Bielli A., de Virgilio M., Frigerio L., Pesca M., et al. (1997). Protein quality control along the route to the plant vacuole. Plant Cell 9 1869–1880 10.1105/tpc.9.10.1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M. (2004). Back to the future with ubiquitin. Cell 116 181–190 10.1016/S0092-8674(03)01074-2 [DOI] [PubMed] [Google Scholar]
- Pilon M., Schekman R., Romisch K. (1997). Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16 4540–4548 10.1093/emboj/16.15.4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R. K., Bohmler S., Bordallo J., Sommer T., Wolf D. H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388 891–895 10.1038/42276 [DOI] [PubMed] [Google Scholar]
- Pollier J., Moses T., Gonzalez-Guzman M., De Geyter N., Lippens S., Vanden Bossche R., et al. (2013). The protein quality control system manages plant defence compound synthesis. Nature 504 148–152 10.1038/nature12685 [DOI] [PubMed] [Google Scholar]
- Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., et al. (2008). Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 32 870–877 10.1016/j.molcel.2008.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S., Wolf D. H. (2007). Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin. Cell Dev. Biol. 18 780–791 10.1016/j.semcdb.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Rancour D. M., Dickey C. E., Park S., Bednarek S. Y. (2002). Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 130 1241–1253 10.1104/pp.011742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour D. M., Park S., Knight S. D., Bednarek S. Y. (2004). Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of Arabidopsis CDC48. J. Biol. Chem. 279 54264–54274 10.1074/jbc.M405498200 [DOI] [PubMed] [Google Scholar]
- Reddy P., Sparvoli A., Fagioli C., Fassina G., Sitia R. (1996). Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled Ig light chains. EMBO J. 15 2077–2085 [PMC free article] [PubMed] [Google Scholar]
- Saijo Y. (2010). ER quality control of immune receptors and regulators in plants. Cell. Microbiol. 12 716–724 10.1111/j.1462-5822.2010.01472.x [DOI] [PubMed] [Google Scholar]
- Saijo Y., Tintor N., Lu X., Rauf P., Pajerowska-Mukhtar K., Haweker H., et al. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28 3439–3449 10.1038/emboj.2009.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C., Buchberger A. (2005). Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 7 999–1006 10.1038/ncb1299 [DOI] [PubMed] [Google Scholar]
- Schuberth C., Buchberger A. (2008). UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 65 2360–2371 10.1007/s00018-008-8072-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. H., Ploegh H. L., Weissman J. S. (2011). Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334 1086–1090 10.1126/science.1209235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O., Cho S. K., Kim E. Y., Kim W. T. (2009). Characterization of three Arabidopsis homologs of human RING membrane anchor E3 ubiquitin ligase. Plant Cell Rep. 28 561–569 10.1007/s00299-009-0680-8 [DOI] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2011). Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 870–875 10.1073/pnas.1013251108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2012). The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol. Plant 5 929–940 10.1093/mp/sss042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D. Y., Vierling E., Guy C. L. (2001). Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 126 789–800 10.1104/pp.126.2.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Park H., Till E. A., Lennarz W. J. (2001). The PUB domain: a putative protein–protein interaction domain implicated in the ubiquitin-proteasome pathway. Biochem. Biophys. Res. Commun. 287 1083–1087 10.1006/bbrc.2001.5688 [DOI] [PubMed] [Google Scholar]
- Ueda T., Matsuda N., Uchimiya H., Nakano A. (2000). Modes of interaction between the Arabidopsis Rab protein, Ara4, and its putative regulator molecules revealed by a yeast expression system. Plant J. 21 341–349 10.1046/j.1365-313x.2000.00681.x [DOI] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. (2004). Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165 41–52 10.1083/jcb.200309132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9 944–957 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Numers N., Survila M., Aalto M., Batoux M., Heino P., Palva E. T., et al. (2010). Requirement of a homolog of glucosidase II β-subunit for EFR-mediated defense signaling in Arabidopsis thaliana. Mol. Plant 3 740–750 10.1093/mp/ssq017 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang W. Z., Song L. F., Zou J. J., Su Z., Wu W. H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148 1201–1211 10.1104/pp.108.126375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D. H., Stolz A. (2012). The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta 1823 117–124 10.1016/j.bbamcr.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Kawanabe M., Hayashi Y., Endo T., Nishikawa S. (2010). A vacuolar carboxypeptidase mutant of Arabidopsis thaliana is degraded by the ERAD pathway independently of its N-glycan. Biochem. Biophys. Res. Commun. 393 384–389 10.1016/j.bbrc.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004). A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429 841–847 10.1038/nature02656 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Tanaka K. (2010). Lectin-like ERAD players in ER and cytosol. Biochim. Biophys. Acta 1800 172–180 10.1016/j.bbagen.2009.07.029 [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., et al. (2006). Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126 571–582 10.1016/j.cell.2006.06.041 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., Boller T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]