Abstract
Carpel margin meristems (CMMs), a pair of meristematic tissues present along the margins of two fused carpel primordia of Arabidopsis thaliana, are essential for the formation of ovules and the septum, two major internal structures of the gynoecium. Although a number of regulatory factors involved in shoot meristem activity are known to be required for the formation of these gynoecial structures, their direct roles in CMM development have yet to be addressed. Here we show that the CUP-SHAPED COTYLEDON genes CUC1 and CUC2, which are essential for shoot meristem initiation, are also required for formation and stable positioning of the CMMs. Early in CMM formation, CUC1 and CUC2 are also required for expression of the SHOOT MERISTEMLESS gene, a central regulator for stem cell maintenance in the shoot meristem. Moreover, plants carrying miR164-resistant forms of CUC1 and CUC2 resulted in extra CMM activity with altered positioning. Our results thus demonstrate that the two regulatory proteins controlling shoot meristem activity also play critical roles in elaboration of the female reproductive organ through the control of meristematic activity.
Keywords: Arabidopsis thaliana, carpel margin meristem, shoot meristem, leaf development, MicroRNA (miRNA), fruit development
Introduction
In the plant shoot, leaves and floral organs are produced from the shoot and floral meristems, respectively. These meristems maintain pluripotent stem cells at the center and differentiate appropriate types of lateral organs at their periphery depending on the developmental context. Although diverse in their shape and function, floral organs (sepals, petals, stamens, and carpels) are considered to be modified leaves and their specific characters are conferred by combinatorial actions of homeotic genes (Krizek and Fletcher, 2005). How unique shapes of individual organ types are generated is a central question in understanding plant shoot development.
The carpel is a component of the gynoecium, a highly complex organ system dedicated to reproduction. Either single or multiple carpel(s) fuse to form the gynoecium and enclose ovules inside. Ovules are formed by meristematic tissues located within or adjacent to carpel primordia (Yamaki et al., 2011) and in Arabidopsis thaliana, they are produced by a pair of meristematic tissues called carpel margin meristems (CMMs; also called medial ridges), which are present along the fused margins of the two carpel primordia (Nole-Wilson et al., 2010). In addition to producing ovules laterally, the CMM pair terminates and fuses with each other along their tip, forming the septum that acts as transmitting tissue for pollen tubes.
Several regulators of shoot meristem activity are involved in carpel margin development (reviewed in Reyes-Olalde et al., 2013). The REPLUMLESS gene (RPL; also known as BLR and PNY) encoding a BELL-type homeodomain protein is expressed in carpel margins and is required for replum development (Roeder et al., 2003). The RPL protein physically interacts with a class I KNOX protein BREVIPEDICELLUS (BP) and the activity of the two proteins counteracts with JAGGED (JAG), FILAMENTOUS FLOWER (FIL), YABBY3 (YAB3), and ASYMMETRIC LEAVES1/2 (AS1 and AS2) genes, which promote the fate of adjacent valve and valve margin tissues (Dinneny et al., 2005; Alonso-Cantabrana et al., 2007; Gonzalez-Reig et al., 2012). Both RPL and BP genes are also expressed in the shoot meristem and affect internode length (Smith and Hake, 2003). The RPL protein interacts with another class I KNOX protein SHOOT MERISTEMLESS (STM) and they act together to maintain stem cells in the shoot meristem (Byrne et al., 2003). Although strong mutant alleles of stm do not produce flowers due to their strong shoot meristem defects, function of STM in carpel development has been accessed using weak stm alleles or inducible RNAi plants, which produce abnormal flowers. In these flowers, carpels often fail to fuse at their margins and develop few ovules, indicating that STM is required for proper formation of carpel margins (Endrizzi et al., 1996; Scofield et al., 2007).
The CUP-SHAPED COTYLEDON genes CUC1 and CUC2 encoding a pair of paralogous NAC transcription factors are required for shoot meristem initiation through promoting STM expression (Hibara et al., 2003). Because of their functional redundancy, seedlings of each single mutant show little morphological phenotype while their double mutants completely lack a shoot meristem and produce severely fused cotyledons. Viable shoots with flowers can be regenerated from double mutant calli and these flowers produce carpels with severe reduction of ovules, septum and replum (Ishida et al., 2000; Figure 1). The Auxin Response Factor MONOPTEROS (MP) is required for CUC1 and CUC2 expression possibly through its direct binding to the gene promoters and the CUC genes in turn affect expression and polarity of the auxin transport protein PIN1 in ovule primordia (Galbiati et al., 2013). In addition, both CUC1 and CUC2 are negatively regulated by the microRNA miR164, which is encoded by three loci in Arabidopsis. Disruption of miR164 encoding genes or that of its target sequences in CUC1 and CUC2 causes misregulation of their expression, resulting in various developmental defects including abnormal carpel development (Mallory et al., 2004; Baker et al., 2005; Nikovics et al., 2006; Sieber et al., 2007; Larue et al., 2009).
Figure 1.
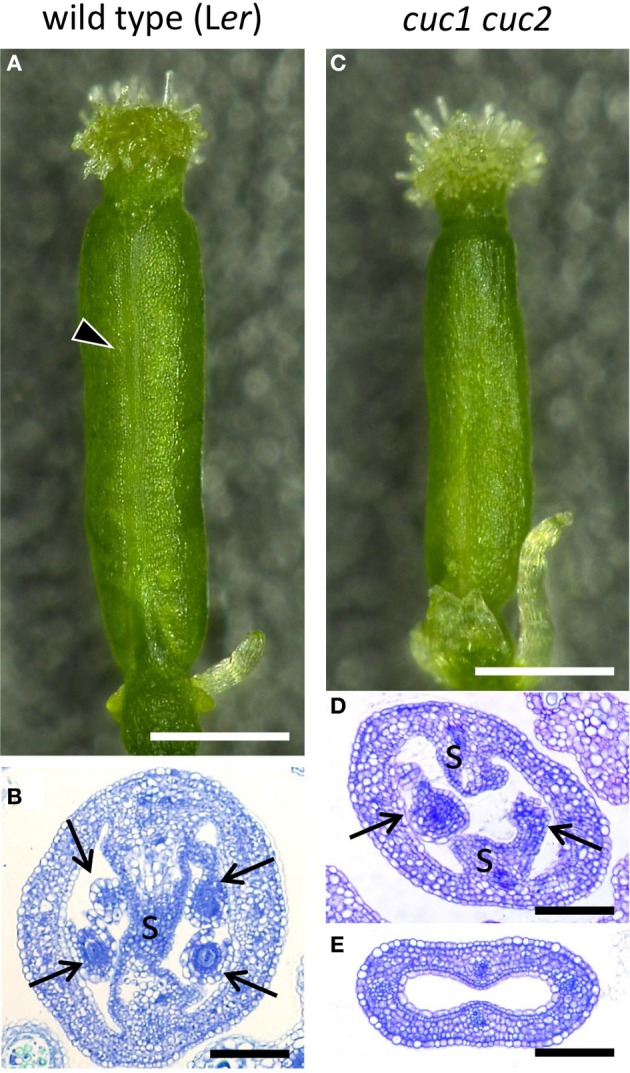
Gynoecium phenotype of cuc1 cuc2. Mature gynoecia from regenerated plants of wild type Ler (A,B) and cuc1 cuc2 (C–E). Whole view (A,C) and transverse sections (B,D,E). Arrowhead in (A) indicates the replum. Arrow, ovule; s, septum. Scale bars are: (A,C), 500 μm; (B,D,E), 100 μm.
The above results point to the importance of CUC1 and CUC2 and in formation of carpel margin structures. However, the developmental basis of the roles for these factors has not been fully investigated. Notably, whether the factors directly affect CMM formation and if so, how they interact during the process remains unknown. Here we investigated the roles for CUC1 and CUC2 in gynoecium development by loss and gain of function approaches. The results demonstrate that the CUC1 and CUC2 genes are critical for normal CMM development.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana accessions Landsberg erecta (Ler) and Columbia (Col) were used as the wild type strains. The cuc1 cuc2 double mutant (cuc1-1 cuc2-1) is in the Ler background (Aida et al., 1997). CUC2g-m4 is in the Col background and was described previously (Nikovics et al., 2006). For construction of CUC1g-m7, the miR164 target sequence (AG CAC GTG TCC TGT TTC TCC A) of CUC1 was replaced by a mutant sequence (AG CAC GTG AGT TGT TTT AGT A), which contains seven silent mutations (underlined). The mutated genomic fragment corresponding to the nucleotides 5108201..5112019 of chromosome 3 (TAIR 10) was cloned into pGreenII 0229 (Hellens et al., 2000) and transformed into Col. A transgenic line displaying extra petal number and reduced sepal growth, a typical phenotype described for miR164 resistant 5mCUC1 (Mallory et al., 2004), was selected and subjected to analysis. This line accumulated CUC1 mRNA ~9.5 fold of the wild-type level in inflorescence apices. Seeds were surface sterilized and sown on MS plates as previously described (Fukaki et al., 1996). After incubation for 2 days at 4°C in the dark, plants were grown in a growth chamber at 23°C under constant white light. Ten- to fourteen-day-old seedlings were transferred onto soil and grown at 23°C under constant white light. Induction of calli from root explants and subsequent shoot regeneration was performed as previously described (Aida et al., 1997). Flower stages were determined as previously described (Smyth et al., 1990). Stage 9 was further subdivided into early, mid and late substages, each corresponding to stages 5, 6, and 7 of anther development (Sanders et al., 1999).
Histological analysis
Histological sections (3 μm) were prepared as previously described(Aida et al., 1997), except that formalin/acetic acid/alcohol (FAA) was used as a fixative. Scanning electron microscopy was carried out as described previously (Aida et al., 1997). In situ hybridization was performed as previously described (Ishida et al., 2000) with following modifications: 6 instead of 8 μm sections were prepared and hybridized at 45°C instead of 42°C. Probes for STM and FIL have been described previously (Long et al., 1996; Sawa et al., 1999). Templates for CUC1 and CUC2 probes were the full-length coding sequences. In the wild type and cuc1 cuc2, coloring reaction was performed for 36 h, with the initial 12 h at room temperature and the remaining at 4°C. In CUC1g-m7 and CUC2g-m4, coloring reaction was carried out for 12 h at room temperature.
Results
CUC1 and CUC2 are required for the initiation of the CMMs
To compare gynoecium development of the wild type (Ler) and cuc1 cuc2, we used inflorescence shoots regenerated from calli of each genotype (Aida et al., 1997; Ishida et al., 2000). Wild-type gynoecia in regenerated plants showed essentially the same morphology as those from non-regenerated plants (Figures 1A,B) and followed normal developmental stages (Figure 2; Smyth et al., 1990). Mature gynoecia of cuc1 cuc2 were somewhat smaller than those of wild type and tended to lose replum tissues most prominently in the apical region of the ovary (compare Figures 1A and 1C; Ishida et al., 2000). In histological sections, the septum and ovules were severely reduced (Figure 1D) or completely missing (Figure 1E).
Figure 2.
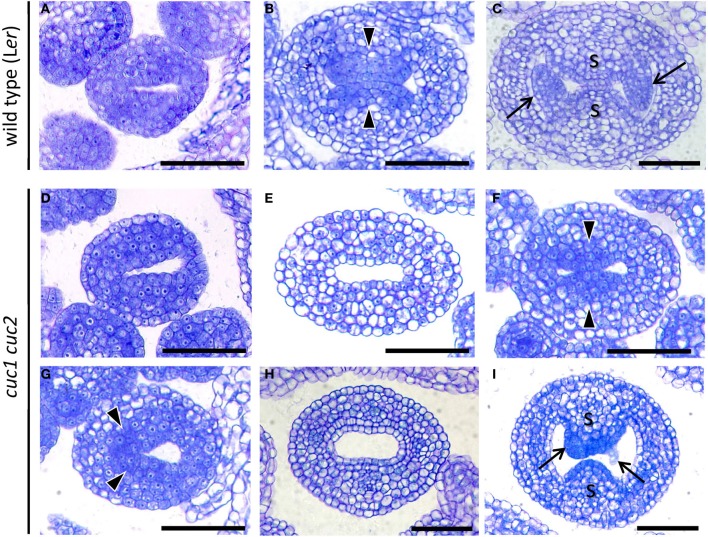
CUC1 and CUC2 are required for formation and stable positioning of CMM. Transverse sections of developing gynoecium at stage 7 (A,D), stage 8 (G), early stage 9 (B,E,F), mid stage 9 (H), and late stage 9 (C,I). Sections are prepared from wild type (A–C) and cuc1 cuc2 (D–I). Arrow, ovule; arrowhead, CMM; s, septum. Scale bars are 50 μm.
Gynoecium primordia of cuc1 cuc2 were indistinguishable from those of the wild type up to stage 7, at which both wild type and cuc1 cuc2 formed a cylindrical primordium consisting mostly of densely cytoplasmic cells (Figures 2A,D). Deviation of the mutant phenotype began at stage 8 to early stage 9, when the wild type initiated two bulges of CMM from the adaxial wall. In the wild-type, cells in the CMMs remained cytoplasmically dense whereas the rest of the cells started vacuolation, which was a sign of cell differentiation (Figure 2B). On the other hand, cuc1 cuc2 frequently failed to form either one or both of the CMMs and cells in the corresponding regions became vacuolated (Figure 2E; Table 1). In some mutant gynoecia, CMMs were formed on both sides, but their size was smaller than that of the wild type (Figure 2F). In addition, the positioning of CMM initiation was often asymmetric (Figure 2G). When the wild type initiated ovule primordia, cells at the contacting surfaces of the two CMMs underwent post-genital fusion to form the septum (Figure 2C). On the other hand, a significant fraction of cuc1 cuc2 gynoecia still failed to initiate CMMs (Figure 2H; Table 1). Some mutant gynoecia developed small bumps at the corresponding positions, but their surfaces often failed to contact (Figure 2I; Table 1). These bumps were likely to retain organogenic activity as indicated by the presence of densely cytoplasmic cells, and might produce ovule-like primordia from their flanks. Taken together, these results show that CUC1 and CUC2 are required for CMM initiation. Occasional CMM formation at asymmetric positions indicates that the CUC gene activities are also required for stable positioning of the CMMs.
Table 1.
Effect of cuc1 cuc2 on CMM formation.
| Genotype | Stage* | CMM formation | CMM contact** | |||
|---|---|---|---|---|---|---|
| Both sides | One side | None | Yes | No | ||
| wild type | 8 | 9 (100%) | 0 | 0 | 8 (89%) | 1 (11%) |
| Early 9 | 14 (100%) | 0 | 0 | 14 (100%) | 0 | |
| Mid 9 | 7 (100%) | 0 | 0 | 7 (100%) | 0 | |
| cuc1 cuc2 | 8 | 3 (33%) | 3 (33%) | 3 (33%) | 3 (100%) | 0 |
| Early 9 | 7 (39%) | 7 (39%) | 4 (22%) | 7 (100%) | 0 | |
| Mid 9 | 6 (55%) | 4 (36%) | 1 (9%) | 3 (50%) | 3 (50%) | |
Expression of CUC1 and CUC2 predicts the sites of CMM initiation
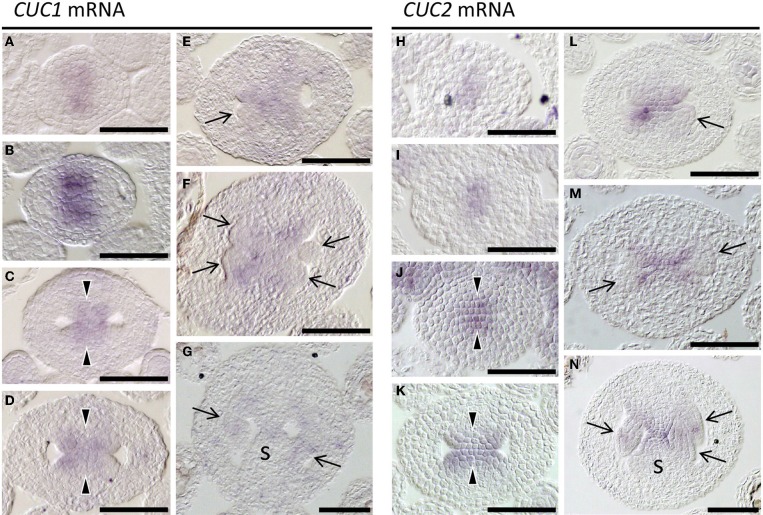
Expression patterns of CUC1 and CUC2 during gynoecium development have been reported only partially (Ishida et al., 2000; Takada et al., 2001; Nahar et al., 2012; Galbiati et al., 2013). We therefore carried out detailed expression analysis. Upon initiation of the gynoecial primordium, their expression was detected at its apical center, where the cleft of the future gynoecium cavity will form (Figures 3A,H). When the primordium became cylindrical, CUC1 and CUC2 expression was detected in the adaxial region of the medial wall, from which the CMM develops (Figures 3B,I). The area of CUC1 expression domain was broader than that of CUC2. When the CMMs began to form, expression of both genes was detected throughout the bulge (Figures 3C,D,J,K). Their expression was missing in the developing ovule primordia but present in the remaining part of the CMMs (Figures 3E,L). Later, transcripts of CUC1 and CUC2 were both detected at the base of ovule primordia, the fused region of the septum, and in ovules (Figures 3F,G,M,N). These results are consistent with the role for CUC1 and CUC2 in CMM formation. Expression patterns from stage 7 to stage 10 are summarized in Supplementary Figure 1 and Supplementary Table 1.
Figure 3.
Expression of CUC1 and CUC2. In situ hybridization on transverse sections of developing wild type (Ler) gynoecia regenerated from calli. Probed with CUC1 (A–G) and CUC2 (H–N). Sections are prepared from gynoecia at stage 6 (A,H), stage 7 (B,I), stage 8 (C,J), early stage 9 (D,K), mid stage 9 (E,L), late stage 9 (F,M), and stage 10 (G,N). Arrow, ovule; arrowhead, CMM; s, septum. Scale bars are 50 μm.
CUC1 and CUC2 are required for STM expression and prevent differentiation of CMM cells
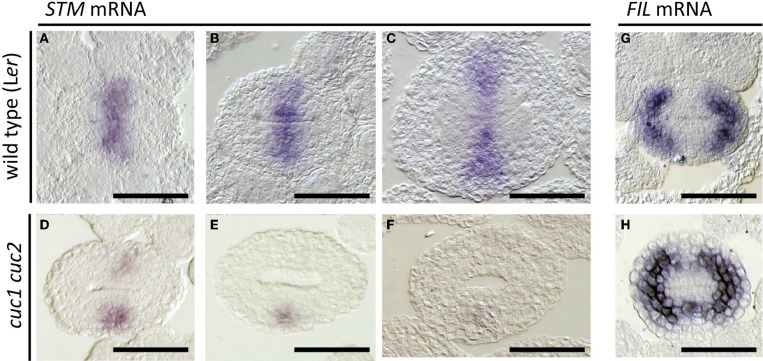
The class I KNOX gene STM plays a critical role in maintaining shoot meristem activity and is also required for proper gynoecium development. As reported previously, STM expression was detected along the carpel margins of early gynoecia (Figure 4A; Long et al., 1996) and continued in the CMM while it was missing in ovule primordia (Figures 4B,C). In cuc1 cuc2, by contrast, STM expression was greatly reduced (Figures 4D–F). Notably, its expression tended to be absent on the adaxial side of the carpel margins while it remained on the abaxial side. These results show that CUC1 and CUC2 are required for STM expression in the CMM.
Figure 4.
Effect of cuc1 cuc2 on STM and FIL expression. In situ hybridization on transverse sections of developing wild type Ler (A–C,G) and cuc1 cuc2 (D–F,H) gynoecia regenerated from calli. Probed with STM (A–F) and FIL (G,H). Sections are prepared from gynoecia at stage 8 (A,D,G,H), early stage 9 (B,E) and mid stage 9 (C,F). Scale bars are 50 μm.
Expression of the FIL gene is detected in the future valve region while it is excluded from the carpel margins (Dinneny et al., 2005; Figure 4G). Together with its close homolog YAB3, it is required for valve development. In cuc1 cuc2, FIL expression extended toward the carpel margins and formed a continuous ring (Figure 4H). These results are consistent with the reduction of carpel margin structures in cuc1 cuc2 and indicate that CUC1 and CUC2 prevent valve differentiation at the carpel margins.
MicroRNA resistant versions of CUC1 and CUC2 genomic fragments cause expansion of the CMMs
We next examined the role of microRNA-dependent regulation of CUC1 and CUC2 in CMM formation. To this end, we used transgenic plants carrying genomic fragments of CUC1 or CUC2 that carry silent mutations in the target sequences of miR164 (CUC1g-m7 and CUC2g-m4, respectively). These plants exhibited expansion of carpel margin structures including the replum (Figures 5A–F) and the septum (Figures 5G–I). In addition, the abaxial surface of carpel margins in CUC2g-m4 was swollen and produced filamentous structures (Figures 5C,F,I; Nikovics et al., 2006).
Figure 5.
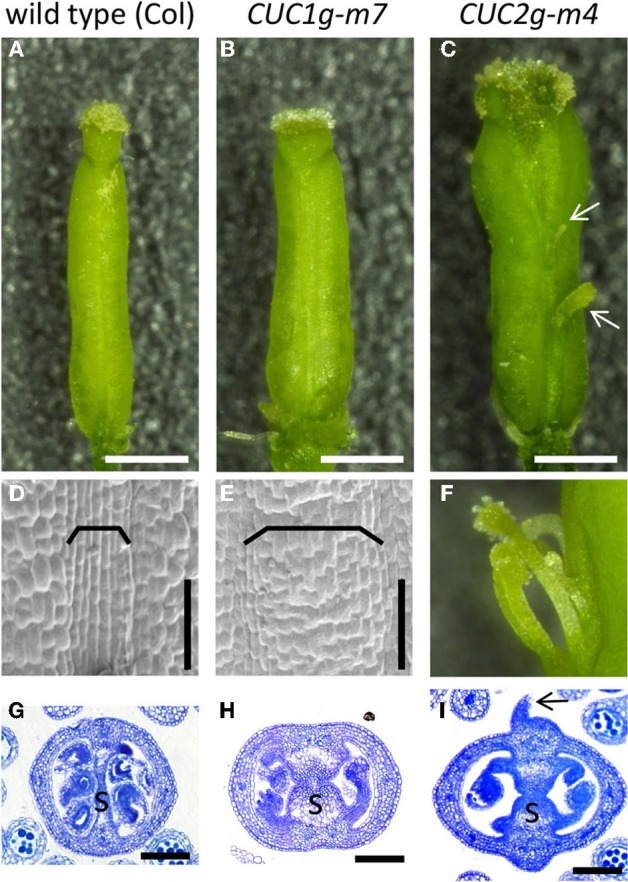
Gynoecium phenotype of CUC1g-m7 and CUC2g-m4. Mature gynoecia of wild type Col (A,D,G), CUC1g-m7 (B,E,H), and CUC2g-m4 (C,F,I). Whole view (A–C), scanning electron micrograph of replum (D,E), close up view of filamentous structures (F) and transverse sections of ovary (G–I). Arrows in (C,I) indicate filamentous structures. Brackets indicate repla. s, septum. Scale bars are: (A–C), 500 μm; (D,E,G–I), 100 μm.
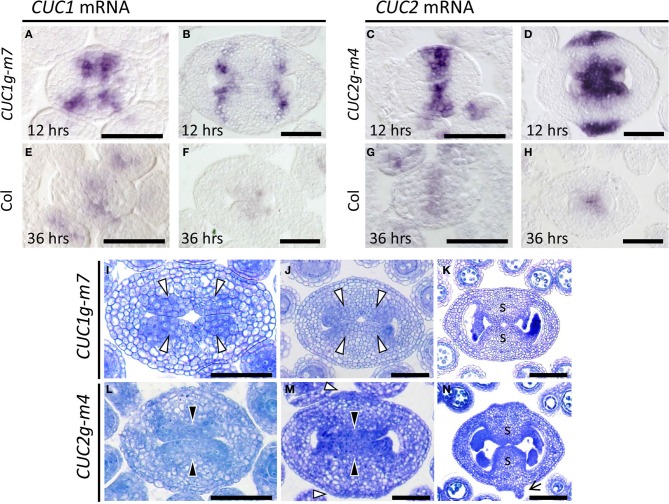
Expression of CUC1 and CUC2 was examined to access the effect of the silent mutations introduced into the transgenes. In CUC1g-m7, CUC1 mRNA initially accumulated in a broad region around the carpel margins with four peaks of staining (Figure 6A), which later dissolved into four discrete spots (Figure 6B). In CUC2g-m4, CUC2 mRNA was first detected broadly throughout the carpel margins (Figure 6C) and later it split into adaxial and abaxial ends (Figure 6D). In both transgenic plants, stronger signals were observed in shorter staining time than in wild type (12 vs. 36 h), indicating that the levels of CUC1 and CUC2 mRNA was significantly elevated (compare Figures 6A–D with 6E–H).
Figure 6.
CMM development in CUC1g-m7 and CUC2g-m4. (A–H) In situ hybridization on transverse sections from developing gynoecia of CUC1g-m7 (A,B), CUC2g-m4 (C,D) and wild type Col (E–H) probed with CUC1 (A,B,E,F) and CUC2 (C,D,G,H). Staining reaction was carried out for 12 h in CUC1g-m7 (A,B) and CUC2g-m4 (C,D), and for 36 h in Col (E–H). Sections are prepared from gynoecia at stage 6 (A,C,E,G) and late stage 9 (B,D,F,H). (I–N) Transverse sections from developing gynoecia of CUC1g-m7 (I–K) and CUC2g-m4 (L–N) at early stage 9 (I,L), late stage 9 (J,M), and stage 11 (K,N). Black arrowheads indicate CMMs at normal positions and white arrowheads indicate those at abnormal positions. Arrow in (N) indicates a primordium of a filamentous structure. s, septum. Scale bars are 50 μm.
We next examined early gynoecium development in these transgenic plants. In CUC1g-m7, CMMs were duplicated and initiated at four positions (Figure 6I, white arrowheads) that corresponded to the peaks of CUC1 mRNA accumulation (Figure 6A). Each pair of the duplicated CMMs grew adaxially as a congenitally fused tissue and contacted each other at the center to undergo post-genital fusion, forming a thicker septum than that of the wild type (Figures 6J,K). The boundary of the fused CMMs was slightly depressed, forming a small central space after the fusion (Figure 6K). In CUC2g-m4, CMMs developed on the adaxial side and were broader compared to the wild type (Figure 6L). In addition, CUC2g-m4 produced ectopic meristematic tissues as indicated by densely cytoplasmic cells on the abaxial side (Figure 6M, white arrowheads). These meristematic tissues further expanded and initiated primordia of filamentous structures on their flanks (Figure 6N, arrow). We interpret these meristematic tissues as ectopic CMMs, although they lack ability to form ovules. Together, the results show that disruption of miR164-mediated regulation of CUC1 and CUC2 strongly affected the size, positioning, and number of the CMMs.
In CUC1g-m7, expression of STM was laterally extended compared to that in wild type (compare Figures 7A with 4B), and four strong staining peaks were found within the expression domain, showing a strong correlation with the pattern of CUC1 expression in this background (compare Figures 7C with 6B). Expression of STM was also broader in CUC2g-m4 than in wild type and was detected throughout the carpel margins including outermost cells on the abaxial side, in which wild type did not accumulate STM transcripts (compare Figures 7B to 4B). This ectopic expression of STM continued in ectopic CMMs on the abaxial side (Figure 7D). On the other hand, FIL expression was not detected in the carpel margins of CUC1g-m5 and CUC2g-m7 as in the wild type (Figures 7E,F). These results indicate that elevated and ectopic levels of CUC gene expression in CUC1g-m7 and CUC2g-m4 cause increased meristematic activity of the CMMs.
Figure 7.
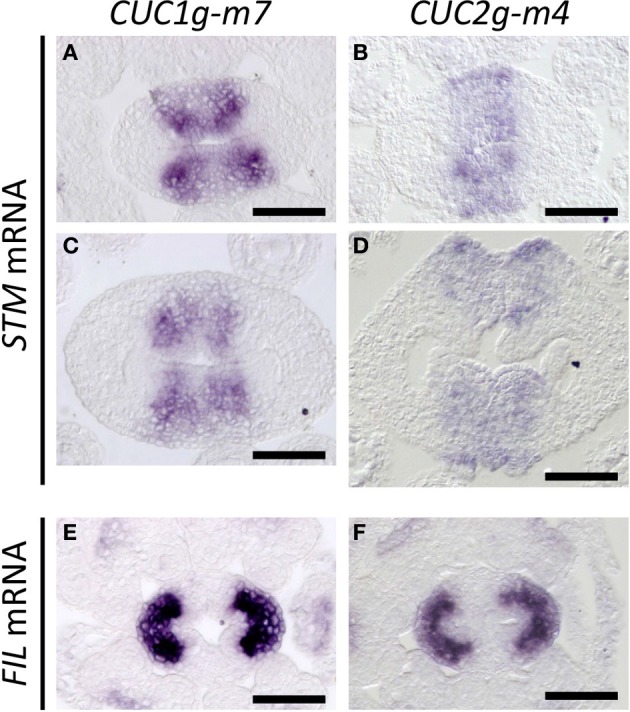
STM and FIL expression in CUC1g-m7 and CUC2g-m4. In situ hybridization on transverse sections from developing gynoecia of CUC1g-m7 (A,C,E) and CUC2g-m4 (B,D,F) probed with STM (A–D) and with FIL (E,F) at early stage 9 (A,B), late stage 9 (C,D) and stage 8 (E,F). Scale bars are 50 μm.
Discussion
Our results demonstrate that CUC1 and CUC2 play critical roles in formation and positioning of CMMs, which are central tissues for generating internal gynoecial organs. The loss of CUC1 and CUC2 activity caused severe reduction and altered positioning of CMMs, indicating that the previously reported defects in ovule and septum formation (Ishida et al., 2000; Galbiati et al., 2013) were due to the failure of forming these meristematic tissues. Moreover, CUC1g-m7 and CUC2g-m4 plants resulted in duplication and expansion of CMMs, and the positions of the duplicated CMMs in each transgenic plant are associated with the peaks of CUC1 or CUC2 transcripts, respectively. These results indicate that CUC1 and CUC2 promote CMM formation and, possively through the interaction with miR164, they are required for correct positioning of the CMMs.
Expression of STM was strictly dependent on CUC1 and CUC2 activities in the CMMs. This result is consistent with the previously reported function of STM in ovule formation (Scofield et al., 2007) and supports that CUC1 and CUC2 act upstream of STM in CMM formation. It has been suggested that CUC1 and CUC2 promote ovule development partly though activating a cytokinin pathway (Galbiati et al., 2013) and STM can promote cytokinin biosynthesis genes in seedling apices (Jasinski et al., 2005; Yanai et al., 2005). Our results are thus consistent with the idea that activation of STM expression by CUC1 and CUC2 promotes cytokinin production, which in turn contributes to ovule formation. Despite significant reduction of STM transcripts in cuc1 cuc2 double mutant gynoecia, they do not show a split carpel phenotype, which has been reported for weak stm mutant alleles (Endrizzi et al., 1996) and inducible STM RNAi plants (Scofield et al., 2007). This aspect of carpel phenotype may reflect earlier function of STM, which is already expressed in the floral meristem before carpel initiation (Long et al., 1996). Alternatively, the remaining STM expression on the abaxial side of cuc1 cuc2 carpel margins (Figures 4D–F) may be sufficient to prevent split of carpels.
In contrast to STM, the area of FIL expressing cells was greatly reduced in cuc1 cuc2, indicating that CUC1 and CUC2 negatively affect FIL expression. This result fits to the model in which factors responsible for carpel margin formation and those responsible for valve/valve margin formation counteract each other, as has been proposed based on interactions among RPL, BP, FIL, JAG, and AS1/2 genes (Alonso-Cantabrana et al., 2007; Gonzalez-Reig et al., 2012). Because the STM protein has shown to physically interact with the carpel margin factor RPL (Byrne et al., 2003; Smith and Hake, 2003), it would be possible that the activation of STM expression by the CUC gene increases the amount of the STM/RPL complex in CMMs, thereby antagonizing the valve/valve margin factors including FIL.
Our results show that activation of the class I KNOX gene STM by CUC1 and CUC2, a critical regulatory step during embryonic shoot meristem formation (Aida et al., 1999; Takada et al., 2001; Hibara et al., 2003), also occurs during CMM formation. The same regulatory relationship is also found in the formation of leaf margin structures (Kawamura et al., 2010) and is conserved among eudicots (Blein et al., 2008). Furthermore, the important roles for auxin and miR164 in regulating expression of CUC genes are also conserved among the processes of shoot meristem, leaf margin and carpel margin formation (Aida et al., 2002; Furutani et al., 2004; Nikovics et al., 2006; Larue et al., 2009; Koyama et al., 2010; Bilsborough et al., 2011; Galbiati et al., 2013). Further investigation on how these common regulatory factors are integrated into each developmental context and their possible relation with context-specific regulatory factors such as floral homeotic genes will be important to understand how unique shapes of different organs are formed.
Author contributors
Yuri Kamiuchi, Masao Tasaka, and Mitsuhiro Aida designed the research. Yuri Kamiuchi, Kayo Yamamoto, Masahiko Furutani, and Mitsuhiro Aida performed the research. Yuri Kamiuchi and Mitsuhiro Aida analyzed the data and wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Patrick Laufs for providing CUC2g-m4 seeds. This work was supported by JSPS KAKENHI Grant Numbers 18770063, 23370023, and MEXT KAKENHI Grant Numbers 21024008, 24114001.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00165/abstract
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Ishida T., Tasaka M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570 [DOI] [PubMed] [Google Scholar]
- Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965–3974 [DOI] [PubMed] [Google Scholar]
- Alonso-Cantabrana H., Ripoll J. J., Ochando I., Vera A., Ferrandiz C., Martinez-Laborda A. (2007). Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134, 2663–2671 10.1242/dev.02864 [DOI] [PubMed] [Google Scholar]
- Baker C. C., Sieber P., Wellmer F., Meyerowitz E. M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15, 303–315 10.1016/j.cub.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Bilsborough G. D., Runions A., Barkoulas M., Jenkins H. W., Hasson A., Galinha C., et al. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U.S.A. 108, 3424–3429 10.1073/pnas.1015162108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., et al. (2008). A conserved molecular framework for compound leaf development. Science 322, 1835–1839 10.1126/science.1166168 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Groover A. T., Fontana J. R., Martienssen R. A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130, 3941–3950 10.1242/dev.00620 [DOI] [PubMed] [Google Scholar]
- Dinneny J. R., Weigel D., Yanofsky M. F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132, 4687–4696 10.1242/dev.02062 [DOI] [PubMed] [Google Scholar]
- Endrizzi K., Moussian B., Haecker A., Levin J. Z., Laux T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 967–979 10.1046/j.1365-313X.1996.10060967.x [DOI] [PubMed] [Google Scholar]
- Fukaki H., Fujisawa H., Tasaka M. (1996). SGR1, SGR2, and SGR3: Novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol. 110, 945–955 10.1104/pp.110.3.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M., Vernoux T., Traas J., Kato T., Tasaka M., Aida M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030 10.1242/dev.01388 [DOI] [PubMed] [Google Scholar]
- Galbiati F., Sinha Roy D., Simonini S., Cucinotta M., Ceccato L., Cuesta C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant J. 76, 446–455 10.1111/tpj.12309 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reig S., Ripoll J. J., Vera A., Yanofsky M. F., Martinez-Laborda A. (2012). Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet. 8:e1003020 10.1371/journal.pgen.1003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 10.1023/A:1006496308160 [DOI] [PubMed] [Google Scholar]
- Hibara K., Takada S., Tasaka M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36, 687–696 10.1046/j.1365-313X.2003.01911.x [DOI] [PubMed] [Google Scholar]
- Ishida T., Aida M., Takada S., Tasaka M. (2000). Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 41, 60–67 10.1093/pcp/41.1.60 [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Kawamura E., Horiguchi G., Tsukaya H. (2010). Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62, 429–441 10.1111/j.1365-313X.2010.04156.x [DOI] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A., Fletcher J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688–698 10.1038/Nrg1675 [DOI] [PubMed] [Google Scholar]
- Larue C. T., Wen J. Q., Walker J. C. (2009). A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J. 58, 450–463 10.1111/j.1365-313X.2009.03796.x [DOI] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 10.1038/379066a0 [DOI] [PubMed] [Google Scholar]
- Mallory A. C., Dugas D. V., Bartel D. P., Bartel B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046 10.1016/j.cub.2004.06.022 [DOI] [PubMed] [Google Scholar]
- Nahar M. A., Ishida T., Smyth D. R., Tasaka M., Aida M. (2012). Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physio.l 53, 1134–1143 10.1093/pcp/pcs057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945 10.1105/tpc.106.045617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S., Azhakanandam S., Franks R. G. (2010). Polar auxin transport together with AINTEGUMENTA and REVOLUTA coordinate early Arabidopsis gynoecium development. Dev. Biol. 346, 181–195 10.1016/j.ydbio.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Reyes-Olalde J. I., Zuniga-Mayo V. M., Chavez Montes R. A., Marsch-Martinez N., De Folter S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18, 644–655 10.1016/j.tplants.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Roeder A. H. K., Ferrandiz C., Yanofsky M. F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13, 1630–1635 10.1016/j.cub.2003.08.027 [DOI] [PubMed] [Google Scholar]
- Sanders P. M., Bui A. Q., Weterings K., McIntire K. N., Hsu Y. C., Lee P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Rep. 11, 297–322 10.1007/s004970050158 [DOI] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Kanaya E., Morita E. H., Okada K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088 10.1101/gad.13.9.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Murray J. A. H. (2007). The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 50, 767–781 10.1111/j.1365-313X.2007.03095.x [DOI] [PubMed] [Google Scholar]
- Sieber P., Wellmer F., Gheyselinck J., Riechmann J. L., Meyerowitz E. M. (2007). Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060 10.1242/dev.02817 [DOI] [PubMed] [Google Scholar]
- Smith H. M., Hake S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15, 1717–1727 10.1105/tpc.012856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Hibara K., Ishida T., Tasaka M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135 [DOI] [PubMed] [Google Scholar]
- Yamaki S., Nagato Y., Kurata N., Nonomura K. I. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev.Biol. 351, 208–216 10.1016/j.ydbio.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15, 1566–1571 10.1016/j.cub.2005.07.060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.