Abstract
Autophagy is a process in which long-lived proteins, damaged cell organelles, and other cellular particles are sequestered and degraded. This process is important for maintaining the cellular microenvironment when the cell is under stress. Many studies have shown that autophagy plays a complex role in human diseases, especially in cancer, where it is known to have paradoxical effects. Namely, autophagy provides the energy for metabolism and tumor growth and leads to cell death that promotes tumor suppression. The link between autophagy and cancer is also evident in that some of the genes that regulate Carcinogenesis, oncogenes and tumor suppressor genes, participate in or impact the autophagy process. Therefore, modulating autophagy will be a valuable topic for cancer therapy. Many studies have shown that autophagy can inhibit the tumor growth when autophagy modulators are combined with radiotherapy and/or chemotherapy. These findings suggest that autophagy may be a potent target for cancer therapy.
Keywords: Autophagy, chemotherapy, chemoresistance, nuclear factor-κB, P53, Bcl-2
In 1955, de Duve et al.[1] first described the term “autophagy” to distinguish lysosomal degradation, or cellular “eating” (phagy) of self (auto), from the breakdown of extracellular material (heterophagy). So-termed autophagy is a genetically programmed, evolutionarily conserved process that degrades long-lived cellular proteins and organelles[2]. Autophagy contributes to the maintenance of the cellular energy homeostasis, clearance of damaged organelles, and adaptation to environmental stress. Although Seglen et al.[3] studied the early and intermediate steps of this process morphologically using electro-injected radioactive probes, significant breakthroughs in understanding the molecular basis of autophagy only occurred following analyses in the genetically facile yeast system. Since Takeshige et al.[4] carried out the first genetic screen for autophagy mutants, there has been a tremendous increase in autophagy research[5].
There are nearly 30 autophagy-related proteins (ATGs) in mammals, and these have been identified based on investigations in yeast[2]. ATGs, through their role in signaling pathways, maintain normal physiological levels of autophagy. Recent studies have demonstrated that defective autophagy has a complicated correlation with many human diseases, including neuronal degeneration[2], immune disorders[6], myopathy, and cancer[7],[8]. For example, the correlation between autophagy and tumorigenesis is ambiguous, and several studies have shown that autophagy regulators alone and in combination with radiotherapy and/or chemotherapy can inhibit tumor growth. Therefore, modulation of autophagy may be a useful strategy for cancer therapy.
The Classification, Mechanisms, and Regulation of Autophagy
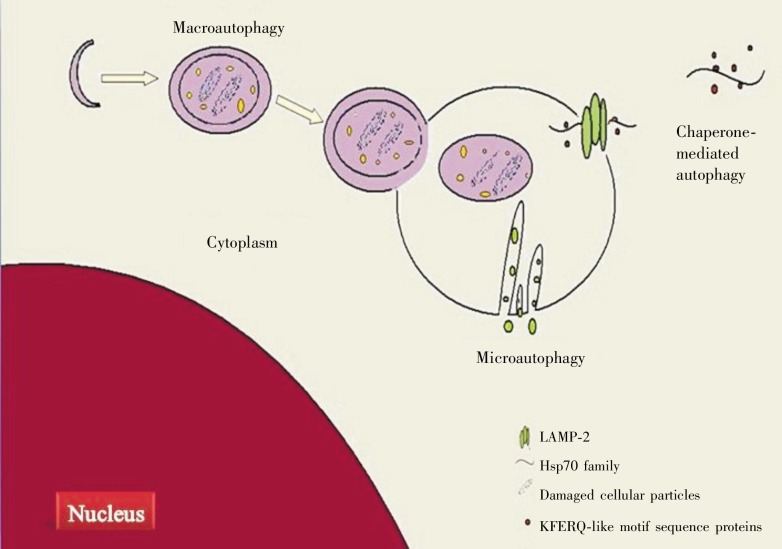
Autophagy is a tightly regulated lysosome-dependent catabolic pathway. During this process, cytosolic particles are sequestered into autophagosomes that subsequently fuse with lysosomes, where their contents are degraded. Also, autophagy can be classified into chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy, based on the pathway of substrate into the lysosome[9] (Figure 1). The type of autophagy that will function depends on the signals, cellular microenvironment, and organs. CMA is only described in mammals and involved in degradation of single, soluble proteins. Delivery of substrate proteins to the lysosome from CMA is not mediated by vesicular transport but executed by a protein translocation pathway through a proteinaceous pore in the lysosomal membrane. The proteins involved in the translocation pathway is a member of the Hsp70 family of chaperones in the cytosol (Hsc70) and the lysosomal lumen (Hsc73) as well as the lysosomal membrane protein LAMP-2A[10]. Cytosolic Hsc70 first recognizes the KFERQ-like motif sequence in proteins and then leads to formation of the Hsc70-cargo complex. In the presence of Hsc73 and other Hsp70 co-chaperones, such as Hsp40, Hsp90, and Hip, the Hsc70-cargo complex is targeted to the lysosomal membrane, where it binds to the cytosolic domain of LAMP-2A, after which degradation occurs. Microautophagy is characterized by the development of tubular invaginations involving the TOR signaling complex, EGO complex (Ego1, Ego3 and the GTPase Gtr2), and vacuolar transport chaperone (VTC) complex (Vtc1, Vtc2, Vtc3 and Vtc4), followed by digestion of small particles[11],[12]. Compared with CMA and microautophagy, macroautophagy is the prevalent autophagy pathway in which long-lived proteins and cellular organelles are enveloped and sequestered in double-membrane vesicles called autophagosomes. Autophagosomes then fuse with lysosomes, where their components are degraded.
Figure 1. The classification of autophagy.
There are three types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). In macroautophagy, damaged cellular particles and long-lived proteins are sequestered and delivered to the lysosome by double-membrane vesicles called autophagosomes. In microautophagy, small particles enter the lysosome through tubular invaginations and undergo digestion. In CMA, substrate proteins are delivered into the lysosome by a member of the Hsp70 family of chaperones in cytosol.
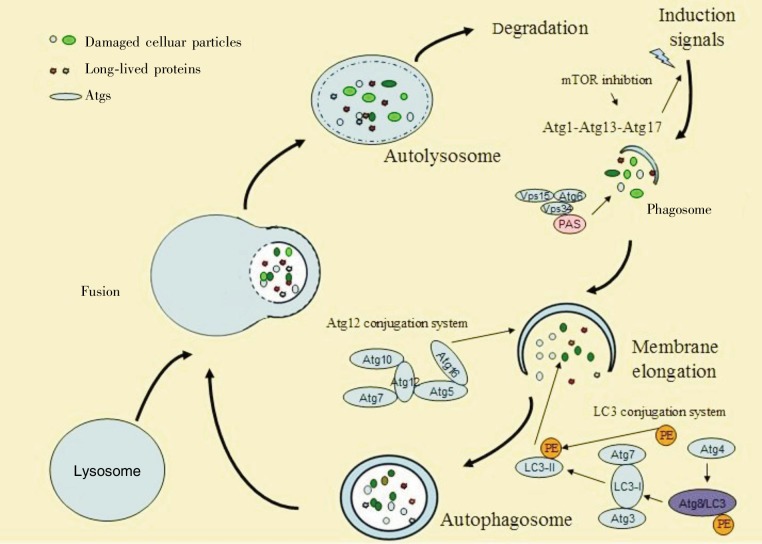
By means of the ATG pathway, autophagy occurs sequentially in several phases: induction; isolation membrane, or phagophore formation; membrane elongation, or autophagosome formation; autolysosome formation; and degradation (Figure 2). The phosphoinositide 3-kinase (PI3K)–protein kinase B (AKT)–mammaliam target of rapamycin (mTOR) axis is involved in autophagy initiation [13]–[15]. Cellular stresses, such as nutrient deficiency, hypoxia, temperature or cell density changes, hormone or growth factor depletion, chemotherapy, and radiotherapy, trigger autophagy through several phosphorylation events within the mTOR complex, which is formed by two distinct complexes termed mTORC1 and mTORC2. mTOR phosphorylation induces formation of the Atg1-Atg13-Atg17 complex and activates autophagy. Recent studies[16],[17] demonstrated that mammalian homologs of ATG1 are Unc-51–like kinases 1 (ULK1) and ULK2. In mammals, the ULK1 complex includes at least three autophagy-related proteins, mammalian ATG13, FIP200 (focal adhesion kinase (FAK) family interacting protein of 200 kDa), and ATG101[18],[19].
Figure 2. The autophagy process and signaling pathway.
Autophagy occurs sequentially in phases: induction, phagophore formation, autophagosome elongation, autolysome formation, and then degradation. Cellular stresses trigger the autophagic pathway by several phosphorylation events within mTOR and drive Atg1-Atg13-Atg17 complex formation. Atgs, such as Vps15, Atg6, and Vps34, then accumulate in the PAS. During the autophagosome elongation phase, two conjugation systems, Atg12 and Atg8/LC, are indispensable for the formation of the autophagosome.
The isolation membrane or so-called phagophore is a crescent-shaped, lipid bilayer membrane characterized by the accumulation of ATGs in pre-autophagosomal structures (PAS), and its generation results from a cascade of reactions induced by the ATG1 complex. In this phase of the autophagy process, the formation of the type III phosphatidylinositol 3-kinase (Ptdlns3K) complex performs a key role in triggering autophagy and its applications in phagophore formation. In mammals, Beclin-1, the homolog of the autophagy-related gene Atg6/vacuolar protein sorting (Vps3), is closely correlated with tumorigenesis; mutations or allelic loss of Beclin-1 are frequently found in breast, ovarian and prostate cancers[20].
Two conjugation systems involved in the elongation phase, ATG12 and ATG8/LC3, are indispensable for autophagosome formation. As the autophagosome matures, the ubiquitin-like protein ATG12 is covalently conjugated to ATG5 through the action of E1- and E2-like proteins ATG7 and ATG10, respectively. Upon the help of ATG7 and ATG10, ATG8 (mammalian homologue LC3) is lipidated by conjugation to phosphatidylethanolamine (PE) [21]. Subsequently, the ATG12-ATG5 dimer and ATG8-PE assemble and are recruited to the autophagosomal membrane via interaction with ATG16. Once the autophagosome is fully expanded, ATG8 is deconjugated from PE through the action of ATG4 and is released back to the cytosol[22]. With this process underway, the autophagosomal membrane becomes lengthened and sequesters cytosolic particles and long-lived proteins further. Through interactions with Rab7 and the SNARE protein Vtilp [23],[24], the outer membrane of the autophagosome fuses to the lysosome to form the autolysosome, where the engulfed cytoplasmic proteins and organelles are degraded by lysosomal hydrolases.
Autophagy occurs at a low level under normal physiological conditions but significantly increases in response to stress, including oxidative stress, chemo-and radiotherapeutic treatments, and nutrient deficiency, suggesting a precise and complicated mechanism in the regulation of autophagy when adapted to the environment change. Although the TOR/mTOR and PI3K/AKT pathways were reported to negatively influence autophagy[5],[25] and the class III PI3K was found to be an efficient, promotive factor[20], the precise mechanisms underlying the autophagy process are still ambiguous.
The Multifaceted and Paradox Correlation between Autophagy and Cancer
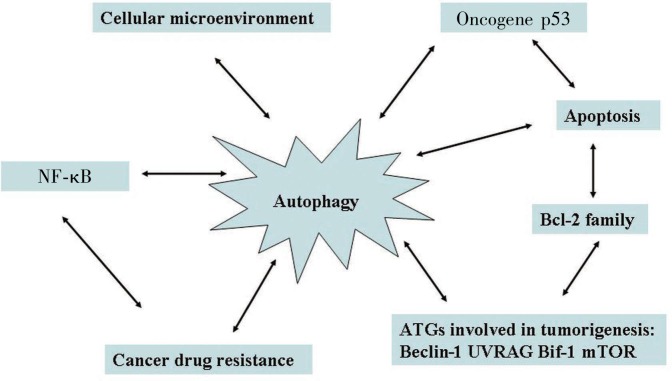
As shown in Figure 3, several interactions between autophagy and other signaling pathways have been recognized to play a functional role in tumorigenesis. Many studies have shown that anti-cancer therapies can induce autophagy in tumor cells, but the role of autophagy in these cells may depend on the type of tumor, the stage of tumorigenesis, and the nature and extent of the insult, suggesting that appropriate modification of autophagy, be it suppression of cell-protective autophagy and enhancement of cell-destructive autophagy, may augment cytotoxicity induced by anti-cancer therapy.
Figure 3. The multifaceted associations between autophagy and tumorigenesis.
Autophagy and tumorigenesis and cancer drug resistance are associated in the following ways: (a) shared signaling pathways such as the PI3K-AKT-mTOR axis[140]; (b) mutual interaction with the cellular microenvironment[141]; (c) mutual regulation of apoptosis via factors such as the Bcl-2 family[142]; and (d) abnormal expression genes (for example, NF-κB[143]), which take part in drug resistance.
The role of the cellular microenvironment and its associated signaling pathways in tumorigenesis
Cell survival, growth, and proliferation require growth factors, abundant nutrients, and oxygen. However, tumor cells are generally deficient in these critical components. For example, when a solid tumor reaches a size of 0.2–2.0 mm in diameter, oxygen, growth factors, and nutrients, including glucose and small molecules like amino acids, cannot efficiently diffuse to cells at the tumor center because of inadequate vascularization[26],[27]. AMP-activated protein kinase (AMPK) is a known key metabolic sensor that monitors the cellular AMP/ATP ratio, then nutrient or energy deprivation trigger AMPK activation. In emerging cancer cells, nutrient or energy deprivation, which is generally characterized as a high AMP or low ATP level, activates AMPK that inhibits mTOR-dependent protein translation through the regulation of the tuberous sclerosis 1 (TSC1)–TSC2 complex or the mTOR-binding protein raptor[28]. mTOR is an autophagy suppressor. Thus, loss of mTOR signaling activates autophagy, which elevates the activities of the cellular catabolic and anabolic processes required to sustain cell survival, growth, and proliferation[29]–[31]. In contrast, defects in autophagy may cause failure of energy homeostasis as well as protein and organelle quality control, leading to accumulation of cellular damage and metabolic stress. Some manifestations of this damage, such as activation of the DNA damage response and generation of genomic instability, may promote tumor initiation and drive cell-autonomous tumor progression. Moreover, in an established tumor, the vasculature is profoundly abnormal and subject to intermittent collapse that similarly creates metabolic stress[32]. In turn, the cells at the tumor center and in sectors of established tumors live in a metabolically stressed microenvironment characterized by hypoxia, pH anomalies, and nutrient deprivation. In these conditions, autophagy eliminates the stress and helps the cells adaption to the harmful signals. However, defects in autophagy impair the survival of tumor cells in these areas, resulting in increased cell death and inflammation. Notably, the cytokine response from inflammation may promote tumor growth and accelerate cell non-autonomous tumor progression[33],[34]. The overarching theme is two-fold. Autophagy protects cells from damaging accumulation under conditions of metabolic stress, allowing efficient stress tolerance and recovery. Furthermore, autophagy is a critical and novel tumor suppressor mechanism.
The hypoxia-inducible factor (HIF) is an evolutionarily conserved transcriptional gene complex consisting of an O2-regulated HIF-1α subunit and a constitutively expressed HIF-1β subunit[35],[36]. HIF-1 activates transcription of the gene encoding the BH3 domain protein BNIP3, which induces selective mitochondrial autophagy by competing with Beclin-1 for binding to Bcl-2, thereby freeing Beclin-1 to trigger autophagy. BNIP3-induced autophagy was originally associated with hypoxic cell death[37]. Recently, however, Bellot et al.[38] demonstrated that the hypoxic microenvironment contributes to survival rather than cell death by inducing autophagy via BNIP3 and BNIP3L.
The involvement of ATGs and their signaling pathway in tumorigenesis
Autophagy is highly regulated and under the control of a number of signaling pathways. The following is a summary of recent research on the regulation of autophagy.
PI3K-AKT-mTOR axis signaling pathway
The PI3K-AKT-mTOR axis, a vital process for initiating the autophagy pathway, regulates several biological events including cell cycle and proliferation and is mutated in many human malignancies[15],[33]. In tumorigenesis, growth signaling constitutively activates receptor tyrosine kinases (RTKs), which then activate Rheb and PI3K. Rheb is a small GTP-binding protein that activates mTOR in its GTP-bound form, whereas Rheb and PI3K converge to activate mTOR to stimulate cell growth and inhibit autophagy[14]. Because most cancers can harbor activating mutations of master regulators, such as TSC1, TSC2, Akt, and ribosomal S6 kinase (RSK)[39], many cancers exhibit enhanced activation of mTOR and inhibit autophagy.
The Vps34 complex signaling pathway
The Vps34 complex, which consists of Vps34, Vps15, ATG6, and ATG14 in yeast and Vp34, Vps15, Beclin-1, the WD domain protein Ambra1, and the endophilin Bif-1 in mammals, is essential for autophagy[40],[41].
Emerging evidence suggests that submembers of Vps34 are involved in tumorigenesis. This is especially true for Beclin-1, a phylogenetically conserved protein that is essential for autophagy. In fact, the first association between autophagy and cancer was the landmark discovery of Beclin-1, which is also a haploinsufficient tumor suppressor[2]. In 1999, Liang et al. [42] used gene transfer techniques in human MCF7 breast carcinoma cells and found that the autophagy promoting activity of Beclin-1 in these cells was associated with inhibition of cellular proliferation. Furthermore, increasing studies show that the Beclin-1 locus (17q21) is frequently subjected to monoallelic deletions in human breast, ovarian, and prostate cancers as well as in brain tumors[43]–[45]. Further study demonstrated that Beclin-mediated tumor suppress involved a Beclin-1 domain. Through this domain, Beclin-1 directly interact with ultraviolet irradiation resistance-associated gene (UVRAG) and this interaction is purported to promote binding of Vps34 to Beclin-1. In addition, Bif-1, also known as SH3GLB1 or endophilin B1, was originally discovered as a Bax-binding protein, interacts with Beclin-1 via UVRAG and promotes Vps34 activation and autophagosome formation. In vivo, UVRAG has been reported to suppress cell proliferation and tumorigenesis, and monoallelic deletions or mutations in UVRAG occur in numerous human malignancies[23],[40]. Furthermore, reduction of Bif-1 expression was observed in gastric carcinomas, invasive urinary bladder, and gallbladder cancers, and mantle cell lymphomas[46]–[48].
Both ATG12 conjugation and LC3 modification (ATG8 lipidation in yeast) have been reported to take part in the dynamic process of autophagosome formation; ATG12 conjugation is essential for the formation of preautophagosomes, and LC3 modification is necessary for the formation of autophagosomes[23]. Recent studies suggest that several key regulators of autophagosome in these two conjugation systems, ATG12 and LC3 conjugations, especially those also associated with apoptosis, are correlated with tumorigenesis. For example, ATG5, playing a role in ATG12 conjugation in the procedure of autophagosome membrane elongation and completion, is reported to interact with Bcl-xL. Kang et al.[49] reported that frameshift mutations of ATG5 in gastric and colorectal cancers may contribute to cancer development by dysregulating the autophagy process. Lee et al.[50] recently demonstrated that cellular and viral FLICE-like inhibitor protein (FLIPs), which protect cells from apoptosis mediated by death receptors, limit the ATG3-mediated step of LC3 modification to regulate autophagosome biogenesis. Nevertheless, the precise mechanism linking autophagy-related conjugations and tumorigenesis is still unknown and requires further investigation.
Autophagy and apoptosis: a complex interaction through the Bcl-2 family and p53 signaling pathways
Bcl-2 family–mediated signaling pathway
The multifunctional Bcl-2 family of proteins contains members that inhibit (Bcl-2, Bcl-xL, Mcl-1, and Bcl-w) or promote (Bax and Bak) apoptosis. The anti-apoptotic members of this family have been reported to suppress autophagy, mainly by interacting with and therefore inhibiting Beclin-1, whereas the pro-apoptotic members of this family and other BH3-only proteins, including Bad, Bik, BNI-P3L, Noxa, Puma, and BimEL, promote autophagy by freeing Beclin-1 from inhibitory interactions with anti-apoptotic Bcl-2–like proteins. Such interactions involve the BH3 domain of Beclin-1 and the so-called “BH3 receptor” domain of anti-apoptotic Bcl-2 family members. Pattingre et al.[51] demonstrated that activation of c-Jun N-terminal kinase 1 (JNK1) by tamoxifen-induced Ceramide required both Bcl-2 phosphorylation and autophagy stimulation, which contributed to the development of anti-estrogen resistance.
p53-mediated signaling pathway
p53, a typical tumor suppressor that is activated by DNA damage- induced stress, Arf activation, and re-expression of p53 in p53-negative tumor cells, has been found to be associated with autophagy [52],[53]. Tasdemir et al.[54],[55] showed that p53 inhibition in enucleated cells increased autophagy, whereas expression of cytoplasmic p53 repressed the enhanced autophagy in p53-null cells. p53 activation can trigger autophagy by up-regulating the transcription of damage-regulated autophagy modulator (DRAM) or by inhibiting mTOR[56]. DRAM1 causes accumulation of autophagosomes, though the underlying mechanism is unknown. Furthermore, because it is specifically located in the lysosome, DRAM1 may regulate the autophagosome-lysosome fusion that is required for the generation of autophagolysosomes during a late stage of the autophagy process. Therefore, it is plausible that DRAM1 regulates the autophagosome-lysosome fusion that is required for the generation of autophagolysosomes.
Autophagy and nuclear factor-kappa B
The nuclear factor-kappa B (NF-κB) system is a critical signaling pathway induced to defend cells from cellular damage and environmental danger[57]. Dysregulation of the NF-κB pathway is associated with cancer development, progression, and drug resistance, in addition to other human conditions, such as inflammatory diseases[58]. Recently, autophagy has been reported to play a role in several cellular functions regulated by NF-κB. Djavaheri-Mergny et al.[59] demonstrated that NF-κB activation mediates autophagy repression through effects on mTOR complex in tumor necrosis factor-alpha (TNF-α)–treated Ewing sarcoma cells. Further, Lee et al.[60] reported that autophagy repression involved suppression of TSC1, which triggered the mTOR pathway. Dan et al.[61] have also demonstrated that activation of mTOR in PTEN-deficient cancer cells involves IκB kinase (IKK)–α, a catalytic subunit of the IKK complex that controls NF-κB activation.
In addition to its role in repressing autophagy, NF-κB has recently been found to activate autophagy. Copetti et al.[62] demonstrated that the NF-κB family member p65/RelA (v-rel reticuloendotheliosis viral Oncogene homolog A) up-regulated Beclin-1 mRNA and protein levels in different cellular systems. Furthermore, they blocked p65 signaling pathway that could led to a decrease in Beclin-1 transcription. Protein Beclin-1 can physically act in association with another protein (PI3KIII/Vps34) as a platform from which to recruit activators and inhibitors of autophagy, then to finely regulate autophagosome formation. Nevertheless, the precise link between NF-κB signaling and autophagic degradation remains to be elucidated.
Autophagy and cancer drug resistance
Acquired drug resistance in cancer cells is the major cause of failure for cancer therapy. Cancer drug resistance is complex, dynamic, and “elusive, ” and it is affected by the following factors: (1) decreased influx and, because of drug transporters, increased efflux of anti-cancer drugs; (2) activation of DNA repair; (3) activation of detoxification, and (4) blockage of apoptosis[63],[64]. Among these factors, current pharmacological approaches aim to restore the efficacy of the standard chemotherapy against drug-resistant cancers by reactivating apoptosis. However, resistance to apoptosis is considered a characteristic of many diverse cancer cells. Recent studies suggest that the autophagy and apoptosis pathways overlap[48]. Furthermore, many studies have shown that molecules that modulate or are involved in the autophagy process, when combined with chemotherapy agents, can reverse drug resistance[65],[66].
5-fluorouracil (5-FU)–based chemotherapy has been reported to be an effective protocol for improving outcome and reducing tumor recurrence rate. Unfortunately, 5-FU resistance is a major cause of failure in colorectal cancer therapy[67]. Recently, Li et al.[68] showed that 5-FU induced apoptosis as well as autophagy, and combination treatment with 5-FU and 3-methyladenine (3-MA), which inhibits autophagy, significantly increased apoptotic cell death. Sasaki et al.[69] reported that combining chloroquine with 5-FU improved efficacy of 5-FU on colon cancer cells. O'Donovan et al.[70] inhibited early autophagy induction using siRNA targeting Beclin-1 and found that ATG7 significantly enhanced the effect of 5-FU and reduced the recovery of drug-treated esophageal cancer cells. These observations suggest that an autophagic response to chemotherapy may function as a survival mechanism that promotes chemoresistance and that selective inhibition of autophagy regulators has the potential to improve chemotherapeutic regimes.
Cancer Therapy and Targeted Autophagy
In recent years, autophagy has been recognized as an important regulator of cancer development and progression and a key factor in determining tumor cell sensitivity to anti-cancer therapy. Numerous conventional and experimental anti-tumor strategies, including combination therapies with autophagy regulators, have shown that targeting autophagy is beneficial in cancer therapy[5],[8] (Table 1). However, autophagy is a complicated process that can have distinct and conflicting consequences for tumor growth depending on given conditions[48].
Table 1. Reports of anti-cancer drugs and chemical compounds and their effect on autophagy in cancer therapy.
| Regulator | Signaling pathway | Targeted cells | Regulation pattern | Synergistic role in combined chemotherapy | Clinical trial | References |
| Single compound | ||||||
| Arsenic trioxide (As2o3) | BNIP | Malignant glioma (MG) | Induction | Cytotoxicity | Yes | [73], [74], [96], [97] |
| Rapamycin and RAD-001 CCI-779 | mTOR | Lung cancer, MG | Induction | Cytotoxicity | Yes | [71], [87], [113] |
| PEITC | Atg5 | Prostate cancer cells | Induction | Cytotoxicity | No | [80], [114] |
| Temozolomide | PTEN | Malignant glioma | Induction | Cytotoxicity | Yes | [76],[101],[104],[115] |
| Dopamine | MAPK-JNK-P38 axis | Neuroblastoma | Induction | Cytotoxicity | No | [116] |
| K5 | Bcl-2-Beclin1 | Endothelial cells | Induction | Cytotoxicity | No | [32], [79] |
| OSU-03012 | ROS | HepG cells | Induction | Cytotoxicity | No | [81], [117] |
| BCG/CWS | TLR2, TLR4 | Colon cancer cells | Induction | Cytotoxicity | No | [84] |
| NVP-BEZ235 | mTOR | Gliomas | Induction | Cytotoxicity | No | [82], [83] |
| Combined compound | ||||||
| Tamoxifen (Ceramide) | Bcl-2-Beclin1 | Breast cancer cells | Induction | Protection | Yes | [85], [118], [119] |
| 3-MA (5-FU) | PI3K | Colon cancer cell | Inhibition | Protection | No | [68], [120] |
| Chloroquine (FK228 (depsipeptide)) | PI3K | Lymphoma model | Inhibition | Protection | No | [121] |
| Bafilomycin A1 (Imatinib) | Vacuolar-ATPase | Colon cancer cells | Inhibition | Protection | No | [110] |
| proteasome inhibitor MG-132 (3-MA) | mTOR pathway | Gastric cancer cell | Induction | Protection | No | [86], [122] |
| HDAC,SAHA (Chloroquine) | HDAC inhibitor | Cervical cancer | Induction | Cytotoxicity | Yes | [123], [124] |
| Pitavastatin (radiation) | NF-κB | Malignant glioma | Induction | Cytotoxicity | Yes | [125] |
| Imatinib(Gleevac) (3-MA) | bcr-abl, c-abl, c-kit, PDGFRa and b | GM, leukaemia, GIST | Induction | Cytotoxicity | Yes | [126]–[128] |
| Evodiamine (5-FU) | ROS | HeLa cells | Induction | Protection | No | [129] |
| Resveratrol (Dihydroceramide) | Ceremide | Ovarian cancer | Induction | Cytotoxicity | Yes | [130]–[132] |
| 5-FU | mTOR | Human colon cancer cell | Induction | Cytotoxicity | Yes | [68], [120] |
| P38 inhibitors (SB202190) | HIF1α-FoxO3A | Ovarian cancer cells | Induction | Cytotoxicity | No | [133]–[135] |
| Vincristine | MD and LD | HeLa cells | Induction | Cytotoxicity | Yes | [136] |
| Siramesine | lysosomal pH | MCF-7 breast carcinoma cells | Induction* | Cytotoxicity | Yes | [137] |
| Synergistic | ||||||
| Oncolytic adenovirus | ? | Glioblastoma | Induction | Cytotoxicity | No | [107], [138], [139] |
| Dasatinib | Akt | Glioblastoma | Induction | Cytotoxicity | No | [108] |
TLR, toll-like receptor; PEITC, phenethyl isothiocyanate; K5, kringle domains of plasminogen (endostatin kringle 5); BCG/CWS, bovis bacillus calmette-guerin; 3-MA, 3-methyladenine; HDAC, SAHA, histone deacetylase inhibitors, suberoylanilide hydroxamic acid; 5-FU, 5-fluorouracil; ROS, reactive oxygen species; MD, microtubule-disturbing; LD, lysosome-destabilizing; GIST, gastrointestinal stromal tumors. Induction*, induce but decreased autophagosome turnover. The question mark (?) indicates that it is still ambiguous or unknown.
Cancer therapy applications of autophagy regulators as single agents
Strategies to use regulators of autophagy as single agents to induce cytotoxicity in tumor cells have been developed in the last year. These agents include rapamycin[71],[72], arsenic trioxide (As2O3) [73]–[75], temozolomide[76]–[78], kringle domains of plasminogen (endostatin kringle 5 ) [79], phenethyl isothiocyanate (PEITC) [80], OSU-03012 (derivative of celecoxib) [81], NVP-BEZ235[82],[83], and cell wall skeleton of Mycobacterium bovis Bacillus Calmette-Guerin (BCG/CWS)[84]. In addition, some autophagic inhibitors, such as 3-MA, chloroquine, bafilomycin A1, tamoxifen, and proteasome inhibitor MG-132[51],[85],[86] (Table 1), also inhibit tumorigenesis by playing a protective role in autophagy.
Utilization of the first prototype of an mTOR inhibitor, rapamycin, is limited due to the poor aqueous solubility and strong immunosuppressive properties[71]. However, its analogs, temsirolimus (CCI-779), everolimus (RAD-001), and deforolimus (AP23573), are used in the clinical and preclinical setting. In particular, temsirolimus was the first mTOR inhibitor approved by the US Food and Drug Administration for cancer treatment, and it is considered the first-line treatment for advanced renal cell carcinoma patients with poor prognostic features[87]–[91]. mTOR inhibitors bind to an intracellular protein, FK506 binding protein 12 (FKBP-12), forming a complex that inhibits the mTOR kinase. Randomized phase III trials have already demonstrated significant clinical benefits of treatment with single-agent temsirolimus in advanced renal cell carcinoma and relapsed and/or refractory mantle cell lymphoma[91]–[93]. Phase I and II trials have also been carried out in other malignancies, such as glioblastoma[94], breast cancer[95], endometrial cancer[93], non-Hodgkin lymphomas[92], and multiple myeloma[93].
In contrast to mTOR inhibitors, As2O3 has an elusive capacity for autophagy induction[73]–[75],[93]. Kanzawa et al.[97],[98] showed that As2O3 triggered autophagic cell death in U-373 malignant glioma cells with BNIP and BNIPL up-regulation. However, autophagy played complex roles in the As2O3-induced death of human acute myeloid leukemia cell line HL60 cells. A study by Yang et al.[74] showed that inhibiting autophagy at an early stage with 3-MA 1 h before As2O3 provoked As2O3-induced death of HL60 cells, whereas 3-MA administered 30 min after As2O3 attenuated As2O3-induced death. Therefore, they concluded that autophagy inhibits As2O3-induced apoptosis during the initiation stage of the autophagy process but amplifies the As2O3-mediated apoptotic program if it is persistently activated. In contrast, Charoensuk et al.[75] reported that As2O3 induced acute promyelocytic leukemia cell death in HL-60 cells via apoptosis but not autophagy. These findings suggest that the As2O3-induced effects depend on the tumor tissue type: it triggers autophagy in solid tumors, whereas it induces cell death by apoptosis in hematological neoplasms. Currently, As2O3 is in clinical trials in gliomas, advanced solid tumors, metastatic liver cancer, and pediatric solid tumors[99],[100].
In addition to the aforementioned agents, new compounds have emerged as promising cancer chemopreventive agents because of their ability to induce autophagic cell death. These compounds include phenethyl isothiocyanate (PEITC), the celecoxib derivative OSU-03012, and kringle domains of plasminogen (endostatin kringle 5, K5). A study by Bommareddy et al.[80] showed that PEITC treatment triggered ATG5-dependent autophagic cell death in human prostate cancer cells, suggesting it may be an effective anti-tumor agent. The mechanism of action of OSU-03012 is presumably 3-phosphoinositide-dependent kinase 1 (PDK1) inhibition. Gao et al.[81] recently demonstrated that OSU-03012 induced autophagic cell death, which is related to the accumulation of reactive oxygen species (ROS) in hepatocellular carcinoma. Ramakrishnan et al.[32] reported that K5, a well-characterized and potential angiogenesis inhibitor, could induce autophagy in endothelial cells with alterations in the Beclin-1–Bcl-2 complex.
Temozolomide (TMZ), a DNA alkylating agent, was reported to reduce the viability of malignant glioma cells in a dose-dependent manner and induce G2/M arrest, and it has been used clinically for the treatment of malignant gliomas[101]. Although the current standard treatment for malignant gliomas is surgical resection followed by adjuvant radiotherapy plus TMZ, this type of cancer continues to maintain a dismal prognosis. O6-alkylguanine-DNA alkyltransferase (AGT) is a DNA repair enzyme that is known to limit the efficacy of TMZ in glioblastoma cells[102]. However, Fu et al.[103] reported that the possible molecular mechanisms underlying TMZ resistance in glioblastoma cells involve down-regulation of autophagy-related proteins in CD133+ cells. In contrast, a recent study carried out by Gao et al.[104] suggested that thalidomide enhanced the cytotoxicity of TMZ by promoting autophagy, which contributed to the up-regulation of PTEN by thalidomide. There is no doubt that more compounds that induce cell death through autophagy will be developed in the near future.
Combined applications of two autophagic regulators to synergistically kill cancer cells
A series of recent studies have reported that the combination of two or more autophagic regulators, especially proautophagic regulators, markedly enhance autophagic cell death. Oncolytic adenoviruses, promising tools in cancer therapy, induce autophagy as a part of their therapeutic effects[105]. Yokoyama et al.[106] jointly used OBP-405, an oncolytic adenovirus regulated by the human telomerase reverse transcriptase promoter with a tropism modification (RGD), as well as TMZ and rapamycin for an inhibition assay in glioblastoma cells. They found that tumor cells were synergistically sensitized to OBP-405 upon stimulation of the autophagic pathway. Ulasov et al.[107] used conditionally replicative adenoviruses (CRAds) with TMZ in the setting of experimental glioma in vivo and in vitro and observed, by Western blotting, increased expression of Bax and p53, elevated levels of Atg5, and decreased expression of Bcl-2. In addition to oncolytic adenoviruses, dasatinib, an ATP-competitive, small-molecule inhibitor, has also been used in combination therapies to induce autophagic cell death. Dasatinib is used to treat drug-resistant tumors expressing mutant BCR-ABL, KIT, or epidermal growth factor receptor (EGFR) by blocking tyrosine phosphorylation sites and inducing a significant increase in autophagic cell death. Milano et al.[108] demonstrated that combination treatment of glioma cells with dasatinib and TMZ resulted in a significant increase in cell cycle disruption and autophagic cell death, which resulted in increased therapeutic efficacy compared to the combination of dasatinib with carboplatin or irinotecan. Therefore, combination of two or more autophagic regulators may be helpful for cancer therapy and/or reverse the cancer drug resistance.
Combined applications of autophagy regulators and chemotherapy in cancer treatment
The idea of inhibiting tumor growth through autophagy blockade was conceived from observations of the role of autophagy and its Cytoprotective effects during chemo- and radiotherapy in tumor cells. As mentioned above, malignant neoplasms are characterized by excessive proliferation, which results in an imbalance in supportive nutrients, hypoxia, and oxidative stress in tumor tissues. In these conditions, tumor cell and tissue survival is dependent upon the initiation of the autophagic process and glycolysis[109]. A number of conventional anti-tumor agents have been assayed and have been found to trigger autophagy as protective mechanism (Table 1).
Many specific autophagy inhibitors, such as 3-MA, chloroquine, bafilomycin A1, represent a general strategy to sensitize cancer cells to conventional chemotherapy[8],[68],[110]. Tamoxifen, the most commonly used anti-estrogen, exerts its pharmacological action by binding to estrogen receptor alpha (ERα) and blocking the growth-promoting action of the estrogen-bound receptor in breast cancer cells. However, the development of anti-estrogen resistance has become a major impediment in the treatment of ERα-positive breast cancer. Samaddar et al.[111] reported that autophagy plays a critical role in the development of anti-estrogen resistance. More specifically, overexpression of Beclin-1 down-regulated estrogenic signaling and growth response and contributed to the development of anti-estrogen resistance. Recently, Pattingre et al.[51],[112] demonstrated that tamoxifen stimulated Ceramide synthesis de novo, which induced dissociation of the Beclin-1–Bcl-2 complex. Further, activation of JNK1 by Ceramide was required both to phosphorylate Bcl-2 and to stimulate autophagy. Several compounds and chemotherapeutic agents have been reported to induce autophagy that protects against chemotherapy-induced cytotoxicity. However, autophagy inhibitors combined with these agents augment their killing capacity.
Conclusions and Perspectives
In summary, autophagy is a ubiquitous process in eukaryotic cells that results in the breakdown of cytoplasm within the lysosome in response to stress conditions, which allows the cell to adapt to environmental and/or developmental changes. However, autophagic cell death can result from excessive autophagy, which is associated with conditions such as neurodegerative diseases, heart diseases, cancer, and others. Therefore, further exploration of the role of autophagy in human disease will be important and promising work. The present research shows that appropriate modification of autophagy, that is, suppression of cell-protective autophagy or enhancement of cell-killing autophagy, can augment cytotoxicity caused by anti-cancer therapy. Hence, modulating autophagy will open new areas for cancer therapy.
Acknowledgments
The authors thank Professor Shou-Rong Ji from the Department of Foreign Language, Wenzhou University, for her help in editing this manuscript.
References
- 1.de Duve C, Pressman BC, Gianetto R, et al. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue [J] Biochem J. 1955;60(4):604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion [J] Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seglen PO, Berg TO, Blankson H, et al. Structural aspects of autophagy [J] Adv Exp Med Biol. 1996;389:103–111. doi: 10.1007/978-1-4613-0335-0_12. [DOI] [PubMed] [Google Scholar]
- 4.Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction [J] J Cell Biol. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease [J] Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu B, Finn OJ. T-cell death and cancer immune tolerance [J] Cell Death Differ. 2008;15(1):70–79. doi: 10.1038/sj.cdd.4402274. [DOI] [PubMed] [Google Scholar]
- 7.Fidzianska A, Bilinska ZT, Walczak E, et al. Autophagy in transition from hypertrophic cardiomyopathy to heart failure [J] J Electron Microsc (Tokyo) 2010;59(2):181–183. doi: 10.1093/jmicro/dfp048. [DOI] [PubMed] [Google Scholar]
- 8.Marx J. Autophagy: is it cancer's friend or foe? [J] Science. 2006;312(5777):1160–1161. doi: 10.1126/science.312.5777.1160. [DOI] [PubMed] [Google Scholar]
- 9.Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease [J] Biochim Biophys Acta. 2009;1792(1):3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Bandyopadhyay U, Cuervo AM. Entering the lysosome through a transient gate by chaperone-mediated autophagy [J] Autophagy. 2008;4(8):1101–1103. doi: 10.4161/auto.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uttenweiler A, Schwarz H, Neumann H, et al. The vacuolar transporter chaperone (VTC) complex is required for microautophagy [J] Mol Biol Cell. 2007;18(1):166–175. doi: 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krick R, Muhe Y, Prick T, et al. Piecemeal microautophagy of the nucleus: genetic and morphological traits [J] Autophagy. 2009;5(2):270–272. doi: 10.4161/auto.5.2.7639. [DOI] [PubMed] [Google Scholar]
- 13.Liu JJ, Lin M, Yu JY, et al. Targeting apoptotic and autophagic pathways for cancer therapeutics [J] Cancer Lett. 2011;300(2):105–114. doi: 10.1016/j.canlet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Ghayad SE, Cohen PA. Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients [J] Recent Pat Anticancer Drug Discov. 2010;5(1):29–57. doi: 10.2174/157489210789702208. [DOI] [PubMed] [Google Scholar]
- 15.Maiuri MC, Tasdemir E, Criollo A, et al. Control of autophagy by oncogenes and tumor suppressor genes [J] Cell Death Differ. 2009;16(1):87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 16.Chan EY, Longatti A, McKnight NC, et al. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism [J] Mol Cell Biol. 2009;29(1):157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan EY, Tooze SA. Evolution of Atg1 function and regulation [J] Autophagy. 2009;5(6):758–765. doi: 10.4161/auto.8709. [DOI] [PubMed] [Google Scholar]
- 18.Ganley IG, Lam du H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy [J] J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy [J] Autophagy. 2009;5(5):649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 20.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34 [J] Biochem J. 2008;410(1):1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation [J] Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 22.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion [J] Cell. 2007;130(1):165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Sun Q, Fan W, Zhong Q. Regulation of Beclin-1 in autophagy [J] Autophagy. 2009;5(5):713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda M, Itoh T. Direct link between Atg protein and small GTPase Rab: Atg16L functions as a potential Rab33 effector in mammals [J] Autophagy. 2008;4(6):824–826. doi: 10.4161/auto.6542. [DOI] [PubMed] [Google Scholar]
- 25.Eisenberg-Lerner A, Bialik S, Simon HU, et al. Life and death partners: apoptosis, autophagy and the cross-talk between them [J] Cell Death Differ. 2009;16(7):966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 26.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth [J] Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bursch W, Karwan A, Mayer M, et al. Cell death and autophagy: cytokines, drugs, and nutritional factors [J] Toxicology. 2008;254(3):147–157. doi: 10.1016/j.tox.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Guan KL. AMP-activated protein kinase and cancer [J] Acta Physiol (Oxf) 2009;196(1):55–63. doi: 10.1111/j.1748-1716.2009.01980.x. [DOI] [PubMed] [Google Scholar]
- 29.Ikenoue T, Hong S, Inoki K. Monitoring mammalian target of rapamycin (mTOR) activity [J] Methods Enzymol. 2009;452:165–180. doi: 10.1016/S0076-6879(08)03611-2. [DOI] [PubMed] [Google Scholar]
- 30.Degtyarev M, De Maziere A, Orr C, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents [J] J Cell Biol. 2008;183(1):101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinojima N, Yokoyama T, Kondo Y, et al. Roles of the Akt/ mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy [J] Autophagy. 2007;3(6):635–637. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan S, Nguyen TM, Subramanian IV, et al. Autophagy and angiogenesis inhibition [J] Autophagy. 2007;3(5):512–515. doi: 10.4161/auto.4734. [DOI] [PubMed] [Google Scholar]
- 33.Morselli E, Galluzzi L, Kepp O, et al. Anti- and pro-tumor functions of autophagy [J] Biochim Biophys Acta. 2009;1793(9):1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Tsuchihara K, Fujii S, Esumi H. Autophagy and cancer: dynamism of the metabolism of tumor cells and tissues [J] Cancer Lett. 2009;278(2):130–138. doi: 10.1016/j.canlet.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 35.Papandreou I, Lim AL, Laderoute K, et al. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L [J] Cell Death Differ. 2008;15(10):1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 36.Mazure NM, Pouyssegur J. Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia [J] Autophagy. 2009;5(6):868–869. doi: 10.4161/auto.9042. [DOI] [PubMed] [Google Scholar]
- 37.Tafani M, Schito L, Anwar T, et al. Induction of autophagic cell death by a novel molecule is increased by hypoxia [J] Autophagy. 2008;4(8):1042–1053. doi: 10.4161/auto.7070. [DOI] [PubMed] [Google Scholar]
- 38.Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains [J] Mol Cell Biol. 2009;29(10):2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoki K, Guan KL. Tuberous sclerosis complex, implication from a rare genetic disease to common cancer treatment [J] Hum Mol Genet. 2009;18(R1):R94–R100. doi: 10.1093/hmg/ddp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy [J] Cell Death Differ. 2009;16(7):947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin-1-PI3K complexes [J] Autophagy. 2009;5(4):534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 42.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by Beclin-1 [J] Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 43.Karantza-Wadsworth V, White E. Role of autophagy in breast cancer [J] Autophagy. 2007;3(6):610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiPaola RS, Dvorzhinski D, Thalasila A, et al. Therapeutic starvation and autophagy in prostate cancer: a new paradigm for targeting metabolism in cancer therapy [J] Prostate. 2008;68(16):1743–1752. doi: 10.1002/pros.20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y, Li DD, Wang LL, et al. Decreased expression of autophagy-related proteins in malignant epithelial ovarian cancer [J] Autophagy. 2008;4(8):1067–1068. doi: 10.4161/auto.6827. [DOI] [PubMed] [Google Scholar]
- 46.Lee JW, Jeong EG, Soung YH, et al. Decreased expression of tumour suppressor Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric carcinomas [J] Pathology. 2006;38(4):312–315. doi: 10.1080/00313020600820880. [DOI] [PubMed] [Google Scholar]
- 47.Kim SY, Oh YL, Kim KM, et al. Decreased expression of Bax-interacting factor-1 (Bif-1) in invasive urinary bladder and gallbladder cancers [J] Pathology. 2008;40(6):553–557. doi: 10.1080/00313020802320440. [DOI] [PubMed] [Google Scholar]
- 48.Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy [J] Apoptosis. 2009;14(4):376–391. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 49.Kang MR, Kim MS, Oh JE, et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability [J] J Pathol. 2009;217(5):702–706. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- 50.Lee JS, Li Q, Lee JY, et al. FLIP-mediated autophagy regulation in cell death control [J] Nat Cell Biol. 2009;11(11):1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattingre S, Bauvy C, Levade T, et al. Ceramide-induced autophagy: to junk or to protect cells? [J] Autophagy. 2009;5(4):558–560. doi: 10.4161/auto.5.4.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bialik S, Kimchi A. Autophagy and tumor suppression: recent advances in understanding the link between autophagic cell death pathways and tumor development [J] Adv Exp Med Biol. 2008;615:177–200. doi: 10.1007/978-1-4020-6554-5_9. [DOI] [PubMed] [Google Scholar]
- 53.Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death [J] Autophagy. 2007;3(1):72–74. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 54.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53 [J] Nat Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tasdemir E, Chiara Maiuri M, Morselli E, et al. A dual role of p53 in the control of autophagy [J] Autophagy. 2008;4(6):810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 56.O'Prey J, Skommer J, Wilkinson S, et al. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy [J] Cell Cycle. 2009;8(14):2260–2265. doi: 10.4161/cc.8.14.9050. [DOI] [PubMed] [Google Scholar]
- 57.Karin M. Nuclear factor-kappaB in cancer development and progression [J] Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 58.Wong ET, Tergaonkar V. Roles of NF-kappaB in health and disease: mechanisms and therapeutic potential [J] Clin Sci (Lond) 2009;116(6):451–465. doi: 10.1042/CS20080502. [DOI] [PubMed] [Google Scholar]
- 59.Djavaheri-Mergny M, Amelotti M, Mathieu J, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy [J] J Biol Chem. 2006;281(41):30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 60.Lee DF, Kuo HP, Chen CT, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway [J] Cell. 2007;130(3):440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 61.Dan HC, Baldwin AS. Differential involvement of IkappaB kinases alpha and beta in cytokine- and insulin-induced mammalian target of rapamycin activation determined by Akt [J] J Immunol. 2008;180(11):7582–7589. doi: 10.4049/jimmunol.180.11.7582. [DOI] [PubMed] [Google Scholar]
- 62.Copetti T, Bertoli C, Dalla E, et al. p65/RelA modulates BECN1 transcription and autophagy [J] Mol Cell Biol. 2009;29(10):2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Connor R. A review of mechanisms of circumvention and modulation of chemotherapeutic drug resistance [J] Curr Cancer Drug Targets. 2009;9(3):273–280. doi: 10.2174/156800909788166583. [DOI] [PubMed] [Google Scholar]
- 64.Mayur YC, Peters GJ, Prasad W, et al. Design of new drug molecules to be used in reversing multidrug resistance in cancer cells [J] Curr Cancer Drug Targets. 2009;9(3):298–306. doi: 10.2174/156800909788166619. [DOI] [PubMed] [Google Scholar]
- 65.Carew JS, Medina EC, Esquivel JA, 2nd, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation [J] J Cell Mol Med. 2010;14(10):2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ondrouskova E, Soucek K, Horvath V, et al. Alternative pathways of programmed cell death are activated in cells with defective caspase-dependent apoptosis [J] Leuk Res. 2008;32(4):599–609. doi: 10.1016/j.leukres.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 67.Segal NH, Saltz LB. Evolving treatment of advanced colon cancer [J] Annu Rev Med. 2009;60:207–219. doi: 10.1146/annurev.med.60.041807.132435. [DOI] [PubMed] [Google Scholar]
- 68.Li J, Hou N, Faried A, et al. Inhibition of autophagy by 3-MA enhances the effect of 5-FU induced apoptosis in colon cancer cells [J] Ann Surg Oncol. 2009;16(3):761–771. doi: 10.1245/s10434-008-0260-0. [DOI] [PubMed] [Google Scholar]
- 69.Sasaki K, Tsuno NH, Sunami E, et al. Chloroquine potentiates the anti-cancer effect of 5-fluorouracil on colon cancer cells [J] BMC Cancer. 2010;10:370. doi: 10.1186/1471-2407-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Donovan TR, O'Sullivan GC, McKenna S. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics [J] Autophagy. 2011;7(6):509–524. doi: 10.4161/auto.7.6.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torguson R, R Waksman. Overview of the 2007 Food and Drug Administration Circulatory System Devices Panel meeting on the Xience V Everolimus-Eluting Coronary Stent [J] Am J Cardiol. 2008;102(12):1624–1630. doi: 10.1016/j.amjcard.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 72.Wu VT, Tan HL, Huang Q, et al. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy [J] Autophagy. 2009;5(6):824–834. doi: 10.4161/auto.9099. [DOI] [PubMed] [Google Scholar]
- 73.Chiu HW, Ho SY, Guo HR, et al. Combination treatment with arsenic trioxide and irradiation enhances autophagic effects in U118-MG cells through increased mitotic arrest and regulation of PI3K/Akt and ERK1/2 signaling pathways [J] Autophagy. 2009;5(4):472–483. doi: 10.4161/auto.5.4.7759. [DOI] [PubMed] [Google Scholar]
- 74.Yang YP, Liang ZQ, Gao B, et al. Dynamic effects of autophagy on arsenic trioxide-induced death of human leukemia cell line HL60 cells [J] Acta Pharmacol Sin. 2008;29(1):123–134. doi: 10.1111/j.1745-7254.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- 75.Charoensuk V, Gati WP, Weinfeld M, et al. Differential cytotoxic effects of arsenic compounds in human acute promyelocytic leukemia cells [J] Toxicol Appl Pharmacol. 2009;239(1):64–70. doi: 10.1016/j.taap.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 76.Zustovich F, Lombardi G, Della Puppa A, et al. A phase II study of cisplatin and temozolomide in heavily pre-treated patients with temozolomide-refractory high-grade malignant glioma [J] Anticancer Res. 2009;29(10):4275–4279. [PubMed] [Google Scholar]
- 77.Pirtoli L, Cevenini G, Tini P, et al. The prognostic role of Beclin-1 protein expression in high-grade gliomas [J] Autophagy. 2009;5(7):930–936. doi: 10.4161/auto.5.7.9227. [DOI] [PubMed] [Google Scholar]
- 78.Addeo R, De Rosa C, Faiola V, et al. Phase 2 trial of temozolomide using protracted low-dose and whole-brain radiotherapy for nonsmall cell lung cancer and breast cancer patients with brain metastases [J] Cancer. 2008;113(9):2524–2531. doi: 10.1002/cncr.23859. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen TM, Subramanian IV, Kelekar A, et al. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells [J] Blood. 2007;109(11):4793–4802. doi: 10.1182/blood-2006-11-059352. [DOI] [PubMed] [Google Scholar]
- 80.Bommareddy A, Hahm ER, Xiao D, et al. Atg5 regulates phenethyl isothiocyanate-induced autophagic and apoptotic cell death in human prostate cancer cells [J] Cancer Res. 2009;69(8):3704–3712. doi: 10.1158/0008-5472.CAN-08-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao M, Yeh PY, Lu YS, et al. OSU-03012, a novel celecoxib derivative, induces reactive oxygen species-related autophagy in hepatocellular carcinoma [J] Cancer Res. 2008;68(22):9348–9357. doi: 10.1158/0008-5472.CAN-08-1642. [DOI] [PubMed] [Google Scholar]
- 82.Vazquez-Martin A, Oliveras-Ferraros C, del Barco S, et al. mTOR inhibitors and the anti-diabetic biguanide metformin: new insights into the molecular management of breast cancer resistance to the HER2 tyrosine kinase inhibitor lapatinib (Tykerb) [J] Clin Transl Oncol. 2009;11(7):455–459. doi: 10.1007/s12094-009-0384-0. [DOI] [PubMed] [Google Scholar]
- 83.Liu TJ, Koul D, LaFortune T, et al. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas [J] Mol Cancer Ther. 2009;8(8):2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuk JM, Shin DM, Song KS, et al. Bacillus calmette-guerin cell wall cytoskeleton enhances colon cancer radiosensitivity through autophagy [J] Autophagy. 2010;6(1):46–60. doi: 10.4161/auto.6.1.10325. [DOI] [PubMed] [Google Scholar]
- 85.Schoenlein PV, Periyasamy-Thandavan S, Samaddar JS, et al. Autophagy facilitates the progression of ERalpha-positive breast cancer cells to antiestrogen resistance [J] Autophagy. 2009;5(3):400–403. doi: 10.4161/auto.5.3.7784. [DOI] [PubMed] [Google Scholar]
- 86.Wu WK, Cho CH, Lee CW, et al. Macroautophagy and ERK phosphorylation counteract the antiproliferative effect of proteasome inhibitor in gastric cancer cells [J] Autophagy. 2010;6(2):228–238. doi: 10.4161/auto.6.2.11042. [DOI] [PubMed] [Google Scholar]
- 87.Malizzia LJ, Hsu A. Temsirolimus, an mTOR inhibitor for treatment of patients with advanced renal cell carcinoma [J] Clin J Oncol Nurs. 2008;12(4):639–646. doi: 10.1188/08.CJON.639-646. [DOI] [PubMed] [Google Scholar]
- 88.Hudes GR. Targeting mTOR in renal cell carcinoma [J] Cancer. 2009;115(10 Suppl):2313–2320. doi: 10.1002/cncr.24239. [DOI] [PubMed] [Google Scholar]
- 89.Konings IR, Verweij J, Wiemer EA, et al. The applicability of mTOR inhibition in solid tumors [J] Curr Cancer Drug Targets. 2009;9(3):439–450. doi: 10.2174/156800909788166556. [DOI] [PubMed] [Google Scholar]
- 90.Coiffier B, Ribrag V. Exploring mammalian target of rapamycin (mTOR) inhibition for treatment of mantle cell lymphoma and other hematologic malignancies [J] Leuk Lymphoma. 2009;50(12):1916–1930. doi: 10.3109/10428190903207548. [DOI] [PubMed] [Google Scholar]
- 91.Wysocki PJ. mTOR in renal cell cancer: modulator of tumor biology and therapeutic target [J] Expert Rev Mol Diagn. 2009;9(3):231–241. doi: 10.1586/erm.09.8. [DOI] [PubMed] [Google Scholar]
- 92.Hess G, Smith SM, Berkenblit A, et al. Temsirolimus in mantle cell lymphoma and other non-Hodgkin lymphoma subtypes [J] Semin Oncol. 2009;36(Suppl 3):S37–S45. doi: 10.1053/j.seminoncol.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 93.Dancey JE, Curiel R, Purvis J. Evaluating temsirolimus activity in multiple tumors: a review of clinical trials [J] Semin Oncol. 2009;36(Suppl 3):S46–S58. doi: 10.1053/j.seminoncol.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 94.Lefranc F, Rynkowski M, DeWitte O, et al. Present and potential future adjuvant issues in high-grade astrocytic glioma treatment [J] Adv Tech Stand Neurosurg. 2009;34:3–35. doi: 10.1007/978-3-211-78741-0_1. [DOI] [PubMed] [Google Scholar]
- 95.Sanchez-Munoz A, Perez-Ruiz E, Jimenez B, et al. Targeted therapy of metastatic breast cancer [J] Clin Transl Oncol. 2009;11(10):643–650. doi: 10.1007/s12094-009-0419-6. [DOI] [PubMed] [Google Scholar]
- 96.Qian W, Liu J, Jin J, et al. Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin-1 [J] Leuk Res. 2007;31(3):329–339. doi: 10.1016/j.leukres.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 97.Kanzawa T, Zhang L, Xiao L, et al. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3 [J] Oncogene. 2005;24(6):980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 98.Kanzawa T, Kondo Y, Ito H, et al. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide [J] Cancer Res. 2003;63(9):2103–2108. [PubMed] [Google Scholar]
- 99.Xu SN, Chen JP, Liu JP, et al. Efficacy of arsenic trioxide for acute promyelocytic leukemia: a systematic review and metaanalysis [J] Zhong Xi Yi Jie He Xue Bao. 2009;7(9):801–808. doi: 10.3736/jcim20090901. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 100.Chen X, Zhang M, Liu LX. The overexpression of multidrug resistance-associated proteins and gankyrin contribute to arsenic trioxide resistance in liver and gastric cancer cells [J] Oncol Rep. 2009;22(1):73–80. [PubMed] [Google Scholar]
- 101.Quick A, Patel D, Hadziahmetovic M, et al. Current therapeutic paradigms in glioblastoma [J] Rev Recent Clin Trials. 2010;5(1):14–27. doi: 10.2174/157488710790820544. [DOI] [PubMed] [Google Scholar]
- 102.Lefranc F, Facchini V, Kiss R. Proautophagic drugs: a novel means to combat apoptosis-resistant cancers, with a special emphasis on glioblastomas [J] Oncologist. 2007;12(12):1395–403. doi: 10.1634/theoncologist.12-12-1395. [DOI] [PubMed] [Google Scholar]
- 103.Fu J, Liu ZG, Liu XM, et al. Glioblastoma stem cells resistant to temozolomide-induced autophagy [J] Chin Med J (Engl) 2009;122(11):1255–1259. [PubMed] [Google Scholar]
- 104.Gao S, Yang XJ, Zhang WG, et al. Mechanism of thalidomide to enhance cytotoxicity of temozolomide in U251-MG glioma cells in vitro [J] Chin Med J (Engl) 2009;122(11):1260–1266. [PubMed] [Google Scholar]
- 105.Tyler MA, Ulasov IV, Lesniak MS. Cancer cell death by design: apoptosis, autophagy and glioma virotherapy [J] Autophagy. 2009;5(6):856–857. doi: 10.4161/auto.8792. [DOI] [PubMed] [Google Scholar]
- 106.Yokoyama T, Iwado E, Kondo Y, et al. Autophagy-inducing agents augment the antitumor effect of telerase-selve oncolytic adenovirus OBP-405 on glioblastoma cells [J] Gene Ther. 2008;15(17):1233–1239. doi: 10.1038/gt.2008.98. [DOI] [PubMed] [Google Scholar]
- 107.Ulasov IV, Sonabend AM, Nandi S, et al. Combination of adenoviral virotherapy and temozolomide chemotherapy eradicates malignant glioma through autophagic and apoptotic cell death in vivo [J] Br J Cancer. 2009;100(7):1154–1164. doi: 10.1038/sj.bjc.6604969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Milano V, Piao Y, LaFortune T, et al. Dasatinib-induced autophagy is enhanced in combination with temozolomide in glioma [J] Mol Cancer Ther. 2009;8(2):394–406. doi: 10.1158/1535-7163.MCT-08-0669. [DOI] [PubMed] [Google Scholar]
- 109.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, et al. Energy metabolism in tumor cells [J] FEBS J. 2007;274(6):1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 110.Wu YC, Wu WK, Li Y, et al. Inhibition of macroautophagy by bafilomycin A1 lowers proliferation and induces apoptosis in colon cancer cells [J] Biochem Biophys Res Commun. 2009;382(2):451–456. doi: 10.1016/j.bbrc.2009.03.051. [DOI] [PubMed] [Google Scholar]
- 111.Samaddar JS, Gaddy VT, Duplantier J, et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance [J] Mol Cancer Ther. 2008;7(9):2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 112.Pattingre S, Bauvy C, Carpentier S, et al. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy [J] J Biol Chem. 2009;284(5):2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wessely R, Kastrati A, Mehilli J, et al. Randomized trial of rapamycin- and paclitaxel-eluting stents with identical biodegradable polymeric coating and design [J] Eur Heart J. 2007;28(22):2720–2725. doi: 10.1093/eurheartj/ehm425. [DOI] [PubMed] [Google Scholar]
- 114.Satyan KS, Swamy N, Dizon DS, et al. Phenethyl isothiocyanate (PEITC) inhibits growth of ovarian cancer cells by inducing apoptosis: role of Caspase and MAPK activation [J] Gynecol Oncol. 2006;103(1):261–270. doi: 10.1016/j.ygyno.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 115.Quinn JA, Jiang SX, Reardon DA, et al. Phase II trial of temozolomide plus o6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma [J] J Clin Oncol. 2009;27(8):1262–1267. doi: 10.1200/JCO.2008.18.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gomez-Santos C, Ferrer I, Santidrian AF, et al. Dopamine induces autophagic cell death and alpha-synuclein increase in human neuroblastoma SH-SY5Y cells [J] J Neurosci Res. 2003;73(3):341–350. doi: 10.1002/jnr.10663. [DOI] [PubMed] [Google Scholar]
- 117.Park MA, Curiel DT, Koumenis C, et al. PERK-dependent regulation of HSP70 expression and the regulation of autophagy [J] Autophagy. 2008;4(3):364–367. doi: 10.4161/auto.5593. [DOI] [PubMed] [Google Scholar]
- 118.Chapman JV, Gouaze-Anderson V, Messner MC, et al. Metabolism of short-chain Ceramide by human cancer cells— implications for therapeutic approaches [J] Biechem Pharmacol. 2010;80(3):308–315. doi: 10.1016/j.bcp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.de Medina P, Silvente-Poirot S, Poirot M. Tamoxifen and AEBS ligands induced apoptosis and autophagy in breast cancer cells through the stimulation of Sterol accumulation [J] Autophagy. 2009;5(7):1066–1067. doi: 10.4161/auto.5.7.9820. [DOI] [PubMed] [Google Scholar]
- 120.Xiong HY, Guo XL, Bu XX, et al. Autophagic cell death induced by 5-FU in Bax or PUMA deficient human colon cancer cell [J] Cancer Lett. 2010;288(1):68–74. doi: 10.1016/j.canlet.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 121.Kim RH, Bold RJ, Kung HJ. ADI, autophagy and apoptosis: metabolic stress as a therapeutic option for prostate cancer [J] Autophagy. 2009;5(4):567–568. doi: 10.4161/auto.5.4.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu WK, Wu YC, Yu L, et al. Induction of autophagy by proteasome inhibitor is associated with proliferative arrest in colon cancer cells [J] Biochem Biophys Res Commun. 2008;374(2):258–263. doi: 10.1016/j.bbrc.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 123.Watanabe M, Adachi S, Matsubara H, et al. Induction of autophagy in malignant rhabdoid tumor cells by the histone deacetylase inhibitor FK228 through AIF translocation [J] Int J Cancer. 2009;124(1):55–67. doi: 10.1002/ijc.23897. [DOI] [PubMed] [Google Scholar]
- 124.Yamaguchi J, Sasaki M, Sato Y, et al. Histone deacetylase inhibitor (SAHA) and repression of EZH2 synergistically inhibit proliferation of gallbladder carcinoma [J] Cancer Sci. 2010;101(2):355–362. doi: 10.1111/j.1349-7006.2009.01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsuboi Y, Kurimoto M, Nagai S, et al. Induction of autophagic cell death and radiosensitization by the pharmacological inhibition of nuclear factor-kappa B activation in human glioma cell lines [J] J Neurosurg. 2009;110(3):594–604. doi: 10.3171/2008.8.JNS17648. [DOI] [PubMed] [Google Scholar]
- 126.Shingu T, Fujiwara K, Bogler O, et al. Stage-specific effect of inhibition of autophagy on chemotherapy-induced cytotoxicity [J] Autophagy. 2009;5(4):537–539. doi: 10.4161/auto.5.4.8164. [DOI] [PubMed] [Google Scholar]
- 127.Shingu T, Fujiwara K, Bogler O, et al. Inhibition of autophagy at a late stage enhances imatinib-induced cytotoxicity in human malignant glioma cells [J] Int J Cancer. 2009;124(5):1060–1071. doi: 10.1002/ijc.24030. [DOI] [PubMed] [Google Scholar]
- 128.Ertmer A, Huber V, Gilch S, et al. The anticancer drug imatinib induces cellular autophagy [J] Leukemia. 2007;21(5):936–942. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- 129.Yang J, Wu LJ, Tashino S, et al. Reactive oxygen species and nitric oxide regulate mitochondria-dependent apoptosis and autophagy in evodiamine-treated human cervix carcinoma HeLa cells [J] Free Radic Res. 2008;42(5):492–504. doi: 10.1080/10715760802112791. [DOI] [PubMed] [Google Scholar]
- 130.Signorelli P, Munoz-Olaya JM, Gagliostro V, et al. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells [J] Cancer Lett. 2009;282(2):238–243. doi: 10.1016/j.canlet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 131.Pervaiz S, Holme AL. Resveratrol: its biologic targets and functional activity [J] Antioxid Redox Signal. 2009;11(11):2851–2897. doi: 10.1089/ars.2008.2412. [DOI] [PubMed] [Google Scholar]
- 132.Ohshiro K, Rayala SK, Kondo S, et al. Identifying the estrogen receptor coactivator PELP1 in autophagosomes [J] Cancer Res. 2007;67(17):8164–8171. doi: 10.1158/0008-5472.CAN-07-0038. [DOI] [PubMed] [Google Scholar]
- 133.Matrone A, Grossi V, Chiacchiera F, et al. p38alpha is required for ovarian cancer cell metabolism and survival [J] Int J Gynecol Cancer. 2010;20(2):203–211. doi: 10.1111/igc.0b013e3181c8ca12. [DOI] [PubMed] [Google Scholar]
- 134.Comes F, Matrone A, Lastella P, et al. A novel cell type-specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells [J] Cell Death Differ. 2007;14(4):693–702. doi: 10.1038/sj.cdd.4402076. [DOI] [PubMed] [Google Scholar]
- 135.Chiacchiera F, Matrone A, Ferrari E, et al. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription [J] Cell Death Differ. 2009;16(9):1203–1214. doi: 10.1038/cdd.2009.36. [DOI] [PubMed] [Google Scholar]
- 136.Groth-Pedersen L, Ostenfeld MS, Hoyer-Hansen M, et al. Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome-destabilizing siramesine [J] Cancer Res. 2007;67(5):2217–2225. doi: 10.1158/0008-5472.CAN-06-3520. [DOI] [PubMed] [Google Scholar]
- 137.Ostenfeld MS, Hoyer-Hansen M, Bastholm L, et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces Cytoprotective autophagosome accumulation [J] Autophagy. 2008;4(4):487–499. doi: 10.4161/auto.5774. [DOI] [PubMed] [Google Scholar]
- 138.Alonso MM, Gomez-Manzano C, Bekele BN, et al. Adenovirus-based strategies overcome temozolomide resistance by silencing the O6-methylguanine-DNA methyltransferase promoter [J] Cancer Res. 2007;67(24):11499–11504. doi: 10.1158/0008-5472.CAN-07-5312. [DOI] [PubMed] [Google Scholar]
- 139.Ryu SY, Kim K, Lee WS, et al. Synergistic growth inhibition by combination of adenovirus mediated p53 transfer and cisplatin in ovarian cancer cell lines [J] J Gynecol Oncol. 2009;20(1):48–54. doi: 10.3802/jgo.2009.20.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen N, Karantza V. Autophagy as a therapeutic target in cancer [J]. Cancer Biol Ther. 2011;11(2):157–168. doi: 10.4161/cbt.11.2.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roy S, Debnath J. Autophagy and tumorigenesis [J] Semin Immunopathol. 2010;32(4):383–396. doi: 10.1007/s00281-010-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis [J] FEBS J. 2011;278(3):403–413. doi: 10.1111/j.1742-4658.2010.07965.x. [DOI] [PubMed] [Google Scholar]
- 143.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer [J] Cell. 2009;137(6):1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]