Abstract
In the mouse, the lineages of cells that give rise to trophectoderm and ICM are generally held to arise between the 8-cell and 16-cell stage of development. This model assumes that all blastomeres have essentially equivalent potential in terms of their fate through the first three rounds of cell division. There is, however, accumulating evidence that the blastomeres of 2-cell stage conceptuses may be compositionally different and contribute unequally to trophectoderm and ICM of the blastocyst. Here, we evaluate these competing points of view relating to when commitment to the trophectoderm lineage occurs in mammals, describe some of the genes that drive trophectoderm specification, and discuss the implications of the two hypotheses in relation to outcomes in commonly used reproductive technologies. Much of what is presently known has been derived from studying the mouse, but where information is available from other species, and particularly from cattle, it has been included.
Keywords: Cattle, Conceptus, Horse, Lineage specification, Mouse
Trophectoderm Lineage Emergence in the Mouse
The general aspects of conceptus development from fertilization up to the time that the blastocyst hatches from the zona pellucida are relatively similar across all eutherian mammals [1, 2], suggesting the process is under some degree of conserved genetic control. It takes about three and a half days for a mouse conceptus to progress from the zygote to the blastocyst stage of development (Fig. 1A). The early blastocyst has about 32 cells, of which one-third to one-quarter are part of the ICM, while the remainder comprise trophectoderm, the precursor cells of trophoblast. The first indication of cell differentiation, however, occurs before the blastocyst stage and begins at the 8-cell stage (Fig. 1A) when the outer cells begin to press against each other, form junctional complexes, and show signs of polarization and surface flattening, a phenomenon known as compaction. By the 16-cell stage, a population of non-polarized, inner cells has emerged [3, 4]. It is now generally accepted that trophectoderm is formed from the outer cell layer of the morula, while the inner cells give rise to the ICM, which subsequently forms the epiblast and primitive endoderm lineages. What remains controversial, however, is whether there is pre-existing information accounting for these cell fate decisions earlier than the 8-cell stage of development, perhaps even as early as the oocyte itself [2, 5, 6].
Fig. 1.
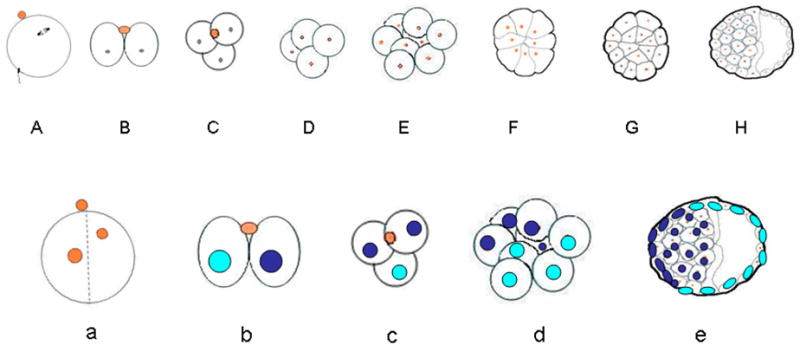
An outline of early development in the mouse. The upper diagram (A), which is based on Fig. 1 of Johnson & McConnell [4] illustrates development from fertilization to the 32-cell expanding blastocyst, during which time the conceptus is enclosed within the zona pellucida (not shown). No net growth occurs during this period so that cells become progressively smaller. A, oocyte at the time of fertilization (0 h), with the 2nd meiotic spindle and the first polar body marking the so-called animal pole of the egg.; B, 2-cell stage conceptus at (~20) h after a meridional division had occurred approximately along the animal-vegetal axis of the zygote (noted in about 75% of cases) with the second polar body positioned between the two blastomeres; C, 3-cell conceptus, which, in the CF1 strain of mouse used in this laboratory, is noted in the majority of conceptuses between 26 and 30 h; D, 4-cell stage (~36 h); E, 8-cell stage (~48 h) before compaction; F, 8-cell stage (~52h) after compaction; G, 16-cell morula (~60h) with an established inner cell population; H, 32-cell blastocyst (84 h; not all cells shown). Note that there is considerable variation in the timing of stages both between and also within strains of mice.
The lower panel (B) illustrates one possible outcome of pre-patterning in early mouse development following a cleavage plane that occurs approximately along the animal vegetal axis of the egg. As shown in a, in about 75% of cases, cleavage occurs within 30° (indicated by the broken lines) of either side of this animal-vegetal pole axis (defined here by the position of the 2nd polar body colored in orange). According to the pre-patterning hypothesis, the two blastomeres at the 2-cell stage are distinct and have become allocated to two separate lineages (illustrated here with blue and aquamarine nuclei). Although these lineages are not completely fixed and depend upon the planes of subsequent cell divisions, in a high proportion of cases, the blastomere that divides first (b & c), will contribute to the embryonic pole of the blastocyst (e), while the one that divides last will be the precursor of mural trophectoderm located towards the abembryonic pole. Thus, the cleavage plane of the zygote defines the eventual clonal boundary between the two lineages.
Tarkowski & Wroblewska postulated that those cells that are entirely enclosed by other blastomeres by the late morula become directed to form ICM, while outer cells receive positional information that drives them to become trophectoderm [7]. A currently more favored version of this model is that cell fate is determined, not in the late morula but earlier [8–11] at the 8-cell stage. In this model, some cells divide in a plane perpendicular to the surface to create additional polarized outer cells, while others divide parallel to the surface to produce the population of inner cells (Fig. 1A). By the 16-cell stage, embryos average about ten outer and six inner cells, although proportions can vary. The strengths of this cell polarity model have been described in detail by Yamanaka et al. [12] and will not be discussed in detail here. Which cell divides in a particular plane, i.e. orthogonal, parallel or obliquely to the outer surface of the conceptus [13], has never been particularly well explained in this model [4], although there is evidence that the blastomeres that reach the 8-cell stage first are primarily the ones most likely to divide equatorially and contribute inner cells [14]. Conversely, those cells with a greatest apical area in contact with the outside of the conceptus [15] and fewest internal contacts [8] (and presumably the ones that reach the 8-cell stage last) appear to be the ones most likely to divide meridionally. A prediction of this model is that all blastomeres at the 8-cell stage will provide progeny to trophectoderm, although some cells will contribute more than others. Many who support the cell polarity model make a second prediction, namely that all eight blastomeres at the early 8-cell stage retain equivalent developmental potential. We return to this issue later.
Other mouse embryologists believe that there are already distinct populations of cells in the 8-cell mouse conceptus whose fates are likely to be quite different from each other (Fig. 1B). Here, the organization of the 8-cell conceptus is considered to stem from differences between the first two blastomeres and most likely from the initial cleavage plane of the zygote, which distributes information already present in the egg to progeny blastomeres unequally. There is consensus among many embryologists that in greater than 75% of cases zygote cleavage occurs along an axis that is within 30° of the placement of the second polar body, thereby roughly bisecting the so-called animal and vegetal poles [5, 16, 17] (Fig. 1B). Some consider this event to separate a blastomere that contributes predominantly to the abembryonic part of the blastocyst, i.e. mural trophoblast, and one that is the precursor of the embryonic end of the blastocyst, i.e. ICM and associated polar trophectoderm [5, 18, 19]. However, not all accept this model, arguing that the position of the second polar body is not a fixed landmark (the “North Pole” of the embryo) but instead moves into the cleavage furrow, thereby providing an illusion of a relatively predetermined division plane [20, 21]. Considerable evidence for and against the pre-patterning model has accumulated. For example, many lineage tracing experiments, in which blastomeres at the 2-cell stage have been “marked” to allow their fate to be followed, have been consistent with this model [17, 18, 22], whereas others appear to contradict it and show a mixing of the progeny of the first two blastomeres at both poles of the conceptus [21, 23–25].
The pre-patterning model predicts that the first two blastomeres are not only likely to contribute unequally to the embryonic and abembryonic poles of the blastocyst, but that they must be compositionally distinct in terms of the information they received from the oocyte. Although compositional differences have yet to be proved, Zernicki-Goetz and colleagues [16, 19, 26] indicate that the blastomere at the 2-cell stage that divides first is the one that contributes most cells to the ICM and polar trophectoderm, while the late dividing one is the main precursor of mural trophectoderm. Importantly, the spatial patterning of cells at the 8-cell stage and the degree of mixing in the blastocyst will be influenced by the orientation, not just of the first, but of each of the cleavage divisions of the early conceptus [26]. For example, if the first dividing 2-cell blastomere subsequently divides equatorially rather than demonstrating the more usual meridional cleavage, axial polarity will probably not be well maintained. These views are consistent with the earlier observations that there is a somewhat fixed pattern of cleavage over the first three rounds of cell division [8] and that the pair of cells to reach the 8-cell stage first contributes disproportionately to the ICM, while the pair that arrived at that stage last is primarily a precursor of trophectoderm [8, 27, 28]. Clearly, such features of preferential contribution to the ICM or trophectoderm are not inconsistent with either the Cell Polarity or Pre-patterning models and could be explained by both.
The concept that the early dividing blastomere and the lagging blastomere of the 2-cell conceptus differ in the manner their progeny are allocated within the blastocyst has received considerable criticism [25]. According to these workers the allocation of cells into the embryonic and abembryonic regions appeared not to be related to the order in which the first two blastomeres cleaved. Indeed, the notion that the polarity of the blastocyst is established by the time the oocyte is fertilized remains an anathema to some and remains the subject of contentious debate [6, 18].
Formation of Trophectoderm in Cattle
The development of the bovine embryo from the zygote stage is similar but not identical to what is observed in the mouse. As in the mouse, cell divisions in the early cleavage stages are not synchronous, as transient 3-cell and 6-cell stages can be observed (unpublished data, this laboratory). With the improvements in technologies for maturing oocytes and, particularly for culturing embryos, the development of the fastest developing one-third of in vitro-produced embryos progresses about as quickly as those recovered from pregnant cows. Most of the embryos likely to progress to blastocyst have cleaved at least once within 24 h after removing the zygotes from fertilization medium and approximately half are at the 8-cell stage by 48 h (72 h after introduction to sperm). Of the latter, about 70% will progress to blastocyst, which begin to emerge late on d 6. Interestingly, the longest cell cycle is the fourth, with a duration greater than 40h [29, 30], whereas in the mouse it is the second, possibly reflecting the relative time of embryonic genome activation in the two species. Compacted morulae form between days 5 and 6 and at least one round of cell divisions later than observed for the mouse.
Accordingly, blastocysts prior to the time they expand and hatch, rather than containing around 30–35 cells as in the mouse, average over 100, of which about one-third are within the ICM, the remainder being trophectoderm [30, 31]. By the time the blastocysts have begun to hatch from day 8 onwards, they comprise over 200 cells[31, 32]. To the authors’ knowledge little has been done in terms of lineage tracing in cattle conceptuses, and so the question of pre-patterning has never been addressed. Clearly, however, manipulations likely to disturb the organization of the cytoskeleton in the oocyte or zygote, are more likely to be damaging if the cytoplasm is organized into zones of different composition. For example it is easy to imagine how removal of the nuclear material and its adherent cytoplasm from an oocyte, followed by fusion with a somatic cell, could be highly disruptive to oocyte organization. Is it possible that the many problems that occur in placental development of cloned animals are at least partially due to disruption of oocyte determinants rather than simply to the aberrant expression of imprinted genes?
Oocyte Asymmetry and the Plane of First Cleavage
Most mammalian oocytes are not radially symmetrical. The nuclear material is usually not centrally placed, and the eggs are often oval in profile. There is a striking unevenness in cytoplasmic texture and in organelle distribution, including the distribution of microtubule organizing centers [33, 34]. Accordingly, the first cleavage division of a zygote, whatever its plane, would be unlikely to create two progeny blastomeres equivalent in composition despite the fact that mitosis must ensure that each daughter blastomere receive sufficient raw materials and organelles to proceed in their development This type of asymmetry is clearly seen in the horse oocyte and zygote, where the localization of fat droplets and mitochondria is polarized (Fig. 2A & 2B). As a result, material is portioned unequally among blastomeres in the early and subsequent cleavage divisions (Fig. 2C & 2D). This situation is somewhat reminiscent of that in the larger yolkier eggs of invertebrates, where the asymmetrical distribution of maternal mRNAs and proteins establish the future embryonic axes [33] and are essential for proper development of the embryo.
Fig. 2.
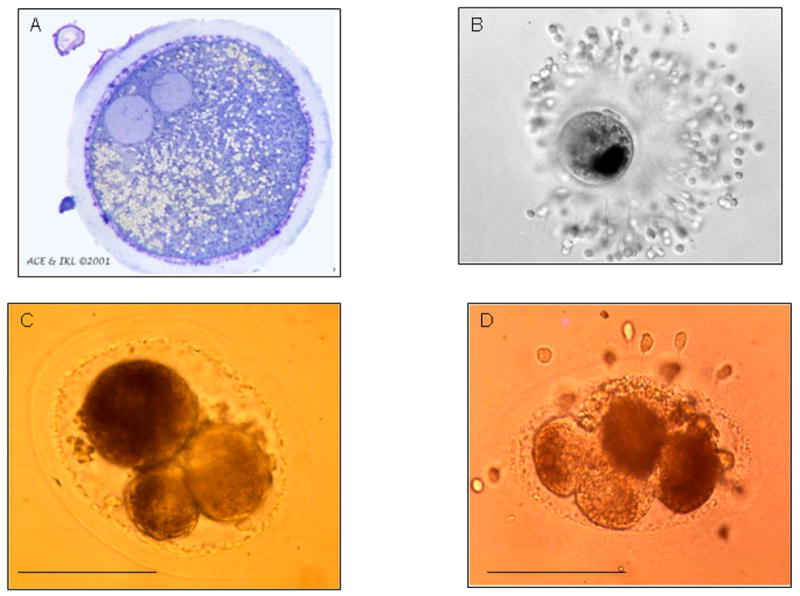
Asymmetry in early horse conceptus development. A, light microscope section (1μm) of a pronuclear stage equine zygote 21 h after insemination. A single cumulus cells remains attached to the zona pellucida. The larger pronuclear profile is probably that of the male pronucleus, since a portion of the midpiece of the fertilizing sperm was found associated with it in an adjacent section viewed under the electron microscope (not shown). The image illustrates the asymmetrical distribution of cytoplasmic particles (lipid bodies and mitochondria) towards one side of the egg; B, zygote (whole mount) just prior to cleavage and recovered from the oviduct of a mare. Note the position of dense cytoplasmic material positioned to the right of the animal-vegetal axis (defined by the position of the polar bodies). C, a fresh, 3-cell stage, horse embryo. Note that the larger blastomere, which presumably has yet to divide, has darker cytoplasm than the two smaller blastomeres; D, a 4-cell stage horse embryo. Here two of the blastomeres have darker cytoplasm than the other two. A, is by courtesy of Dr. A.C. Enders, University of California-Davis; B is an unpublished image acquired by Dr. J. Bézard and provided by Dr. Keith J. Betteridge (University of Guelph, Canada) from a study by King et al [84]. C & D (Bar 100 μm) correspond to plates 5 and 3, respectively, of Betteridge et al [85], with permission.
Although the mouse oocyte is often regarded as if it were a radially symmetrical structure, it clearly is not, either at maturity or during its development [34]. Whether, the cytoplasm is organized into distinct zones containing molecules with different informational content that provide cell lineage guidance through subsequent development, as occurs in amphibians and insects, remains a divisive topic [4, 33, 35]. There are reports that certain gene products, including leptin and STAT3, are localized to the cortical region near the animal pole of mouse oocytes and then become asymmetrically distributed at zygote cleavage [36], but this observation has not been confirmed [4]. If such selective distribution of information to progeny blastomeres does in fact occur, much depends upon how the first cleavage plane is established. If, for example, cleavage of the oocyte is purely random [37],, pre-patterning based on asymmetric distribution of oocyte information would be improbable. On the other hand, a more predictable cleavage plane, such as the one illustrated in Fig. 1B, might ensure unequal loading of the first two blastomeres and a blueprint that could conceivably be maintained through subsequent cleavage divisions.
Hiiragi, Solter and colleagues [6, 21] take strong exception to the notion that the first cleavage plane determines the embryonic/abembryonic axis of the blastocyst and resist the notion that polarity specification might arise in the egg. They assert that the mouse zygote divides roughly perpendicularly to a line drawn between the two pronuclei as they approach each other just prior to the time that the cleavage furrow begins to form [6] and that the second polar body is mobile and navigates its way into the cleavage plane. In this model, cleavage is not a random event, although the division plane is clearly not positionally predictable prior to fertilization, as it is in the one proposed by those who support pre-patterning [18]. According to Hiiragi & Solter [6] the very unpredictability of the cleavage plane ensures that the blastomeres at the 2-cell stage must be essentially developmentally equivalent in potential. Another argument against pre-patterning, is that attempts to disturb oocyte cytoplasmic organization, e.g. by centrifugation, stirring [38] or partial removal of zygote cytoplasm from either of its poles [39] should disturb subsequent development. But is such a conclusion necessarily true? Could informational macromolecules occupying particular zones within the oocyte, become preferentially loaded into one blastomere and not into the other, thereby setting the stage for lineage separation and ultimately commitment? It might be argued that as long as there is continuity between the microtubule network that orientates the first mitotic division, the microtubule organizing centers, and the cytoskeletal system that confines maternal mRNAs and proteins to particular cytoplasmic zones of the oocyte, unequal and directed loading might be possible. The degree to which cytoskeleton, associated with information macromolecules, would be obliged to re-orientate prior to zygote cleavage would depend upon the angle of the cell division plane. More systematic cell lineage tracing studies, especially on models other than the mouse, are needed to address whether asymmetries in the egg persist during development to the blastocyst.
The model described above could also be reconciled with the one proposed by Piotrowska & Zernicka-Goetz [40], namely that sperm entry position and the accompanying fertilization cone influences the orientation of the cleavage plane and ear-marks the blastomere that will subsequently divide last at the 2-cell stage. In the case of ascidians and amphibians, for example, the egg loses its primary animal-vegetal axis as the sperm penetrates, and the cytoplasm becomes reorganized through directed movement of its cytoskeleton to define the embryonic axes [41–43].
Blastomere Equivalence and Totipotency
One repeated argument against the concept that mammalian early development is prepatterned in the egg and polarity established early is the capacity of isolated blastomeres to be incorporated into all three germ layers of chimeras, and to develop into normal pups. Integration into chimeras is relatively easily explained even if some degree of pre-patterning exists. Blastomeres on track to be incorporated into the ICM would presumably be expected to demonstrate pluripotency and hence be integrated into chimeric mice. Those destined for trophectoderm, if rigidly committed, would possibly fail to demonstrate pluripotency, but since the efficiency of such manipulations is generally low, such failures would likely be overlooked. By constructing chimeras, Tarkowski and colleagues [44] calculated how many cells of a 4-cell blastomere are capable of contributing cells to the body of a mouse. Interestingly, they concluded that only two of the blastomeres of a 4-cell stage embryo have that potential. This result would appear to be inconsistent with the notion of equivalence among the blastomeres. On the other hand, how easily are individual blastomeres able to be progenitors of live young, i.e. demonstrate totipotency? In the mouse, such experiments are not clear cut. Fertile adults [45] and even twins have certainly been derived from blastomeres isolated at the 2-cell stage of development [45–49], even though relative cell numbers in the trophectoderm and ICM are significantly perturbed. Individual blastomeres from later stages have been successfully incorporated into chimeras [50], but appear not to have the potential to give rise to pups unless they are mixed with additional cells [51–53], which may provide the missing trophoblast component. Such isolated blastomeres have been noted to give rise to blastocysts, but these usually have either a small ICM or lack an ICM completely [54]. To the authors’ knowledge, there is no instance where each blastomere from a dissociated mouse conceptus beyond the 2-cell stage has provided an individual pup. The usual explanation has been that there is simply insufficient cytoplasm and cells to allow proper development to occur. Interestingly, however, a recent attempt to derive embryonic stem (ES) cells from individual blastomeres of 8-cell stage embryos produced five ES cell lines and seven trophoblast stem cell lines [55], suggesting that these blastomeres were already committed to a particular fate.
On the other hand, the production of live young from individual blastomeres after blastomere separation from conceptuses beyond the 2-cell stage has been achieved in species other than the mouse, including rabbits, sheep and cattle, even without the addition of trophoblast support cells [56–58]. There is one remarkable example of a four-cell stage bovine embryo providing quadruplets [58]. Whether these differences reflect greater plasticity of the developmental program of some species relative to others or whether additional factors, e.g. the timing of embryonic genome activation are responsible, remains unclear. Each of the species listed above activate their genomes later than in the mouse. Conceivably, lineage commitment might not be locked in until maternal gene products of oocyte origin activate a quorum of necessary embryonic genes.
Genes Required for Initial Trophectoderm Specification
Gene ablation experiments in the mouse have revealed many genes are required to form a fully functional placenta [59–64]. Only a few of these transcription factors have been examined in the bovine trophoblast. Hand1 [65] and Mash2 [66, 67], both essential genes for mouse placental development, are expressed in bovine blastocysts, but no detailed study on their cellular distributions have been performed. Although it seems likely that these transcription factors are important in establishing trophectoderm functional differentiation in bovine as well as mouse embryos, no such role has yet been proved. RNAi knockdown of these and other genes is likely to be an invaluable means of studying these transcription factors in species, such as cattle, where gene knock-outs are impractical.
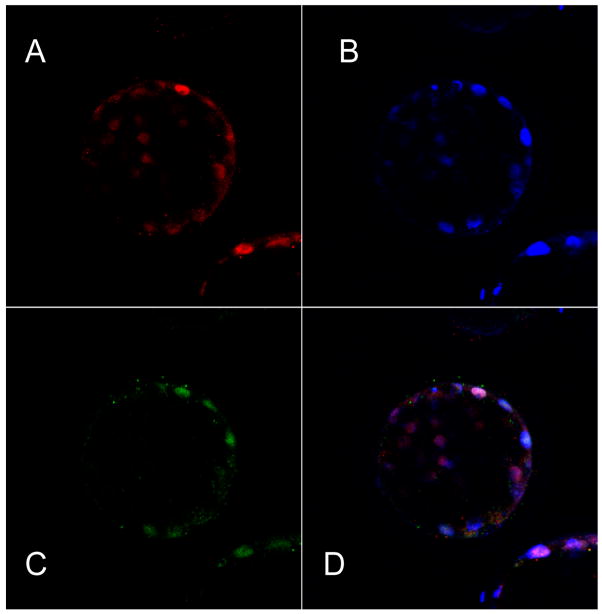
Deletion of some genes in the mouse leads to developmental failure before or at implantation, suggesting that they are required in trophectoderm specification and not simply in its functional differentiation. For example, deletion of the gene Pou5f, which encodes the POU-transcription factor, Oct4, provides conceptuses that lacked markers for an ICM and appeared to consist largely if not entirely of trophectoderm [68]. Accordingly, trophectoderm only seems to differentiate in the mouse when Pou5f1 becomes down-regulated. A similar situation occurs in murine embryonic stem cells, which default to trophoblast when Pou5f1 is silenced [69]. The generality that Oct4 protein is absent from functional trophectoderm does not apply across all species, however. In bovine embryos, Oct4 can be detected in trophectoderm until day 10, two to three days after the blastocyst first forms[70, 71]. In trophectoderm from d8 blastocysts it is co-expressed with the trophectoderm-specific marker, Cdx2 (Fig. 3), and Oct4 staining in the ICM can be quite faint compared with some cells in trophectoderm. Indeed, it may be that the mouse is the exception rather than the rule, since Oct4 is also expressed in early human trophectoderm[72]. These observations indicate that Oct4 may not be a binary off-on switch, but that its dosage relative to other transcription factors may be critical.
Fig. 3.
Confocal section of a day 8 bovine blastocyst immunostained to compare the localization of Oct4 (A, red), Cdx 2 (C, green) and nuclear material (B, Blue). D is a merged image. Bovine blastocysts were fixed in 2% paraformaldehyde in PBS for 15 min at room temperature, transferred to PBS/0.1% bovine serum albumin (BSA)at 4 C for 10 min, and permeabilized in 0.1% Triton X-100 in PBS/BSA for 15 min at room temperature. Samples were then placed in blocking solution (5% goat serum in PBS/BSA) for 30 min, then incubated with primary antibody (anti-Cdx2 mouse monoclonal) from BioGenex diluted 1:100 in 0.1% Triton X-100 in PBS/BSA for 2 h at room temperature. For control experiments with anti-Cdx2 (not shown), blastocysts were incubated with secondary antibody alone. The embryos were then washed, incubated for 2 h at room temperature with Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) diluted 1:500 in 0.1% Triton X-100 in PBS, and counter stained with DAPI (Sigma) They were then mounted in PBS/BSA drops suspended between glass slides under a cover well imaging chamber gasket (20 mm × 0.5 mm- Invitrogen). Optical sections were collected with a Zeiss LSM 510 two photon, confocal system, with excitation and emission settings corresponding to the fluorophores conjugated to the secondary antibodies and DAPI (blue). Oct-4 staining was performed as described earlier) [86].
The formation of trophectoderm is not, therefore, simply a default pathway initiated by the down-regulation of Pou5f1. In the mouse, it is becoming evident that it is a directed event requiring specific transcription factors. One of these is the caudal-related gene, Cdx2 [73, 74] while another is the T-box gene Eomes [75, 76]. Cdx2 shows a reciprocal pattern of expression to Oct4 at the blastocyst stage in the mouse, i.e. it is absent from the ICM but expressed in trophectoderm. Cdx2 knock out conceptuses also fail to implant, but do form a rudimentary blastocoel cavity [74, 77]. Eomes, by contrast, is expressed in ICM as well as trophectoderm [78] and, when deleted, the embryos form blastocysts that fail only after placentation has been initiated [76]. Neither kind of mutant embryo forms trophoblast outgrowths when cultured, and Cdx2 -/- embryos cannot be coaxed to produce trophectoderm stem cells [64]. Eomes may be necessary for proliferation of cytotrophoblast, but possibly not in its earliest specification. Cdx2, however, may be a true specification gene. It remains unclear when Cdx2 is first expressed in bovine embryos, although it is clearly confined to trophectoderm by the time the blastocyst first forms (Fig. 3).
Two recent reviews have made a strong case that the specification of murine trophectoderm requires the down-regulation of Pou5f1 accompanied by the up-regulation of Cdx2 [12, 59]. According to this hypothesis, by the time the compacted morula is about to make the transition to blastocyst, the future trophectoderm cells are largely Cdx2-positive and Oct4-negative, although both transcription factors may be expressed together as the developmental transition of the outer cells is being initiated, with each repressing the other’s target genes [79]. Indeed, the reciprocal nature of the relationship between these two transcription factors may be key to proper trophectoderm specification and differentiation, both in mouse embryos and embryonic stem cells [80]. Gain of Cdx2 function in murine ES cells, for example, down regulates Pou5f1 and promotes trophoblast differentiation [77, 79].
Concluding Remarks
The controversy over whether axis formation first evident in the blastocyst is directed by the organization of the oocyte and early embryo is likely to remain controversial. The fact that normal looking blastocysts and fertile offspring can be derived from conceptuses that have been “damaged” by removal of blastomeres or that have been created by technologies expected to perturb the organization of the egg ooplasm, such as ICSI or nuclear transfer, emphasizes the plasticity of early embryo development in mammals. This regulative behavior in itself is a major intuitive argument against the concept of pre-patterning. To many developmental biologists working with invertebrates, however, the reluctance to accept the concept that the mammalian oocyte is pre-patterned is puzzling, as the phenomenon has been so clearly confirmed at the morphological, biochemical and genetic levels in other phyla, including ascidians (sea squirts), insects, nematodes, and amphibians [33]. In these animal groups, the establishment of the main embryonic axes is clearly an outcome of the relative partitioning and regional mobilization of informational macromolecules from the cytoplasm of the oocyte [42, 43, 81–83]. Even in sea urchins, where cell-cell interactions are important for lineage derivation, some degree of pre-patterning occurs. Of course, it can be argued that care must be taken in drawing analogies between eutherian mammals, which possess a placenta, and other animals that do not, since the establishment of the embryonic axes may only become necessary once the ICM has formed. On the other hand, it would seem a radical evolutionary departure if a core specification process be so summarily abandoned in mammals. As pointed out by Gardner [38], information residing in the egg may help establish the general guidelines for initiating cleavage patterns, but other regulative mechanisms, such as cell position, may then intervene to keep development on course.
Acknowledgments
The research was supported by NIH Grant HD 21896. Steven Smith is a trainee on Grant 5 T32 GM008396-15 entitled “Molecular basis of gene expression and signal processing from NIH/NIGMS. The authors gratefully acknowledge the help of Norma McCormack for editorial assistance and re-creating Fig. 1.
References
- 1.Wimsatt WA. Some comparative aspects of implantation. Biol Reprod. 1975;12:1–40. doi: 10.1095/biolreprod12.1.1. [DOI] [PubMed] [Google Scholar]
- 2.McLaren A. The embryo. Cambridge: Cambridge University Press; 1972. [Google Scholar]
- 3.Fleming JA, Choi Y, Johnson GA, Spencer TE, Bazer FW. Cloning of the ovine estrogen receptor-alpha promoter and functional regulation by ovine interferon-tau. Endocrinology. 2001;142:2879–2887. doi: 10.1210/endo.142.7.8245. [DOI] [PubMed] [Google Scholar]
- 4.Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–597. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Gardner RL. Specification of embryonic axes begins before cleavage in normal mouse development. Development. 2001;128:839–847. doi: 10.1242/dev.128.6.839. [DOI] [PubMed] [Google Scholar]
- 6.Hiiragi T, Solter D. Mechanism of First Cleavage Specification in the Mouse Egg: Is Our Body Plan Set at Day 0? Cell Cycle. 2005:4. doi: 10.4161/cc.4.5.1680. [DOI] [PubMed] [Google Scholar]
- 7.Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18:155–180. [PubMed] [Google Scholar]
- 8.Graham CF, Deussen ZA. Features of cell lineage in preimplantation mouse development. J Embryol Exp Morphol. 1978;48:53–72. [PubMed] [Google Scholar]
- 9.Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- 10.Wiley LM, Kidder GM, Watson AJ. Cell polarity and development of the first epithelium. Bioessays. 1990;12:67–73. doi: 10.1002/bies.950120204. [DOI] [PubMed] [Google Scholar]
- 11.Rossant J. Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Semin Cell Dev Biol. 2004;15:573–581. doi: 10.1016/j.semcdb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006 doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland AE, Speed TP, Calarco PG. Inner cell allocation in the mouse morula: the role of oriented division during fourth cleavage. Dev Biol. 1990;137:13–25. doi: 10.1016/0012-1606(90)90003-2. [DOI] [PubMed] [Google Scholar]
- 14.Garbutt CL, Johnson MH, George MA. When and how does cell division order influence cell allocation to the inner cell mass of the mouse blastocyst? Development. 1987;100:325–332. doi: 10.1242/dev.100.2.325. [DOI] [PubMed] [Google Scholar]
- 15.Pickering SJ, Maro B, Johnson MH, Skepper JN. The influence of cell contact on the division of mouse 8-cell blastomeres. Development. 1988;103:353–363. doi: 10.1242/dev.103.2.353. [DOI] [PubMed] [Google Scholar]
- 16.Piotrowska K, Zernicka-Goetz M. Role for sperm in spatial patterning of the early mouse embryo. Nature. 2001;409:517–521. doi: 10.1038/35054069. [DOI] [PubMed] [Google Scholar]
- 17.Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development. 2003;130:5113–5122. doi: 10.1242/dev.00725. [DOI] [PubMed] [Google Scholar]
- 18.Gardner RL. The case for prepatterning in the mouse. Birth Defects Res C Embryo Today. 2005;75:142–150. doi: 10.1002/bdrc.20038. [DOI] [PubMed] [Google Scholar]
- 19.Zernicka-Goetz M. Developmental cell biology: cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6:919–928. doi: 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- 20.Hiiragi T, Solter D. First cleavage plane of the mouse egg is not predetermined but defined by the topology of the two apposing pronuclei. Nature. 2004;430:360–364. doi: 10.1038/nature02595. [DOI] [PubMed] [Google Scholar]
- 21.Motosugi N, Bauer T, Polanski Z, Solter D, Hiiragi T. Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev. 2005;19:1081–1092. doi: 10.1101/gad.1304805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–3748. doi: 10.1242/dev.128.19.3739. [DOI] [PubMed] [Google Scholar]
- 23.Caamano JN, Ryoo ZY, Youngs CR. Promotion of development of bovine embryos produced in vitro by addition of cysteine and beta-mercaptoethanol to a chemically defined culture system. J Dairy Sci. 1993;81:369–374. doi: 10.3168/jds.S0022-0302(98)75586-9. [DOI] [PubMed] [Google Scholar]
- 24.Chroscicka A, Komorowski S, Maleszewski M. Both blastomeres of the mouse 2-cell embryo contribute to the embryonic portion of the blastocyst. Mol Reprod Dev. 2004;68:308–312. doi: 10.1002/mrd.20081. [DOI] [PubMed] [Google Scholar]
- 25.Waksmundzka M, Wisniewska A, Maleszewski M. Allocation of Cells in Mouse Blastocyst Is Not Determined by the Order of Cleavage of the First Two Blastomeres. Biol Reprod. 2006 doi: 10.1095/biolreprod.106.053165. [DOI] [PubMed] [Google Scholar]
- 26.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132:479–490. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- 27.Kelly SJ, Mulnard JG, Graham CF. Cell division and cell allocation in early mouse development. J Embryol Exp Morphol. 1978;48:37–51. [PubMed] [Google Scholar]
- 28.Surani MA, Barton SC. Spatial distribution of blastomeres is dependent on cell division order and interactions in mouse morulae. Dev Biol. 1984;102:335–343. doi: 10.1016/0012-1606(84)90198-2. [DOI] [PubMed] [Google Scholar]
- 29.Holm P, Booth PJ, Callesen H. Kinetics of early in vitro development of bovine in vivo- and in vitro-derived zygotes produced and/or cultured in chemically defined or serum-containing media. Reproduction. 2002;123:553–565. [PubMed] [Google Scholar]
- 30.Lequarre AS, Marchandise J, Moreau B, Massip A, Donnay I. Cell cycle duration at the time of maternal zygotic transition for in vitro produced bovine embryos: effect of oxygen tension and transcription inhibition. Biol Reprod. 2003;69:1707–1713. doi: 10.1095/biolreprod.103.017178. [DOI] [PubMed] [Google Scholar]
- 31.Van Soom A, Boerjan ML, Bols PE, Vanroose G, Lein A, Coryn M, de Kruif A. Timing of compaction and inner cell allocation in bovine embryos produced in vivo after superovulation. Biol Reprod. 1997;57:1041–1049. doi: 10.1095/biolreprod57.5.1041. [DOI] [PubMed] [Google Scholar]
- 32.Knijn HM, Gjorret JO, Vos PL, Hendriksen PJ, van der Weijden BC, Maddox-Hyttel P, Dieleman SJ. Consequences of in vivo development and subsequent culture on apoptosis, cell number, and blastocyst formation in bovine embryos. Biol Reprod. 2003;69:1371–1378. doi: 10.1095/biolreprod.103.017251. [DOI] [PubMed] [Google Scholar]
- 33.Edwards RG, Beard HK. Oocyte polarity and cell determination in early mammalian embryos. Mol Hum Reprod. 1997;3:863–905. doi: 10.1093/molehr/3.10.863. [DOI] [PubMed] [Google Scholar]
- 34.Albertini DF, Barrett SL. The developmental origins of mammalian oocyte polarity. Semin Cell Dev Biol. 2004;15:599–606. doi: 10.1016/j.semcdb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Johnson MH. Mammalian development: axes in the egg? Curr Biol. 2001;11:R281–284. doi: 10.1016/s0960-9822(01)00139-7. [DOI] [PubMed] [Google Scholar]
- 36.Antczak M, Van Blerkom J. Oocyte influences on early development: the regulatory proteins leptin and STAT3 are polarized in mouse and human oocytes and differentially distributed within the cells of the preimplantation stage embryo. Mol Hum Reprod. 1997;3:1067–1086. doi: 10.1093/molehr/3.12.1067. [DOI] [PubMed] [Google Scholar]
- 37.Louvet-Vallee S, Vinot S, Maro B. Mitotic spindles and cleavage planes are oriented randomly in the two-cell mouse embryo. Curr Biol. 2005;15:464–469. doi: 10.1016/j.cub.2004.12.078. [DOI] [PubMed] [Google Scholar]
- 38.Gardner RL. Scrambled or bisected mouse eggs and the basis of patterning in mammals. Bioessays. 1999;21:271–274. doi: 10.1002/(SICI)1521-1878(199904)21:4<271::AID-BIES2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 39.Zernicka-Goetz M. Fertile offspring derived from mammalian eggs lacking either animal or vegetal poles. Development. 1998;125:4803–4808. doi: 10.1242/dev.125.23.4803. [DOI] [PubMed] [Google Scholar]
- 40.Piotrowska K, Zernicka-Goetz M. Early patterning of the mouse embryo--contributions of sperm and egg. Development. 2002;129:5803–5813. doi: 10.1242/dev.00170. [DOI] [PubMed] [Google Scholar]
- 41.Davidson EH, Cameron RA, Ransick A. Specification of cell fate in the sea urchin embryo: summary and some proposed mechanisms. Development. 1998;125:3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 42.Sardet C, Prodon F, Pruliere G, Chenevert J. [Polarization of eggs and embryos: some common principles] Med Sci (Paris) 2004;20:414–423. doi: 10.1051/medsci/2004204414. [DOI] [PubMed] [Google Scholar]
- 43.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 44.Tarkowski AK, Ozdzenski W, Czolowska R. How many blastomeres of the 4-cell embryo contribute cells to the mouse body? Int J Dev Biol. 2001;45:811–816. [PubMed] [Google Scholar]
- 45.Tarkowski AK. Experiments on the development of isolated blastomers of mouse eggs. Nature. 1959;184:1286–1287. doi: 10.1038/1841286a0. [DOI] [PubMed] [Google Scholar]
- 46.Tsunoda Y, McLaren A. Effect of various procedures on the viability of mouse embryos containing half the normal number of blastomeres. J Reprod Fertil. 1983;69:315–322. doi: 10.1530/jrf.0.0690315. [DOI] [PubMed] [Google Scholar]
- 47.Rands GF. Size regulation in the mouse embryo. II The development of half embryos. J Embryol Exp Morphol. 1986;98:209–217. [PubMed] [Google Scholar]
- 48.Papaioannou VE, Mkandawire J, Biggers JD. Development and phenotypic variability of genetically identical half mouse embryos. Development. 1989;106:817–827. doi: 10.1242/dev.106.4.817. [DOI] [PubMed] [Google Scholar]
- 49.Illmensee K, Kaskar K, Zavos PM. Efficient blastomere biopsy for mouse embryo splitting for future applications in human assisted reproduction. Reprod Biomed Online. 2005;11:716–725. doi: 10.1016/s1472-6483(10)61690-2. [DOI] [PubMed] [Google Scholar]
- 50.Tarkowski AK. Mouse chimaeras revisited: recollections and reflections. Int J Dev Biol. 1998;42:903–908. [PubMed] [Google Scholar]
- 51.Kelly SJ. Studies of the potency of the early cleavage blastomeres of the mouse. In: Balls M, Wild AE, editors. The Early Development of Mammals. Cambridge, London, New York, Mebourne: Cambridge University Press; 1975. pp. 97–105. [Google Scholar]
- 52.Kelly SJ. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J Exp Zool. 1977;200:365–376. doi: 10.1002/jez.1402000307. [DOI] [PubMed] [Google Scholar]
- 53.Tarkowski AK, Ozdzenski W, Czolowska R. Identical triplets and twins developed from isolated blastomeres of 8- and 16-cell mouse embryos supported with tetraploid blastomeres. Int J Dev Biol. 2005;49:825–832. doi: 10.1387/ijdb.052018at. [DOI] [PubMed] [Google Scholar]
- 54.Rossant J. Investigation of inner cell mass determination by aggregation of isolated rat inner cell masses with mouse morulae. J Embryol Exp Morphol. 1976;36:163–174. [PubMed] [Google Scholar]
- 55.Chung Y, Klimanskaya I, Becker S, Marh J, Lu SJ, Johnson J, Meisner L, Lanza R. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature. 2006;439:216–219. doi: 10.1038/nature04277. [DOI] [PubMed] [Google Scholar]
- 56.Moore NW, Adams CE, Rowson LE. Developmental potential of single blastomeres of the rabbit egg. J Reprod Fertil. 1968;17:527–531. doi: 10.1530/jrf.0.0170527. [DOI] [PubMed] [Google Scholar]
- 57.Willadsen SM. The development capacity of blastomeres from 4- and 8-cell sheep embryos. J Embryol Exp Morphol. 1981;65:165–172. [PubMed] [Google Scholar]
- 58.Johnson WH, Loskutoff NM, Plante Y, Betteridge KJ. Production of four identical calves by the separation of blastomeres from an in vitro derived four-cell embryo. Vet Rec. 1995;137:15–16. doi: 10.1136/vr.137.1.15. [DOI] [PubMed] [Google Scholar]
- 59.Rossant J, Chazaud C, Yamanaka Y. Lineage allocation and asymmetries in the early mouse embryo. Philos Trans R Soc Lond B Biol Sci. 2003;358:1341–1348. 1349. doi: 10.1098/rstb.2003.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cross JC, Anson-Cartwright L, Scott IC. Transcription factors underlying the development and endocrine functions of the placenta. Recent Prog Horm Res. 2002;57:221–234. doi: 10.1210/rp.57.1.221. [DOI] [PubMed] [Google Scholar]
- 61.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 62.Hemberger M, Cross JC. Genes governing placental development. Trends Endocrinol Metab. 2001;12:162–168. doi: 10.1016/s1043-2760(01)00375-7. [DOI] [PubMed] [Google Scholar]
- 63.Knofler M, Vasicek R, Schreiber M. Key regulatory transcription factors involved in placental trophoblast development--a review. Placenta. 2001;22(Suppl A):S83–92. doi: 10.1053/plac.2001.0648. [DOI] [PubMed] [Google Scholar]
- 64.Rossant J. Stem Cells from the Mammalian blastocyst. Stem Cells. 2001;19:477–482. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- 65.Degrelle SA, Campion E, Cabau C, Piumi F, Reinaud P, Richard C, Renard JP, Hue I. Molecular evidence for a critical period in mural trophoblast development in bovine blastocysts. Dev Biol. 2005;288:448–460. doi: 10.1016/j.ydbio.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 66.Jang G, Jeon HY, Ko KH, Park HJ, Kang SK, Lee BC, Hwang WS. Developmental competence and gene expression in preimplantation bovine embryos derived from somatic cell nuclear transfer using different donor cells. Zygote. 2005;13:187–195. doi: 10.1017/s0967199405003217. [DOI] [PubMed] [Google Scholar]
- 67.Arnold DR, Lefebvre R, Smith LC. Characterization of the Placenta Specific Bovine Mammalian Achaete Scute-like Homologue 2 (Mash2) Gene. Placenta. 2006 doi: 10.1016/j.placenta.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 69.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells [see comments] Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 70.van Eijk MJ, van Rooijen MA, Modina S, Scesi L, Folkers G, van Tol HT, Bevers MM, Fisher SR, Lewin HA, Rakacolli D, Galli C, de Vaureix C, Trounson AO, Mummery CL, Gandolfi F. Molecular cloning, genetic mapping, and developmental expression of bovine POU5F1. Biol Reprod. 1999;60:1093–1103. doi: 10.1095/biolreprod60.5.1093. [DOI] [PubMed] [Google Scholar]
- 71.Kirchhof N, Carnwath JW, Lemme E, Anastassiadis K, Scholer H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. 2000;63:1698–1705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- 72.Hansis C, Grifo JA, Krey LC. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol Hum Reprod. 2000;6:999–1004. doi: 10.1093/molehr/6.11.999. [DOI] [PubMed] [Google Scholar]
- 73.Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- 74.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 75.Hancock SN, Agulnik SI, Silver LM, Papaioannou VE. Mapping and expression analysis of the mouse ortholog of Xenopus Eomesodermin. Mech Dev. 1999;81:205–208. doi: 10.1016/s0925-4773(98)00244-5. [DOI] [PubMed] [Google Scholar]
- 76.Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 77.Tolkunova E, Cavaleri F, Eckardt S, Reinbold R, Christenson LK, Scholer HR, Tomilin A. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem Cells. 2006;24:139–144. doi: 10.1634/stemcells.2005-0240. [DOI] [PubMed] [Google Scholar]
- 78.McConnell J, Petrie L, Stennard F, Ryan K, Nichols J. Eomesodermin is expressed in mouse oocytes and pre-implantation embryos. Mol Reprod Dev. 2005;71:399–404. doi: 10.1002/mrd.20318. [DOI] [PubMed] [Google Scholar]
- 79.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 80.Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. 2005;68:106–112. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- 81.Davidson EH. Lineage-specific gene expression and the regulative capacities of the sea urchin embryo: a proposed mechanism. Development. 1989;105:421–445. doi: 10.1242/dev.105.3.421. [DOI] [PubMed] [Google Scholar]
- 82.Nusslein-Volhard C. Determination of the embryonic axes of Drosophila. Dev Suppl. 1991;1:1–10. [PubMed] [Google Scholar]
- 83.Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- 84.King WA, Bezard J, Bousquet D, Palmer E, Betteridge KJ. The meiotic stage of preovulatory oocytes in mares. Genome. 1987;29:679–682. doi: 10.1139/g87-114. [DOI] [PubMed] [Google Scholar]
- 85.Betteridge KJ, Eaglesome MD, Mitchell D, Flood PF, Beriault R. Development of horse embryos up to twenty two days after ovulation: observations on fresh specimens. J Anat. 1982;135:191–209. [PMC free article] [PubMed] [Google Scholar]
- 86.Roberts RM, Ezashi T, Das P. Trophoblast gene expression: Transcription factors in the specification of early trophoblast. Reproductive Biology and Endocrinology. 2004;2:47. doi: 10.1186/1477-7827-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]