Abstract
TRIM (tripartite motif) proteins primarily function as ubiquitin E3 ligases that regulate the innate immune response to infection. TRIM25 [also known as Efp (oestrogen-responsive finger protein)] has been implicated in the regulation of oestrogen receptor α signalling and in the regulation of innate immune signalling via RIG-I (retinoic acid-inducible gene-I). RIG-I senses cytosolic viral RNA and is subsequently ubiquitinated by TRIM25 at its N-terminal CARDs (caspase recruitment domains), leading to type I interferon production. The interaction with RIG-I is dependent on the TRIM25 B30.2 domain, a protein-interaction domain composed of the PRY and SPRY tandem sequence motifs. In the present study we describe the 1.8 Å crystal structure of the TRIM25 B30.2 domain, which exhibits a typical B30.2/SPRY domain fold comprising two N-terminal α-helices, thirteen β-strands arranged into two β-sheets and loop regions of varying lengths. A comparison with other B30.2/SPRY structures and an analysis of the loop regions identified a putative binding pocket, which is likely to be involved in binding target proteins. This was supported by mutagenesis and functional analyses, which identified two key residues (Asp488 and Trp621) in the TRIM25 B30.2 domain as being critical for binding to the RIG-I CARDs.
Keywords: B30.2, E3 ligase, interferon, oestrogen-responsive finger protein (Efp), retinoic acid-inducible gene-I (RIG-I), tripartite motif (TRIM), tripartite motif-containing 25 (TRIM25)
INTRODUCTION
The innate immune system presents the first line of defence against invading microbial pathogens. Detection of bacterial or viral components is mediated by host cell pattern-recognition receptors including the transmembrane TLRs (Toll-like receptors) [1], the cytoplasmic RLRs [RIG-I (retinoic acid-inducible gene-I)-like receptors] and NLRs [NOD (nucleotide-binding oligomerization domain)-like receptors] [1–3]. Recognition of viral RNAs by TLRs and RLRs activates signalling pathways that result in the expression of pro-inflammatory mediators, including type I IFNs (interferons), to establish an antiviral state in infected and surrounding cells [2–4]. Increasingly, TRIM (tripartite motif; also known as RBCC) proteins, such as pyrin, TRIM5α, TRIM21, TRIM22, TRIM25, TRIM27, TRIM30α and Riplet, are being found to play key roles in restricting viral infection, with many regulating TLR, RLR and NLR signalling cascades [5–12].
TRIM proteins are characterized by an N-terminal zinc finger RING domain, one or two B-box domains, and a CCD (coiled-coil domain). The RING domain confers ubiquitin E3 ligase activity, the function of the B-box domain is largely unknown [13] and the CCD is implicated in multimerization of TRIM proteins [14]. Approximately 50 %of human TRIM proteins also contain a B30.2 domain at the C-terminus, and it is this domain that is thought to recruit substrate proteins as targets for RING E3 ligase activity [14]. Named after the B30.2 exon found within the MHC class I region [15], the B30.2 domain was originally defined by the presence of three highly conserved sequence motifs (LDP, WEVE and LDYE), and is only found in vertebrates with an adaptive immune system [16,17]. The SPRY domain was identified based on a sequence repeat in the dual-specificity kinase spore lysis A and in the Ca2+-release channel ryanodine receptors [18]. The B30.2 domain consists of a ‘SPRY’ region preceded by a conserved N-terminal extension, known as the ‘PRY’ region [17]. The solution of several B30.2 and SPRY domain structures has revealed a characteristic β-sandwich fold [19–21].
TRIM25 [also known as Efp (oestrogen-responsive finger protein)] was originally identified as an oestrogen-responsive gene and plays a key role in the development of the uterus. Expression of TRIM25 has also been associated with ovarian and breast cancer, and is thought to positively promote cell growth by targeting the cell-cycle regulator 14-3-3σ for proteasomal degradation [22–26]. However, more recently TRIM25 has been identified as a key component of the RIG-I signalling pathway. The RIG-I receptor is activated by RNA viruses, such as influenza and hepatitis C virus [3], and this initiates a signalling cascade that results in the activation of NF-κB (nuclear factor κB) and interferon regulatory factors, and the production of IFNα/β [27–29]. Specifically, binding of viral RNA to the RIG-I CTD/RD (C-terminal repressor domain) [30] and the hydrolysis of ATP is thought to induce a conformational change in RIG-I, which exposes the first CARD (caspase recruitment domain) of RIG-I for interaction with TRIM25 [31,32], and results in the attachment of Lys63-linked polyubiquitin chains to the second RIG-I CARD [7,33]. RIG-I then translocates to the mitochondrial surface where it interacts with the transmembrane adaptor protein MAVS [mitochondrial antiviral signalling protein; also known as IPS-1 (IFNβ promoter stimulator 1)/Cardif (CARD adaptor inducing IFNβ)/VISA (virus-induced signalling adaptor)]. TRIM25 acts as a positive regulator of the type I IFN production pathway by activating RIG-I in the initial stages of viral infection. In addition, it has been shown that TRIM25 leads to MAVS degradation, which releases kinase-containing complexes (IKK [IκB (inhibitor of nuclear factor κB) kinase] and TBK1 {TANK [TRAF (tumour-necrosis-factor-receptor-associated factor)-associated nuclear factor-κB activator]-binding kinase 1}) to activate transcription and, finally, type I IFN production [8,34].
The TRIM25 interaction with RIG-I is negatively regulated by a LUBAC (linear ubiquitin assembly complex) containing the E3 ligases HOIL-1L (haem-oxidized IRP2 ubiquitin ligase 1) and HOIP (HOIL-1-interacting protein) [35]. The C-terminal RBR (ring-in-between-RING) domains of HOIL-1L and HOIP interact with the TRIM25 B30.2 domain to induce the ubiquitination and proteasomal degradation of TRIM25 [35]. The importance of TRIM25 in the RIG-I-mediated response to infection has been demonstrated in Trim25−/− mouse embryonic fibroblasts which, following infection with Sendai or vesicular stomatitis virus, were unable to produce IFNβ or restrict viral replication [7,36]. TRIM25 is itself up-regulated in response to IFN in a positive-feedback loop that further augments the antiviral response [37].
As a complement to the RIG-I crystal structures [31,32], and as an initial step towards understanding how TRIM25 interacts with multiple target proteins to regulate innate anti-viral signalling and oestrogen responses, we present the first crystal structure of the TRIM25 B30.2 domain. By comparison with the binding interface of previously published B30.2/SPRY structures in complex with ligands, we further suggest the TRIM25 loop regions and surface pockets that are likely to be involved in binding, and using mutagenesis identify two key residues which are critical for binding to RIG-I.
EXPERIMENTAL
Sample preparation
The TRIM25 B30.2 domain was amplified from full-length murine Trim25 cDNA (GenBank® accession number NM_009546.2; Open Biosystems, Thermo Scientific) by PCR with primers containing AscI and EcoRI restriction sites (Geneworks). The construct was cloned into an in-house pGEX-4T bacterial expression vector. DNA sequencing confirmed the integrity of the resulting plasmid, which was then transformed into Escherichia coli BL21 (DE3) cells. Protein expression was induced upon addition of 0.5 mM IPTG at an D600 of 0.8. After a 4 h induction at 30 °C, the cells were harvested by centrifugation and frozen at −80 °C. The GST (glutathione transferase) fusion protein was purified from clarified cell lysates using glutathione–Sepharose 4B (GE Healthcare), then cleaved overnight at 4 °C with thrombin precision protease (Roche). Cleaved protein was eluted in 50 mM Tris/HCl, 150 mM NaCl and 2 mM DTT (pH 7.5) and further purified by gel-filtration chromatography (Superdex 200 16/60, GE Healthcare), and anion-exchange chromatography (Mono Q, GE Healthcare).
Crystallization
Purified protein was concentrated to 10 mg/ml in 50 mM MES, 75 mM NaCl and 2 mM DTT (pH 6.0), using Amicon 10 kDa molecular-mass cut-off centrifugal concentrators (Millipore). Crystallization screens using the sitting-drop/vapour diffusion method at 281 K were performed at the Bio21 Collaborative Crystallization Centre (C3), Parkville, Australia. Crystals were obtained under multiple conditions and were optimized by mixing 1 μl of protein solution and 1 μl of reservoir solution containing 20–24 % PEG 3350, 0.2 M NaCl and 0.1 M Tris/HCl (pH 8.5). Prior to data collection, crystals were transferred to a cryoprotectant solution consisting of reservoir solution supplemented with 20 %ethylene glycol, then mounted in a nylon loop and flash-frozen in liquid nitrogen.
Data collection and structure determination
Diffraction data were collected on beamline MX2 at the Australian Synchrotron. Data were integrated using XDS [38] and scaled using SCALA [39] from the CCP4 suite [40]. A molecular replacement solution was found using PHASER [41], and the pyrin PRYSPRY domain (PDB code 2WL1, chain A) as a model. Building and refinement were performed in COOT [42] and PHENIX [43] respectively. Refinement converged with R = 0.1734 and Rfree = 0.1944 for data to 1.77 Å, with deviations from ideality of 0.007 Å for bonds and 0.998° for angles. Further data collection and refinement statistics are reported in Table 1. Figures were generated using PyMOL (http://www.pymol.org).
Table 1.
Data collection and refinement statistics (molecular replacement)
| Parameter | Value |
|---|---|
| PDB code | 4B8E |
| Data collection | |
| Protein | TRIM25440–634 |
| Space group | P221 21 |
| Cell dimensions | |
| a, b, c (Å) | 58.149, 73.561 91.009 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 0.953690 |
| Resolution (Å) | 91.01–1.78 (1.88–1.78) |
| Rmerge (%) | 5.44 (60.7) |
| I/σI | 20.9 (3.5) |
| Completeness (%) | 99.8 (99.2) |
| Redundancy | 7.2 (7.2) |
| Refinement | |
| Resolution (Å) | 91.01–1.78 |
| Number of reflections | 274645 |
| Unique reflections | 38142 |
| Rwork /Rfree (%) | 17.34/19.44 |
| Number of atoms | |
| Protein | 3024 |
| Ligand | 10 |
| Solvent | 224 |
| B-factors | |
| Protein | 32.20 |
| Chain A | 24.68 |
| Chain B | 40.09 |
| Solvent | 34.60 |
| RMSD | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.000 |
| Ramachandran plot statistics | |
| Ramachandran favoured (%) | 98 |
| Ramachandran outliers (%) | 0 |
Values in parentheses are for the highest-resolution shell.
Plasmid construction
GST-mRIG-I(2CARD) has been described previously [44]. Myc-tagged mRIG-I(2CARD) was subcloned into pEF-IRES-Puro encoding a C-terminal Myc tag between AflII and NotI. Point mutatations in the mouse TRIM25 B30.2 domain (residues 440–630) were generated using PCR, splicing by overlap extension [45]. Constructs encoding proteins with a C-terminal FLAG epitope tag (DYKDDDDK) were generated by PCR to give fragments with in-frame AscI and MluI restriction enzyme sites at the N- and C-termini respectively and subcloned into the mammalian expression vector pEF-Flag-I, a derivative of pEFBOS [46]. TRIM25 full-length mutants were generated by PCR site-directed mutagenesis using pEF-mTRIM25-Flag as a template. All constructs were fully sequenced to confirm the mutagenesis.
Co-immunoprecipitation and immunoblot analysis
HEK (human embryonic kidney)-293T cells were lysed in NP-40 (Nonidet P40) buffer [50 mM Hepes (pH 7.4), 150 mM NaCl, 1 % (v/v) NP-40 and protease inhibitor cocktail; Sigma], followed by centrifugation at 16 060 g for 20 min at 4 °C. For co-immunoprecipitation, 0.75 ml of post-centrifuged lysates were incubated with ~2.0 μg of antibody at 4 °C overnight, followed by incubation with a 50 %slurry of Protein A/G agarose (Amersham) for 2 h at 4 °C. Immunoprecipitated proteins were extensively washed with lysis buffer and eluted with SDS/Laemmli buffer by boiling for 5 min.
For immunoblot analysis, proteins were resolved by SDS/PAGE (12 % gel) and transferred on to PVDF membranes. The following primary antibodies were used: anti-FLAG (M2; 1:2000 dilution; Sigma), anti-Myc (1:2000 dilution; Covance), anti-GST (1:2000 dilution; Sigma) and anti-ubiquitin (P4D1; 1:500 dilution; Santa Cruz Biotechnology). The proteins were visualized by an enhanced chemiluminescence reagent (Pierce) and detected by a Luminescent imaging system (Fuji LAS-4000).
Luciferase reporter assay
Luciferase assays were performed as described previously [47]. Briefly, HEK-293T cells, seeded into 12-well plates, were transfected with 200 ng of IFNβ luciferase construct and 300 ng of β-gal (β-galactosidase)-expressing pGK-β-gal. In addition, 15 ng of plasmid encoding vector or pEF-IRES-mRIG-I(2CARD)-Myc and 200 ng of pEF-Flag-mTRIM25 WT (wild-type) or mutants were transfected. At 48 h post-transfection, cells were lysed and subjected to a luciferase assay (Promega) according to the manufacturer’s instructions. Luminescence and absorbance were measured, and luciferase values were normalized to β-galactosidase to correct for transfection efficiency.
RESULTS
Crystal structure of the TRIM25 B30.2 domain
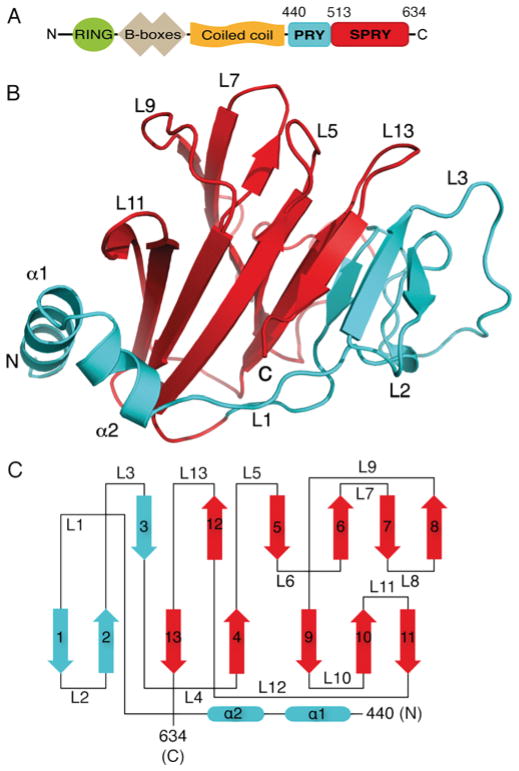
Full-length TRIM25 is a 634 residue 70.6 kDa protein, which is comprised of a RING domain, two B-boxes, a coiled-coil and a B30.2 (PRYSPRY) domain (Figure 1A). Residues 440–634 corresponding to the mouse B30.2 domain were expressed in E. coli cells as a GST-fusion protein, and purified using standard procedures [48]. The TRIM25B30.2 protein crystallized in 25 % (w/v) PEG 3350, 0.2 M NaCl and 0.1 M Tris/HCl (pH 8.5), with two molecules in the asymmetric unit. Phases were obtained via molecular replacement using the pyrin B30.2 domain (PDB [49] code 2WL1 [50]) and refined at 1.8 Å, with R and Rfree values of 0.1734 and 0.1944 respectively (Table 1).
Figure 1. The TRIM25 B30.2 domain structure.
(A) Domain architecture of the full-length TRIM25 molecule highlighting the PRY (blue) and SPRY (red) tandem sequence motifs that comprise the B30.2 domain. (B) Cartoon representation of the three-dimensional structure of the TRIM25 B30.2 domain with the PRY and SPRY regions coloured blue and red respectively. α-Helices and key loops are annotated. (C) Topology diagram of the TRIM25 B30.2 domain with the structural elements coloured according to PRY (blue) and SPRY (red). α-Helices and β-strands are numbered and shown as rounded rectangles and arrows respectively. Loop regions are numbered L1–L13.
The structure consists of a hydrophobic core comprising thirteen β-strands that form two antiparallel β-sheets in a bent β-sandwich configuration. Two α-helices at the N-terminus pack against one face of the β-sheet core with α1 running almost parallel and α2 running perpendicular to the direction of the β-strands (Figures 1B and 1C). The PRY (residues 440–512) and SPRY (residues 513–633) regions of the TRIM25 B30.2 domain form a single structural module (Figure 1B), and exhibit a similar topology to the B30.2 domain structures of pyrin, TRIM21 and TRIM72 [5,50–52]. The TRIM25 structure resolved eight more residues at the N-terminus of the PRY region than previous B30.2 structures. These residues reveal an extra α-helix (α1), which forms hydrophobic interactions with the adjacent β-sheet, while presenting polar residues to the solvent-exposed surface. Specifically, Leu446, Phe449 and Leu450 from α1 are involved in forming hydrophobic interactions with Phe592, Ile594 and Phe596 from β10, and Leu604 and Lys607 from β11. Therefore α1 shields a hydrophobic patch on the underlying β-sheet from solvent exposure. The total solvent-accessible surface area presented by our three-dimensional structure of the TRIM25 B30.2 domain is 8850 Å2 (Figure 1B).
Structural conservation suggests a conserved binding site in B30.2 domains
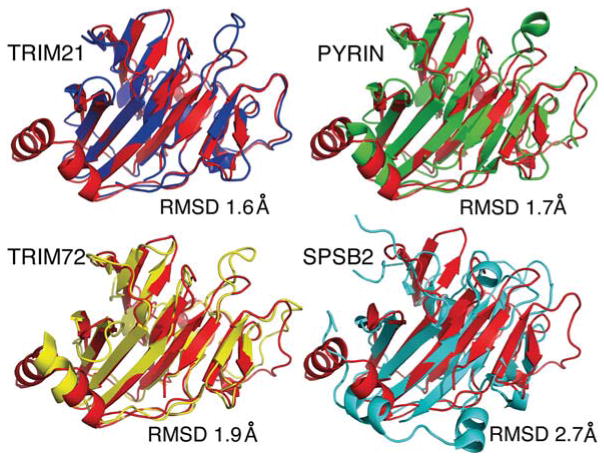
To assess the level of structural conservation between the murine TRIM25 B30.2 domain and other known B30.2/SPRY domain structures, the former was aligned with the X-ray crystal structures of mouse TRIM21 (PDB code 2VOK [51]), mouse SPSB2 (SPRY domain-containing suppressor of cytokine signalling box2; PDB code 3EK9 [53]), human pyrin (PDB code 2WL1 [50]) and human TRIM72 (PDB code 3KB5 [52]). The RMSD values were calculated from structural alignments conducted using DALI [54]. The TRIM25 B30.2 domain shows the highest level of structural conservation with the TRIM21 B30.2 domain, (RMSD of 1.6 Å), followed by human pyrin and TRIM72 (RMSD of 1.7 Å and 1.9 Å respectively). These are comparatively lower than the RMSD value of 2.7 Å obtained for the mouse SPSB2 SPRY domain (Figure 2).
Figure 2. Structural similarity of the TRIM25 B30.2 domain with other B30.2/SPRY domains.
Cartoon representations of the TRIM25 B30.2 domain (red) aligned with the B30.2 domain crystal structures of TRIM21 (blue; PDB code 2VOK), pyrin (green; PDB code 2WL1) and TRIM72 (yellow; PDB code 3KB5), and the N-terminus and SPRY domain structure of SPSB2 (light blue; PDB code 3EK9). All structural alignments are shown in the same orientation with the corresponding RMSD values.
In the three available structures of B30.2/SPRY domains bound to ligand, TRIM21–IgG Fc [5,51], SPSB1–VASA peptide and SPSB2–VASA peptide [55], ligand binding is mediated by a similar overall surface (Figures 3A and 3B). In TRIM21, this region is centred on the top of one face of the β-sandwich, and includes loops 5 and 13, which link the two sheets together (Figure 3B). In TRIM25, this region contains three structurally adjacent solvent-exposed phenylalanine residues in loops 5, 7 and 13 (Phe528, Phe559 and Phe623 respectively), which are raised considerably above the plane of the β-sandwich (Figures 3C and 3D, left-hand panel). At the base of this motif lies an interesting ladder of parallel tryptophan and arginine residue side chains [Trp621 (β12), Arg546 (loop 6), Arg549 (loop 6) and Trp66 (β7)] arranged into a W-R-R-W π-stack (Figures 3C and 3D, left-hand panel). Taken together, the highly solvent-exposed phenylalanine residues and the adjacent W-R-R-W stack form an obvious potential ligand-binding surface that is composed of both hydrophobic and hydrogen bonding capable side chains (Figure 3D, left-hand panel). In addition, as the analogous region is involved in TRIM21 ligand binding, we hypothesize that it will form at least part of the TRIM25 interaction surface.
Figure 3. Known interaction sites for TRIM21 and SPSB2 suggest a putative binding pocket on the TRIM25 B30.2 domain.
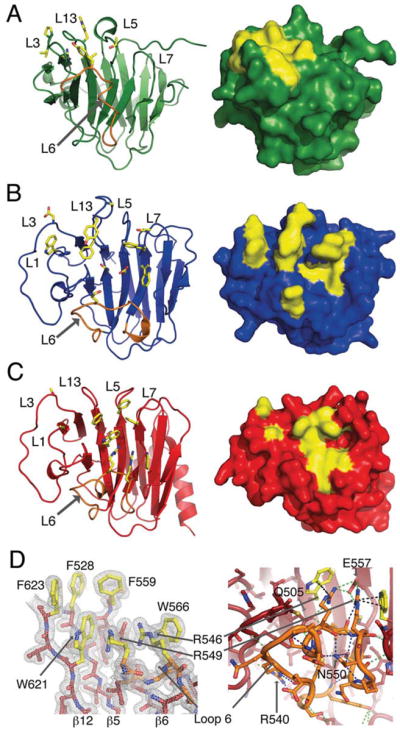
Key residues involved in the SPSB2–VASA peptide interaction (PDB code 3EMW) (A) and TRIM21 interaction with the Fc region of IgG (PDB code 2VOL) (B) are marked in yellow. (C) Homologous loops in the TRIM25 B30.2 domain suggest three phenylalanine residues and a W-R-R-W stack (yellow) that could be involved in binding. (A–C) Cartoon (left-hand panels) and surface (right-hand panels) representations are shown. Loop 6 is highlighted (orange) in the cartoon representations. (D) Electron density of the key residues likely to be involved in binding is shown in the left-hand panel. In the right-hand panel, a stick representation of loop 6 is shown (orange), with the polar contacts within the loop (blue), the contacts to residues in other parts of the domain (green) and the bonds within the W-R-R-W π-stack (black) shown as broken lines.
In TRIM21, the ligand-binding epitope also extends into the PRY region of the domain, to include Trp295 in loop 1 and Asp308 in loop 3 [51] (Figure 3B). A similar section of the region preceding the SPRY domain is also used in SPSB1 and SPSB2 to co-ordinate ligand binding [55] (Figure 3A). TRIM25 residues that structurally align with these are highlighted in Figure 3(C). The SPSB1 and SPSB2 structures show binding to a short linear peptide motif, whereas TRIM21 binds in a conformation-dependent manner to a more extensive ligand [51,55]. In fact, most of the surface used by TRIM21 is occluded in SPSB1 and SPSB2 by the very long (22 residue) loop 6, which blocks the majority of one face of the β-sandwich, leaving only a small surface available for ligand binding (Figure 3A). In TRIM25, loop 6 is packed against the β-sheet, sandwiched tightly between loops 3 and 8. It is held in a highly twisted, yet rigid, conformation by numerous hydrogen bonds both within the loop as well as to the β-sheet proper (Figure 3D, right-hand panel). For example, between the β-sheet and loop 6, Glu557 in β6 forms salt bridges with both Arg546 and Arg549 (which are part of the W-R-R-W motif), whereas the side chain of Gln505 in β3 acts as a hydrogen-bond donor and acceptor to the backbone of Ser545 and Leu547 respectively (Figure 3D, right-hand panel). The side chain of Arg540 forms hydrogen bonds both with the backbone carbonyl of Gln498 (loop 3), and with Gly536 and Met538 within loop 6. Also within loop 6, the side chain of Asn550 makes no less than four hydrogen bonds to other residues with the loop (Glu544, Arg546, Asn552 and Ser553). The B-factors right across loop 6 in both chains are low and are, in fact, similar to the core of the β-sandwich. We hypothesize that the complicated conformation and rigidity of loop 6 functions to shield parts of the underlying β-sheet not involved in ligand binding, as well as directly forming part of the ligand-binding site. When compared with previously published B30.2/SPRY structures, loop 6 varies dramatically in length and conformation suggesting that it may be a key feature in dictating the binding specificity of the B30.2/SPRY domain (Figures 3A–3C).
Sequence conservation of key residues within a putative binding site on the TRIM25 B30.2 domain
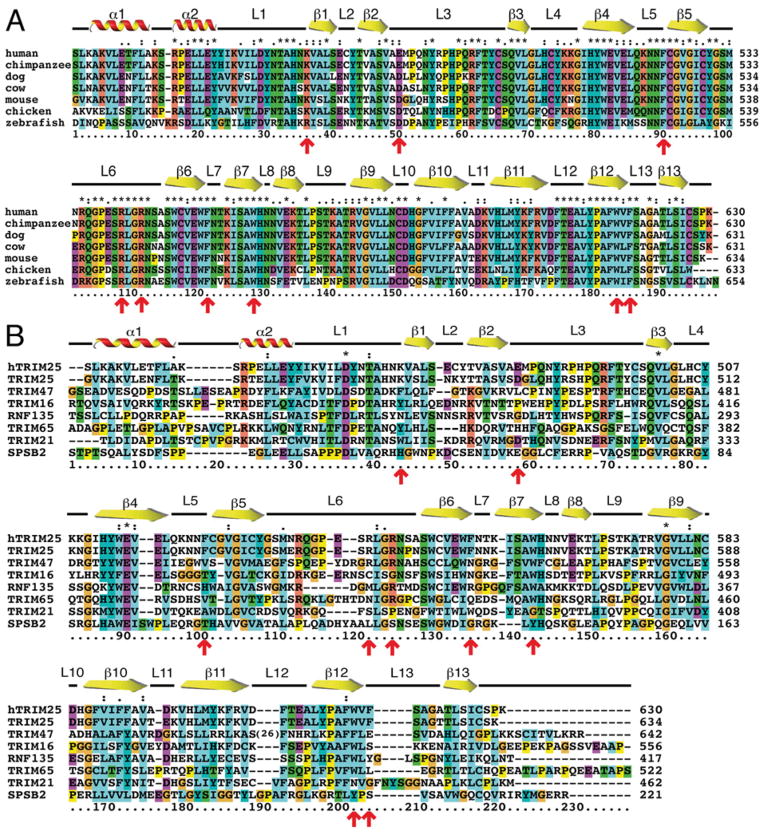
Alignment of the TRIM25 B30.2 sequences across vertebrate species reveals the expected sequence conservation of structural elements such as β-strands and α-helices, but also conservation of residues within loop regions (Figure 4A), suggesting that TRIM25 B30.2 domain function is likely to be conserved across these species. Alignment of human and mouse TRIM25 B30.2 domains with other related murine B30.2/SPRY domains confirms that regions with relative conservation across the different family members correspond to the secondary structural elements (Figure 4B), and this is consistent with the similarity of the TRIM25 B30.2 structure to known B30.2/SPRY domain structures (Figure 2A). Residues Lys474 (loop 1) and Asp488 (loop 3) exhibit high sequence similarity across species (Figure 4A), whereas Asp488 is conserved in TRIM21, but not in other related B30.2 domains (Figure 4B). As concerns our predicted binding site, the three phenylalanine residues within loops 5 (Phe528), 7 (Phe559) and 13 (Phe623) are unique to TRIM25, where they are highly conserved across species (Figure 4A). Similarly, the W-R-R-W motif is also conserved in all vertebrate TRIM25 molecules (Figure 4A); however, only TRIM47 is predicted to contain a similarly π-stacked (W-R-R-F) motif at the same position (Figure 4B). Finally, loop 6 is extremely variable in sequence between different B30.2 domains, yet once again is highly conserved within TRIM25 across species (Figure 4). Therefore a combination of residues, including Asp488, Lys474, Arg546 and Trp566, and loop 6 appeared likely to contribute to TRIM25 B30.2 domain-binding specificity.
Figure 4. Mouse TRIM25 B30.2 domain sequence is conserved across species, but not across other closely related mouse B30.2 domains.
(A) Alignment of murine TRIM25 B30.2 domain against the same domain sequence from other species. (B) Alignment of human (hTRIM25) and mouse TRIM25 B30.2 domain with closely related mouse B30.2/SPRY domains. Sequences were sourced from GenBank® and aligned using ClustalX 2.0.12 [62]. Secondary structural elements positioned above the alignment correspond to residues from the mouse TRIM25 B30.2 domain crystal structure. Red arrows indicate key residues in TRIM25, which have been tested by mutagenesis for binding to the RIG-I CARDs.
Asp488 and Trp621 are critical for binding of the TRIM25 B30.2 domain to RIG-I
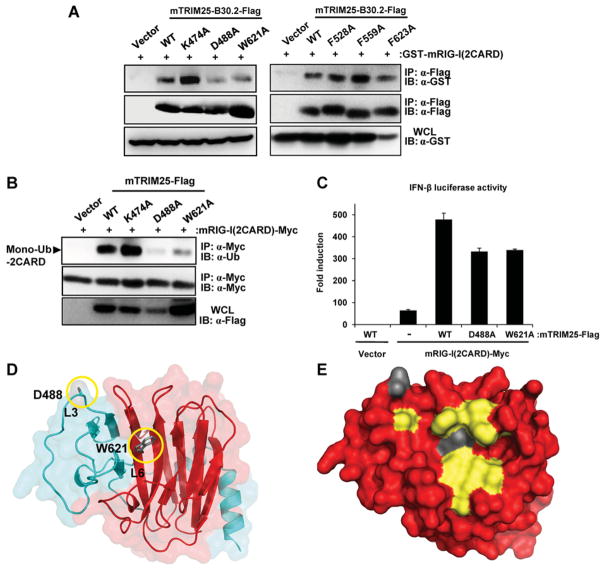
To test whether the residues identified by structural comparison and sequence alignment formed part of a putative binding site, we mutated the key residues to alanine and tested the constructs for binding to a known TRIM25-interacting protein, the RIG-I CARD domains. Expression constructs for FLAG-tagged mouse TRIM25 B30.2 domain WT or various mutants and the mouse RIG-I CARDs fused to GST [GST–mRIG-I(2CARD)], were transiently transfected into HEK-293T cells and binding was then assessed by co-immunoprecipitation and Western blotting. Mutation within the TRIM25 B30.2 domain converting Lys474, Phe528, Phe559 or Phe623 into an alanine residue did not have an appreciable effect on binding to GST–mRIG-I(2CARD). In contrast, mutation of Asp488 or Trp621 dramatically reduced binding to the RIG-I CARDs (Figure 5A). Mutation of Arg546, Arg549 or Trp566 to an alanine residue abrogated protein expression, suggesting that these mutations disrupted the tertiary structure of the TRIM25 B30.2 domain (results not shown).
Figure 5. The mutations D488A and W621A decreases binding of the TRIM25 B30.2 domain to the RIG-I CARDs and result in decreased RIG-I CARD ubiquitination and signalling.
(A) HEK-293T cells were transfected with GST–mRIG-I(2CARD) together with either empty vector, Flag-tagged mTRIM25-B30.2 WT or mutants. At 48 h post-transfection, whole-cell lysates (WCLs) were immunoprecipitated (IP) using an anti-FLAG antibody, followed by immunoblot (IB) analysis using an anti-GST or anti-FLAG antibody. Expression of GST–mRIG-I(2CARD) was determined in the whole-cell lysates using an anti-GST antibody. (B) HEK-293T cells were transfected with mRIG-I(2CARD)–Myc together with either empty vector or FLAG-tagged mTRIM25 WT or mutants. At 48 h post-transfection, cells were lysed and whole-cell lysates subjected to immunoprecipitation using an anti-Myc antibody, followed by immunoblotting using an anti-ubiquitin (Ub) or anti-Myc antibody. Whole-cell lysates were further used for immunoblotting with an anti-FLAG antibody to determine the expression of mTRIM25 WT and mutants. The arrow indicates the mono-ubiquitinated form of mRIG-I(2CARD)–Myc. (C) HEK-293T cells were transfected with IFNβ luciferase and constitutive β-gal-expressing pGK-β-gal constructs together with either empty vector and FLAG–mTRIM25 WT, mRIG-I(2CARD)–Myc and vector, or mRIG-I(2CARD)–Myc together with FLAG-tagged mTRIM25 WT or mutants. At 48 h post-transfection, the luciferase and β-gal values were determined. The results, expressed as means ± S.D. (n = 2), are representative of three independent experiments. (D) Cartoon representation of the TRIM25 B30.2 domain highlighting the location of Asp488 and Trp621 in relation to the PRY (blue) and SPRY (red) regions. Loops 3 and 6 are indicated. (E) Surface representation of the TRIM25 B30.2 domain depicting the location of Asp488 and Trp621 in the context of the residues identified as part of a putative binding site.
Binding of TRIM25 to RIG-I results in ubiquitination of the second CARD of RIG-I, facilitating an interaction with MAVS and the production of IFNβ [7]. To determine whether mutation of Asp488 or Trp621 was sufficient to alter the ability of TRIM25 to induce ubiquitination-dependent RIG-I activation, the mutants were generated in the context of full-length mouse TRIM25 and tested for their ability to ubiquitinate RIG-I and promote RIG-I downstream signalling. Consistent with the binding data, mutation of either Asp488 or Trp621 almost completely abolished the ability of TRIM25 to ubiquitinate the RIG-I CARDs (Figure 5B). Similarly, using an IFNβ reporter assay, mutation of either Asp488 or Trp621 reduced the ability of TRIM25 to enhance RIG-I 2CARD-mediated IFNβ induction by 31 % (Figure 5C).
In conclusion, two residues (Asp488 and Trp621) were identified as being critical for TRIM25 B30.2 binding to the RIG-I CARDs, strongly suggesting that they form part of the binding interface (Figures 5D and 5E).
DISCUSSION
Previous studies have shown that TRIM25 has an important role in the innate immune response to viral infection [7], and may function as an oncogene in breast cancer leading to poorer patient outcomes [25,26]. The primary mechanism of action is thought to be recruitment of a target protein by the C-terminal B30.2 protein-interaction domain, followed by the transfer of ubiquitin from the RING-bound E2-ubiquitin conjugate to a lysine residue in the target protein. TRIM25 has been reported to synthesize both Lys63- and Lys48-linked polyubiquitin chains, leading respectively to the activation of RIG-I [7] and degradation of MAVS [8]. Although B-box domains are thought to co-ordinate zinc ions, their specific role within the context of TRIM25 remains unknown [56]. The CCD is required for multimerization of TRIM25, which is important for RIG-I activation and downstream signalling [57]. In the present study we have solved the crystal structure of the TRIM25 B30.2 domain and identified a putative binding surface; the structure of the RING, B-box and CCD domains are yet to be elucidated.
The previously published B30.2 domain structure of TRIM72 has 192 residues (62 PRY) resolved, whereas pyrin has 190 (62 PRY) and TRIM21 has 192 (52 PRY) residues resolved [5,50–52]. Among SPRY-only proteins, SPSB1 has 203 (64 in the N-terminal region), SPSB4 has 201 (64 N-terminal) and SPSB2 has 199 (66 N-terminal) residues resolved [53,55]. The TRIM25 B30.2 domain structure has a total of 193 (79 PRY) residues resolved, and therefore may include more of the PRY region than previous B30.2 structures. In addition to the characteristic bent β-sandwich, the TRIM25 B30.2 crystal structure revealed two α-helices in the PRY region of the B30.2 domain. Structural alignments with previously published B30.2/SPRY domains show that the TRIM25 B30.2 domain structure contains an additional α-helix at the N-terminus (α1), which forms non-polar interactions with hydrophobic residues on the adjacent β-sheet, thereby shielding it from solvent exposure. Analysis of other B30.2 domain structures, revealed a lack of similarly hydrophobic residues on the β-sheet (results not shown), suggesting that the additional α-helix may be unique to TRIM25. Interestingly, this region appears more similar in topology to the N-terminal region of the SPRY-only proteins, SPSB1, SPSB2 and SPSB4, which also contain two α-helices (Figure 2B). The structure of the TRIM25 B30.2 domain therefore supports earlier suggestions that the PRY region is related to the N-terminal region preceding the SPRY motif in SPRY-only proteins, despite the low sequence similarity [21,53,58].
It is obvious from the TRIM25 B30.2 crystal structure that together the PRY and SPRY tandem motifs form a single modular domain, similar to that seen in previously published B30.2/SPRY domain structures. The structure of murine and human TRIM21 in complex with the Fc region of IgG revealed key residues from the PRY and SPRY regions that are involved in binding [5,51]. Similarly, residues from the N-terminus and SPRY region of SPSB1 and SPSB2 are involved in binding the DINNN peptide motif found in VASA and inducible nitric oxide synthase [21,55,59]. We have utilized structural and sequence comparisons to extrapolate from these known interactions, to suggest residues that form a putative binding pocket in TRIM25 and which might be involved in interacting with target proteins such as RIG-I or MAVS. We observed several interesting features within this putative binding site, such as three highly solvent-exposed phenylalanine residues sitting above a quartet of residues (W-R-R-W) involved in a π-stack.
When compared with previous B30.2 structures, the TRIM25 structure highlights the diversity of loop 6. In TRIM25, it has a very complicated conformation, constrained by numerous intra-and inter-loop hydrogen bonds, salt-bridges and van der Waals contacts. We suggest that by covering or revealing part of the β-sandwich, loop 6 will play a major role in determining binding specificity by both blocking parts of the β-sheet surface not required for ligand binding, as well as providing residues that directly contribute to ligand binding.
The TRIM25 B30.2 domain has been shown to be both necessary and sufficient for the interaction with the RIG-I CARDs in cell lysates [7,60], and experiments in vitro have shown this to be a direct interaction [61]. We have used mutagenesis to explore a putative binding site, which we have identified by structural and sequence comparisons, identifying two residues (Asp488 in loop 3 and Trp621 in loop 6) that appear to be critical for the interaction of the TRIM25 B30.2 domain with the RIG-I CARDs. Although mutation of other conserved residues (Lys474, Phe528, Phe559 or Phe623) did not alter binding to RIG-I it is still conceivable that these residues will contribute to the binding interface, but that mutation to an alanine residue was not sufficient to have an impact on binding in an overexpression system. Mutation of Arg546, Arg549 or Trp566 resulted in significantly reduced protein expression (results not shown), suggesting that these mutations reduced protein stability and are likely to disrupt the π-stack which anchors loop 6 in place. The location of these two key residues, Asp488 in the PRY and Trp621 in the SPRY region of the TRIM25 B30.2 domain (Figure 5D), lends further support to the notion that the PRY and SPRY regions do indeed form a single modular domain. Interestingly, Asp488 is conserved in TRIM21 (Figures 2 and 4B) and the analogous residue (Asp308) has been shown to form hydrogen bonds with residues in the Fc region of IgG [5] (Figure 3B), suggesting that there is a common modality of binding across these two B30.2 domains.
As the crystal structure of mallard duck RIG-I contained the CARD domains in an inactive conformation, it is difficult to ascertain complementary residues on the first CARD of RIG-I that are likely to be involved in binding TRIM25. Furthermore, although mutational analyses revealed residues in human RIG-I that were important for binding TRIM25 (Thr55) and ubiquitination (Lys172) [60], neither residue is conserved in the mallard duck.
The structure of a mouse or human TRIM25 B30.2–RIG-I CARD complex is required to fully understand the binding interface. This information, together with the current mutagenesis data will inform future experiments designed to test the involvement of the TRIM25 B30.2 domain in binding target proteins, and facilitate crystallization of TRIM25 in complex with those binding partners.
A number of other potential TRIM25-binding partners have been identified. For instance, the TRIM25 B30.2 domain has been shown to interact with the RBR domains of HOIL-1L and HOIP, which reportedly target TRIM25 for proteasomal degradation [35]. However, a direct molecular interaction between the RBR of HOIL-1L/HOIP and the TRIM25 B30.2 domain was not shown, and the residues involved in binding remain to be elucidated. Targeting this interaction has the potential to augment TRIM25-mediated activation of RIG-I, leading to an increased IFN response in immunocompromised patients. TRIM25 has also been shown to interact with MAVS in cells [8], but a direct interaction was not shown and it remains unclear whether the B30.2 domain is necessary and sufficient for binding to MAVS.
B30.2/SPRY domain proteins are becoming increasingly important in innate immune signalling. The role of TRIM25 as an activator of RIG-I signalling makes it an important target for modulating the immune response to RNA viruses, such as measles, vesicular stomatitis, influenza and hepatitis C [3]. Our analysis of the crystal structure of the TRIM25 B30.2 domain identifies key residues and a putative binding pocket (Figure 5E) that may be involved in binding substrate proteins. Importantly, small molecules that target the TRIM25 B30.2 domain may offer greater potential for specificity, as opposed to generic inhibitors of the ubiquitin–proteasome cascade or therapeutics targeting the RING domain, which may have an impact on multiple RING E3 ligases. The structure of the TRIM25 B30.2 domain as described in the present paper will facilitate a detailed understanding of how this domain interacts with its putative target proteins.
Acknowledgments
The authors thank the staff at the MX2 Beamline, Australian Synchrotron, Dr J. Newman at the Bio21 Collaborative Crystallization Centre (C3), and Dr J. Zhang, Dr J. Murphy, Dr R. Lewis and Dr P. Czabotar from the Walter and Eliza Hall Institute of Medical Research for assistance and valuable discussions.
FUNDING
This work was supported, in part, by the National Health and Medical Research Council (NHMRC) Australia [grant numbers 461219, 637348, 487922 and 361646] and fellowships to S.E.N. and N.A.N., the Victorian State Government Operational Infrastructure Scheme grant, the U.S. National Institutes of Health [grant number R01 AI087846 (to M.U.G.)], the Australian Postgraduate Awards (to A.A.D.) and the Australian Research Council via a Future Fellowship to J.J.B.
Abbreviations used
- CARD
caspase recruitment domain
- CCD
coiled-coil domain
- β-gal
β-galactosidase
- GST
glutathione transferase
- HEK
human embryonic kidney
- HOIL-1L
haem-oxidized IRP2 ubiquitin ligase 1
- HOIP
HOIL-1-interacting protein
- IFN
interferon
- MAVS
mitochondrial antiviral signalling protein
- NLR
NOD (nucleotide-binding oligomerization domain)-like receptor
- NP-40
Nonidet P40
- RBR
ring-in-between-RING
- RIG-I
retinoic acid-inducible gene-I
- RLR
RIG-I-like receptor
- SPSB
SPRY domain-containing suppressor of cytokine signalling box
- TLR
Toll-like receptor
- TRIM
tripartite motif
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Akshay D’Cruz designed and performed experiments and wrote the paper. Nadia Kershaw solved the crystal structure. Jessica Chiang and May Wang performed experiments. Nicos Nicola, Jeffrey Babon, Michaela Gack and Sandra Nicholson designed and supervised the experiments and contributed to writing of the paper.
References
- 1.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 2.Creagh EM, O’Neill LAJ. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 5.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 8.Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012;10:44. doi: 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, et al. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 10.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 11.Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, Mechti N, Mothes W, Poli G, Luban J, Vicenzi E. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-κB-responsive long terminal repeat elements. J Virol. 2011;85:5183–5196. doi: 10.1128/JVI.02302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3:513–527. doi: 10.1002/emmm.201100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 14.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vernet C, Boretto J, Mattei MG, Takahashi M, Jack LJ, Mather IH, Rouquier S, Pontarotti P. Evolutionary study of multigenic families mapping close to the human MHC class I region. J Mol Evol. 1993;37:600–612. doi: 10.1007/BF00182746. [DOI] [PubMed] [Google Scholar]
- 16.Henry J, Ribouchon MT, Offer C, Pontarotti P. B30.2-like domain proteins: a growing family. Biochem Biophys Res Commun. 1997;235:162–165. doi: 10.1006/bbrc.1997.6751. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DA, de Bono B, Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116:411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponting C, Schultz J, Bork P. SPRY domains in ryanodine receptors (Ca2+-release channels) Trends Biochem Sci. 1997;22:193–194. doi: 10.1016/s0968-0004(97)01049-9. [DOI] [PubMed] [Google Scholar]
- 19.Grutter C, Briand C, Capitani G, Mittl PR, Papin S, Tschopp J, Grutter MG. Structure of the PRYSPRY-domain: implications for autoinflammatory diseases. FEBS Lett. 2006;580:99–106. doi: 10.1016/j.febslet.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 20.Masters SL, Yao S, Willson TA, Zhang JG, Palmer KR, Smith BJ, Babon JJ, Nicola NA, Norton RS, Nicholson SE. The SPRY domain of SSB-2 adopts a novel fold that presents conserved Par-4-binding residues. Nat Struct Mol Biol. 2006;13:77–84. doi: 10.1038/nsmb1034. [DOI] [PubMed] [Google Scholar]
- 21.Woo JS, Imm JH, Min CK, Kim KJ, Cha SS, Oh BH. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orimo A, Inoue S, Minowa O, Tominaga N, Tomioka Y, Sato M, Kuno J, Hiroi H, Shimizu Y, Suzuki M, et al. Underdeveloped uterus and reduced estrogen responsiveness in mice with disruption of the estrogen-responsive finger protein gene, which is a direct target of estrogen receptor alpha. Proc Natl Acad Sci USA. 1999;96:12027–12032. doi: 10.1073/pnas.96.21.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda K, Orimo A, Higashi Y, Muramatsu M, Inoue S. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett. 2000;472:9–13. doi: 10.1016/s0014-5793(00)01421-6. [DOI] [PubMed] [Google Scholar]
- 24.Sakuma M, Akahira J, Suzuki T, Inoue S, Ito K, Moriya T, Sasano H, Okamura K, Yaegashi N. Expression of estrogen-responsive finger protein (Efp) is associated with advanced disease in human epithelial ovarian cancer. Gynecol Oncol. 2005;99:664–670. doi: 10.1016/j.ygyno.2005.07.103. [DOI] [PubMed] [Google Scholar]
- 25.Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. Efp targets 14-3-3σ for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T, Urano T, Tsukui T, Horie-Inoue K, Moriya T, Ishida T, Muramatsu M, Ouchi Y, Sasano H, Inoue S. Estrogen-responsive finger protein as a new potential biomarker for breast cancer. Clin Cancer Res. 2005;11:6148–6154. doi: 10.1158/1078-0432.CCR-05-0040. [DOI] [PubMed] [Google Scholar]
- 27.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 28.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 32.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 35.Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 37.Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. Xds. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabsch W. SCALA. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 40.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 46.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao S, Masters SL, Zhang JG, Palmer KR, Babon JJ, Nicola NA, Nicholson SE, Norton RS. Backbone 1H, 13C and 15N assignments of the 25 kDa SPRY domain-containing SOCS box protein 2 (SSB-2) J Biomol NMR. 2005;31:69–70. doi: 10.1007/s10858-004-6924-6. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur J Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 50.Weinert C, Grutter C, Roschitzki-Voser H, Mittl PR, Grutter MG. The crystal structure of human pyrin b30.2 domain: implications for mutations associated with familial Mediterranean fever. J Mol Biol. 2009;394:226–236. doi: 10.1016/j.jmb.2009.08.059. [DOI] [PubMed] [Google Scholar]
- 51.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci USA. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park EY, Kwon OB, Jeong BC, Yi JS, Lee CS, Ko YG, Song HK. Crystal structure of PRY-SPRY domain of human TRIM72. Proteins. 2010;78:790–795. doi: 10.1002/prot.22647. [DOI] [PubMed] [Google Scholar]
- 53.Kuang Z, Yao S, Xu Y, Lewis RS, Low A, Masters SL, Willson TA, Kolesnik TB, Nicholson SE, Garrett TJ, Norton RS. SPRY domain-containing SOCS box protein 2: crystal structure and residues critical for protein binding. J Mol Biol. 2009;386:662–674. doi: 10.1016/j.jmb.2008.12.078. [DOI] [PubMed] [Google Scholar]
- 54.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filippakopoulos P, Low A, Sharpe TD, Uppenberg J, Yao S, Kuang Z, Savitsky P, Lewis RS, Nicholson SE, Norton RS, Bullock AN. Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4. J Mol Biol. 2010;401:389–402. doi: 10.1016/j.jmb.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borden KL. RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 57.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Cruz AA, Babon JJ, Norton RS, Nicola NA, Nicholson SE. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 2013;22:1–10. doi: 10.1002/pro.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuang Z, Lewis RS, Curtis JM, Zhan Y, Saunders BM, Babon JJ, Kolesnik TB, Low A, Masters SL, Willson TA, et al. The SPRY domain-containing SOCS box protein SPSB2 targets iNOS for proteasomal degradation. J Cell Biol. 2010;190:129–141. doi: 10.1083/jcb.200912087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci USA. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng M, Ding Z, Xu L, Kong L, Wang W, Jiao S, Shi Z, Greene MI, Cong Y, Zhou Z. Structural and biochemical studies of RIG-I antiviral signaling. Protein Cell. 2013;4:142–154. doi: 10.1007/s13238-012-2088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]