Abstract
We investigated the genome-wide distribution of CNVs in the Alzheimer's disease (AD) Neuroimaging Initiative (ADNI) sample (146 with AD, 313 with Mild Cognitive Impairment (MCI), and 181 controls). Comparison of single CNVs between cases (MCI and AD) and controls shows overrepresentation of large heterozygous deletions in cases (p-value < 0.0001). The analysis of CNV-Regions identifies 44 copy number variable loci of heterozygous deletions, with more CNV-Regions among affected than controls (p = 0.005). Seven of the 44 CNV-Regions are nominally significant for association with cognitive impairment. We validated and confirmed our main findings with genome re-sequencing of selected patients and controls. The functional pathway analysis of the genes putatively affected by deletions of CNV-Regions reveals enrichment of genes implicated in axonal guidance, cell–cell adhesion, neuronal morphogenesis and differentiation. Our findings support the role of CNVs in AD, and suggest an association between large deletions and the development of cognitive impairment
Keywords: Alzheimer's disease, Copy Number Variable Regions (CNV-Regions), Copy Number Variations (CNVs), Genome-wide scan, Next Generation Sequencing (NGS)
1. Introduction
Copy number variations (CNVs) represent an important source of genetic diversity that can affect biological functions. With the advent of genome-wide tools, CNV mapping on a genomic scale has proven to be crucially relevant to integrate the information provided by SNP microarray technologies in studies of complex traits [1]. The recent availability of high-throughput genotyping technologies has propelled an increasing number of genome-wide association studies (GWAS) of Alzheimer's disease (AD) producing a progressive number of candidate genes (http://www.alzgene.org/). Nevertheless, the risk factors identified in AD patients collectively explain a relatively modest amount of the total genetic risk for the disease. CNV may be an important piece to complete the puzzle of the “missing heritability” [2] in the study of cognitive impairment and AD. Recent studies supported a substantial role of chromosomal structural variations in the pathogenesis of several neurological disorders [3]. This evidence is consistent with the hypothesis that genetic susceptibility to late onset complex disorders such as Alzheimer's disease (AD), Amyotrophic Lateral Sclerosis (ALS) and Parkinson's disease (PD) depends on dosage-sensitive loci directly affected by copy number variations [4]. CNV position and dose-related effects may have a strong indirect influence on gene expression, as supported by recent studies on CNV's global role in shaping the human transcriptome [5]. These effects may be further enhanced by the increased action of environmental age-related factors [6].
Three recent studies screened for an effect of CNVs in three different samples of late-onset AD [7–9] using both whole genome and candidate gene approaches. Despite the lack of statistically significant results, several genes were deemed interesting for further investigations since they were modestly enriched in CNVs in cases compared to controls. Whole genome SNP-microarray CNV studies may generally fail to reach statistical significance in case–control study designs because of the rarity of the events. A significant methodological challenge is identifying a common pattern of variation across different subjects when structural events have different sizes. Large structural events partially overlapping with previously identified smaller CNVs in the general population are usually not considered novel, but the effect of a very large CNV may be totally different from those of much smaller CNVs. To address this concern, Redon et al. [10] introduced the concept of loci-encompassing CNV-Regions. CNV-Regions can provide a more realistic representation of the distribution of structural rearrangements across subjects than single copy number events [11]. The effect size of a CNV may explain only a small proportion of the whole genetic variability underlying common disorders, since rarely a single CNV has a high frequency in the general population [12]. On the contrary, a combination of different CNVs in different individuals, can alter biological functions in the same important pathway and result in a much larger effect, although identifying the correct pattern of jointly acting CNVs is complex. In this context the proximity of CNVs to functional genetic variants and regulatory elements is likely to be critical. Recent papers have described how CNVs are strongly involved in controlling overall gene expression [5]: their genome-wide distribution is non-random, and is strongly correlated with genomic features like exons, segmental duplications, repetitive elements (e.g. Alu elements) [1] and microRNAs [13]. Interestingly, CNVs can shape the expression level of a gene not only by gene dosage, but also by inducing a variety of epigenetic modifications some of which can act on genes located more than half a megabase from the physical location of the CNV [5,14].
A further challenge is developing a copy number detection algorithm that is not biased toward the specific probe-content of different available commercial array platforms [15]. Different algorithms and parameter optimization can lead to substantial differences in CNV detection [16,17]. Another critical aspect of a SNP microarray approach is the difficulty in resolving the breakpoints of CNVs, whose assignment is based on estimates driven from array intensities that are dependent on SNPs density [18]. To address these limitations, we opted for a strategy that integrates the SNP microarray information with deep sequencing validation of the most promising results [15,19,20].
In this study, we present the genome-wide distribution of Copy Number Variations (CNVs) and Copy-Number-Variable Regions (CNV-Regions) using SNP microarrays in the Alzheimer's Disease Neuro-imaging Initiative (ADNI) cohort for which we have previously described genome-wide association analyses of individual SNPs [21,22].We looked for duplications and deletions using intensity data from SNP microarrays in the ADNI cohort composed by 640 subjects, namely 181 healthy controls, 313 mild cognitive impairment (MCI) and 146 Alzheimer's disease (AD) patients. The affected cases (e.g. MCI and AD) include individuals with mild to moderate cognitive impairment. We investigated the association of CNVs and CNV-Regions with cognitive impairment as expressed in MCI and late-onset AD patients. We created CNV-Region profiles to investigate the potential consequences of these structural variations on gene function using a pathway-based framework. Finally, we confirmed our CNV results comparing individual calls with a different algorithm and validated our main findings by re-sequencing our CNV-Regions in selected patients and controls.
2. Results
2.1. Quality control
After removing subjects that did not pass the quality score threshold for the raw intensity data (QS > 0.2), the ADNI sample contained 640 subjects subdivided in 146 AD patients, 313 MCI patients and 181 matched healthy controls. The Nexus CNV calling algorithm identified 11,058 CNVs from the raw data across the whole sample. In 85 subjects we identified and excluded from the analysis 373 “overlapping” CNVs that can be grouped in 3 categories: 1) 74 segments called as a contiguous deletion and duplication (33 subjects), 2) 89 segments in which heterozygous and homozygous deletions could not be distinguished (40 cases), 3) 1 case where a heterozygous duplication and a homozygous duplication could not be distinguished. There were 208 segments with contiguous heterozygous deletions (92 cases) that were analyzed as 92 deletions after merging two or more segments for each of the 92 subjects. This latter phenomenon seems to be due to slight differences in the intensities data that are detected by the Nexus CNV-calling algorithm as different events. We performed this merge procedure since the “call fragmentation” issue is known to possibly affect the assessment of large CNVs [16]. In total, 116 segments of the group of 208were removed from the final list of CNVs along with the CNVs listed in categories 1 (74 segments), 2 (89 segments), and 3 (2 segments), leaving 10,777 CNVs.
2.2. Genome-wide map of CNVs detected in the ADNI sample
Our first aim was to evaluate the frequencies of CNVs detected in the ADNI population defined by deletions, duplications, size and diagnosis as reported in Table 2. Of the 10,777 CNVs, we identified 8443 heterozygous deletions, 806 homozygous deletions, and 1528 duplications, including 5 multi-allelic variants (i.e. high copy gain). The median size of CNVs was 230 kb, ranging from 2.5 kb to 72 Mb. Homozygous deletions tended to be smaller than heterozygous deletions and were similar in size and number in patients and controls, as were duplications.
Table 2.
Characteristics of CNVs detected using microarray data in the ADNI sample.
| CNV size ranges |
Homozygous deletions | Heterozygous deletions | Duplications | High copy gain | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTRL | MCI | AD | Total | CTRL | MCI | AD | Total | CTRL | MCI | AD | Total | CTRL | MCI | AD | Total | ||
| 0–20 kb | Number of CNV detected | 41 | 96 | 33 | 170 | 46 | 72 | 33 | 151 | 2 | 18 | 6 | 26 | 1 | 1 | – | 2 |
| Mean CNV size kb (SD) | 11,382 (4807) | 10,522 (4206) | 11,130 (3797) | 10,847 (4277) | 14,560 (3116) | 14,249 (3220) | 14,235 (3534) | 14,341 (3241) | 15,439 (2099) | 15,496 (2880) | 16,244 (3004) | 15,664 (2780) | 16,054 (0) | 16,054 (0) | – | 16,054 | |
| 20–100 kb | Number of CNV detected | 120 | 170 | 77 | 367 | 468 | 777 | 369 | 1614 | 91 | 199 | 84 | 374 | 0 | 1 | – | 1 |
| Mean CNV size kb (SD) | 45,912 (20,436) | 47,134 (21,647) | 44,442 (20,671) | 46,169 (21,023) | 62,431 (22,446) | 63,201 (21,922) | 62,080 (21,978) | 62,721 (22,079) | 58,457 (18,978) | 58,685 (22,608) | 59,282 (22,812) | 58,764 (21,774) | – | 46,259 (0) | – | 46,259 | |
| 100 kb–450 kb | Number of CNV detected | 77 | 127 | 64 | 268 | 1092 | 2084 | 915 | 4091 | 212 | 380 | 168 | 760 | 2 | 0 | – | 2 |
| Mean CNV size kb (SD) | 153,417 (47,905) | 153,644 (40,175) | 156,993 (40,218) | 154,379 (42,412) | 255,886 (116,761) | 263,490 (117,022) | 262,689 (116,642) | 261,281 (116,884) | 242,418 (106,611) | 248,233 (103,951) | 238,330 (102,066) | 244,422 (104,231) | 148,958 (32,608) | – | – | 148,958 (32,608) | |
| 450 kb–2 Mb | Number of CNV detected | 0 | 1 | 0 | 1 | 473 | 1001 | 432 | 1906 | 99 | 162 | 89 | 350 | – | – | – | – |
| Mean CNV size kb (SD) | – | 527,566 (0) | – | 527,566 (0) | 873,373 (353,832) | 913,183 (386,963) | 875,095 (356,450) | 894,671 (372,491) | 857,426 (306,587) | 881,915 (344,363) | 866,698 (316,327) | 871,119 (326,257) | – | – | – | – | |
| 2 Mb–10 Mb | Number of CNV detected | – | – | – | – | 100 | 445 | 72 | 617 | 1 | 9 | 3 | 13 | – | – | – | – |
| Mean CNV size kb (SD) | – | – | – | – | 3,487,306 (1,616,961) | 4,223,484 (1,933,931) | 3,641,255 (1,812,325) | 4,036,226 (1,893,946) | 2,268,253 (0) | 2,673,911 (539,851) | 2,948,826 (642,019) | 2,706,148 (542,653) | – | – | – | – | |
| > 10 Mb | Number of CNV detected | – | – | – | – | 6 | 46 | 12 | 64 | – | – | – | – | – | – | – | – |
| Mean CNV size kb (SD) | – | – | – | – | 13,032,082 (3,996,942) | 13,562,461 (9,103,163) | 14,729,801 (5,023,062) | 13,731,614 (8,069,805) | – | – | – | – | – | – | – | – | |
| All | Number of CNV detected | 238 | 394 | 174 | 806 | 2185 | 4425 | 1833 | 8443 | 405 | 768 | 350 | 1523 | 3 | 2 | 5 | |
| Mean CNV size kb (SD) | 74,744 (63,843) | 73,764 (67,772) | 79,522 (66,631) | 75,297 (66,344) | 526,015 (1,065,102) | 907,721 (2,088,887) | 589,584 (1,458,033) | 739,869 (1,752,991) | 355,300 (356,042) | 355,757 (431,596) | 374,569 (431,851) | 359,958 (412,733) | 104,656 (80,121) | 31,156 (21,358) | 75,256 (70,317) | ||
The number of CNVs per individual did not differ significantly between cases and healthy controls. We observed a slightly higher number of the total copy number events in MCI subjects (mean #-of-calls = 18.1) compared to either AD patients (mean #-of-calls = 15.5) or healthy controls (mean #-of-calls = 15.6) (see Table 2 for details).
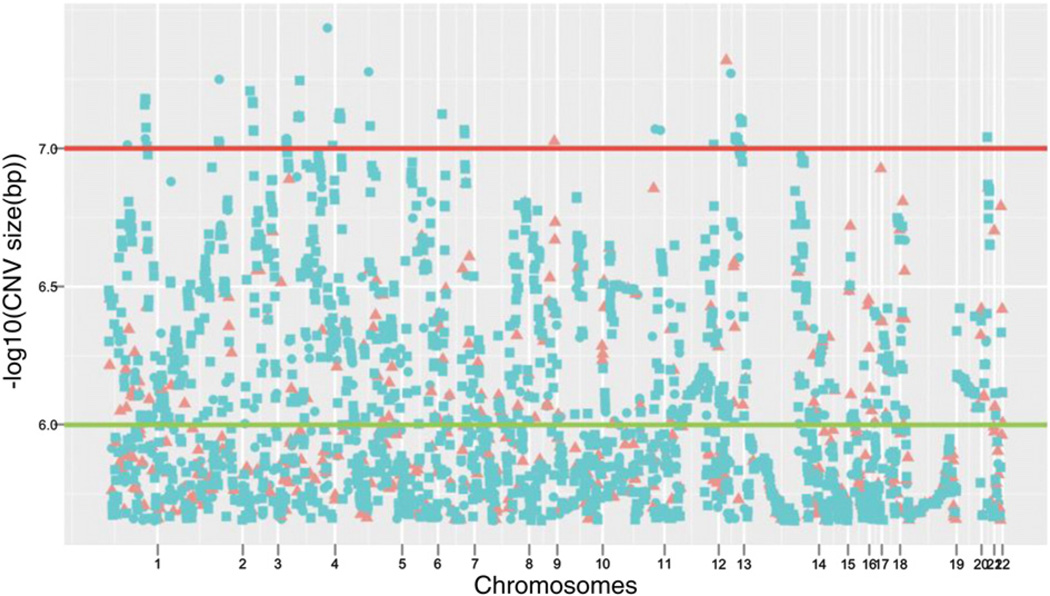
Although the differences in the total number of CNVs was not significant, we observed a greater number of copy number events with size >450 kb in AD and MCI, as well a larger mean size of these events: AD (mean size = 590 kb), MCI (mean size = 908 kb) versus healthy controls (mean size = 526 kb).When analyzed by CNV type, heterozygous deletions were significantly larger in MCI and AD patients than in healthy controls (χ2 = 136.92, p-value < 0.0001). Fig. 1 compares the genomic distributions of heterozygous deletions larger than 450 kb in cases and healthy controls in an adapted version of the typical Manhattan plot, where instead of the p-values per SNP by chromosome we used the CNV size per subject by chromosome.
Fig. 1.
Manhattan plot of the CNV size distribution for heterozygous deletions >450 kb in the ADNI sample. Each dot represents the CNV size in logarithmic scale of each deletion for each chromosome. The distance between ordered CNVs is relative and does not accurately depict the physical location of the deletions on the genome. Blue circles and squares represent CNVs belonging to MCI and AD subjects respectively. Red triangles represent CNVs belonging to healthy control subjects. The horizontal lines display two different size levels: green line for size 1 Mb and red line for size 10 Mb.
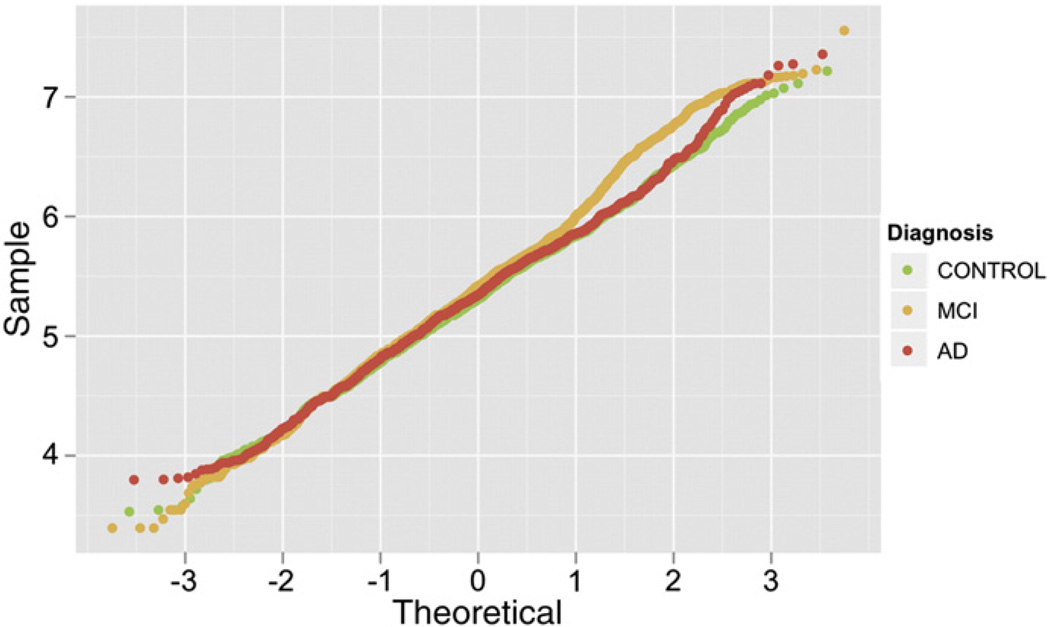
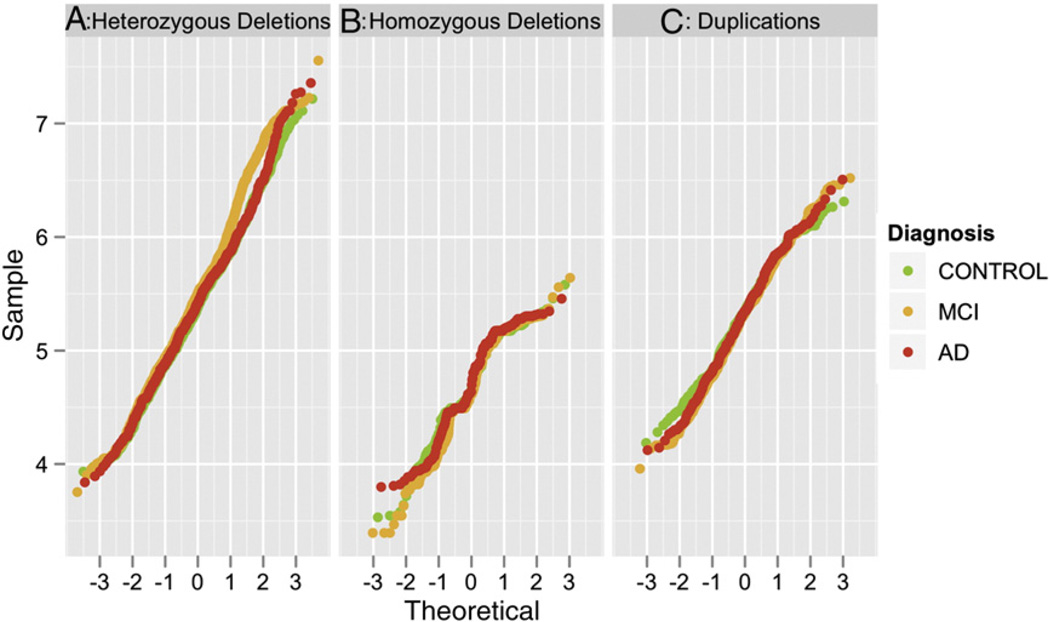
A Q–Q (quantile–quantile) plot revealed departures from the expected distribution of CNV size values (Fig. 2). The distribution of CNV size differed among the three diagnostic groups. In particular, the MCI CNV size significantly deviates from the reference line starting from a value of approximately 450 kb. The distribution of CNV size of the AD patients deviates from the normality starting at a value of approximately 4 Mb. The departure from the reference line increases as the CNV size increases for both MCI and AD cases but not healthy controls, with a greater departure for MCI. The deviations in the Q–Q plot are primarily driven by the distribution of heterozygous deletion events (Fig. 3).
Fig. 2.
Q–Q plot of the size distribution of CNVs detected in the ADNI sample by diagnostic group (healthy controls in green, MCI in orange and AD in red). The size of the CNVs are in logarithmic scale, for example the value = 4 in the table corresponds to CNVs with size = 10,000 bp, or the value = 7 to CNVs size equal to 10,000,000 bp.
Fig. 3.
Q–Q plot of the size distribution of CNVs detected in the ADNI sample represented by CNV type (panel A, heterozygous deletions; panel B, homozygous deletions; panel C, duplications) and diagnostic group (healthy controls in green, MCI in orange and AD in red). The size of the CNVs are in logarithmic scale, for example the value = 4 in the table corresponds to CNVs with size = 10,000 bp, or the value = 7 to CNVs size equal to 10,000,000 bp.
2.3. Identification of CNV-Regions with rare large deletions
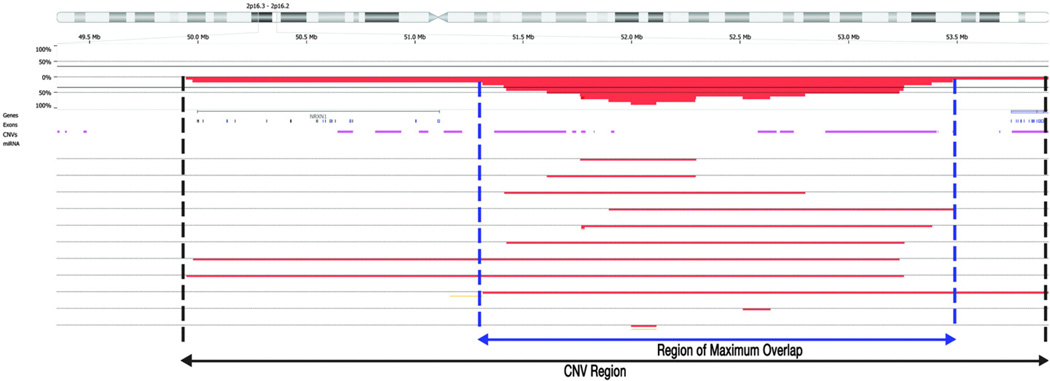
Comparing the CNVs detected in our sample with those previously reported in reference control populations, like the Toronto's Database of Genomic Variants (DGV), requires consideration of the size of the CNV. For example, very large deletions are likely to encompass regions that harbor smaller copy number events reported in control reference populations and the biological consequences of larger deletions can be different and more functionally relevant. Consequently, we carefully evaluated the size and location of CNVs in order to distinguish potentially pathogenic from benign CNVs, as discussed by Conrad et al. [23]. CNVs are common among healthy individuals, and the strategy to identify potentially pathogenic candidates is key. Thus, following Conrad et al. [23], we consider CNVs as potentially pathogenic if the overlapping segments are at least 30% larger than those reported in the reference controls (Fig. 4) and are shared by four or more patients (at least 1% of our sample). Fig. 4 shows an example of the relationship between CNVs described in the DGV reference population and the CNV-Regions observed in our sample on chromosome 2p16.3-p16.2. We used the intensity data from the SNP microarray to estimate the breakpoint of each CNV (Conrad et al. [23]). Applying these criteria we identified 44 independent heterozygous deleted CNV-Regions (see Table S1).
Fig. 4.
CNV detection and CNV-Regions (Copy-Number-Variable Regions) boundaries. The physical map of chromosome 2p16.3-p16.2 is depicted. The percentages of duplications and deletions size are indicated by horizontal percentage reference line for the region. The CNVs from the reference population DGV are indicated by horizontal line (purple). Each horizontal line indicates a study subject and the red indicates the extent of the deletion. The vertical blue lines indicate the area of maximum overlap (95%) of deletions for the study subjects.
To provide a first level of confirmation of these 44 CNV-Regions, we used an alternative algorithm for CNV identification from single nucleotide polymorphism arrays, i.e. PennCNV [24]. Although Nexus and PennCNV provide very different CNV calls both in terms of overall number and size [18], we opted for PennCNV since it is currently the most widely used algorithm. As expected, we found that 43% of deletions and 55% of duplications identified by Nexus were also called by PennCNV. Then, we focused specifically on CNV calls by PennCNV overlapping with the previously defined 44 CNV-Regions. We found that 42 over 44 CNV-Regions harbored CNVs detected by PennCNV, 29 of those with deletions larger than 450 kb (Table S2). As expected, PennCNV calls had a smaller size, but the overall coverage of deletions within each CNV-Region confirms the pattern of disruption potentially impacting the same genes and regulatory elements. Despite the differences between the algorithms, these data confirm the presence of regions of putative altered functional activity within the boundaries of the identified CNV-Regions.
2.4. CNV-Regions association analysis
To better investigate the increased number of heterozygous deletions evidenced by the Q–Q plot we evaluated the distribution of the 44 CNV–Regionswith a logistic regression approach, since these regions may have more explanatory power than CNVs alone. Overall, the number of affected subjects with more than one CNV-Region deletion is significantly greater in cases than controls (χ2 = 14.79, p-value = 0.005), with the highest contribution due to a larger proportion of MCI subjects having more than 5 CNV-Regions than healthy controls (Table 3). To further unravel the role of each single CNV-Region in determining the strength of the overall association, we tested each CNV-Region for association with cognitive impairment. We observed that seven of the forty-four CNV-Regions have nominal p-value < 0.05: CNV-Region 7, CNV-Region 14, CNV-Region 23, CNV-Region 28, CNV-Region 38, CNV-Region 48 and CNV-Region 70. These large deletions were present only in cases (MCI and AD patients) with the exception of one healthy control (CNV-Regions 28 and 70).
Table 3.
Co-occurrence of large deletions at more than 5 CNV-Regions.
| Diagnostic group | Number of Individuals with CNV-Regions | ||||
|---|---|---|---|---|---|
| No CNVRs |
1–5 CNVRs |
>5 CNVRs |
Total | ||
| Healthy (n = 181) | Observed frequency | 162 (89.5%) | 18 (9.9%) | 1 (0.84%) | 181 |
| Expected frequency | 147.9 (81.7%) | 29.4 (16.24%) | 3.7 (2%) | ||
| MCI (n = 313) | Observed frequency | 239 (76.36%) | 64 (20.44%) | 10 (3.2%) | 313 |
| Expected frequency | 255.8 (81.72%) | 50.9 (16.26%) | 6.4 (2.04%) | ||
| AD (n = 146) | Observed frequency | 122 (83.56%) | 22 (7.03%) | 2 (1.37%) | 146 |
| Expected frequency | 119.3 (81.71%) | 23.7 (16.23%) | 3 (2.05%) | ||
| Total | 523 | 104 | 13 | ||
Pearson chi2 (4) = 14.79 p-value = 0.005
2.5. CNVs validation with next generation sequencing
To provide an initial validation of our findings, we used low-coverage whole genome re-sequencing to confirm the detection of CNVs in the seven CNV-Regions associated with cognitive impairment. We selected six subjects: two patients that presented CNVs in almost all the seven CNV-Regions, two healthy controls who did not report CNV calls (negative controls), and two additional patients who presented two and three CNVs, both called by Nexus and PennCNV. As reported in the 1000 genome study [25], low-coverage whole genome re-sequencing proved to be valid for CNV identification and provided higher resolution compared with microarrays. With sequencing, we confirmed the CNV-Regions, as well as corroborating the reliability of the longer Nexus algorithm. On average, 87.3% of the sequencing reads passed quality filters, and approximately 85% align to the reference sequence (see Supplementary Table S3 for further details). Ninety-nine percent of the bases of the aligned reads match the reference with an average error rate of 0.006. The average coverage genome wide was ~ 1.0×. Alignment metrics are reported in Supplementary Table S3.
Of the two sequenced patients with 7 CNV-Regions with nominally significant p-values, one had CNV calls in 5 of the 7 CNV-Regions that were identified by PennCNV. The second patient had CNV calls in all the seven CNV-Regions, 5 of which were identified by PennCNV. The remaining two CNV-Regions that were not called by PennCNV have nonetheless been validated by NGS (for details on CNV calls boundaries across Nexus and the two validation methods see Table S5). As expected, the two healthy subjects did not present any NGS-based CNV in the seven CNV-Regions.
Of the other two patients selected for having CNVs called by both Nexus and PennCNV, one presented two CNV calls on chromosomes 3 and 14. On chromosome 3, Nexus called a CNV of 146 kb (35,991,950–36,138,541) and PennCNV a CNV of 118 kb (36,027,069–36,145,613). Both segments were cross-validated by the NGS-based CNV of 921 kb (35,394,001–36,316,000) with additional refinement of the boundaries of the actual deletion. On chromosome 14 Nexus called a CNV of 711 kb (21,308,832–22,020,471) and PennCNV called a CNV of 70 kb (21,701,518–21,771,960). The NGS-based CNV size of 855 kb confirms the deletion at the locus and is in accord with Nexus overall size of the deletion. On chromosomes 5, 6, and 22: Nexus called three CNVs with sizes of 206 kb, 72 kb, and 199 kb, respectively. The CNVs called by PennCNV had sizes of 108 kb, 60 kb, and 2 kb, respectively. The sequencing cross-validated CNVs on chromosomes 5, and 6 with sizes of 72 kb and 68 kb, but not on chromosome 22.
2.6. CNV-Region genes: functional classification (DAVID)
We analyzed the 231 genes that map within the boundaries of our CNV-Regions with DAVID, to identify related biological processes and molecular functions enriched in our dataset. The gene functional classification analysis identified 10 clusters of genes that share annotation terms for the same biological function. Among them only four have enrichments score > 1.3, a value equivalent to a p-value = 0.05, generally considered an adequate significance threshold for annotation enrichment analyses. Table 4 presents the results for the top four gene functional clusters along with the top functional annotation terms reported for each cluster and their enrichment fold value relative to the entire human genome (DAVID Background: Homo Sapiens). The functional analysis shows enrichment in our data for four different gene families: genes that encode for trans-membrane proteins, immunoglobulin-like domains found in several diverse protein families, the semaphorin protein family and leucine-repeat-rich (LRR) proteins.
Table 4.
Functional gene cluster classifications using the Database for Annotation, Visualization and Integrated Discovery (DAVID) for enrichment scores (ES) > 1.3.
| Gene functional classification | Top 3 associated annotation terms | ||||
|---|---|---|---|---|---|
| Gene cluster | N. of genes | Enrichment score | Annotation category | Annotation term | Enrichment fold |
| 1 | 37 | 1.91 | UP_SEQ_FEATURE | transmembrane region | 3.79 |
| SP_PIR_KEYWORDS | transmembrane | 3.76 | |||
| SP_PIR_KEYWORDS | membrane | 2.99 | |||
| 2 | 8 | 1.78 | SP_PIR_KEYWORDS | Immunoglobulin domain | 35.81 |
| INTERPRO | IPR007110:Immunoglobulin-like | 29.10 | |||
| INTERPRO | IPR003598:Immunoglobulin subtype 2 | 60.95 | |||
| 3 | 4 | 1.70 | UP_SEQ_FEATURE | domain:Sema | 462.41 |
| PIR_SUPERFAMILY | PIRSF005526:semaphorin | 396.21 | |||
| INTERPRO | IPR001627:Semaphorin/CD100 antigen | 403.04 | |||
| 4 | 6 | 1.56 | UP_SEQ_FEATURE | repeat:LRR 10 | 136.13 |
| UP_SEQ_FEATURE | repeat:LRR 9 | 117.98 | |||
| UP_SEQ_FEATURE | repeat:LRR 8 | 106.90 | |||
We also performed a functional cluster analysis annotation, which allows including even those genes that might not exert identical biological function but that are likely to co-function in the context of the same biological network. This analysis identified 7 of the 71 total clusters above the significant threshold of ES = 1.3 (Supplementary Table S6). The overall biological function of the two top clusters, Clusters 1 and 2 (ES = 2.03 and 1.71 respectively) is associated with genes implicated in glycosylation, the posttranslational modification process of integral membrane proteins like glycoproteins. Cluster 3 relates to axon guidance, neurogenesis and differentiation and includes the genes coding for the semaphorin protein family (e.g. SEMA3A, -3C, -3D, -3E) and the axon guidance receptors (e.g. ROBO1 and ROBO2) as well as transcription factor involved in the differentiation of retinal ganglion cells (e.g. ATOH7). Interestingly, the two most significant annotation terms (p-value < 0.01 corrected for multiple testing) both belong to Cluster 3: the UniProt classification of the superfamily of semaphorin proteins, obtained from the Protein Information Resource SuperFamily (PIRSF) site, and the axon guidance pathway as described by KEGG [26]. Cluster 4 is enriched for annotation terms related to proteins containing immunoglobulin-like domains and includes genes coding for neural cell adhesion molecules (e.g., NCAM2 and JAM2) as well as genes that regulate cell surface interactions during nervous system development (e.g. CNTN5). Cluster 5 relates to biological processes implicated in the modulation of the assembly that allows the fusion of transport vesicles and the plasma membrane in the cytoplasm (e.g. STXBP5) and in synaptic vesicle trafficking to cytoplasmic vesicles (e.g. PLCO). Cluster 6 reveals the enrichment of annotation terms related to neuron development, differentiation and more specifically neuron projection morphogenesis and axonogenesis. Besides the above mentioned semaphorin protein family and axon guidance receptors, the biological functions enriched in this cluster are associated with genes such as SLITRK-1, -5 and -6, responsible for enhancing neuronal dendrite outgrowth, and FOXG1, a transcription repression factor which plays an important role in brain development. Finally, cluster 7 relates to cell–cell adhesion and includes genes coding for cadherins, glycoproteins involved in Ca2+-mediated cell–cell adhesion (e.g. CDH7, CDH19, and PCDH7) as well as for proteins implicated in cellular migration (e.g. ROBO1 and ROBO2).
3. Discussion
CNV mapping has proven to be highly relevant in increasing our understanding of genetic susceptibility to complex traits. We focused our investigations on CNV-Regions rather than CNVs, mostly because CNV-Regions are more likely to capture the extent of loci disrupted by deletions or duplications. Based on Redon el al. [10] a CNV-Region is defined as the union of juxtaposed or overlapping unique CNVs all of which may impact the same biological function. The assessment of the enrichment of biological signatures of the functional sequences that fall within a CNVR (i.e. genes, regulatory elements, non-coding RNAs) may inform on the functional genomic impact of the CNVs. Recently, several studies have attempted to study the functional impact of CNVs by integrating CNVs and gene expression data by reconstructing the functional CNV-Region networks [5,27,28]. The overall number of subjects with CNV-Regions is low, a finding compatible with CNV-Regions being rare events. We hypothesize that multiple rare CNV-Regions characterize a profile that confer susceptibility to cognitive impairment by acting synergistically in combination with other genetic, epigenetic or environmental factors. We found a statistically significant overrepresentation of subjects with more than one CNV-Region in affected cases compared to healthy controls and we found that very large deletions >450 kb are associated with MCI and/or AD. Recent studies supported the role of the overall load of large deletions in many complex traits [29,30]. Our findings are consistent in particular with the hypothesis that links large copy number events and disease susceptibility as previously described in the context of large CNV studies of schizophrenia, autism and other psychiatric disorders [31,32]
In addition, several studies provided evidence for a substantial role of chromosomal structural variations in the pathogenesis of neurological disorders [3,33–35]. For AD, three papers [7–9] reported the results of an initial CNVs analysis with both genome-wide and candidate gene strategies, evaluating the association of single CNVs to AD and MCI. Heinzen and colleagues [7] reported a duplication in the CHRNA7 gene, although not significantly overrepresented in cases, and the possibility of large heterozygous deletions in cases. Swaminathan and colleagues identified some potential candidate genes enriched in CNVs in cases (CSMD1, SLC35F2, HNRNPCL1 as well as the candidate gene CHRFAM7A), although none met the conventional significance (p-value < .05) after correction for multiple testing. Recently, the same authors replicated these findings in an independent sample from the NIA-LOAD/NCRAD Family Study, with the identification of a new candidate gene (IMMP2L) possibly involved in AD susceptibility [9]. These previous observations of single CNVs enriched in MCI and AD subjects within candidate genes may have failed to reach statistical significance in a traditional case–control study design because of the rarity of the events. Our focus on CNV-Regions shifts the attention from a single event to the region of overlap of events characterized by different sizes, but likely affecting the same underlying functional biology of the deleted or disrupted gene(s). This is consistent with the biological plausibility of the previous findings, despite the lack of statistical significance. Furthermore, the interpretation of the clinical significance of single CNVs, especially for small events < 500 kb, is challenging since their pathogenicity is modulated by many factors. We also applied ab initio a stringent filtering procedure to ultimately pull out a set of rare candidate CNVs, excluding all the events quite common throughout the healthy population [36]. Our approach should be seen as a complementary methodology to single CNVs that leverages the power of genome-wide CNV-Region profiling to overcome the limitation of incomplete penetrance and variable expressivity of single CNVs
Our findings of increased number and size of heterozygous deletions associated with late-onset cognitive impairment are consistent with the neurobiology of late-onset diseases. The presence of CNVs in a coding region can alter the abundance of the corresponding transcripts affecting the amount of protein product that may influence cell differentiation [3]. Excessive protein production may lead to age-dependent protein misfolding with implied disruption of protein transport, mitochondrial dysfunction and apoptosis [37]. On the other hand, also CNVs present in the vicinity of genes may influence their expression through a variety of epigenetic mechanisms [5]. An advantage of CNVs and particularly CNV-Regions is that they identify structural changes within DNA that have the potential to affect gene function. To further elucidate the clinical relevance of our CNV-Regions, we analyzed the gene functions or pathways these CNV-Regions might affect using DAVID. The deletions within these CNV-Regions occur in genes implicated in the biological pathways of axonal guidance, neuronal morphogenesis and differentiation, cell–cell adhesion and glycoprotein glycosilation [38].
We established the reliability of our CNV calls using two different algorithms implemented in Nexus and PennCNV, and confirmed our most promising findings by assessing CNV consensus calls and CNV-Region boundaries using a sequencing strategy. A NGS-based CNV detection approach provides the highest sensitivity currently available and allows refining the CNV boundaries, as well as identifying events that cannot be detected by the most sensitive array technologies [39,40]. While deep-coverage (≥25×) whole-genome sequencing costs are significantly dropping, low-coverage sequencing (1–6× base coverage) is still the most feasible option [41]. Low and high sequencing both provide data comparable to CGH-based (comparative genomics hybridization) data [42]. The better resolution of NGS-based CNV detection identifies additional CNVs in the AD and in MCI patients compared to SNP microarray calls, strengthening our results. An increasing number of algorithms that interrogate deep sequencing data for CNV discovery are becoming available, although there is not yet a consensus on a “gold standard” method and analysis strategy. A weak point common to many NGS-based discovery approaches is the requirement for a paired reference sequenced genome since they were originally developed for the detection of cancer CNVs where the paired reference genome for a tumor is the related normal tissue [43]. There is no current consensus yet on the criteria to build a reference genome for CNV calling for complex diseases. Here, we opted to use ERDS, a read-depth NGS-based CNV discovery approach that relies on depth-of-coverage (DOC) (i.e. the density of reads mapping to the region) and detects changes in copy number by comparing the observed DOC within a sliding window of the genome to a reference genome [44–46]. This is currently the only sequencing-based CNV discovery method that allows for the accurate prediction of absolute copy numbers [44,47]. The greatest advantage of ERDS, and DOC algorithms in general, is the ability to detect broader range of CNV events, with best reliability for large events and better breakpoint resolution [48].
The overall high rate of consensus calls across approaches, both cross-algorithm reliabilities using PennCNV and NGS-based confirmation, supports the accuracy of Nexus CNV detection algorithm [49,50]. Recently, Dellinger et al. reported that Nexus may be affected by an overcall of CNVs, especially with more relaxed analysis parameters [18]. Our comparative analysis however supports Nexus reliability, provided that the analysis is set up with conservative parameters to find a good trade-off between sensitivity and specificity in CNVs detection.
Our findings of large deletions suggest a link between chromosomal structural alterations and the development of MCI and AD. The CNV-Region strategy captures much more realistic information than the simple description of a catalog of single CNV events, enhancing the genomic relevance of CNVs into a more clinical translational perspective [38]. The higher prevalence in AD and MCI subjects of large deletions in genes involved in neurodevelopment and brain functions makes them good candidates for the definition of predictive profiles for the disease evolution as possible indicators of progressive brain dysfunction. The putative clinical significance of the large deletions reported here is based on several factors including the lack of complete overlapping with benign losses spanning these genomic regions (as reported in DGV), gene function and in most cases tissue specific expression. Although these structural alterations warrant future molecular investigations to fully understand their functional role, it is possible that disruption of the gene regulatory networks is the final common pathway [51]. We are aware that CNV analysis of DNA from blood has limitations, including the difficulty of directly studying the consequence of the identified CNV-Regions on the neuronal activity, particularly in the heterozygous state. There is considerable evidence about the presence of CNVs in normal brains [52,53], although the extent of these structural variations is unknown as is their role in brain functioning. Future research plans include the analysis of post-mortem brain from healthy, MCI and AD subjects to verify the neuronal presence of the CNV-Regions we detected, and possibly their effect on overall brain gene expression and pathways. It is very likely that the dysfunction of specific neuronal pathways underlying AD and MCI depends on additional genetic and/or epigenetic mechanisms to manifest a particular phenotype. Studies of characteristics that are quantitative, change over time, or vary across clinical disorders like cognitive impairment, offer a great opportunity to deepen our understanding of the role of genetic variation on human behavior and diseases. This can perhaps be best accomplished by determining the correspondence between ‘dimensional’ phenotypic and genomic variation data.
4. Material and methods
4.1. Ethics statement
Study subjects gave written informed consent at the time of enrollment for imaging and genetic sample collection and completed clinical symptom assessments approved by each participating sites' Institutional Review Board (IRB).
4.2. ADNI
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu\ADNI). For up-to-date information see www.adni-info.org.
4.3. Participants
All subjects were part of the ADNI longitudinal multi-site observational study that included AD, mild cognitive impairment (MCI), and elderly individuals with normal cognition. All subjects have been assessed with clinical and cognitive measures at the time of collection, including ADAS-Cog, CDR-SB and MMSE, MRI and PET scans (FDG and 11C PIB) and blood and CNS biomarkers. Brain imaging, biological samples, and clinical assessments are longitudinally collected for a target of 200 healthy controls, 400 MCI, and 200 AD subjects. All AD patients included in the study are sporadic cases of mild AD that met NINCDS/ADRDA criteria for probable AD [54–56], between the ages of 55–90, with an MMSE score of 20–26 inclusive and having an MRI consistent with the diagnosis of AD (Table 1). Further details about the inclusion and exclusion criteria can be found in the ADNI protocols [57]. For CNV whole-genome screening analysis, we downloaded the entire ADNI genotyping dataset publicly available at the following link: http://www.loni.ucla.edu/ADNI/Data/. Following CNV quality control measures (described below), data from a total of 146 AD, 313 MCI subjects and 181 healthy controls were included in the analysis based on the diagnostic information collected at baseline.
Table 1.
Clinical and demographic characteristics of healthy controls, MCI and mild AD subjects at baseline assessment.
| Control | MCI | AD | |
|---|---|---|---|
| # subjects | 229 | 398 | 193 |
| Mean Age | 75.93 ± 5.0 | 74.83 ± 7.48 | 75.33 ± 7.47 |
| Gender (male/female) | 119/110 | 257/141 | 101/91 |
| Smoker/non-smoker | 84/145 | 163/235 | 75/118 |
| Handedness (right/left) | 211/18 | 362/36 | 181/12 |
| Ethnicity (Hispanic/non-Hispanic/unknown) | 2/224/3 | 14/380/4 | 4/187/2 |
| Race (American Indian or Alaskan Native/Asian/African American/White/more than one race) | 0/3/16/210/0 | 1/9/15/372/1/ | 0/2/8/181/2 |
| Mean years of education | 16.04 ± 2.86 | 15.67 ± 3.04 | 14.70 ± 3.1 |
| MMSE | 29.11 ± 0.99 | 27.03 ± 1.77 | 23.32 ± 2.06 |
| CDR global (0–0.5–1) # subjects in each category and mean values | 229–0–0 | 2–396–0 | 0–100–93 |
| 0.0 | 0.49 ± 0.035 | 0.74 ± 0.25 | |
| ADAS-cog | 6.20 ± 2.91 | 11.49 ± 4.42 | 18.36 ± 6.67 |
| APOE (ε2/ε3/ε4) | 38/354/66 | 28/509/259 | 9/214/163 |
4.4. Genotyping
The ADNI sample was genotyped using the Human-610 Quad BeadChip with a total of 620,901 markers, including 21,890 intensity-only probes specifically designed to improve CNV detection and 27,635 additional probes in SNPs desert genomic regions to enrich CNV coverage. DNA collection, genotyping and relative quality control analysis details are provided in Potkin et al. [21]. The intensity data were analyzed with Illumina GenomeStudio and are publicly available on the LONI website (www.loni.ucla.edu/ADNI/Data). For the purpose of this study we confined the analysis to autosomal chromosomes.
4.5. CNV segmentation algorithm
We used Nexus v5 (Biodiscovery Inc., El Segundo, CA, USA) to produce CNV calls. Nexus calling algorithm SNPrank Segmentation is based on the Circular Binary Segmentation model [58]. To detect CNVs and allelic ratio anomalies, it relies on the normalized measure of the total signal intensity for the two alleles of a SNP, defined as the Log R ratio (LRR) and the normalized measure of the allelic intensity ratio of the two alleles, defined as the B allele frequencies (BAF). Both LRR and BAF were computed from the array intensity data with Illumina Genome Studio v1.0.2 software using the manufacturer's cluster file as a reference [21]. SNPrank Segmentation classifies three types of CNVs: 1) CN gain, corresponding to duplications (CN = 3 or 4 copies); 2) CN Loss, corresponding to single copy deletions (CN = 1 copy); 3) homozygous copy loss corresponding to a complete deletion event (CN = 0). We used standard calling parameters recommended by the manufacturer for Illumina array data: 1) minimum number of probes per segment of 5, 2) max contiguous probe spacing of 1000 kbp, and 3) significance threshold of p-value = 1 × 10−6 for CNV calling. The combination of these measures is particularly indicated for the detection of CNV in regions scarcely covered, while simultaneously accounting for the risk of false positive calls (e.g. centromeric and telomeric CNV, or CNV calls merely due to a background waviness of the LRR that involves few adjacent probes). We also ran PennCNV to evaluate CNV calls that had a minimum of three SNPs on autosomes in addition to standard default quality control parameters for CNV calling.
4.6. Statistical analysis
We used Nexus a specific pre-processing quality control parameter (QCscore) to check for the quality of the individuals' intensity raw data to minimize the risk of false positive CNV calls and we excluded samples exceeding QCscore of 0.2, a threshold value empirically determined for Illumina array data by the manufacturer. We removed overlapping CNVs within the same subject blind to diagnostic group where the overlapping refers to either a situation where more than one deletion or duplication were called for the same subject in the same region, or the same event was specified twice for the same subject using PLINK software package [59] (http://pngu.mgh.harvard.edu/purcell/plink), release v1.07. Proportion of overlap concordance of Nexus CNV calls with the PennCNV calls using PLINK was also calculated to provide validation across different CNV algorithms. We performed descriptive statistics of single CNV distributions by size and type using chi-square and Fisher exact test. To identify copy number variable loci (i.e. CNV-Regions for partially overlapping CNVs), we used the “union overlap” tool of PLINK that uses the ratio of the number of base-pairs intersected between different CNVs and the length of the CNV as a denominator in calculating the proportion overlap. The “union overlap” tool allowed to both 1) select segments that were 95% overlapping between them in a region with boundaries defined by our own data and 2) exclude segments that were overlapping for more than 70% with previously described CNVs. To investigate the effect of CNV-Regions on cognitive impairment, we tested for association with diagnoses and MMSE score using a logistic regression model. All descriptive and association analyses of single CNVs and CNV-Regions were performed with STATA11 (StataCorp Stata Statistical Software: Release11. 200×. College Station, TX: StataCorp LP).
4.7. Bioinformatics and in-silico functional pathway databases
We used the Database for Annotation, Visualization and Integrated Discovery (DAVID), release 6.7 [60] to screen the genes that appeared to be affected by CNV-Region variants. DAVID classifies the genes into functional groups based on annotation similarity criteria. DAVID calculates an enrichment score that, relative to the ADNI dataset, ranks the relevance of the annotation terms that describe the genes included in a functional cluster. To be included in the final list, we required a gene to harbor a copy number variant in at least 3 subjects and to map in the region of maximum overlapping within the boundaries of a CNV-Region. The list of genes affected by CNV-Region variants we submitted for the gene functional analysis had 231 entries. We used the fuzzy clustering algorithm implemented in the “gene functional classification” tool of DAVID to classify functionally related genes into groups based on co-occurrence of “motifs” underlying shared biological modules. Then, we refined our results with the “functional annotation clustering” analysis to cluster the annotation terms associated with the genes in our list. For the analysis of annotation enrichment, we set options to the default “medium” stringency criteria values provided by DAVID.
4.8. Graphics (Nexus and R-ggplot2)
All plots and histograms were created using the plotting system “ggplot2” of R (http://had.co.nz/ggplot2/). All images related to CNVs and CNV-Regions have been created within the graphical framework provided by Nexus v5.
4.9. Whole genome sequencing
After sonication of genomic DNA using a Covaris S2 to an average size of 250–300 bp, libraries were constructed manually using the Wellcome Trust protocol and reagents [61]. Each library was then sequenced to depth of ~4–7× coverage on a Illumina HiSeq2000 DNA sequencer using v5 kits, with pair-end reads of 75–100 bp flow cell. The Illumina pipeline (v1.7–1.8) was then used to convert digital images into base pair calls (with quality scores). The number of raw reads generated per patient is reported in Supplementary Table S3.
Sequencing reads were aligned to the NCBI36 human reference genome (Ensembl hg18 release 50: ftp://ftp.ensembl.org/pub/current/fasta/homo_sapiens/dna/) using BWA software v0.5.9 (http://bio-bwa.sourceforge.net/) [62]. We used the software package SAMtools v0.7.1 (http://samtools.sourceforge.net/) to generate the SAM/BAM format files after screening the alignment data for duplicate reads, sorting and indexing procedures. SummaryAlignmentmetrics were calculated using the Collect Alignment Summary Metrics program implemented in the PICARD Java-based command-line software package v2.6.21 (http://picard.sourceforge.net/). The CNV calling was performed with “Estimation by Read Depth with Single Nucleotide Variants” (ERDS) software v1.02 (http://web.duke.edu/~mz34/erds.htm), a Hidden Markov Model (HMM) based approach that relies on depth-of-coverage (DOC) to infer the copy number state. It represents an extensions to the methods described in [63]. The algorithm has been described more in detail in [64]. A deletion was called when its average read depth was below 0.7 * expected read depth (corresponding to copy number < 1.4), and a duplication is called when its average read depth is above 1.3 * expected read depth (corresponding to copy number >2.6). Expected read depth is calculated using the expectation maximization (EM) approach and corrected by GC bias. Sequence Variant Analyzer (SVA, http://www.svaproject.org) and Integrative Genome Viewer (IGV, http://www.broadinstitute.org/software/igv/) were used to visually inspect and annotate the CNV-Regions.
Supplementary Material
Acknowledgments
The genotyping was performed by Jennifer Webster and Drs. David Craig and Matt Huentelman of TGen. We want to acknowledge the support and numerous contributions by the ADNI investigators and the ADNI Industry Advisory Board (Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories), and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration and the Foundation for the National Institutes of Health. We gratefully acknowledge the participation of the ADNI subjects and their family members.
Genotyping was performed at The Translational Genomics Institute, Phoenix AZ, by Jennifer Webster and Drs. David Craig and Matt Huentelman. Sample processing, storage and distribution were provided by the NIA-sponsored National Cell Repository for Alzheimer's Disease by Dr. Tatiana Foroud and Kelley Faber. Sample verification and quality control bioinformatics were provided by Drs. Li Shen, Sungeun Kim and Kwangsik Nho of the Indiana University Center for Neuroimaging, Nathan Pankratz of the IU Dept of Medical and Molecular Genetics, Drs. Guia Guffanti and Anita Lakatos of UC Irvine, and Bryan DeChairo of Pfizer, Inc.
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu\ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principle Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California — San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada.
This analysis was supported by grants from the Transdisciplinary Imaging Genetics Center (TIGC-P20 RR020837-01), the Alzheimer's Disease Neuroimaging Initiative (ADNI U01 AG024904-01, and supplement 3U01AG024904-03S5), the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIH), the Function Imaging Biomedical Informatics Research Network (FBIRN U24-RR021992, National Center for Research Resources), commercial support from Vanda Pharmaceuticals, and private support from an anonymous Foundation and anonymous donations. Additional contributions made through the Foundation for the NIH from Merck & Co. Inc., Pfizer, Inc., and Gene Network Sciences, Inc. partially supported the genotyping results reported here. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant and supplement). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to thank Soheil Shams and Ziwei Che (Biodiscovery Inc., El Segundo, CA) for assistance and troubleshooting in CNV analysis with Nexus software.
Footnotes
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu\ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at www.loni.ucla.edu\ADNI\Collaboration\ADNI_Manuscript_Citations.pdf.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygeno.2013.04.004.
Contributor Information
Guia Guffanti, Email: Guffant@nyspi.columbia.edu.
Federica Torri, Email: ftorri@uci.edu.
Jerod Rasmussen, Email: rasmussj@uci.edu.
Andrew P. Clark, Email: clarkap@usc.edu.
Anita Lakatos, Email: alakatos@uci.edu.
Jessica A. Turner, Email: jturner@mrn.org.
Andrew J. Saykin, Email: asaykin@iupui.edu.
Michael Weiner, Email: Michael.Weiner@ucsf.edu.
Marquis P. Vawter, Email: mvawter@uci.edu.
James A. Knowles, Email: knowles@med.usc.edu.
Steven G. Potkin, Email: sgpotkin@uci.edu.
Fabio Macciardi, Email: fmacciar@uci.edu.
References
- 1.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalman B, Vitale E. Structural chromosomal variations in neurological diseases. Neurologist. 2009;15:245–253. doi: 10.1097/NRL.0b013e3181963cef. [DOI] [PubMed] [Google Scholar]
- 4.Blauw HM, Veldink JH, van Es MA, van Vught PW, Saris CG, van der Zwaag B, Franke L, Burbach JP, Wokke JH, Ophoff RA, van den Berg LH. Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen. Lancet Neurol. 2008;7:319–326. doi: 10.1016/S1474-4422(08)70048-6. [DOI] [PubMed] [Google Scholar]
- 5.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum. Mol. Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 6.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 7.Heinzen EL, Need AC, Hayden KM, Chiba-Falek O, Roses AD, Strittmatter WJ, Burke JR, Hulette CM, Welsh-Bohmer KA, Goldstein DB. Genome-wide scan of copy number variation in late-onset Alzheimer's disease. J. Alzheimers Dis. 2010;19:69–77. doi: 10.3233/JAD-2010-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaminathan S, Kim S, Shen L, Risacher SL, Foroud T, Pankratz N, Potkin SG, Huentelman MJ, Craig DW, Weiner MW, Saykin AJ. The Alzheimer's disease neuroimaging initiative, genomic copy number analysis in Alzheimer's disease and mild cognitive impairment: an ADNI study. Int J Alzheimers Dis. 2011;2011:729478. doi: 10.4061/2011/729478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swaminathan S, Shen L, Kim S, Inlow M, West JD, Faber KM, Foroud T, Mayeux R, Saykin AJ. Analysis of copy number variation in Alzheimer's disease: the NIALOAD/NCRAD Family Study. Curr. Alzheimer Res. 2012;9:801–814. doi: 10.2174/156720512802455331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogler C, Gschwind L, Rothlisberger B, Huber A, Filges I, Miny P, Auschra B, Stetak A, Demougin P, Vukojevic V, Kolassa IT, Elbert T, de Quervain DJ, Papassotiropoulos A. Microarray-based maps of copy-number variant regions in European and sub-Saharan populations. PLoS One. 2010;5:e15246. doi: 10.1371/journal.pone.0015246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 13.Marcinkowska M, Szymanski M, Krzyzosiak WJ, Kozlowski P. Copy number variation of microRNA genes in the human genome. BMC Genomics. 2011;12:183. doi: 10.1186/1471-2164-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 15.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, Macdonald JR, Mills R, Prasad A, Noonan K, Gribble S, Prigmore E, Donahoe PK, Smith RS, Park JH, Hurles ME, Carter NP, Lee C, Scherer SW, Feuk L. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat. Biotechnol. 2011;29:512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genomic. Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 18.Dellinger AE, Saw SM, Goh LK, Seielstad M, Young TL, Li YJ. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res. 2010;38:e105. doi: 10.1093/nar/gkq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 21.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr, Weiner MW, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Grzeda KR, Stewart C, Grubert F, Urban AE, Snyder MP, Marth GT. Copy Number Variation detection from 1000 Genomes project exon capture sequencing data. BMC Bioinformatics. 2012;13:305. doi: 10.1186/1471-2105-13-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C, Ahn J, Yoon Y, Park S. Identification of functional CNV region networks using a CNV-gene mapping algorithm in a genome-wide scale. Bioinformatics. 2012;28:2045–2051. doi: 10.1093/bioinformatics/bts318. [DOI] [PubMed] [Google Scholar]
- 29.Yeo RA, Gangestad SW, Gasparovic C, Liu J, Calhoun VD, Thoma RJ, Mayer AR, Kalyanam R, Hutchison KE. Rare copy number deletions predict individual variation in human brain metabolite concentrations in individuals with alcohol use disorders. Biol. Psychiatry. 2011;70:537–544. doi: 10.1016/j.biopsych.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo RA, Gangestad SW, Liu J, Calhoun VD, Hutchison KE. Rare copy number deletions predict individual variation in intelligence. PLoS One. 2011;6:e16339. doi: 10.1371/journal.pone.0016339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ISC. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 33.Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat. Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupski JR. Brain copy number variants and neuropsychiatric traits. Biol. Psychiatry. 2012;72:617–619. doi: 10.1016/j.biopsych.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, Nezarati MM, Asamoah A, Jackson KE, Gowans GC, Martin JA, Carmany EP, Stockton DW, Schnur RE, Penney LS, Martin DM, Raskin S, Leppig K, Thiese H, Smith R, Aberg E, Niyazov DM, Escobar LF, El-Khechen D, Johnson KD, Lebel RR, Siefkas K, Ball S, Shur N, McGuire M, Brasington CK, Spence JE, Martin LS, Clericuzio C, Ballif BC, Shaffer LG, Eichler EE. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 37.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 38.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 39.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HY, Leng J, Li R, Li Y, Lin CY, Luo R, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi X, Stromberg MP, Stutz AM, Urban AE, Walker JA, Wu J, Zhang Y, Zhang ZD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park H, Kim JI, Ju YS, Gokcumen O, Mills RE, Kim S, Lee S, Suh D, Hong D, Kang HP, Yoo YJ, Shin JY, Kim HJ, Yavartanoo M, Chang YW, Ha JS, Chong W, Hwang GR, Darvishi K, Kim H, Yang SJ, Yang KS, Kim H, Hurles ME, Scherer SW, Carter NP, Tyler-Smith C, Lee C, Seo JS. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat. Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood HM, Belvedere O, Conway C, Daly C, Chalkley R, Bickerdike M, McKinley C, Egan P, Ross L, Hayward B, Morgan J, Davidson L, MacLennan K, Ong TK, Papagiannopoulos K, Cook I, Adams DJ, Taylor GR, Rabbitts P. Using next-generation sequencing for high resolution multiplex analysis of copy number variation from nanogram quantities of DNA from formalin-fixed paraffin-embedded specimens. Nucleic Acids Res. 2010;38:e151. doi: 10.1093/nar/gkq510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Brown C, Durbin R, Hurles ME, Edwards PA, Bignell GR, Stratton MR, Futreal PA. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, Sahinalp SC, Gibbs RA, Eichler EE. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, Frazer KA. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darvishi K. Application of Nexus copy number software for CNV detection and analysis. Curr. Protoc. Hum. Genet. 2010;Chapter 4(Unit 4):14, 11–28. doi: 10.1002/0471142905.hg0414s65. [DOI] [PubMed] [Google Scholar]
- 50.Matsuzaki H, Wang PH, Hu J, Rava R, Fu GK. High resolution discovery and confirmation of copy number variants in 90 Yoruba Nigerians. Genome Biol. 2009;10:R125. doi: 10.1186/gb-2009-10-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potkin SG, Macciardi F, Guffanti G, Fallon JH, Wang Q, Turner JA, Lakatos A, Miles MF, Lander A, Vawter MP, Xie X. Identifying gene regulatory networks in schizophrenia. Neuroimage. 2010;53:839–847. doi: 10.1016/j.neuroimage.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S, Chun J. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J. Comp. Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J. Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, Delacourte A, Galasko D, Gauthier S, Jicha G, Meguro K, O'Brien J, Pasquier F, Robert P, Rossor M, Salloway S, Stern Y, Visser PJ, Scheltens P. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 55.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 56.Storandt M, Grant EA, Miller JP, Morris JC. Rates of progression in mild cognitive impairment and early Alzheimer's disease. Neurology. 2002;59:1034–1041. doi: 10.1212/wnl.59.7.1034. [DOI] [PubMed] [Google Scholar]
- 57.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 59.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 61.Quail MA, Swerdlow H, Turner DJ. Improved protocols for the illumina genome analyzer sequencing system. Curr. Protoc. Hum. Genet. 2009;Chapter 18(Unit 18):12. doi: 10.1002/0471142905.hg1802s62. (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, Cirulli ET, Fellay J, Dickson SP, Gumbs CE, Heinzen EL, Need AC, Ruzzo EK, Singh A, Campbell CR, Hong LK, Lornsen KA, McKenzie AM, Sobreira NL, Hoover-Fong JE, Milner JD, Ottman R, Haynes BF, Goedert JJ, Goldstein DB. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.