Abstract
Purpose of Review
Description of the immunologic components needed for autoimmune diabetes.
Recent Findings
The MHC class II molecules are the primary susceptibility genes for many autoimmune diseases, including type 1 diabetes. Understanding of the structural interaction between MHC molecules, antigenic peptides, and T cell receptors (the three components of the trimolecular complex) has increased greatly over the last several years. The components of the anti-insulin trimolecular complex and findings that insulin is a key autoantigen in type 1 diabetes are reviewed.
Summary
The anti-insulin trimolecular complex is well defined in the non-obese diabetic mouse model. Insulin and specifically, the amino acid sequence 9 to 23 of the insulin B chain, represents a primary antigenic target for islet autoimmunity in the non-obese diabetic mouse model of type 1 diabetes with a specific mutation of this peptide preventing all diabetes. Initial studies suggest the human homologues of the anti-insulin trimolecular complex may be relevant in human disease.
Keywords: diabetes, autoimmunity, insulin, human leukocyte antigen, T cell receptor
Introduction
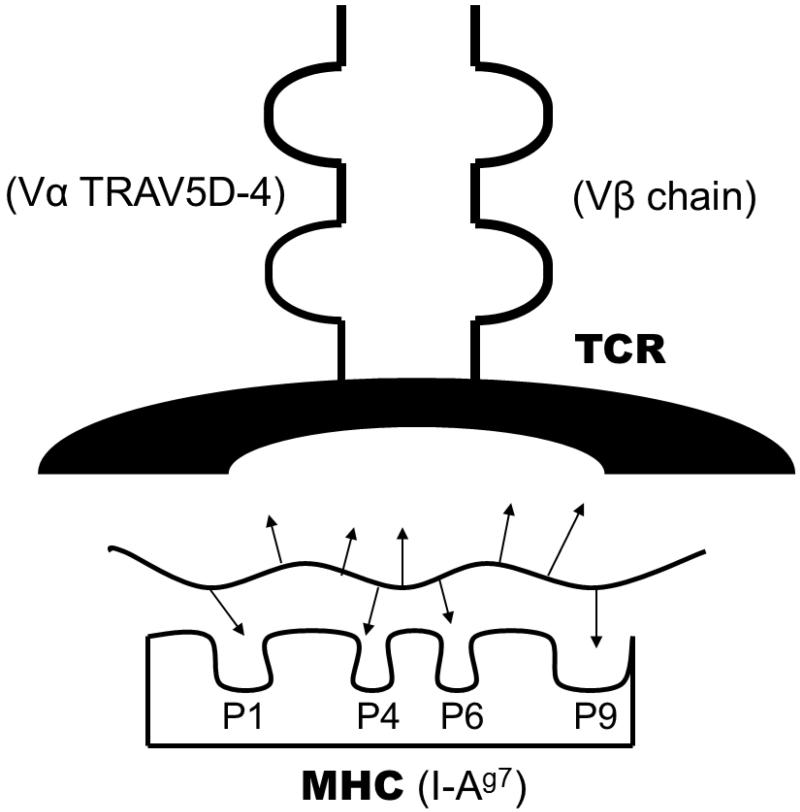
Type 1A diabetes (T1D) is the immune mediated form of diabetes. The incidence of T1D is increasing dramatically, doubling approximately every 20 years (1). The immunologic mediators of the disease are becoming more defined. The trimolecular complex consists of antigen presenting cells, peptide fragments, and specific T cell receptors that recognize peptide bound to antigen presenting cells, and this trimolecular complex leads to tissue specific targeting in autoimmune diseases. The trimolecular complex for insulin is now well characterized for the non-obese diabetic mouse model of T1D (figure 1). The components of the anti-insulin trimolecular complex in mice and humans will be reviewed.
Figure 1.
The anti-insulin trimolecular complex is composed of a T cell receptor on CD4 T cells, a peptide, and major histocompatability complex class II molecule. The MHC is composed of a binding groove with pockets that can accommodate amino acid side chains (represented as arrows) of the peptide. The MHC class II binding groove can accompany nine amino acid peptides and contains distinct pockets that interact with peptide side chains. Peptides can have varying affinities for MHC class II molecules depending on the amino acid sequence and alignment in the binding groove. Those peptide side chains not binding to the MHC molecule can interact with the T cell receptor. The components for the non-obese diabetic mouse are denoted in parentheses. MHC = major histocompatability complex, P1-P9 denote pockets, and TCR = T cell receptor.
The Major Histocompatability Complex
The major determinant of genetic susceptibility to T1D is conferred by genes in the human leukocyte antigen (HLA) complex, which is divided into three regions: class I, II, and III. Alleles of the class II genes, DQ and DR (and to a lesser extent DP), are the most important determinants of type 1 diabetes. These class II molecules are mainly expressed on antigen presenting cells (macrophages, dendritic cells, and B cells) and present antigens to CD4+ T lymphocytes. Haplotypes (combinations of DR and DQ alleles on the same chromosome) are strongly associated with T1D. Each unique amino acid sequence of DR and DQ is given a number. Since DRA does not vary, haplotypes can be defined by specific DRB, DQA, and DQB alleles. The DR3 and DR4 haplotypes are present in more than 90% of people with T1D possessing one or both of these haplotypes versus 40% of the US population (2). The DR3 haplotype contains the alleles DQA1*0501 and DQB1*0201, while high risk DR4 contains both DQA1*0301 and DQB1*0302. The most extensively studied allele DQ8 (DQA1*0301 and DQB1*0302), present in approximately 60% of type 1 diabetics, has been crystallized accommodating an insulin peptide (3). A polymorphism in the beta chain of DQ8 (substitution of β57 aspartic acid for valine, leucine, or alanine) leads to changes in the peptide binding groove of the molecule (4;5). Subtle structural changes to the binding groove of MHC class II molecules likely leads to diabetes susceptibility by decreasing the affinity by which autoantigens are presented or changing the register of binding (6;7).
Heterozygous DR3/4 individuals have a much higher risk for diabetes development than homozygous DR3/3 and DR4/4 individuals. Recent work by Erlich and coworkers helps explain this phenomenon by analyzing the large family data set of the Type 1 Diabetes Genetics Consortium (8). The T1D risk of DR3/4 individuals is the result of a trans-encoded DQ molecule (DQA and DQB encoded by different chromosomes) that form in DR3/4 individuals, namely DQA1*0501 and DQB1*0302. Trans-encoded DQ8 molecules differ in peptide presentation and T cell recognition of peptides compared to cis-encoded molecules (9;10). Work is being done to identify additional genes contributing to diabetes risk within or linked to the MHC region such as HLA-A24 (11), HLA-B39 (12), and a locus telomeric of the UBD gene several million base pairs away from DR and DQ alleles (13). We believe there are genes to be identified within the MHC that predispose to diabetes risk.
In addition to HLA genes, many genetic loci contributing to diabetes risk have been implicated through genome wide association studies (14), which involves analyzing thousands of single nucleotide polymorphisms from large populations to find alleles associated with a particular disease. While HLA alleles confer the highest risk, multiple non-HLA genetic polymorphisms modify disease risk. One of these non-HLA genes, the group of longer variable number of tandem nucleotide repeats (VNTR) 5′ of the insulin gene, protects against diabetes. The decreased diabetes risk is associated with greater insulin message in the thymus and resultant deletion of autoreactive T cells in the thymus (15;16). Maturing T cells in the thymus that react with autoantigens are deleted and the more insulin that is present in the thymus leads to less insulin reactive T cells.
Insulin as an autoantigen
Insulin has been focused on as a T1D autoantigen for decades since autoantibodies to this molecule were discovered in patients having T1D, (17) and often precedes the development of other islet autoantibodies (18). Numerous studies have implicated insulin as an autoantigen in human T1D. Pugliese and coworkers demonstrated that polymorphisms upstream of the insulin gene (VNTR) which are associated with levels of insulin expression in the thymus correlate with the risk of T1D development (15). Alleva and colleagues detected T cells that react with the amino acid sequence 9-23 of the insulin B chain (B:9-23) in the peripheral blood of T1D patients but not in HLA-matched normal subjects (19). More recently, Kent and coworkers identified CD4 T cells in the pancreatic lymph nodes of T1D patients that react with the amino acid sequence 1-15 of the insulin A chain (20). Subsequent studies further support that such insulin A:1-15 reactive CD4 T cells are detected in peripheral blood in T1D patients (21;22). In addition, multiple investigators have reported CD8 T cells that react with preproinsulin epitopes (23-25), some of which have been shown to kill beta cells in vitro (26) and in vivo using a humanized mouse model (27). Given the above evidence, it is likely that the T cell response to insulin or preproinsulin contribute to T1D development of humans.
Much of what we know about autoantigens in T1D comes from the study of the animal model of T1D. The non-obese diabetic (NOD) mouse develops diabetes spontaneously and shares many similarities to the human disease. Wegmann and coworkers first established CD4 T cell clones that react with insulin from pre-diabetic NOD islets (28) and found that the majority of them responds to the insulin B:9-23 peptide, which has an identical amino acid sequence in mouse and human insulin. Also, the structure of human DQ8 is very similar to the mouse MHC class II presenting molecule, I-Ag7. Both MHC molecules share a polymorphism, the substitution of β57 aspartic acid for a small hydrophobic amino acid. Aspartic acid has a negatively charged side chain that forms a salt bridge with α76 arginine forming pocket 9 of the MHC class II binding groove; disrupting this salt bridge leads to a very basic pocket along the binding groove implicated in diabetes susceptibility (4;6). Several investigators confirmed the existence of CD4 T cells reacting with the insulin B:9-23 in lymphocytes infiltrating the NOD pancreatic islets although the frequency of such B:9-23 reactive T cells in the islets differs by studies (29;30). These insulin B:9-23-reactive T cell clones are capable of inducing diabetes when adoptively transferred or transgenically introduced into immuno-compromised recipients (31-33). Furthermore, the administration of the insulin B:9-23 peptide or the altered form of this peptide prevents and delays diabetes onset in NOD mice (34-36). This evidence indicates that the CD4 T cell response to the insulin B:9-23 peptide contributes to islet autoimmunity in NOD mice.
It is still a fundamental question whether insulin is essential for human T1D development. In the NOD mouse model, elimination of the majority of T cells that react with insulin (37;38) but not with glutamic acid decarboxyalse (GAD) (39) or insulin glucose related phosphatase (IGRP) (40) results in protection from diabetes. Conversely, lack of insulin expression in thymic epithelial cells results in aggressive diabetes development even in a diabetes-resistant mouse strain (41). We demonstrated that NOD mice lacking native insulin genes but transgenic for insulin with an alanine mutation at B:16 (B16:A insulin) are completely protected from diabetes development (42;43). In addition, Krishnamurthy and coworkers demonstrated that the response to IGRP is downstream of that to insulin (40). Thus, it is quite likely that insulin is required to drive anti-islet autoimmunity in the NOD mouse, although this does not necessarily imply that insulin is the only single autoantigen essential for islet cell autoimmunity. What insulin or preproinsulin epitopes are essential for islet autoimmunity? As described above, B:9-23 is the most studied peptide for NOD diabetes. The complete prevention of insulin autoantibodies in insulin knockout NOD mice with B16:A insulin is abrogated when the alanine mutation is converted back to tyrosine which is the native insulin amino acid sequence (44). Further adoptive transfer experiments with CD4 T cells suggest insulin B:9-23 presented by I-Ag7 is an essential epitope contained in the preproinsulin molecule. Other peptides (e.g. insulin A:7-21 (34), insulin C:15-30 (29), B:2-17 (29), insulin B:24-C:36 (45;46), and preproinsulin:47-64 (47)) demonstrated as CD4 spontaneous insulin epitopes for NOD diabetes are also being investigated.
Why is the insulin B:9-23 peptide pathogenic? T cells are “educated” not to attack self antigens in the thymus (16;41;48) and also in the peripheral immune organs (49;50). Recently, Unanue and colleagues proposed and demonstrated a novel concept of autoantigen presentation. Namely, the B:9-23 peptide binds to I-Ag7 with a low affinity, and T cells reacting with such poor binding peptides may escape negative selection in the thymus (47;51). I-Ag7 presents other autoantigens in an unorthodox manner. Recent work by Haskins and colleagues elucidated a natural epitope of highly diabetogenic T cell clones (BDC-2.5, BDC-10.1, and BDC-5.10.3)(52), a chromogranin peptide, binds to I-Ag7 in an extremely unusual fashion (53). Another idea is that antigens presented by MHC molecules in a targeted organ (i.e. pancreatic islets) are slightly modified and are different from such “educating” tissues, and as a result, T cells reacting with modified antigens are not eliminated or become tolerant. These antigen modifications include alternative splicing of antigens in the thymus (54) and post-translational modification of antigens in the pancreas (21). Autoantigens including the insulin B:9-23 may rarely be expressed in the proper fashion to eliminate or silence T cells in the thymus and peripheral immune organs, which may facilitate T cell differentiation toward autoreactivity.
T cell receptors recognizing the insulin peptide-MHC complex
Identifying the types of T cells and the specific T cell receptor (TCR) sequences provides insight into the pathogenesis of the immune destruction of beta cells. Just as the NOD model provided significant insights into insulin as an autoantigen in T1D, the model provides an avenue to understand how TCRs recognize insulin-MHC complexes. T cells recognize antigens via an interaction between a TCR, comprised of a TCR alpha and beta chain, and peptide-MHC complex. Compared to insulin peptides and MHC molecules, we know relatively little about the association of TCRs with T1D. However, cumulative evidence suggests that skewed TCR sequences preferentially respond to specific autoantigens including B:9-23. Alpha and beta chain genes encode up to 100 different variable (TRAV, TRBV) and junctional (TRAJ, TRBJ) segments. Individual T cells assemble TCRs with multiple nucleotides (N region) between these two segments. Millions of TCR sequences are theoretically possible, and approximately 70% of TCRs reacting with B:9-23 in NOD mice utilize a conserved TCR alpha chain sequences containing identical variable (TRAV5D-4) and junctional (TRAJ53/42) regions (29;55). Two alpha chains that contain these conserved regions but with different N regions can induce insulin autoantibodies in vivo when introduced into NOD mice lacking native alpha chains, whereas a beta chain derived from an insulin B:9-23 reactive TCR (12-4.1) does not (56). The current evidence suggests that the conserved alpha chain sequence recognizes the insulin peptide-I-Ag7 complex by paring with multiple different beta chains, which may predispose to islet autoimmunity. In humans, skewed TCR usage for T cells recognizing insulin A:1-15 and other potential islet antigens have been observed (20;57;58).
A basic question is what makes conserved TCRs target self antigens. Analyzing crystallography of the trimolecular complex is the direct way to understand how TCRs recognize peptide/MHC complexes. Crystallization of a TCR-insulin-MHC complex is yet to be completed. However, five autoimmune CD4 TCRs that react with myelin basic protein (autoantigen for multiple sclerosis) have been solved. All of these structures reveal low binding affinity either between the TCR and peptide-MHC complex or between the peptide and MHC (59-62). Two of the TCRs that have a poor affinity for the peptide-MHC complex bind in an unusual fashion. These binding motifs with low affinity may allow autoreactive T cells to escape selection in the thymus and lead to autoimmune disorders. Spontaneously occurring insulin B:9-23-reactive T cells can be classified into two subsets (33). One subset can only recognize the specific B:9-23 peptide but not insulin, which suggests that the two types of T cells recognize B:9-23 in a different fashion. Thus, both structural and functional studies indicate that unusual and/or weak binding to peptide-MHC complexes may allow T cells to escape thymic selection and become autoreactive T cells.
Are there skewed TCR sequences in the pancreas that target beta cells? A recent study by Vignali and coworkers demonstrates that only T cells recognizing beta cell antigens infiltrate pancreatic islets (63). With the difficulty in accessing the human pancreas, several studies have examined TCR sequences in the peripheral blood. However, Tisch and coworkers demonstrated that TCR sequences recognizing pancreatic antigens in the peripheral blood, which were not diabetogenic, are different from those in pancreatic islets (64). Sequencing TCRs infiltrating pancreatic islets is important. T cells and TCRs from human pancreata and lymph nodes are less well studied than the organs from animal models. The retroperitoneal location of the pancreas makes it one of the most inaccessible organs in the body. Coupled with the facts that islets make up a very small percentage of cells in the pancreas and are surrounded by exocrine cells with proteolytic enzymes, it is difficult to obtain tissue specimens for analysis. The recent creation of a Network for Pancreatic Organ Donors with Diabetes (nPOD) has been established through the Juvenile Diabetes Research Foundation to obtain pancreata and related tissue specimens from cadaveric organ donors with T1D and those who are autoantibody positive but without clinical disease. This project will help answer many remaining questions. How many T cell clones infiltrate the pancreas? What are the TCR sequences of those clones? Does islet cell antigen presentation occur in peripheral lymphoid organs? Do T cells in the peripheral blood correlate with T cells invading the pancreas? Some of these questions have been already studied in the animal models, but answers to these questions will increase our understanding of the immunopathogenesis of human T1D, provide avenues for T cell specific immunotherapies, and improve assays to detect and measure the functionality of autoreactive T cells in peripheral blood.
Conclusion
The anti-insulin trimolecular complex has been defined and well studied in animals. The human counterpart is likely critical for diabetes development but further research needs to be done to define the insulin autoantigens and T cell receptors responsible for islet autoimmunity. With increased understanding of this basic immunology, we are hopeful that type 1 diabetes can be prevented and ultimately cured.
Reference List
- (1)**.Harjutsalo V, Sjoberg L, Tuomilehto J. Time trends in the incidence of type 1 diabetes in Finnish children: a cohort study. Lancet. 2008 May 24;371(9626):1777–82. doi: 10.1016/S0140-6736(08)60765-5. This study showed that the incidence of type 1 diabetes in Finnish children is increasing dramatically, and the number of new cases diagnosed at or before 14 years of age will double in the next 15 years and the age of onset will be younger (0-4 years).
- (2).Lie BA, Ronningen KS, Akselsen HE, Thorsby E, Undlien DE. Application and interpretation of transmission/disequilibrium tests: transmission of HLA-DQ haplotypes to unaffected siblings in 526 families with type 1 diabetes. Am J Hum Genet. 2000 Feb;66(2):740–3. doi: 10.1086/302780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide/HLA-DQ8 complex and susceptibility to type 1 diabetes. Nature Immunology. 2001;2:501–7. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- (4).Corper AL, Stratmann T, Apostolopoulos V, Scott CA, Garcia KC, Kang AS, Wilson IA, Teyton L. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science. 2000 Apr 21;288(5465):505–11. doi: 10.1126/science.288.5465.505. [DOI] [PubMed] [Google Scholar]
- (5).Suri A, Vidavsky I, van der DK, Kanagawa O, Gross ML, Unanue ER. In APCs, the autologous peptides selected by the diabetogenic I-Ag7 molecule are unique and determined by the amino acid changes in the P9 pocket. J Immunol. 2002;168:1235–43. doi: 10.4049/jimmunol.168.3.1235. [DOI] [PubMed] [Google Scholar]
- (6).Suri A, Walters JJ, Gross ML, Unanue ER. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J Clin Invest. 2005 Aug;115(8):2268–76. doi: 10.1172/JCI25350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Suri A, Walters JJ, Rohrs HW, Gross ML, Unanue ER. First signature of islet beta-cell-derived naturally processed peptides selected by diabetogenic class II MHC molecules. J Immunol. 2008 Mar 15;180(6):3849–56. doi: 10.4049/jimmunol.180.6.3849. [DOI] [PubMed] [Google Scholar]
- (8)**.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, Lavant E, Louey A, Moonsamy P. HLA DR-DQ Haplotypes and Genotypes and Type 1 Diabetes Risk: Analysis of the Type 1 Diabetes Genetics Consortium Families. Diabetes. 2008 Feb 5;57:1084–92. doi: 10.2337/db07-1331. This study showed that the trans-complementing heterodimer encoded by DQA1*0501 and DQB1*0302 confers a very high risk for type 1A diabetes.
- (9).Reichstetter S, Kwok WW, Nepom GT. Impaired binding of a DQ2 and DQ8-binding HSV VP16 peptide to a DQA1*0501/DQB1*0302 trans class II heterodimer. Tissue Antigens. 1999 Jan;53(1):101–5. doi: 10.1034/j.1399-0039.1999.530111.x. [DOI] [PubMed] [Google Scholar]
- (10).Tollefsen S, Arentz-Hansen H, Fleckenstein B, Molberg O, Raki M, Kwok WW, Jung G, Lundin KE, Sollid LM. HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J Clin Invest. 2006 Aug;116(8):2226–36. doi: 10.1172/JCI27620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nakanishi K, Inoko H. Combination of HLA-A24, -DQA1*03, and -DR9 contributes to acute-onset and early complete beta-cell destruction in type 1 diabetes: longitudinal study of residual beta-cell function. diab. 2006 Jun;55(6):1862–8. doi: 10.2337/db05-1049. [DOI] [PubMed] [Google Scholar]
- (12).Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, Todd JA. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007 Dec 6;450(7171):887–92. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Aly TA, Baschal EE, Jahromi MM, Fernando MS, Babu SR, Fingerlin TE, Kretowski A, Erlich HA, Fain PR, Rewers MJ, Eisenbarth GS. Analysis of Single Nucleotide Polymorphisms Identifies Major Type 1A Diabetes Locus Telomeric of the Major Histocompatibility Complex. Diabetes. 2008;57(3):770–6. doi: 10.2337/db07-0900. [DOI] [PubMed] [Google Scholar]
- (14).Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007 Jul;39(7):857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pugliese A, Zeller M, Fernandez A, Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD. The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type I diabetes. Nat Genet. 1997;15(3):293–7. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- (16).Chentoufi AA, Polychronakos C. Insulin expression levels in the thymus modulate insulin-specific autoreactive T-cell tolerance: the mechanism by which the IDDM2 locus may predispose to diabetes. diab. 2002 May;51(5):1383–90. doi: 10.2337/diabetes.51.5.1383. [DOI] [PubMed] [Google Scholar]
- (17).Palmer JP, Asplin CM, Clemons P, Lyen K, Tatpati O, Raghu PK, Paquette TL. Insulin antibodies in insulin-dependent diabetics before insulin treatment. Science. 1983;222(4630):1337–9. doi: 10.1126/science.6362005. [DOI] [PubMed] [Google Scholar]
- (18).Achenbach P, Koczwara K, Knopff A, Naserke H, Ziegler AG, Bonifacio E. Mature high-affinity immune responses to (pro)insulin anticipate the autoimmune cascade that leads to type 1 diabetes. J Clin Invest. 2004 Aug;114(4):589–97. doi: 10.1172/JCI21307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Alleva DG, Crowe PD, Jin L, Kwok WW, Ling N, Gottschalk M, Conlon PJ, Gottlieb PA, Putnam AL, Gaur A. A disease-associated cellular immune response in type 1 diabetics to an immunodominant epitope of insulin. J Clin Invest. 2001 Jan;107(2):173–80. doi: 10.1172/JCI8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005 May 20;435(7039):224–8. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- (21).Mannering SI, Harrison LC, Williamson NA, Morris JS, Thearle DJ, Jensen KP, Kay TW, Rossjohn J, Falk BA, Nepom GT, Purcell AW. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med. 2005 Nov 7;202(9):1191–7. doi: 10.1084/jem.20051251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mannering SI, Pang SH, Williamson NA, Naselli G, Reynolds EC, O’Brien-Simpson NM, Purcell AW, Harrison LC. The A-chain of insulin is a hot-spot for CD4+ T cell epitopes in human type 1 diabetes. Clin Exp Immunol. 2009 May;156(2):226–31. doi: 10.1111/j.1365-2249.2009.03907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pinkse GG, Tysma OH, Bergen CA, Kester MG, Ossendorp F, van Veelen PA, Keymeulen B, Pipeleers D, Drijfhout JW, Roep BO. Autoreactive CD8 T cells associated with {beta} cell destruction in type 1 diabetes. Proc Natl Acad Sci U S A. 2005 Dec;102(51):18425–30. doi: 10.1073/pnas.0508621102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hassainya Y, Garcia-Pons F, Kratzer R, Lindo V, Greer F, Lemonnier FA, Niedermann G, Van Endert PM. Identification of naturally processed HLA-A2 - Restricted proinsulin epitopes by reverse immunology. diab. 2005;54(7):2053–9. doi: 10.2337/diabetes.54.7.2053. [DOI] [PubMed] [Google Scholar]
- (25).Toma A, Haddouk S, Briand JP, Camoin L, Gahery H, Connan F, LaForgue D, Caillat-Zucman S, Guillet JG, Carel JC, Muller S, Choppin J, Boitard C. Recognition of a subregion of human proinsulin by class I-restricted T cells in type 1 diabetic patients. Proc Natl Acad Sci U S A. 2005 Jul 26;102(30):10581–6. doi: 10.1073/pnas.0504230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26)**.Skowera A, Ellis RJ, Varela-Calvino R, Arif S, Huang GC, Van-Krinks C, Zaremba A, Rackham C, Allen JS, Tree TI, Zhao M, Dayan CM, Sewell AK, Unger W, Drijfhout JW, Ossendorp F, Roep BO, Peakman M. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J Clin Invest. 2008 Oct;118(10):3390–402. doi: 10.1172/JCI35449. This study provides direct evidence that autoreactive cytotoxic CD8 T cells that can kill beta cells are present in the circulation of patients with type 1 diabetes.
- (27).Jarchum I, DiLorenzo TP. Ins2 deficiency augments spontaneous HLA-A*0201-restricted T cell responses to insulin. J Immunol. 2010 Jan 15;184(2):658–65. doi: 10.4049/jimmunol.0903414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wegmann DR, Norbury-Glaser M, Daniel D. Insulin-specific T cells are a predominant component of islet infiltrates in pre-diabetic NOD mice. Eur J Immunol. 1994;24(8):1853–7. doi: 10.1002/eji.1830240820. [DOI] [PubMed] [Google Scholar]
- (29).Halbout P, Briand JP, Becourt C, Muller S, Boitard C. T cell response to preproinsulin I and II in the nonobese diabetic mouse. J Immunol. 2002 Sep 1;169(5):2436–43. doi: 10.4049/jimmunol.169.5.2436. [DOI] [PubMed] [Google Scholar]
- (30).Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J Immunol. 2007 May 15;178(10):6051–7. doi: 10.4049/jimmunol.178.10.6051. [DOI] [PubMed] [Google Scholar]
- (31).Daniel D, Gill RG, Schloot N, Wegmann D. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur J Immunol. 1995;25(4):1056–62. doi: 10.1002/eji.1830250430. [DOI] [PubMed] [Google Scholar]
- (32).Jasinski JM, Yu L, Nakayama M, Li MM, Lipes MA, Eisenbarth GS, Liu E. Transgenic insulin (B:9-23) T-cell receptor mice develop autoimmune diabetes dependent upon RAG genotype, H-2g7 homozygosity, and insulin 2 gene knockout. Diabetes. 2006 Jul;55(7):1978–84. doi: 10.2337/db06-0058. [DOI] [PubMed] [Google Scholar]
- (33)**.Mohan JF, Levisetti MG, Calderon B, Herzog JW, Petzold SJ, Unanue ER. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat Immunol. 2010 Feb 28; doi: 10.1038/ni.1850. Authors demonstrated that two subsets of B:9-23-reactive T cells, which differs in antigen recognition, infiltrate pancreatic islets in the NOD mouse. One subset can only recognize the specific B:9-23 peptide but not insulin. This study also suggests that antigen processing in the targeted cells may be critical to stimulate autoreactive T cells.
- (34).Daniel D, Wegmann DR. Protection of nonobese diabetic mice from diabetes by intranasal or subcutaneous administration of insulin peptide B-(9-23) Proc Natl Acad Sci USA. 1996;93(2):956–60. doi: 10.1073/pnas.93.2.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Alleva DG, Gaur A, Jin L, Wegmann D, Gottlieb PA, Pahuja A, Johnson EB, Motheral T, Putnam A, Crowe PD, Ling N, Boehme SA, Conlon PJ. Immunological Characterization and Therapeutic Activity of an Altered-Peptide Ligand, NBI-6024, Based on the Immunodominant Type 1 Diabetes Autoantigen Insulin B-Chain (9-23) Peptide. diab. 2002 Jul;51(7):2126–34. doi: 10.2337/diabetes.51.7.2126. [DOI] [PubMed] [Google Scholar]
- (36).Kobayashi M, Abiru N, Arakawa T, Fukushima K, Zhou H, Kawasaki E, Yamasaki H, Liu E, Miao D, Wong FS, Eisenbarth GS, Eguchi K. Altered B:9 23 insulin, when administered intranasally with cholera toxin adjuvant, suppresses the expression of insulin autoantibodies and prevents diabetes. J Immunol. 2007 Aug 15;179(4):2082–8. doi: 10.4049/jimmunol.179.4.2082. [DOI] [PubMed] [Google Scholar]
- (37).French MB, Allison J, Cram DS, Thomas HE, Dempsey-Collier M, Silva A, Georgiou HM, Kay TW, Harrison LC, Lew AM. Transgenic expression of mouse proinsulin II prevents diabetes in nonobese diabetic mice. diab. 1996;46:34–9. doi: 10.2337/diab.46.1.34. [DOI] [PubMed] [Google Scholar]
- (38).Jaeckel E, Lipes MA, von Boehmer H. Recessive tolerance to preproinsulin 2 reduces but does not abolish type 1 diabetes. Nat Immunol. 2004 Oct;5(10):1028–35. doi: 10.1038/ni1120. [DOI] [PubMed] [Google Scholar]
- (39).Jaeckel E, Klein L, Martin-Orozco N, von Boehmer H. Normal incidence of diabetes in NOD mice tolerant to glutamic acid decarboxylase. J Exp Med. 2003 Jun 16;197(12):1635–44. doi: 10.1084/jem.20030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Krishnamurthy B, Dudek NL, McKenzie MD, Purcell AW, Brooks AG, Gellert S, Colman PG, Harrison LC, Lew AM, Thomas HE, Kay TW. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J Clin Invest. 2006 Dec 1;116(12):3258–65. doi: 10.1172/JCI29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41)**.Fan Y, Rudert WA, Grupillo M, He J, Sisino G, Trucco M. Thymus-specific deletion of insulin induces autoimmune diabetes. EMBO J. 2009 Sep 16;28(18):2812–24. doi: 10.1038/emboj.2009.212. This study demonstrates insulin expression in thymic epithelial cells is essential to be protected from diabetes development.
- (42).Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005 May 12;435(7039):220–3. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Nakayama M, Babaya N, Miao D, Sikora K, Elliott JF, Eisenbarth GS. Thymic expression of mutated B16:A preproinsulin messenger RNA does not reverse acceleration of NOD diabetes associated with insulin 2 (thymic expressed insulin) knockout. J Autoimmun. 2005 Nov;25(3):193–8. doi: 10.1016/j.jaut.2005.09.014. [DOI] [PubMed] [Google Scholar]
- (44).Nakayama M, Beilke JN, Jasinski JM, Kobayashi M, Miao D, Li M, Coulombe MG, Liu E, Elliott JF, Gill RG, Eisenbarth GS. Priming and effector dependence on insulin B:9-23 peptide in NOD islet autoimmunity. J Clin Invest. 2007 Jul 2;117(7):1835–43. doi: 10.1172/JCI31368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chen W, Bergerot I, Elliott JF, Harrison LC, Abiru N, Eisenbarth GS, Delovitch TL. Evidence that a peptide spanning the B-C junction of proinsulin is an early autoantigen epitope in the pathogenesis of type 1 diabetes. J Immunol. 2001 Nov 1;167(9):4926–35. doi: 10.4049/jimmunol.167.9.4926. [DOI] [PubMed] [Google Scholar]
- (46).Bresson D, Togher L, Rodrigo E, Chen YL, Bluestone JA, Herold KC, von Herrath M. Anti-CD3 and nasal proinsulin combination therapy enhances remission from recent-onset autoimmune diabetes by inducing Tregs. Journal of Clinical Investigation. 2006;116(5):1371–81. doi: 10.1172/JCI27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Levisetti MG, Lewis DM, Suri A, Unanue ER. Weak proinsulin peptide-major histocompatibility complexes are targeted in autoimmune diabetes in mice. diab. 2008 Jul;57(7):1852–60. doi: 10.2337/db08-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002 Nov 15;298(5597):1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- (49).Gardner JM, DeVoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, Johannes KP, Su MA, Chang HY, Krummel MF, Anderson MS. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008 Aug 8;321(5890):843–7. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Yip L, Su L, Sheng D, Chang P, Atkinson M, Czesak M, Albert PR, Collier AR, Turley SJ, Fathman CG, Creusot RJ. Deaf1 isoforms control the expression of genes encoding peripheral tissue antigens in the pancreatic lymph nodes during type 1 diabetes. Nat Immunol. 2009 Sep;10(9):1026–33. doi: 10.1038/ni.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J Immunol. 2007 May 15;178(10):6051–7. doi: 10.4049/jimmunol.178.10.6051. [DOI] [PubMed] [Google Scholar]
- (52).Haskins K. Pathogenic T-cell clones in autoimmune diabetes: more lessons from the NOD mouse. Adv Immunol. 2005;87:123–62. 123–62. doi: 10.1016/S0065-2776(05)87004-X. [DOI] [PubMed] [Google Scholar]
- (53)**.Stadinski BD, Delong T, Reisdorph N, Reisdorph R, Powell RL, Armstrong M, Piganelli JD, Barbour G, Bradley B, Crawford F, Marrack P, Mahata SK, Kappler JW, Haskins K. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol. 2010 Mar;11(3):225–31. doi: 10.1038/ni.1844. This study identified a novel autoantigen for type 1 diabetes of the NOD mouse model. A series of highly diabetogenic CD4 T cell clones (BDC clones) recognize a chromogranin A peptide presented by I-Ag7 in an extremely unusual fashion.
- (54).Dogra RS, Vaidyanathan P, Prabakar KR, Marshall KE, Hutton JC, Pugliese A. Alternative splicing of G6PC2, the gene coding for the islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP), results in differential expression in human thymus and spleen compared with pancreas. diabetol. 2006;49(5):953–7. doi: 10.1007/s00125-006-0185-8. [DOI] [PubMed] [Google Scholar]
- (55).Simone E, Daniel D, Schloot N, Gottlieb P, Babu S, Kawasaki E, Wegmann D, Eisenbarth GS. T cell receptor restriction of diabetogenic autoimmune NOD T cells. Proc Natl Acad Sci USA. 1997;94(6):2518–21. doi: 10.1073/pnas.94.6.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56)**.Kobayashi M, Jasinski J, Liu E, Li M, Miao D, Zhang L, Yu L, Nakayama M, Eisenbarth GS. Conserved T cell receptor alpha-chain induces insulin autoantibodies. Proc Natl Acad Sci U S A. 2008 Jul 22;105(29):10090–4. doi: 10.1073/pnas.0801648105. This study shows that a skewed TCR repertoire with a conserved TCR alpha chain sequence is sufficient to induce insulin autoimmunity.
- (57).Reijonen H, Mallone R, Heninger AK, Laughlin EM, Kochik SA, Falk B, Kwok WW, Greenbaum C, Nepom GT. GAD65-Specific CD4+ T-Cells with High Antigen Avidity Are Prevalent in Peripheral Blood of Patients With Type 1 Diabetes. Diabetes. 2004 Aug;53(8):1987–94. doi: 10.2337/diabetes.53.8.1987. [DOI] [PubMed] [Google Scholar]
- (58).Gebe JA, Yue BB, Unrath KA, Falk BA, Nepom GT. Restricted autoantigen recognition associated with deletional and adaptive regulatory mechanisms. J Immunol. 2009 Jul 1;183(1):59–65. doi: 10.4049/jimmunol.0804046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005 May;6(5):490–6. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005 Sep 7;24(17):2968–79. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005 Jan;22(1):81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- (62).Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007 Sep;8(9):975–83. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- (63)**.Lennon GP, Bettini M, Burton AR, Vincent E, Arnold PY, Santamaria P, Vignali DA. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity. 2009 Oct 16;31(4):643–53. doi: 10.1016/j.immuni.2009.07.008. This study shows that the ability of T cells to infiltrate or accumulate in the pancreatic islets is cell autonomous, antigen specific, and tightly regulated.
- (64)**.Li L, He Q, Garland A, Yi Z, Aybar LT, Kepler TB, Frelinger JA, Wang B, Tisch R. beta cell-specific CD4+ T cell clonotypes in peripheral blood and the pancreatic islets are distinct. J Immunol. 2009 Dec 1;183(11):7585–91. doi: 10.4049/jimmunol.0901587. This study demonstrates that CD4 T cells in the peripheral blood and in the islets specific for beta cell antigens may differ regarding pathogenicity and TCR repertoire.