Abstract
Endogenous short RNAs and the conserved plant homeodomain (PHD) zinc-finger protein ZFP-1/AF10 regulate overlapping sets of genes in Caenorhabditis elegans, which suggests that they control common biological pathways. We have shown recently that the RNAi factor RDE-4 and ZFP-1 negatively modulate transcription of the insulin/PI3 signaling-dependent kinase PDK-1 to promote C. elegans fitness. Moreover, we have demonstrated that the insulin/IGF-1-PI3K-signaling pathway regulates the activity of the DAF-16/FOXO transcription factor in the hypodermis to nonautonomously promote the anterior migrations of the hermaphrodite-specific neurons (HSNs) during embryogenesis of C. elegans. In this study, we implicate the PHD-containing isoform of ZFP-1 and endogenous RNAi in the regulation of HSN migration. ZFP-1 affects HSN migration in part through its negative effect on pdk-1 transcription and modulation of downstream DAF-16 activity. We also identify a novel role for ZFP-1 and RNAi pathway components, including RDE-4, in the regulation of HSN migration in parallel with DAF-16. Therefore, the coordinated activities of DAF-16, ZFP-1, and endogenous RNAi contribute to gene regulation during development to ensure proper neuronal positioning.
Keywords: cell migration, insulin/IGF-1 signaling, RNAi, ZFP-1/AF10/MLLT10, RDE-4
WITH the identification of endogenous short interfering RNAs (endo-siRNAs) in many organisms, including plants, fungi, flies, nematodes, and mammals (reviewed in Li and Liu 2011), has come the challenge of identifying their biological and physiological roles. Caenorhabditis elegans endo-siRNAs that are generated by RNA-dependent RNA polymerases (RdRP) on messenger RNA (mRNA) templates (Ruby et al. 2006; Aoki et al. 2007; Pak and Fire 2007) have been termed 22G-RNAs due to the fact that they preferentially begin with guanosine and are 22 nucleotides in length (Gu et al. 2009). There are two main classes of 22G-RNAs, which are designated according to their interacting Argonaute protein. The first class includes 22G-RNAs antisense to protein-coding genes, which are found in complex with the Argonaute CSR-1 (Chromosome Segregation and RNAi-deficient) (Claycomb et al. 2009; Gu et al. 2009). 22G-RNAs of the second class, which are present in complex with the WAGO family of Argonautes, are antisense to transposons, pseudogenes, cryptic loci, and some coding genes (Gu et al. 2009).
RDE-4 is a double-stranded RNA (dsRNA)-binding protein and a member of the Dicer complex required to initiate silencing in response to exogenous RNAi (Parrish and Fire 2001; Tabara et al. 2002) and for the production of some endo-siRNAs (Lee et al. 2006; Vasale et al. 2010). We have previously demonstrated that RDE-4 cooperates with the RNAi-promoting factor Zinc-Finger Protein 1 (ZFP-1) (Dudley et al. 2002; Grishok et al. 2005; Kim et al. 2005) in modulating insulin/IGF-1 signaling (IIS) through its negative effect on transcription of the conserved 3-phosphoinositide-dependent kinase-1 (pdk-1) gene (Mansisidor et al. 2011). The upregulation of pdk-1 in rde-4(ne299) and zfp-1(ok554) mutants was shown to be responsible for the increased sensitivity to stress and shortened life span observed in these animals (Mansisidor et al. 2011). We have recently discovered a novel function of the IIS pathway in the hypodermis, which serves to nonautonomously regulate hermaphrodite-specific neuron (HSN) migration in a DAF-16-dependent manner (Kennedy et al. 2013). Therefore, we were intrigued by the possibility that RNAi may also contribute to the regulation of IIS during embryogenesis to influence neuronal migration. Indeed, here we describe a role for both RNAi factors and the chromatin-binding protein ZFP-1 in the regulation of HSN migration.
ZFP-1 is a homolog of mammalian AF10 (Acute Lymphoblastic Leukemia 1-Fused gene from chromosome 10) and is best known for its role in leukemia caused by its fusion to the mixed lineage leukemia (MLL) gene, which creates the MLL-AF10 oncogene (Chaplin et al. 1995). However, the normal developmental roles of AF10 remain elusive; therefore, studying its homolog ZFP-1 in C. elegans may help advance our understanding of this protein. In fact, recently, the highly conserved N-terminal plant homeodomain (PHD) fingers of ZFP-1 were shown to be essential for C. elegans viability (Avgousti et al. 2013). The PHD-finger domain of ZFP-1 interacts with methylated lysine 4 of histone H3 (H3K4me3) and promotes the localization of the protein to the promoters of active genes (including pdk-1) enriched in H3K4me3 during embryogenesis (Avgousti et al. 2013). Moreover, ZFP-1 was recently shown to be required for the recruitment of DOT-1.1, a histone H3 lysine 79 methyltransferase, to the promoters of highly expressed genes throughout development and to function, along with DOT-1.1, in the negative modulation of their transcription in a negative feedback mechanism (Cecere et al. 2013).
Here, we demonstrate that the long isoform of ZFP-1, which contains the N-terminal PHD fingers, is required for HSN migration. Combining epistasis analysis with the examination of DAF-16 subcellular localization, we show that the upregulation of pdk-1 in zfp-1 loss-of-function mutants contributes to the observed HSN undermigration phenotype by limiting DAF-16 nuclear activity. Furthermore, we implicate a number of RNAi factors, including the dsRNA-binding protein RDE-4, the dicer-related helicase DRH-3, and the Argonaute CSR-1, in the regulation of HSN migration. However, these genes are likely to function in parallel to DAF-16. Our study expands the limited understanding of the normal developmental roles of both ZFP-1/AF10 and endogenous RNAi by highlighting their requirement in establishing proper neuronal positioning during development.
Materials and Methods
C. elegans strains
Strains were maintained at 20° using standard methods (Brenner 1974). Bristol N2 was the wild-type strain used. The following strains were provided by the Hobert lab: SK4013 zdIs13 (tph-1::gfp)IV (Clark and Chiu 2003) and OH8482 otIs225 (cat-4::gfp)II; him-8IV. The following strain was provided by the Hengartner lab: zfp-1(op481)III. The following strains were provided by the Jose lab: AMJ217 rde-4(ne301)III; jamEx52 (Punc-54::rde-4(+)&3′ UTR plus Punc-54::gfp&3′ UTR) and AMJ188 rde-4(ne301)III; jamEx3 (Pf25b3.3::rde-4(+)&3′ UTR plus Pf25b3.3::gfp&3′ UTR). The following mutant and transgenic strains were obtained through the Caenorhabditis Genetics Center: LGI—CF1038 daf-16(mu86), WM153 C04F12.1(tm1637), WM206 drh-3(ne4253), WM30 rde-3(ne298), NL2098 rrf-1(pk1417), EL476 rrf-2(ok210); LGII—TJ1052 age-1(hx546), NL2099 rrf-3(pk1426); LGIII—RB774 zfp-1(ok554), CB4681 nDf17/qC1 dpy-19(e1259) glp-1(q339); LGIV—WM193 {csr-1(tm892)IV; neIs20 [pie-1::3xFLAG::csr-1 + unc-119(+)]}; LGV—WM158 ergo-1(tm1860), rde-1(ne300); LGX—JT709 pdk-1(sa709), VC446 alg-1(gk214). Compound mutant strains and transgenes used are as follows: AGK82—zfp-1(ok554)III; tph-1::gfpIV, AGK153—zfp-1(op481)III; tph-1::gfpIV, AGK154—{zfp-1(ok554) unc-119(ed3)III; tph-1::gfpIV; armIs5 [ZFP-1::FLAG unc-119(+)]}, AGK133—{zfp-1(ok554) unc-119(ed3)III; tph-1::gfpIV; armEx5 [ZFP-1::GFP unc-119(+)]}, AGK156—{zfp-1(ok554) unc-119(ed3)III; tph-1::gfpIV; armEx14 [PHD1-PHD2::FLAG ZFP-1 short unc-119(+)]}, AGK211—{zfp-1(ok554) unc-119(ed3)III; tph-1::gfpIV; armIs2 [unc-119(+)]}, AGK343—{zfp-1(ok554)III; tph-1::gfpIV; armEx91 [punc-86::zfp-1(long isoform)::gfp pPD118.33 (myo-2::gfp)]} (line 1 in figure S2A of this paper), AGK335—{zfp-1(ok554)III; tph-1::gfpIV; armEx85 [punc-86::zfp-1(long isoform)::gfp pPD118.33 (myo-2::gfp)]} (line 2 in figure S2A of this paper), AGK336—{zfp-1(ok554)III; tph-1::gfpIV; armEx86 [punc-86::zfp-1(long isoform)::gfp pPD118.33 (myo-2::gfp)]} (line 3 in figure S2A of this paper), AGK339—{zfp-1(ok554)III; tph-1::gfpIV; armEx89 [punc-86::zfp-1(long isoform)::gfp pPD118.33 (myo-2::gfp)]} (line 4 in figure S2A of this paper), AGK580—daf-16(mu86)I; tph-1::gfpIV, AGK236: pdk-1(sa709)X; tph-1::gfpIV, AGK155: zfp-1(ok554)III; pdk-1(sa709)X; tph-1::gfpIV, AGK470: age-1(hx546)II; tph-1::gfpIV, AGK285: age-1(hx546)II; zfp-1(ok554)III; tph-1::gfpIV, AGK277: daf-16(mu86)I; age-1(hx546)II; zfp-1(ok554)III; tph-1::gfp IV, AGK98: rde-4(ne299)III; tph-1::gfpIV, AGK246—rde-4(ne299)III; pdk-1(sa709)X; tph-1::gfpIV, AGK268—age-1(hx546)II; rde-4(ne299)III; tph-1::gfpIV, AGK124—alg-1(gk214)X; tph-1::gfpIV, AGK81—C04F12.1(tm1637)I; tph-1::gfpIV, AGK208—{csr-1(tm892)IV; tph-1::gfpIV; neIs20 [pie-1::3xFLAG::csr-1 + unc-119(+)]}, AGK654—drh-3(ne4253)I; tph-1::gfpIV, AGK146: tph-1::gfpIV; ergo-1(tm1860)V, AGK144—nrde-3(gg066); tph-1::gfpIV, AGK218: tph-1::gfpIV; rde-1(ne300)V, AGK194: rde-3(ne298)I; tph-1::gfpIV, AGK123: rrf-1(pk1417)I; tph-1::gfpIV, AGK121: rrf-2(ok210)I; tph-1::gfpIV, AGK145: rrf-3(pk1426)II; tph-1::gfpIV, AGK710—{zfp-1(ok554)III; tph-1::gfpIV; armEx257 [pdpy-7::daf-16b::tagrfp pPD118.33 (myo-2::gfp)]}, AGK711—{tph-1::gfpIV; armEx257 [pdpy-7::daf-16b::tagrfp pPD118.33 (myo-2::gfp)]}, AGK724—{pdk-1(sa709)X; armEx257 [pdpy-7::daf-16b::tagrfp pPD118.33 (myo-2::gfp)]}, AGK725—{zfp-1(ok554)III; pdk-1(sa709)X; armEx257 [pdpy-7::daf-16b::tagrfp pPD118.33 (myo-2::gfp)]}, AGK 721—{rde-4(ne299)III; tph-1::gfpIV; armEx257 [pdpy-7::daf-16b::tagrfp pPD118.33 (myo-2::gfp)]}, AGK688—{rde-4(ne299)III; tph-1::gfpIV; armEx273 [pdpy-7::rde-4::tagrfp pPD118.33 (myo-2::gfp)]} (line 1 in figure 5B of this paper), AGK689—{rde-4(ne299)III; tph-1::gfpIV; armEx274 [pdpy-7::rde-4::tagrfp pPD118.33 (myo-2::gfp)]} (line 2 in figure 5B of this paper), AGK690—{rde-4(ne299)III; tph-1::gfpIV; armEx275 [pdpy-7::rde-4::tagrfp pPD118.33 (myo-2::gfp)]} (line 3 in figure 5B of this paper).
Molecular biology and transgenic lines
Standard molecular biology techniques were used to construct transgenes. Germline transformation was performed by direct injection of various plasmid DNAs into the gonads of adult wild-type animals as described (Mello et al. 1991). A zfp-1(long isoform) complementary DNA (cDNA) was obtained by PCR with N2 cDNA as a template with the forward primer containing the BalI site 5′- ATGAAGAAGTGGCCAATGAAGGAGATGGTAGGTGGATGC-3′ and the reverse primer containing the AgeI site 5′-CAAGTTATTGGTTACCGGTCCTTTTCCATTCGGAGTTGCAGATG-3.′ A 5.1-kb fragment upstream of the unc-86 transcriptional start site was isolated by PCR from genomic DNA with the forward primer containing the SphI site 5′-CGTGACACTGCATGCTTCAAAAACTGTCAACTAACAAGAT-3′ and the reverse primer containing the SalI site 5′-CGGATGCGGTTGTCGACTCATTCAATTTCACTTTTTCATTCG-3.′ The zfp-1 cDNA and the unc-86 promoter were subcloned into the Fire Kit GFP vector pPD95.75. The resulting punc86::zfp-1::gfp transgene was injected at 0.5 ng/μl with the co-injection marker pPD118.33 (myo-2::gfp) at 4 ng/μl. The rde-4 cDNA was obtained by PCR with N2 cDNA as a template with the forward primer containing the BamHI site 5′-TCACGTGGATCCATGGATTTAACCAAACTAACGTTTG-3′ and the reverse primer containing the KpnI site 5′-TACTCAGGTACCCCATCAAATCATAGGTGTTGA-3′. The dpy-7 (Gilleard et al. 1997) promoter was obtained by PCR with N2 genomic DNA as a template. The dpy-7 promoter was amplified with the forward primer containing the SphI site 5′-GTTATTGCATGCTCCACGATTTCTCGCAACACATCCC-3′ and the reverse primer containing the SalI site 5′-GCGTCGGTCGACAAAGAACAGGGTGTGATAAATGAAT-3.′ DNA was inserted into a modified Fire Kit vector pPD95.75, in which we replaced the GFP with a TAGRFP sequence using the KpnI/EcoRI sites. Pdpy-7::rde-4::tagrfp was generated by inserting a SphI/SalI dpy-7 fragment and BamHI/KpnI rde-4 cDNA fragment into our modified pPD95.75 vector. The resulting pdpy-7::rde-4::tagrfp transgene was injected at 2.5 ng/μl with the co-injection marker pPD118.33 (myo-2::gfp) at 2.5 ng/μl. Tissue-specific GFP or TagRFP expression was confirmed using Nomarski and fluorescence microscopy.
Visualization of the HSN neurons
The HSN neurons were detected by staining adult hermaphrodites with rabbit anti-serotonin (Sigma) as previously described (Garriga et al. 1993) or by using the zdIs13[tph-1::gfp] or otIs225[cat-4::gfp] transcriptional reporters.
Microscopy and quantification
Live animals or embryos were mounted on 2% agarose pads and immobilized with 25 mM sodium azide (Sigma). Worms were examined using a Zeiss AxioImager Z1. All phenotypes were scored as the percentage of animals with at least one undermigrated HSN, and results are presented as stacked bar graphs in Figure 1, Figure 2, Figure 4, and Figure 5 to represent the proportion of HSNs in positions along the anterior–posterior (A–P) body axis in these animals. When one or both HSN cell bodies were located posterior to their normal position near the vulva in adult animals, the animal was scored as a mutant. The colored stacks within each column on the graphs presented in Figure 1, Figure 2, Figure 4, and Figure 5 represent the severity of migration of HSNs along the A–P axis within only those animals containing at least one undermigrated HSN. Thus, since there are two HSNs within each animal and not every HSN is undermigrated, a colored region could represent a HSN that remains unaffected in the animal, i.e., the pink color in the graphs, while the other HSN within the same animal is undermigrated (one HSN must be affected for an animal to be included in the graph) and therefore represented by the reddish/purple, green, or blue color. Statistical significance was calculated using the z-test.
RNA extraction and RT-PCR
RNA extraction and reverse transcriptase PCR (RT-PCR) was carried out as described previously (Mansisidor et al. 2011). The following primers were used to detect the pdk-1 transcript: 5′-CCTACAGCCAGGTATTCCG-3′ (forward) and 5′-GATCACGAAATAAATTCTAGCCTGG-3′ (reverse).
Results
C. elegans ZFP-1/AF10 regulates the migration of the HSNs
In our recent study (Kennedy et al. 2013), we showed that insulin/IGF-1-PI3K signaling modulates the activity of the DAF-16/FOXO transcription factor in the hypodermis to nonautonomously regulate the anterior migrations of the two bilaterally symmetric HSNs during embryogenesis of C. elegans. With the discovery that ZFP-1 negatively regulates pdk-1, a component of the IIS pathway, via direct repression of pdk-1 transcription (Mansisidor et al. 2011), we were prompted to investigate the connection between ZFP-1 and IIS during HSN migration. We found that the zfp-1(ok554) loss-of-function mutant (Cui et al. 2006; Avgousti et al. 2013; Cecere et al. 2013) and the zfp-1(op481) mutant (Gysi et al. 2013)—both of which lead to the generation of mRNAs that contain premature stop codons after the PHD fingers—display HSN undermigration defects (Figure 1, A–C). We used a tryptophan hydroxylase GFP reporter (tph-1::gfp) that marks the HSNs in adult animals for scoring the migration phenotype.
Figure 1.
HSN undermigration defects in zfp-1 mutants. (A) Epifluorescent images of the HSN in adults of wild-type (A) and zfp-1(ok554) mutant animals (B), visualized with a tryptophan hydroxylase GFP reporter (tph-1::gfp). Images are oriented with the posterior of the animal to the right. Asterisks indicate the vulva and the arrowhead in A denotes the position of properly migrated HSN(s). The arrow in B illustrates a HSN that has failed to migrate the full distance in the zfp-1(ok554) mutant animal. Bars, 20 μm. (C) Quantification of the percentage of animals with an undermigrated HSN from a minimum of two pooled independent experiments per strain in control tph-1::gfp (n = 177) vs. zfp-1(ok554) (n = 172) and zfp-1(op481) (n = 130) mutants. (D) The long isoform of ZFP-1 (867 aa) has two predicted PHD zinc fingers in the N terminus and an octapeptide motif and leucine zipper in the C terminus. The short isoform of ZFP-1 (635 aa) lacks the PHD fingers but retains the OM-LZ motifs. zfp-1 fosmid constructs used to rescue the HSN undermigration defects present in the zfp-1(ok554) mutant are illustrated below the ZFP-1 proteins. Expression data for these ZFP-1 transgenes can be found in Avgousti et al. (2013). (E) Two different ZFP-1 fosmid constructs rescue the HSN undermigration defects in the zfp-1(ok554) mutants. The ZFP-1 fosmid construct expressing only an intact short isoform did not rescue the HSN undermigration defects present in the zfp-1 mutant. Quantification of the percentage of animals with an undermigrated HSN from a minimum of two pooled independent experiments per strain in the (−) control armIs2 Unc-119 (+); zfp-1(ok554) unc-119 (ed3) (n = 106) vs. the addition of the armEx5 zfp-1::gfp (n = 128), armIs5 zfp-1::flag (n = 172) or armEx14 PHD1-PHD2::flag (n = 135) fosmids into the (−) control background. HSNs were visualized with a tph-1::gfp reporter. The stacked bars show information about the position of all HSNs in the affected worms (see the description below). Worm schematic legend (located at the bottom center of the figure): Stacked bars represent the proportion of HSNs at different positions along the A–P body axis within only those animals containing at least one undermigrated HSN. Thus, since there are two HSNs within each animal and not every HSN is affected, one region (light pink) represents the wild-type HSN that remains unaffected in animals containing a second undermigrated HSN, which is represented by either the reddish/purple, green, or blue region. ***P < 0.001, N.S., not significant (z-test). For all figures, error bars represent standard error of the proportion (SEP) for the entire stacked column.
The zfp-1(op481) allele was originally isolated in a mutagenesis screen performed to identify genetic enhancers of a null mutation in the heparan sulfate proteoglycan core protein SDN-1, which is involved in D-type motor axon guidance (Gysi et al. 2013). Moreover, the op481 allele was shown to exhibit an identical axon guidance phenotype as the ok554 allele of zfp-1 (Gysi et al. 2013). Surprisingly, however, the axon guidance phenotype in the zfp-1(ok554); sdn-1(zh20) double mutant was dependent upon the presence of the integrated transgene used to visualize the axon guidance defects (Gysi et al. 2013). Therefore, to exclude the possibility that the tph-1::gfp transgene was enhancing or causing the zfp-1 mutant undermigration phenotype, we stained wild-type worms and zfp-1 mutant worms with an anti-serotonin antibody (Garriga et al. 1993). The extent of HSN undermigration was similar in stained and transgenic worms, thereby ruling out the above possibility (Supporting Information, Figure S1, A–C). These results also suggest that ZFP-1 specifically affects the process of HSN migration, rather than cell fate, since all undermigrated HSNs showed expression of the neurotransmitter serotonin, a late step in HSN maturation (Desai et al. 1988). Therefore, the phenotype of the zfp-1 loss-of-function mutant is similar to the HSN defects that we observed in daf-16 null mutants (Kennedy et al. 2013).
Long isoform of ZFP-1 is required for HSN migration
There are two isoforms of the ZFP-1 protein (Figure 1D): a long isoform that contains two PHD fingers, an octapeptide motif (OM) and a leucine zipper motif (LZ), and a shorter isoform that lacks the PHD fingers but retains the OM and LZ (Avgousti et al. 2013). The OM-LZ motifs present in both the long and the short isoforms have been shown to be required for MLL-AF10-mediated oncogenesis (DiMartino et al. 2002), whereas the PHD fingers present in the long isoform of ZFP-1 have recently been shown to be required for early development of C. elegans independent of the OM-LZ motifs (Avgousti et al. 2013). Moreover, it was demonstrated that the long isoform of ZFP-1 is preferentially expressed in the germline and embryos and that the short isoform is highly expressed at larval stages (Avgousti et al. 2013). Also, there is evidence that the long isoform of ZFP-1 binds to the pdk-1 promoter during embryogenesis, when HSN migration takes place (Avgousti et al. 2013). Therefore, we sought to determine if the long isoform of ZFP-1 was required for HSN migration.
First, we made use of two different zfp-1 fosmid constructs that express recombinant ZFP-1 protein isoforms tagged with either GFP or FLAG at the C-terminal portions of the proteins (Mansisidor et al. 2011) (Figure 1D) to rescue the zfp-1(ok554) HSN undermigration phenotype. These constructs contain all the regulatory sequences present at the zfp-1 locus and recapitulate endogenous patterns of expression of ZFP-1 isoforms (Avgousti et al. 2013). We found that both fosmid constructs significantly rescued the HSN undermigration defects present in the zfp-1(ok554) mutant (Figure 1E). Next, to clarify which isoform(s) were required for rescue, we made use of another zfp-1 fosmid that introduces a FLAG tag downstream of the PHD fingers followed by a stop codon, thereby leaving only the larva-enriched short isoform intact (Avgousti et al. 2013) (Figure 1D). In contrast to the fosmids that express both isoforms, this construct was unable to rescue the HSN undermigration defects present in the zfp-1 mutant (Figure 1E). Moreover, overexpressing the N-terminal fragment of the ZFP-1 long isoform (PHD1-PHD2::FLAG), which is identical to the truncated fragment retained in the zfp-1(ok554) mutant (Figure 1D) (Avgousti et al. 2013), in the zfp-1(ok554) mutant background does not affect the penetrance of the HSN undermigration phenotype (Figure 1E). This result indicates that the residual fragment expressed in the zfp-1(ok554) mutant does not have neomorphic properties responsible for the HSN migration phenotype observed in the zfp-1(ok554) mutant because increased dosage of the fragment does not lead to increased severity of the phenotype. On the contrary, previously reported data demonstrate that reducing the dosage of the PHD-finger domain is lethal (Avgousti et al. 2013). Importantly, it was also previously shown that the lethality of animals containing one copy of the zfp-1(ok554) allele placed in trans to a deficiency chromosome (nDf17) was rescued by the fosmid expressing the PHD finger region (PHD1-PHD2::FLAG) (Avgousti et al. 2013), which indicates that the transgenic construct PHD1-PHD2::FLAG is functional. Combined, these results indicate that the short isoform or the increased dosage of the PHD1-PHD2 portion of the long isoform are not sufficient to rescue the HSN migration defects present in the zfp-1(ok554) mutant and that the presence of the long isoform containing both the PHD fingers and the C terminus of the protein is required for proper HSN migration.
ZFP-1 works both parallel to and through DAF-16 to control HSN migration
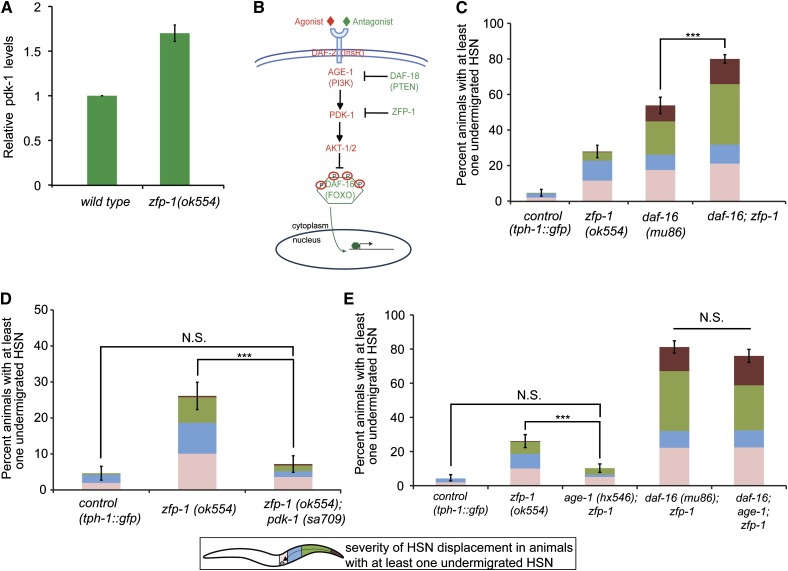
ZFP-1 has been shown to negatively regulate pdk-1 expression at multiple developmental stages (Mansisidor et al. 2011), including the embryo (Figure 2A). To determine whether the HSN undermigration phenotype present in zfp-1 loss-of-function mutants was due to a de-repression of pdk-1 and increased IIS leading to decreased nuclear DAF-16 activity (Figure 2B), we performed an epistasis analysis between zfp-1 and IIS pathway components. First, we created a double-mutant strain between zfp-1(ok554) and the null allele daf-16(mu86) (Lin et al. 1997) to establish whether DAF-16 is the sole output of ZFP-1 involved in HSN migration. We observed a significant enhancement of the daf-16(mu86) HSN undermigration phenotype, indicating that the undermigration phenotype caused by a loss of zfp-1 activity may not be due exclusively to decreased DAF-16 activity (Figure 2C).
Figure 2.
ZFP-1 functions through DAF-16 and in parallel to DAF-16 to regulate HSN migration. (A) Relative pdk-1 mRNA expression in mixed embryos. mRNA expression of pdk-1 in zfp-1(ok554) and wild-type worms was measured by RT-quantitative PCR and normalized to wild type. Actin (act-3) was used as an internal control. Results of two biological replicas are shown; error bars represent standard deviation. (B) Schematic of the IIS pathway affected by ZFP-1. When pathway components in red are active, DAF-16 is phosphorylated and inactive due to its retention in the cytoplasm. When pathway components in green are active, DAF-16 is no longer phosphorylated and can translocate into the nucleus to regulate its target genes. (C) The zfp-1(ok554) mutant enhances the daf-16(mu86) null mutant phenotype. Quantification of the percentage of animals with an undermigrated HSN from at least two pooled independent experiments per strain in control tph-1::gfp (n = 129) vs. zfp-1(ok554) (n = 165), daf-16(mu86) (n = 117), and daf-16(mu86); zfp-1(ok554) (n = 275) mutant animals. (D) A pdk-1 loss-of-function mutant suppresses the HSN undermigration defects in zfp-1 mutant animals. Quantification of the percentage of animals with an undermigrated HSN from two pooled independent experiments per strain in control tph-1::gfp (n = 129), zfp-1(ok554) (n = 134), and zfp-1(ok554); pdk-1(sa709) (n = 125) mutant animals. Note that pdk-1(sa709) animals do not exhibit HSN migration defects at 20 °C (not shown). (E) An age-1(hx546) loss-of-function mutant suppresses the HSN undermigration defects in zfp-1 mutant animals in a DAF-16-dependent manner. Quantification of the percentage of animals with an undermigrated HSN from two pooled independent experiments per strain in control tph-1::gfp (n = 129) vs. zfp-1(ok554) (n = 134), age-1(hx546); zfp-1(ok554) (n = 146), and daf-16(mu86); age-1(hx546); zfp-1(ok554) (n = 125). Note that age-1(hx546) animals do not exhibit HSN migration defects at 20 °C (not shown). HSNs were visualized with a tph-1::gfp reporter. See Figure 1 for detailed description of worm schematic legend. ***P <0.001, N.S., not significant (z-test). Error bars represent SEP.
Next, we asked whether or not the HSN undermigration defects in zfp-1(ok554) could be contributed, in part, to reduced DAF-16 activity due to increased pdk-1 expression. We performed epistasis analysis between the pdk-1(sa709) loss-of-function mutant (Paradis et al. 1999; Mansisidor et al. 2011) and the zfp-1(ok554) mutant to determine whether decreasing pdk-1 expression in the zfp-1 mutant background could suppress the HSN undermigration defects. We observed full suppression of the HSN undermigration defects in the double mutant (Figure 2D), suggesting that repression of pdk-1 by ZFP-1 plays a significant role in regulating HSN migration.
To further demonstrate that enhanced IIS in the zfp-1(ok554) mutant is responsible for inhibiting HSN migration, we also performed epistasis analysis between zfp-1(ok554) and age-1(hx546)/PI3K reduction-of-function mutants (Friedman and Johnson 1988; Tissenbaum and Ruvkun 1998). Similar to our analysis of pdk-1 and zfp-1 mutants, we also observed a full suppression of HSN undermigration defects in age-1; zfp-1 double mutants (Figure 2E). To determine whether the suppression of the zfp-1 mutant phenotype in IIS pathway mutants was due to a release of DAF-16 inhibition, we created a triple mutant: daf-16(mu86); age-1(hx546); zfp-1(ok554) to remove daf-16 activity from the age-1; zfp-1 double mutant. We determined that a loss of daf-16 activity in the age-1; zfp-1 double-mutant background now prevented the suppression of the zfp-1(ok554) mutant phenotype by age-1(hx546) (Figure 2E). Together, these results suggest that the negative regulation of the IIS pathway by ZFP-1 contributes to the control of HSN migration in a DAF-16-dependent manner.
Since the phenotype of the zfp-1(ok554) allele is fully suppressed by mutations in pdk-1 and age-1 (Figure 2, D and E), we can attribute the HSN undermigration seen in the zfp-1(ok554) mutant solely to reduced DAF-16 activity. However, the genetic enhancement observed in the daf-16; zfp-1 double mutant reveals an additional function for ZFP-1 in parallel with DAF-16, which becomes more essential for proper HSN migration in the absence of DAF-16. Given that both DAF-16 and ZFP-1 regulate transcription, it is likely that they are involved in the regulation of common downstream target genes, which ultimately affect HSN migration.
zfp-1(ok554) limits DAF-16 nuclear localization in the embryonic hypodermis
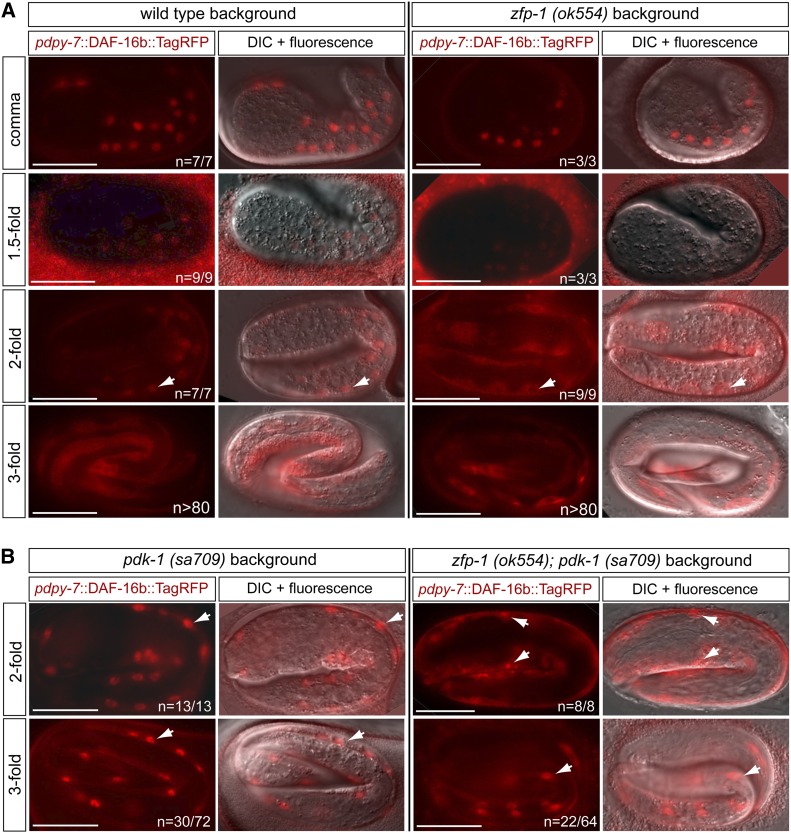
To investigate the effect of the negative regulation of pdk-1 by ZFP-1 on DAF-16 subcellular localization during HSN migration, we examined expression of the DAF-16b::TagRFP protein from a transgene driven by the dpy-7 hypodermal promoter (Kennedy et al. 2013) in the zfp-1(ok554) mutant background. Previously, we showed that DAF-16 localizes to the hypodermal nuclei in the embryo to promote HSN migration (Kennedy et al. 2013). Therefore, we reasoned that if negative regulation of pdk-1 by ZFP-1 during HSN migration contributes to inhibition of IIS to promote nuclear DAF-16 activity, then, in the zfp-1 mutant, nuclear localization of DAF-16 should be affected. We find that, in the wild-type background, hypodermally expressed DAF-16 is predominantly nuclear during the comma, 1.5-, and 2-fold stages, when HSN migration is occurring (Sulston et al. 1983; Pan et al. 2006), and then it becomes cytoplasmically and/or perinuclearly localized by the 3-fold stage (Figure 3A, left panels). In fact, we have recently reported the finding that dynamic nuclear localization of DAF-16 takes place in the embryonic hypodermal tissue during normal development (Kennedy et al. 2013). In the zfp-1(ok554) mutant background, although hypodermally expressed DAF-16 is initially nuclearly localized during the comma and 1.5-fold stage (Figure 3A, right panels), it becomes restricted from the nucleus prematurely by the 2-fold stage (Figure 3A, right panels). This result suggests that the change in DAF-16 nuclear localization at the 2-fold stage is responsible for the HSN migration defects observed in the zfp-1(ok554) mutant.
Figure 3.
Mislocalization of hypodermally expressed DAF-16 in the zfp-1(ok554) mutant is partially restored by loss of pdk-1. (A) In wild-type animals, DAF-16b::TagRFP expressed from a hypodermal promoter is localized to the hypodermal nuclei during the comma, 1.5-, and 2-fold embryonic stages when HSN migration is occurring and is mostly cytoplasmic or perinuclear during the 3-fold stage (left panels). In the zfp-1(ok554) mutant, DAF-16b::TagRFP fails to persist in the nuclei during the 2-fold stage when HSN migration is still occurring (right panels). Arrows in the 2-fold panels point to hypodermal nuclei to illustrate that DAF-16b::TagRFP is nuclearly localized at this stage in wild-type embryos but not in zfp-1(ok554) mutant embryos. (B) In the pdk-1(sa709) loss-of-function mutant, DAF-16b::TagRFP persists in the hypodermal nuclei from the 2-fold stage into the 3-fold stage in 30 of 72 embryos (left panels). The pdk-1(sa709) mutation partially suppresses the absence of DAF-16 nuclear location in zfp-1(ok554) mutant embryos during the 2-fold stage (right panels). Arrows in the 2-fold panels point to hypodermal nuclei to illustrate that DAF-16b::TagRFP is nuclearly localized in the pdk-1(sa709) mutant embryos and shows significant nuclear localization in the zfp-1(ok554); pdk-1(sa709) mutant embryos. Arrows in the 3-fold panels highlight examples of hypodermal nuclei in embryos in which DAF-16b::TagRFP persists after HSN migration has ended. Bars, 20 μm.
As predicted by our epistasis analysis between pdk-1 and zfp-1 for the HSN undermigration phenotype (Figure 2D), the pdk-1(sa709) loss-of-function mutant also suppresses the change in DAF-16 nuclear localization in zfp-1(ok554) mutant embryos (Figure 3, A and B, right panels). First, we confirmed that, consistent with a release of DAF-16 nuclear inhibition under conditions of reduced insulin/IGF-1 signaling, the DAF-16 nuclear localization is more prominent in the pdk-1 (sa709) loss-of-function mutant compared to wild-type embryos (Figure 3, A and B, left panels). Specifically, we find that DAF-16 remains nuclear localized throughout the threefold stage in 30 of 72 pdk-1(sa709) embryos (42%) (Figure 3B, left panels) compared to 0 of >80 wild-type embryos (Figure 3A, left panels). Next, we determined that in zfp-1(ok554); pdk-1(sa709) double-mutant two- and threefold embryos that the subcellular localization of DAF-16 is shifted toward greater nuclear localization when compared to the zfp-1(ok554) single mutant (Figure 3, A and B, right panels). The loss-of-function pdk-1 mutant does not fully suppress the absence of DAF-16 nuclear localization observed in zfp-1(ok554) twofold embryos (Figure 3A, right panels) when compared to localization in wild-type embryos at this stage (Figure 3A, left panels) since both nuclear and cytoplasmic localization is often observed in the same embryo in zfp-1(ok554); pdk-1(sa709). However, we reason that even partial restoration of nuclear localization of DAF-16 at the twofold stage in the zfp-1(ok554) mutant is sufficient to activate downstream DAF-16 target genes required for HSN migration. We also note that a significant number (33%) of zfp-1(ok554); pdk-1(sa709) embryos exhibit the persistence of nuclear DAF-16 at the threefold stage (Figure 3B, right), which is consistent with pdk-1(sa709) being epistatic to zfp-1(ok554).
In total, these results suggest that ZFP-1 functions nonautonomously in the hypodermis to negatively regulate IIS and modulate DAF-16 nuclear localization during HSN migration. Consistently, we also find that the cDNA of the long isoform of zfp-1 fused to GFP driven by the unc-86 promoter, which is expressed in a subset of neurons including the HSN (Baumeister et al. 1996), does not rescue the HSN undermigration defects seen in zfp-1(ok554) mutants (Figure S2, A and B). Interestingly, a conserved microRNA has also been recently reported to function nonautonomously in the hypodermis to regulate genes required for proteoglycan biosynthesis to ensure proper HSN migration (Pedersen et al. 2013). Overall, the action of ZFP-1 in controlling DAF-16 dynamics in the hypodermis that we describe here complements well our earlier findings of the nonautonomous roles of both DAF-16 and DAF-18 (PTEN) in regulating HSN migration (Figure 2B) (Kennedy et al. 2013).
RNAi factors RDE-4, DRH-3, and CSR-1 regulate HSN migration
A genome-wide expression study has revealed that ZFP-1 and RDE-4, a dsRNA-binding protein required for the initiation of exogenous RNAi (Parrish and Fire 2001; Tabara et al. 2002), regulate close to 250 overlapping genes (Grishok et al. 2008). Consistent with this finding, both RDE-4 and ZFP-1 have been shown to affect longevity and the stress response by negatively regulating the conserved IIS kinase pdk-1 (Mansisidor et al. 2011). Therefore, we examined a null allele of rde-4 (Tabara et al. 2002), as well as mutants in 11 other RNAi components that represent multiple small RNA pathways [i.e., CSR-1, WAGO, ERI (enhanced RNA1), microRNA, and nuclear and exogenous RNAi (reviewed in Grishok 2013)] for defects in HSN migration (Table 1). We crossed RNAi mutants to a tph-1::gfp reporter strain and identified three mutants that display HSN migration defects: the rde-4(ne299) null mutant (Tabara et al. 2002), the drh-3(ne4253) loss-of-function mutant (Gu et al. 2009), and a csr-1(tm892) partially rescued strain (Claycomb et al. 2009) (Table 1 and Figure 4, A and B). Strikingly, the highest percentage of undermigration defects was observed in rde-4 null mutants, with almost all affected animals exhibiting migration defects in both HSNs, as shown by the relatively small proportion of HSNs in the wild-type position (Figure 4B, pink in stack). Also, in addition to the undermigration defect observed in 16% of the drh-3(ne4253) mutant animals (Figure 4B), we also noted a smaller percentage of animals with an overmigration defect (∼9%). drh-3 encodes a dicer-related helicase, which participates in multiple endogenous RNAi pathways in C. elegans (Gu et al. 2009), and csr-1 encodes an Argonaute, which binds 22G-RNAs antisense to protein-coding genes (Claycomb et al. 2009; Gu et al. 2009). Consistent with only a reduction of function in the drh-3 and csr-1 mutants, fewer animals exhibited HSN undermigration defects compared to the rde-4 null mutant (Figure 4B). We used a drh-3 loss-of-function allele since null mutants are infertile (Gu et al. 2009). Since csr-1(tm892) mutants are also sterile, we used a partially rescued transgenic strain, where CSR-1 is expressed in the germline to produce 38% viable progeny (Claycomb et al. 2009) that we could analyze for migration defects.
Table 1. RNAi mutants surveyed for HSN migration defect.
| RNAi pathway gene | Class | Nature of mutation | HSN migration defect? |
|---|---|---|---|
| alg-1 (gk214) | Argonaute | Loss of functiona | No |
| C04f12.1 (tm892) | Argonaute | Loss of functionb | No |
| csr-1 (tm892) | Nuclear Argonaute | Partially rescued mutantc | Yes |
| drh-3 (ne4253) | Helicase | Loss of functiond | Yes |
| ergo-1 (tm1860) | Argonaute | Loss of functionb | No |
| nrde-3 (gg066) | Nuclear Argonaute | Nulle | No |
| rde-1 (ne300) | Argonaute | Putative nullf | No |
| rde-3 (ne298) | Nucleotidyl-transferase | Loss of functionf | No |
| rde-4 (ne299) | dsRNA binding | Nullf | Yes |
| rrf-1 (pk1417) | RdRP | Nullg | No |
| rrf-2 (ok210) | RdRP | Nullh | No |
| rrf-3 (pk1426) | RdRP | Nullg | No |
HSNs were visualized by crossing a tph-1::gfp reporter into individual mutant strains.
Deletion allele provided by the C. elegans Gene Knockout Project. The deletion covers part of the expected promoter region and the first two exons.
Figure 4.
RNAi pathway genes rde-4, drh-3, and csr-1 affect HSN migration. (A) Differential interference contrast image of adult hermaphrodite (top) and corresponding epifluorescent image of the HSN (bottom) in an adult rde-4(ne299) mutant animal. HSNs are visualized with a tryptophan hydroxylase GFP reporter (tph-1::gfp). Image is oriented with the posterior of the animal to the right. Asterisk indicates the vulva and the arrowhead denotes the position of a properly migrated HSN. The arrow illustrates a HSN that has failed to migrate the full distance from its birthplace in the tail. (B) Quantification of the percentage of animals with an undermigrated HSN from a minimum of three pooled independent experiments per strain in tph-1::gfp (n = 127) and cat-4::gfp (n = 127) controls vs. rde-4(ne299) (n = 211), drh-3(ne4253) (n = 376), and the partially rescued csr-1(tm982) (n = 342) mutant animals. Note that the cat-4::gfp reporter was used to visualize the HSNs in the partially rescued csr-1(tm892) strain. (C) Placing rde-4(ne299) over a deficiency allele, nDf17, also resulted in HSN undermigration defects. Quantification of the percentage of animals with an undermigrated HSN in control animals [rde-4(ne299)/qC1] (n = 22) vs. rde-4(ne299)/nDf17 animals (n = 26). (D) RDE-4 works in parallel to DAF-16 to regulate HSN migration. Quantification of the percentage of animals with an undermigrated HSN from a minimum of two pooled independent experiments per strain in control tph-1::gfp (n = 127) vs. daf-16(mu86) (n = 150), rde-4(ne299) (n = 211) and daf-16(mu86); rde-4(ne299) (n = 270) mutant animals. (E) A pdk-1 loss-of-function mutant does not suppress rde-4 null HSN undermigration defects. Quantification of the percentage of animals with an undermigrated HSN in control tph-1::gfp (n = 64) vs. rde-4(ne299) (n = 50), rde-4(ne299); pdk-1(sa709) (n = 60). (F) An age-1 loss-of-function mutant does not suppress rde-4 null HSN undermigration defects. Quantification of the percentage of animals with an undermigrated HSN in control tph-1::gfp (n = 50) vs. rde-4(ne299) (n = 50) and age-1(hx546); rde-4(ne299) mutant animals. HSNs were visualized with a tph-1::gfp reporter. See Figure 1 for detailed description of worm schematic legend. ***P < 0.001, N.S., not significant (z-test). Error bars represent SEP.
To further confirm that the HSN undermigration defect that we observed in rde-4 null mutants is due to mutations in rde-4 and not to the background of the strain, we placed the rde-4(ne299) allele over a deficiency allele, nDf17, which is balanced by chromosome qC1 [dpy-19(e1259) glp-1(q339)] (Edgley et al. 2006). We crossed rde-4(ne299) males to nDf17/qC1 animals and scored F1 animals for HSN migration defects. We determined the genotypes of the F1 animals by the presence or absence of the qC1 phenotypes, which are dumpy and sterile, segregating in the F2 generation. Only F1 animals carrying the deficiency allele nDf17 displayed penetrant HSN undermigration defects (Figure 4C). Moreover, we also stained rde-4(ne299) worms with an anti-serotonin antibody (Garriga et al. 1993) to further exclude the possibility that the HSN undermigration defects observed in rde-4 null animals were the result of transgene background effects. We observed HSN undermigration defects in stained worms without the tph-1::gfp transgene (Figure S3, A and B). Similar to zfp-1 mutants, rde-4 mutants are not affected in the production of serotonin since undermigrated HSNs contain the neurotransmitter.
RDE-4 works in parallel to DAF-16 and in the neuronal tissue to regulate HSN migration
Since RDE-4 was shown to inhibit pdk-1 transcription (Mansisidor et al. 2011), we analyzed epistatic relationships between the rde-4 mutant and IIS pathway components to determine if RDE-4, like ZFP-1, also regulates HSN migration in part by inhibiting IIS to promote DAF-16 nuclear localization. First, we combined the rde-4(ne299) null allele with the daf-16(mu86) null allele and observed a significant enhancement of the daf-16 null phenotype (Figure 4D). This result suggests that rde-4 and daf-16 are working in parallel genetic pathways to affect HSN migration.
Since we found that the zfp-1 mutant phenotype can be suppressed by a reduction in pdk-1 and age-1 expression, we also performed epistasis analysis using rde-4(ne299) and either the pdk-1(sa709) mutant or the age-1(hx546) mutant to determine if the HSN undermigration phenotype seen in rde-4(ne299) could also be suppressed by reduced IIS. In contrast to the epistasis experiments with zfp-1, we observed no suppression of the HSN undermigration defects in either double-mutant combination (Figure 4, E and F).
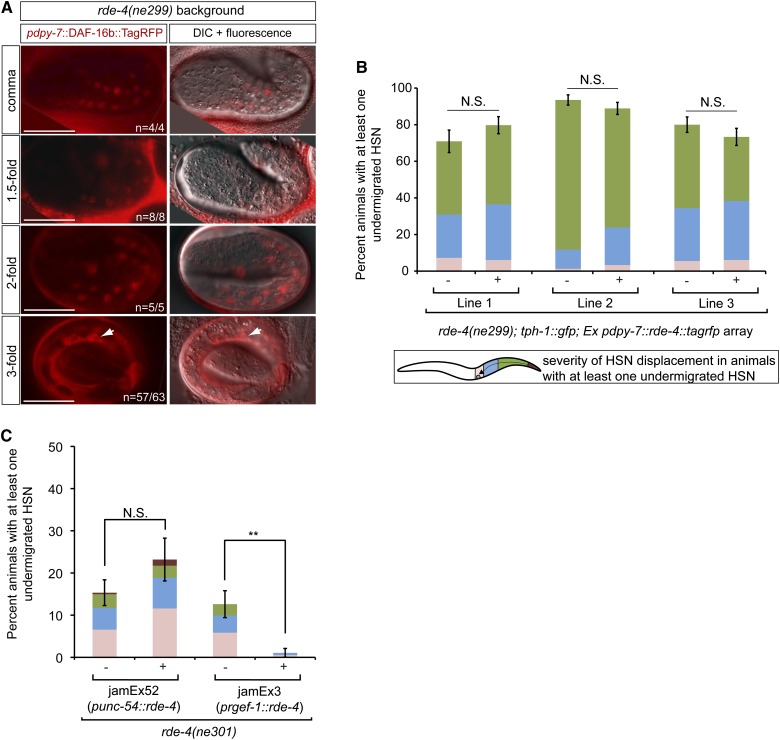
In addition, we have analyzed the cellular distribution of hypodermally expressed DAF-16b::TagRFP in rde-4(ne299) null mutant embryos (Figure 5A) and found it to be similar to that observed in the wild-type background (Figure 3A, left panels): DAF-16 is predominantly nuclear during the comma, 1.5-, and 2-fold stages when HSN migration is occurring before becoming more restricted to the cytoplasm (Figure 5A). The absence of any alterations in DAF-16 nuclear localization during HSN migration in the rde-4 mutant background compared to wild type is consistent with the lack of suppression that we observe for the HSN undermigration defect in the rde-4(ne299); pdk-1(sa709) double mutant (Figure 4E). Combined, these results indicate that RDE-4 does not contribute to the regulation of HSN migration by promoting hypodermal DAF-16 nuclear localization during embryogenesis. Consistently, we also find that the cDNA of rde-4 fused to TagRFP driven by the hypodermal promoter dpy-7 fails to rescue the HSN undermigration defects seen in the rde-4 null mutant (Figure 5B).
Figure 5.
RDE-4 functions in neurons to promote HSN migration. (A) In rde-4(ne299) null animals, the distribution of DAF-16b::TagRFP expressed from a hypodermal promoter is unchanged when compared to wild-type embryos (see Figure 3A, left panels). The distribution remains localized to the hypodermal nuclei during the comma, 1.5-, and 2-fold embryonic stages when HSN migration is occurring and is mostly cytoplasmic or perinuclear during the 3-fold stage. Note that in a few 3-fold embryos (6/63), we observed persistent nuclear localization of DAF-16b::TagRFP. The arrows in the 3-fold panels highlight the cytoplasmic/perinuclear localization of DAF-16b::TagRFP in the remaining 57 of the 63 embryos. Bars, 20 μm. (B) Expression of RDE-4 driven by the dpy-7 promoter does not rescue the HSN undermigration defect in rde-4(ne299) mutants. The rde-4(ne299) mutants that had the rescuing array (+) or their siblings that had lost the array (−) were scored. Number of animals scored per line are the following: line 1, (−) n = 55 and (+) n = 74; line 2, (−) n = 77 and (+) = 90; line 3, (−) n = 90 and (+) = 90. (C) Expression of RDE-4 driven by a pan-neuronal promoter (rgef-1) but not a body-wall muscle promoter (unc-54) rescues the HSN migration defect of rde-4(ne301) mutants. The rde-4(ne301) mutants that carried either the punc-54::rde-4 or prgef-1::rde-4 array (+) and their corresponding siblings that had lost the array (−) were scored. Number of animals scored for the punc-54::rde-4-containing strain are the following: (+) n = 69 and (−) n = 137. Number of animals scored for the prgef-1::rde-4-containing strain are the following: (+) n = 93 and (−) n = 111. HSNs were visualized by anti-serotonin staining. See Figure 1 for detailed description of worm schematic legend. **P <0.01, N.S., not significant (z-test). Error bars represent SEP.
To determine the site of action of RDE-4 in HSN migration, we utilized two different functional tissue-specific rde-4 transgenes (A. Jose, personal communication). The rde-4 cDNA was driven by either a body-wall muscle promoter, unc-54, or a pan-neuronal promoter, rgef-1, in the rde-4(ne301) null mutant background (strains were generously provided by the A. Jose lab). Expression of rde-4 cDNA in the neuronal tissue but not in the body-wall muscle rescued the HSN migration defects of rde-4 mutants (Figure 5C). This result suggests that RDE-4 functions cell-autonomously to control HSN migration and is in agreement with our findings that RDE-4 does not function in the same genetic pathway or tissue as DAF-16.
We observed that the HSN undermigration defect is less severe in the genetic background of the two rde-4 mutant strains expressing the extrachromosomal muscle- and neuronal-specific transgenes (Figure 5C) compared to the strains used throughout the rest of this study. We have used strains containing the rde-4(ne299) allele while the strains from the Jose lab contain the rde-4(ne301) allele. Although these two alleles contain an identical lesion and were isolated from a clonal population during a screen for RNAi-defective mutants (Tabara et al. 1999), they have been maintained and outcrossed independently. As a result, they may have slightly different genetic backgrounds accounting for the variability in HSN undermigration defects observed between the two rde-4 alleles.
Discussion
Our genetic analysis has identified the conserved chromatin-binding factor ZFP-1/AF10, the dsRNA-binding protein RDE-4, the dicer-related helicase DRH-3, and the Argonaute CSR-1 as regulators of neuronal migration during C. elegans embryogenesis. We demonstrate that ZFP-1 facilitates proper HSN migration in part through its negative regulation of the conserved IIS kinase gene pdk-1 (Mansisidor et al. 2011) and the resulting activation of DAF-16/FOXO, which we have previously shown to act in the hypodermis to promote HSN migration (Kennedy et al. 2013). We also show that both ZFP-1 and RDE-4 work in parallel to DAF-16 to stimulate HSN migration during embryogenesis.
Recently, the mammalian homolog of ZFP-1, AF10, was shown to exist in a complex with the H3K79 histone methyltransferase DOT1L, which was called DotCom (Mohan et al. 2010). Interestingly, β-catenin, an important downstream mediator of Wnt signaling, was shown to co-immunoprecipitate with DOT1L complex components in HEK293T cells (Mohan et al. 2010). Moreover, reduced expression of Wingless/Wnt targets was observed after knockdown of the fly AF10 ortholog Alhambra (or Dalf), as well as after the knockdown of other DotCom orthologs, in Drosophila larvae (Mohan et al. 2010). Additionally, AF10 was independently found in a complex with the downstream effectors of Wnt signaling TCF4 and β-catenin in colorectal cancer cells and implicated in the regulation of Wnt target genes in these cells as well as in zebrafish (Mahmoudi et al. 2010).
In C. elegans, it is well known that Wnt signaling controls HSN migration in a β-catenin-independent manner (Harris et al. 1996; Forrester et al. 2004) by acting as a repellent to guide the HSN out of the embryonic tail and into the mid-body of the animal (Pan et al. 2006). However, the connection between AF10 and canonical Wnt signaling (β-catenin-dependent) in vertebrates highlights the intriguing possibility that the interaction between ZFP-1/AF10 and Wnt pathway components may be conserved in C. elegans. Based on our observations that the negative regulation of pdk-1 by ZFP-1 affects DAF-16 hypodermal nuclear localization during HSN migration and that an HSN-specific promoter driving the expression of a ZFP-1::GFP transgene fails to rescue the HSN undermigration defects in zfp-1 mutant animals, it is likely that ZFP-1 functions nonautonomously to promote HSN migration. One possibility is that ZFP-1, independently or in combination with DAF-16, regulates the transcription of the Wnt’s, which affect HSN migration nonautonomously (Pan et al. 2006). Another possibility is that ZFP-1 regulates the transcription of negative regulators of Wnt signaling, such as CAM-1, that can function as sinks to limit extracellular Wnt’s and impede HSN migration (Kim and Forrester 2003; Forrester et al. 2004; Green et al. 2007). Future studies will elucidate these potential roles of ZFP-1 in the regulation of Wnt signaling during HSN migration.
There are multiple endo-siRNA pathways in C. elegans, including the CSR-1 RNAi pathway (Claycomb et al. 2009; Gu et al. 2009), the WAGO RNAi pathway (Gu et al. 2009), and the ERI RNAi pathway (Lee et al. 2006; Gent et al. 2009; Han et al. 2009; Vasale et al. 2010). The biological roles of the endo-RNAi pathways in C. elegans remain largely uncharacterized. A major known role of the WAGO endo-RNAi pathway is in genome surveillance: the silencing of transposable elements, aberrant transcripts (Gu et al. 2009), and gene duplications (Vasale et al. 2010; Fischer et al. 2011). The ERI pathway has been implicated in regulating genes required for spermatogenesis and fertility (Gent et al. 2009; Han et al. 2009; Conine et al. 2010), whereas the CSR-1 RNAi pathway has been recently shown to positively regulate histone mRNA expression (Avgousti et al. 2012).
Mutations in rde-4 have been associated with decreased life span (Welker et al. 2007; Mansisidor et al. 2011) and increased sensitivity to oxidative stress (Mansisidor et al. 2011) and temperature (Blanchard et al. 2011). An increase in stress sensitivity and reduced life span of rde-4 mutants has been attributed to the upregulation of pdk-1 (Mansisidor et al. 2011). Here, we describe a novel role for RDE-4 in embryonic neuronal migration, which is consistent with previous observations that the generation of some somatic 22G-RNAs is dependent on RDE-4 (Lee et al. 2006; Gu et al. 2009). Interestingly, RDE-4 has also been recently shown to participate in the production of endogenous 22G-RNAs required to mediate odor adaptation in adult animals (Juang et al. 2013).
The majority of 22G-RNAs that are in the complex with the Argonaute CSR-1 are antisense to protein-coding genes (Claycomb et al. 2009), suggesting that multiple genes and genetic pathways may be regulated by these endo-siRNAs. Similar to the regulation of embryonic HSN migration described here, both CSR-1 and DRH-3 have been implicated in another embryonic process: the specification of the excretory duct cell, a component of the worm’s renal system (Rocheleau et al. 2008). A genome-wide RNAi screen identified these RNAi pathway genes to act redundantly with the KSR-1 (kinase suppressor of Ras) scaffolding protein in Ras-mediated excretory duct cell specification (Rocheleau et al. 2008). Since DRH-3 is a component of the RdRP complex (Gu et al. 2009; Thivierge et al. 2012) required for the biogenesis of all 22G-RNAs, including those present in the complex with CSR-1 (Gu et al. 2009), it is most likely that DRH-3-dependent CSR-1-bound endo-siRNAs regulate a number of genes affecting developmental events, such as HSN migration and excretory duct specification.
Furthermore, RDE-4 and Dicer have been implicated in the production of 26G-RNAs, which subsequently promote RdRP-dependent, Dicer-independent biogenesis of some WAGO-bound 22G-RNAs (Vasale et al. 2010). Therefore, it is possible that RDE-4 works upstream of some CSR-1 22G-RNAs that regulate genes affecting HSN migration. Surprisingly, none of the RdRPs implicated in 26G-RNA and 22G-RNA production (Ruby et al. 2006; Aoki et al. 2007; Pak and Fire 2007; Vasale et al. 2010) displayed an HSN migration phenotype when examined as null mutants (Table 1). One possible explanation for this is redundancy among the RdRPs required for HSN migration.
In summary, our work has highlighted a novel biological role for the conserved chromatin-binding factor ZFP-1/AF10 and the endo-siRNA pathway in neuronal migration in C. elegans.
Supplementary Material
Acknowledgments
We thank I. Greenwald, O. Hobert, and members of the Grishok lab for helpful discussions; G. Cecere and A. Mansisidor for generating the fosmid-based transgenic strains; M. Hengartner and S. Gysi for providing the zfp-1(op481) allele and for sharing data before publication; A. Jose for providing rde-4 transgenic strains; X. Zhou for help in generating plasmid-based transgenic lines; and I. Greenwald for a generous offer of technical assistance by her staff. The zfp-1(ok554) and rrf-2(ok210) strains were provided by the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation, which is part of the International C. elegans Gene Knockout Consortium. Some strains were provided by the Caenhorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 0D010440). The ergo-1(tm1860), C04F12.1(tm1637), and csr-1(tm892) alleles were generated by the National Bioresource Project (Japan). This work was supported by Hormones: Biochemistry and Molecular Biology Training Grant T32 DK0007328 and Genetics and Development Training Grant T32 GM007088 (to L.M.K.) and NIH Director’s New Innovator Award (1DP2OD006412-01) and the Irma T. Hirschl Career Scientist Award (to A.G.).
Footnotes
Communicating editor: P. Sengupta
Literature Cited
- Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H., 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26: 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti D. C., Palani S., Sherman Y., Grishok A., 2012. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 31: 3821–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti D. C., G Cecere, and A Grishok, 2013. The conserved PHD1-PHD2 domain of ZFP-1/AF10 is a discrete functional module essential for viability in Caenorhabditis elegans. Mol. Cell. Biol. 33: 999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister R., Liu Y., Ruvkun G., 1996. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes Dev. 10: 1395–1410. [DOI] [PubMed] [Google Scholar]
- Blanchard D., Parameswaran P., Lopez-Molina J., Gent J., Saynuk J. F., et al. , 2011. On the nature of in vivo requirements for rde-4 in RNAi and developmental pathways in C. elegans. RNA Biol. 8: 458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G., Hoersch S., Jensen M. B., Dixit S., Grishok A., 2013. The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol. Cell 50: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin T., Ayton P., Bernard O. A., Saha V., Della Valle V., et al. , 1995. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood 85: 1435–1441. [PubMed] [Google Scholar]
- Clark S. G., Chiu C., 2003. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130: 3781–3794. [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Batista P. J., Gu W., Claycomb J. M., Chaves D. A., et al. , 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107: 3588–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Kim E., Han M., 2006. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C., Garriga G., McIntire S. L., Horvitz H. R., 1988. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature 336: 638–646. [DOI] [PubMed] [Google Scholar]
- DiMartino J. F., Ayton P. M., Chen E. H., Naftzger C. C., Young B. D., et al. , 2002. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood 99: 3780–3785. [DOI] [PubMed] [Google Scholar]
- Dudley N. R., Labbe J. C., Goldstein B., 2002. Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA 99: 4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M. L., D. L. Baillie, D. L. Riddle, and A. M. Rose, 2006 Genetic balancers (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.89.1, http//www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Fischer S. E., Montgomery T. A., Zhang C., Fahlgren N., Breen P. C., et al. , 2011. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 7: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Kim C., Garriga G., 2004. The Caenorhabditis elegans Ror RTK CAM-1 inhibits EGL-20/Wnt signaling in cell migration. Genetics 168: 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, D. B., and T. EJohnson, 1988. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga G., Desai C., Horvitz H. R., 1993. Cell interactions control the direction of outgrowth, branching and fasciculation of the HSN axons of Caenorhabditis elegans. Development 117: 1071–1087. [DOI] [PubMed] [Google Scholar]
- Gent J., Schvarzstein M., Villeneuve A. M., Gu S. G., Jantsch V., et al. , 2009. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics 183: 1297–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard J. S., J. D Barry, and I. L Johnstone, 1997. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. L., Inoue T., Sternberg P. W., 2007. The C. elegans ROR receptor tyrosine kinase, CAM-1, non-autonomously inhibits the Wnt pathway. Development 134: 4053–4062. [DOI] [PubMed] [Google Scholar]
- Grishok A., 2013. Biology and mechanisms of short RNAs in Caenorhabditis elegans. Adv. Genet. 83: 1–69. [DOI] [PubMed] [Google Scholar]
- Grishok A., Sinskey J. L., Sharp P. A., 2005. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 19: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Hoersch S., Sharp P. A., 2008. RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105: 20386–20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Vasale J., Batista P. J., et al. , 2009. Distinct Argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Pavelec D. M., Burkhart K. B., Harding S., et al. , 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysi S., Rhiner C., Flibotte S., Moerman D. G., Hengartner M. O., 2013. A network of HSPG core proteins and HS modifying enzymes regulates Netrin-dependent guidance of D-type motor neurons in Caenorhabditis elegans. PLoS ONE 8: e74908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T., Manoharan A. P., Harkins T. T., Bouffard P., Fitzpatrick C., et al. , 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 106: 18674–18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J., Honigberg L., Robinson N., Kenyon C., 1996. Neuronal cell migration in C. elegans: regulation of Hox gene expression and cell position. Development 122: 3117–3131. [DOI] [PubMed] [Google Scholar]
- Juang B. T., Gu C., Starnes L., Palladino F., Goga A., et al. , 2013. Endogenous Nuclear RNAi mediates behavioral adaptation to odor. Cell 154: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy L. M., Pham S. C., Grishok A., 2013. Nonautonomous regulation of neuronal migration by insulin signaling, DAF-16/FOXO, and PAK-1. Cell Rep. 4: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Forrester W. C., 2003. Functional analysis of the domains of the C elegans Ror receptor tyrosine kinase CAM-1. Dev. Biol. 264: 376–390. [DOI] [PubMed] [Google Scholar]
- Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A., et al. , 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Hammell C. M., Ambros V., 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu Y., 2011. Diverse small non-coding RNAs in RNA interference pathways. Methods Mol. Biol. 764: 169–182. [DOI] [PubMed] [Google Scholar]
- Lin K., Dorman J. B., Rodan A., Kenyon C., 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T., Boj S. F., Hatzis P., Li V. S., Taouatas N., et al. , 2010. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/beta-catenin coactivators essential for intestinal homeostasis. PLoS Biol. 8: e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansisidor A. R., Cecere G., Hoersch S., Jensen M. B., Kawli T., et al. , 2011. A conserved PHD finger protein and endogenous RNAi modulate insulin signaling in Caenorhabditis elegans. PLoS Genet. 7: e1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M., Herz H. M., Takahashi Y. H., Lin C., Lai K. C., et al. , 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Fire A., 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244. [DOI] [PubMed] [Google Scholar]
- Pan C. L., Howell J. E., Clark S. G., Hilliard M., Cordes S., et al. , 2006. Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10: 367–377. [DOI] [PubMed] [Google Scholar]
- Paradis S., Ailion M., Toker A., Thomas J. H., Ruvkun G., 1999. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Fire A., 2001. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7: 1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Pedersen M. E., Snieckute G., Kagias K., Nehammer C., Multhaupt H. A., et al. , 2013. An epidermal microRNA regulates neuronal migration through control of the cellular glycosylation state. Science 341: 1404–1408. [DOI] [PubMed] [Google Scholar]
- Rocheleau C., Cullison K., Huang K., Bernstein Y., Spilker A., et al. , 2008. The Caenorhabditis elegans ekl (Enhancer of ksr-1 Lethality) genes include putative components of a germline small RNA pathway. Genetics 178: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Jan C., Player C., Axtell M. J., Lee W., et al. , 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207. [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., et al. , 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., E Schierenberg, J. G White,, and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W. G., Fleenor J., Grishok A., et al. , 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit E., Siomi H., Mello C. C., 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861–871. [DOI] [PubMed] [Google Scholar]
- Thivierge C., Makil N., Flamand M., Vasale J. J., Mello C. C., et al. , 2012. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat. Struct. Mol. Biol. 19: 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H. A., Ruvkun G., 1998. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics 148: 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale J. J., Gu W., Thivierge C., Batista P. J., Claycomb J. M., et al. , 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker N. C., Habig J. W., Bass B. L., 2007. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA 13: 1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C. C., et al. , 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.