Abstract
We develop a maximum penalized-likelihood (MPL) method to estimate the fitnesses of amino acids and the distribution of selection coefficients (S = 2Ns) in protein-coding genes from phylogenetic data. This improves on a previous maximum-likelihood method. Various penalty functions are used to penalize extreme estimates of the fitnesses, thus correcting overfitting by the previous method. Using a combination of computer simulation and real data analysis, we evaluate the effect of the various penalties on the estimation of the fitnesses and the distribution of S. We show the new method regularizes the estimates of the fitnesses for small, relatively uninformative data sets, but it can still recover the large proportion of deleterious mutations when present in simulated data. Computer simulations indicate that as the number of taxa in the phylogeny or the level of sequence divergence increases, the distribution of S can be more accurately estimated. Furthermore, the strength of the penalty can be varied to study how informative a particular data set is about the distribution of S. We analyze three protein-coding genes (the chloroplast rubisco protein, mammal mitochondrial proteins, and an influenza virus polymerase) and show the new method recovers a large proportion of deleterious mutations in these data, even under strong penalties, confirming the distribution of S is bimodal in these real data. We recommend the use of the new MPL approach for the estimation of the distribution of S in species phylogenies of protein-coding genes.
Keywords: fitness effects, selection coefficient, penalized likelihood, mitochondria, chloroplast, influenza
ESTIMATION of the distribution of selection coefficients (S = 2Ns) of new mutations in protein-coding genes is of much interest (Eyre-Walker and Keightley 2007). Theoretical considerations (Akashi 1999, figure 1) and empirical observations of mutation experiments (e.g., Wloch et al. 2001; Sanjuan 2010; Hietpas et al. 2011) find the distribution of S is bimodal: one mode centered around nearly neutral mutations (−2 ≤ S ≤ 2) and the other mode centered around highly deleterious mutations (S ≪ −10). However, phylogenetic-based methods to estimate the distribution of S have failed to recover the bimodal distribution and, in particular, have not detected the large proportion of highly deleterious mutants observed in mutation experiments (Thorne et al. 2007; Yang and Nielsen 2008; Rodrigue et al. 2010; Rodrigue 2013).
In a previous article (Tamuri et al. 2012) we examined a maximum-likelihood (ML) method to estimate the distribution of S over amino acids in protein-coding genes from phylogenetic data. The method is based on the model of Halpern and Bruno (1998) and uses a multiple-sequence alignment of several species to estimate the fitness F of each amino acid at each codon location in a protein-coding gene. We refer to this model as the site-wise mutation–selection (swMutSel) model. The method ignores polymorphism and treats differences among sequences as fixed differences among species; it is thus not suitable for population data. Analysis of computer simulations (Tamuri et al. 2012) showed the method can recover the distribution of S when the distribution is bimodal. Furthermore, analysis of two real data sets (Tamuri et al. 2012) indicated the distribution of S among new mutations is bimodal in the protein-coding genes studied.
The swMutSel model is highly parameterized. For a protein-coding gene of length Lc codons, 19 × Lc site-specific fitnesses are estimated. The large dimension of the estimation problem is challenging, and extreme estimates of the fitnesses (i.e., F = −∞) of some amino acids at some locations are common. Considering this, Rodrigue (2013) argued that the large proportion of deleterious mutants estimated for real data using the swMutSel model is the result of overfitting by the ML method. Rodrigue (2013) suggested a hierarchical Bayesian approach is preferable to estimate parameters in such high-dimensional problems. However, analysis of various protein-coding genes using a mixture model version of swMutSel under such a hierarchical Bayesian approach again did not detect the large proportion of deleterious mutations expected (Rodrigue et al. 2010; Rodrigue 2013). It is unclear whether a strong prior on the fitnesses (for example, a unimodal prior with concentrated probability mass around 0) or a limited number of categories in the mixture model is responsible for the underestimation of the proportion of deleterious mutants.
A limitation of the Bayesian method suggested by Rodrigue (2013) is that the posterior distribution of fitnesses cannot be calculated analytically, and computationally expensive MCMC sampling is necessary to calculate the distribution, therefore limiting the size of analyzed data sets. A fast approach is to use numerical optimization to find the highest mode of the posterior distribution to obtain the maximum a posteriori estimates of the fitnesses. The method is essentially the same as maximum penalized-likelihood (MPL) estimation when the penalty function is a probability density (the prior) on the parameters (Cox and O’Sullivan 1990). Because the MPL method is fast, it can be used on long sequence alignments and phylogenies of hundreds to thousands of species. It incorporates one of the advantages of the Bayesian method, the penalization of extreme estimates, thus correcting overfitting (Cox and O’Sullivan 1990). Furthermore, the strength of the penalty function can be varied to assess how informative a particular data set is about the parameter values. Several phylogenetic problems have been addressed using penalized likelihood (e.g., Nielsen 1997; Sanderson 2002).
Here we develop an MPL approach to estimate the distribution of S under the swMutSel model. Using a combination of computer simulations and real data analysis, we evaluate the effect of various penalty functions on the estimation of the fitnesses and we assess how reliable the estimated distribution of S is for different penalty strengths. We show the new method regularizes the estimates of fitnesses for small, uninformative data sets (i.e., it penalizes extreme fitness estimates), but can still recover a large proportion of deleterious mutations when these are present in simulated data. Furthermore, we study the effects of taxon sampling and sequence divergence on the accuracy of the estimation of the distribution of S. We find accuracy of estimates increases with the number of sequences in the phylogeny or the level of sequence divergence. Results for real data indicate high proportions of deleterious mutations as predicted by theory and as seen in mutation experiments. We recommend the use of the new MPL swMutSel approach for the analysis of real data.
Theory
The site-wise mutation–selection model
For full details of the model and the population genetics assumptions used see Tamuri et al. (2012) (pp. 1103–1104). We assume a protein-coding gene evolving in a Fisher–Wright population with effective gene number N (i.e., the population number is N for haploid and N/2 for diploid organisms). Imagine a new mutant allele in the population with scaled Malthusian fitness F = 2Nf. Selection and random drift will act on the new allele and the mutant will eventually become fixed in the population or lost. If the allele becomes fixed, we say the population has been substituted. We model the substitution process at the codon level in the protein-coding gene. The substitution rate from codon i to j at location k in the gene is
| (1) |
where Sij,k = Fj,k − Fi,k is the selection coefficient for the i to j mutation, and Fi,k and Fj,k are the fitnesses of the two codons. Parameter μij is the neutral mutation rate from i to j, which can be constructed from any standard nucleotide substitution model (e.g., see Yang and Nielsen 2008; Tamuri et al. 2012). The effect of selection is thus to accelerate or slow down the substitution rate with respect to the rate of a neutral mutation. That is, when Sij,k > 0, Sij,k = 0, or Sij,k < 0, then qij,k > μij, qij,k = μij, and qij,k < μij, respectively. Note that Fij,k (and thus Sij,k and qij,k) vary over locations in the protein, while μij is the same for all locations.
We assume codon substitution is a continuous-time Markov process. The qij,k thus form the off-diagonal elements of a rate matrix Qk. The transition probability matrix Pk(t) = exp(tQk) can then be used to calculate the likelihood of a sequence alignment under a given tree topology, using standard methods (Yang 2006). We assume no selection on codon usage, so Fi,k = Fj,k if i and j code for the same amino acid. We use the HKY85 nucleotide substitution model (Hasegawa et al. 1985) to construct μij, so six additional global parameters are necessary: a multiple substitution factor τ, the transition–transversion ratio κ, three nucleotide frequency parameters (π*), and a branch scaling parameter c (see Tamuri et al. 2012 for details). The tree topology and the branch lengths are assumed known (i.e., they can be estimated using quicker methods and fixed during estimation of fitnesses with the swMutSel model).
The equilibrium frequency of codon j at location k is given by
| (2) |
where , and are the equilibrium frequencies of the nucleotides at the three positions of codon j in the absence of selection, and We assume STOP codons are lethal within protein-coding genes (i.e., FSTOP = −∞), and therefore πSTOP = 0.
Let aai be the amino acid encoded by i. At equilibrium, the proportion of nonsynonymous i to j mutations at location k among all nonsynonymous mutations for all locations is
| (3) |
where the sum is over all pairs i ≠ j and the indicator function aai≠aaj = 1 if aai ≠ aaj and = 0 otherwise. The distribution of S among nonsynonymous mutations in the protein-coding gene is given by the distribution of Sij,k values weighted by their corresponding mij,k proportions. We can represent the distribution in histogram form. Writing wI for the width of the Ith histogram bin, the proportion of mutations in the Ith bin (centered on the value SI) is
| (4) |
where the indicator function a<S≤b = 1 if a < S ≤ b and = 0 otherwise.
There has been much discussion in the literature about the relative proportions of deleterious, neutral, and advantageous mutations in protein evolution (e.g., Kimura 1983; Ohta 1992; Akashi 1999), so we study these proportions in detail here. We define an i to j mutation at location k as deleterious if Sij,k < −2, as nearly neutral if −2 ≤ Sij,k < 2, and as advantageous if 2 ≤ Sij,k (Li 1978). The proportions of the three types of mutations are written p−, p0, and p+, respectively. For example, the proportion of advantageous mutations is
| (5) |
We note a few points about estimation of the distribution of S in this work. First, we are interested only in the distribution of S for nonsynonymous mutations. Equations 3 and 4 can be used to calculate the distribution among all mutations if we include synonymous i to j codon mutations in the calculation. However, because we assume synonymous mutations are neutral, a large peak of neutral mutations would be obtained when calculating the distribution (e.g., Tamuri et al. 2012, figure 2). Second, the distribution can also be calculated among substitutions (i.e., those mutations becoming fixed in the population) by using πi,kqij,k instead of πi,kμij in Equation 3. The distribution of S among substitutions is symmetrical; i.e., h(SI) = h(−SI), because the substitution model of Equation 1 is reversible and there is a detailed balance of slightly advantageous/deleterious mutations becoming fixed and lost in the population. We do not consider the distribution of S among substitutions in this work. Third, mutations with S < −10 and those with S > 10 are binned together in the calculation of h(S), so we calculate the distribution of S between −10 and 10. Fourth, we consider mutations toward STOP codons to be nonsynonymous, and they are included in the calculation of h(−10) and p−.
Penalized likelihood
The penalized-likelihood function is
| (6) |
where P(θ) is a penalty function, L(θ) is the likelihood function, and θ are the model parameters. Taking the logarithm on both sides gives the penalized log-likelihood
| (7) |
where p(θ) = log P(θ) and ℓ(θ) = log L(θ). The penalized likelihood is defined up to a proportionality constant, so any constant factors of P(θ) or L(θ) can be ignored. If P(θ) is a probability density on θ, then Equation 6 has a Bayesian interpretation, and finding the maximum penalized-likelihood estimates (MPLE) is equivalent to finding the highest mode of the posterior distribution. Using log-penalty functions of the form p(θ) = λf(θ) is desirable, where λ (>0) is called the regularization parameter. When λ = 0, the problem is reduced to standard maximum-likelihood estimation. Large values of λ regularize the estimates of θ; that is, the estimates become concentrated around the mode of P(θ) and extreme estimates are penalized (Cox and O’Sullivan 1990).
In this work, ℓ(θ) is the log-likelihood of a sequence alignment calculated on a phylogeny using the swMutSel model, and θ are the substitution model parameters. The penalty function depends only on the site-wise fitnesses, Fk = (F1,k, … , F20,k) and the same penalty function is applied to all locations. As only the fitness differences (Sij,k = Fj,k − Fi,k) enter the likelihood function, we fix one of the fitnesses to zero, and thus only 19 fitnesses are estimated for site k. We drop the k subindex and simply write F for the site-specific fitness vector. Below we describe two penalty functions on F, the first based on the multivariate normal distribution and the second based on the Dirichlet distribution.
Multivariate normal penalty:
We use a penalty function proportional to a MVN density
| (8) |
where , and Σ is a 19 × 19 covariance matrix. Ignoring constant factors, we define the log-penalty function as
| (9) |
Note that λ = 1/σ2 is the regularization parameter.
Equation 9 penalizes fitness values as they move away from the fitness mean . With this parameterization we obtain the same penalty value for whichever fitness we decide to fix to zero (which is not true for other MVN penalties). Some tedious algebraic manipulation of the exponent of Equation 8 shows the density has mean vector 0, equal variances v = 2σ2 (the diagonal of Σ), and correlation of 0.5 between any Fi and Fj (the off-diagonal elements of Σ are all 0.5v). Because the mode of the density is at F = 0, extreme estimates of F (toward ∞ or −∞) are penalized.
Dirichlet-based penalty:
We use a penalty function proportional to a transformed Dirichlet density with parameter α (>0),
| (10) |
where is the inverse multivariate logit transform of the fitnesses (with 0 ≤ θi ≤ 1 and ), and J = |∂(θi, … , θ19)/∂(F1, … , F19)| is the Jacobian of the transform. In the Appendix we show The log-penalty function is then
| (11) |
and the regularization parameter is λ = α. Because of the Jacobian, the Dirichlet-based density of Equation 10 has a single mode at F = 0 for any α > 0. As with the MVN penalty, extreme estimates of F (toward ∞ or −∞) are penalized.
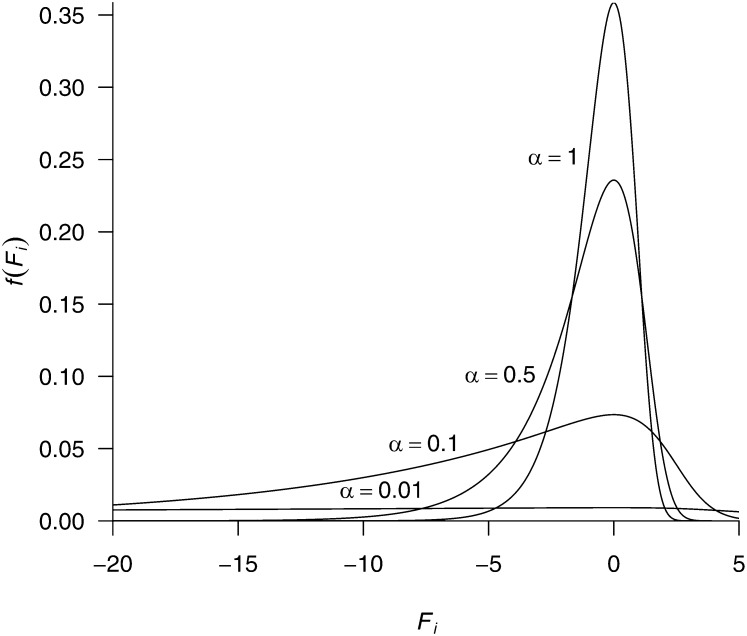
The Dirichlet distribution is useful to construct a penalty on a set of parameters with the constraint 0 ≤ θi ≤ 1 and However, the Dirichlet cannot be used directly to construct a penalty on the fitnesses because −∞ < Fi < ∞. Therefore, we use the multivariate logit transform, mapping the fitnesses from the real line to the (0, 1) interval and enforcing the constraint (which is mathematically equivalent to fixing one of the fitnesses to an arbitrary constant value, say F20 = 0). It follows from standard probability theory that if the probability density on θ is Dirichlet, then the density on its transform F is Dirichlet times the Jacobian. The Jacobian guarantees the probability mass is preserved during the transformation. Figure 1 shows the marginal density of Fi for various values of α. Note that a seemingly uninformative Dirichlet density with α = 1 is rather informative about Fi (Figure 1). We implement the Dirichlet-based penalty on the fitnesses because it is equivalent to the Dirichlet-based prior used by Rodrigue et al. (2010) in their hierarchical Bayesian framework to estimate the distribution of S; it is also equivalent to the use of amino acid pseudocounts to estimate the fitnesses as done by Halpern and Bruno (1998).
Figure 1.
Marginal distribution density of Fi when θ = (θi) ∼ Dirichlet(θ | α). The marginal density of θi is f(θi) = Beta(θi | α, αk − α), and the marginal density of Fi is f(Fi) = f(θi) × J = Beta(θi | α, αk − α) × θi(1 − θi), with θi = exp Fi/(k − 1 + exp Fi). A Dirichlet distribution with α = 1 is very informative on the transformed parameter space on F: The 95% equal-tail range of θi is (0.00133, 0.176), corresponding to a 95% range for Fi of (−3.68, 1.41).
Selection of λ and Kullback–Leibler divergence:
The value of λ must be set by the user before statistical inference is carried out. Selection of λ is subjective and an important issue in MPL estimation. Some authors use cross-validation (sequential removal of data and reestimation of parameters) to choose the value of λ (see Sanderson 2002, for a phylogenetic example). Cross-validation would be computationally expensive with our approach. We suggest the distribution of S should be estimated under various values of λ, and the estimated distributions can be compared to assess how informative a particular data set is about the true distribution.
The Kullback–Leibler (KL) divergence can be used to compare two probability distributions. Writing h0(S) and h1(S) for two estimates of the distribution of S (the histograms, Equation 4) estimated using different values of the regularization parameter (λ = λ0, λ1), then the KL divergence of h1(S) from h0(S) is
| (12) |
If the estimated distributions are identical [i.e., h0(SI) = h1(SI) for all I], then DKL = 0. Large values of DKL indicate dissimilar distributions (DKL is always nonnegative). An informative data set should produce similar estimates of the distribution of S (low DKL) for different values of λ. The KL divergence is asymmetrical; i.e., DKL(h0, h1) ≠ DKL(h1, h0). Here we set h0 to be the histogram estimated with the weaker penalty (λ0 < λ1).
Equation 12 can also be used to measure the divergence of an estimated distribution from the true distribution (e.g., in a simulation study). In this case h0(S) is the histogram for the true distribution and h1(S) the histogram for the estimate.
Materials and Methods
Analysis of simulated data sets
We used computer simulations to study the impact of taxon sampling, level of sequence divergence, and penalty function on the MPL estimation of the distribution of S, when (1) the true distribution is centered around neutral mutations and (2) the true distribution is strongly bimodal with a large proportion of deleterious mutations. We assess the effect of the value of the regularization parameter λ on the estimated distribution of S and its effect on the estimate of the proportion of deleterious mutations, p−. Because the penalty functions used (Equations 9 and 11) penalize extreme fitness estimates, strong penalties (large λ) are expected to lead to underestimates of p− and p+ and overestimates of p0. We are interested in assessing whether increasing the number of taxa leads to more accurate estimates of the distribution of S, in particular when the distribution contains a large proportion of deleterious mutations (large p−). We also examine the effects of levels of divergence between taxa.
Sequence alignments can be simulated on a phylogeny using the swMutSel model with standard methods (Yang 2006). Sequence alignments of length Lc = 1000 codons were simulated with mutational parameters κ = 2, π* = (0.25), and τ = 0 on the unimodal or bimodal distributions of S for various simulation conditions as described in full below.
Unimodal and bimodal distributions of S:
Two types of data sets were simulated. First, we simulated data under a distribution of S centered around nearly neutral mutations. For each location (sequence position), one amino acid was randomly selected to have F = 0 and the remaining 19 fitnesses were drawn from a normal distribution with mean 0 and variance 1, F ∼ N(0, 1). Once the fitnesses were sampled for all 1000 locations, the true distribution of S was calculated using Equation 4. We assume STOP codons have F = −∞, so the true histogram of S has a small mode at −10, corresponding to mutations toward STOP codons. Second, we simulated data sets under a bimodal distribution of S, with a large proportion of deleterious mutations. For each location, one amino acid was randomly selected to have F = 0. The fitnesses of 10 randomly selected amino acids were drawn from the normal distribution F ∼ N(0, 1), and then the remaining 9 fitnesses were drawn from F ∼ N(−10, 1), producing a distribution of S with a large proportion of deleterious mutations.
Taxon sampling:
We simulated sequence alignments on six phylogenies with different numbers of taxa. We started with a rooted, symmetrical, bifurcating 64-taxon tree with branch lengths of 0.25, for a tree height of 1.5 and tree length (sum of branch lengths) of 31.5. We then constructed a 128-taxon tree by replacing every leaf in the 64-taxon tree with a bifurcating node with branch lengths of 0.25 leading to two new leaves, increasing the tree height to 1.75 and the tree length to 63.5. The same procedure was used to construct 256-, 512-, 1024-, and 4096-taxon trees. In the swMutSel model branch lengths are given as neutral substitutions per site, which can be scaled to the usual substitutions per site as explained by Tamuri et al. (2012).
Level of sequence divergence:
In addition to examining how taxon sampling affects estimation of S, we studied the effect of varying levels of divergence. Using the 4096-taxon tree, we set all branch lengths to 1/1024, to produce a tree with height of 3/256. We then doubled the length of every branch to 1/512, resulting in a tree with height of 3/128. We continued this doubling procedure until reaching a tree having branch lengths of 8.0 and height of 96. In total we generated 14 trees. Unimodal and bimodal data sets were simulated on each tree, using the fitnesses described above.
MPL estimation:
Fitnesses for all data sets were estimated using the MPL method, fixing the tree topology, branch lengths, and mutational parameters to their true values. At each site, one amino acid has fitness fixed to zero and 19 fitnesses are estimated. We performed MPL with (1) the multivariate normal (MVN) penalty with σ = 1, 10, 100, and 1000; (2) the Dirichlet penalty with α = 2.0, 1.0, 0.1, and 0.01; and (3) without penalty (λ = 0; i.e., σ = ∞ or α = 0). Penalties with α > 0.1 or σ < 10 are very informative, having a single mode with the probability mass being very concentrated around F = 0. They are used here to study the effect of informative penalties on the estimation of the distribution of S. Numerical optimization of the parameters was repeated three times with different random starting values to test for convergence and obtain reliable results. Given the fitness estimates, we then calculated the estimated distribution of S for each data set and the corresponding proportions of deleterious (p−), neutral (p0), and advantageous (p+) nonsynonymous mutations. The Kullback-Leibler divergence DKL between the true and the estimated distribution of S was calculated using Equation 12 with h0(S) set as the true distribution.
Analysis of real data sets
We estimated the distribution of S for three real data sets. Two data sets (mitochondrial proteins and influenza polymerase) were analyzed by Tamuri et al. (2012), using the ML method, and showed estimated distributions of S with large proportions of deleterious mutations. We reanalyzed these two data sets with the MPL method to assess whether the estimates of the distribution of S and in particular of p− are robust or the result of overfitting by the ML method. The third data set is a chloroplast protein-coding gene alignment of thousands of species, analyzed here to assess the effect of a large phylogeny on the estimated distribution of S.
For each data set, the fitnesses were estimated using the MPL method with the MVN penalty (σ = 10 and 100), with the Dirichlet penalty (α = 0.01 and 0.1), and without penalty (λ = 0). To reduce computation time, we estimated branch lengths on a fixed tree topology with the FMutSel0 model (Yang and Nielsen 2008), using the CODEML program from the PAML package (Yang 2007). The FMutSel0 model is similar to the model of Equation 1, but fitnesses are equal for all locations (i.e., only 19 fitnesses are estimated for the entire alignment). Branch lengths in the swMutSel analyses are fixed to the FMutSel0 estimates. The mutational parameters (κ, π*, c, and τ) were estimated using the swMutSel model with no penalty (λ = 0), using the FMutSel0 estimates as starting values.
Mammal mitochondrial proteins:
We analyzed the 12 protein-coding genes on the heavy strand of the mitochondrial genome of 244 placental mammals. The 12 genes have similar base compositions and substitution pattern (Yang and Rannala 2006) and are treated here as a single gene. The concatenated alignment is 3598 codons long. The tree topology and alignment are from Tamuri et al. (2012).
Influenza PB2 protein:
We analyzed the pb2 polymerase gene of 401 influenza viruses isolated from 80 human and 321 avian hosts. The alignment is 759 codons long. The PB2 polymerase is involved in the replication of the virus within the host cell, and it has been suggested to be involved in host adaptation (Boivin et al. 2010). The tree topology and alignment are from Tamuri et al. (2009).
Plant chloroplast rbcL:
We analyzed the highly conserved rbcL gene of 3490 Eudicots (a group of flowering plants). The alignment, which is 457 codons long, is from Stamatakis et al. (2010). We estimated the tree topology using RAxML (Stamatakis et al. 2005), using the GTR + Γ substitution model. The rbcL gene encodes the large subunit of the Rubisco enzyme, one of the most abundant proteins on Earth, responsible for the photosynthetic fixation of CO2 from the atmosphere into organic compounds.
Results
Analysis of simulated data sets
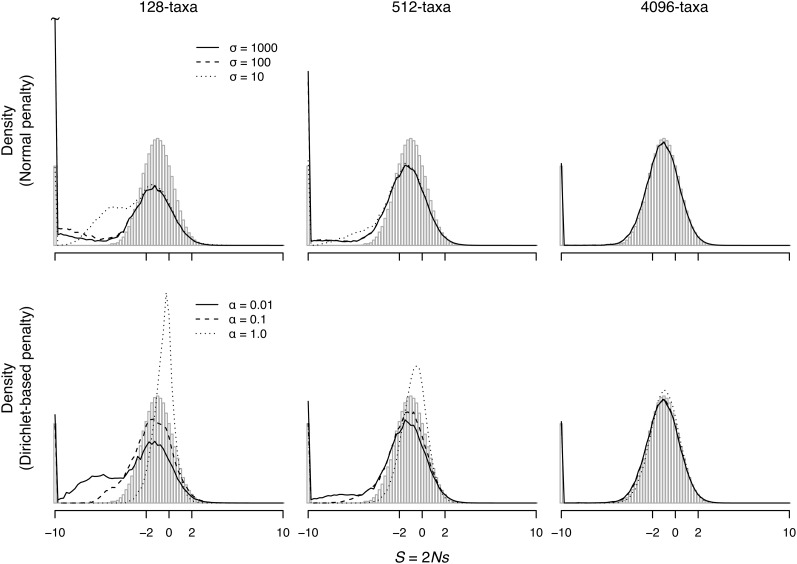
Table 1 lists the true and estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations for the simulated data sets when the true distribution of fitnesses is unimodal. Both the number of taxa and the strength of the penalty influence the proportions. The model overestimates the proportions of deleterious mutations for smaller trees and weak penalties, but it underestimates the proportion under the stronger penalties. For the largest 4096-taxon tree, the estimated proportions are close to the true proportions for all penalties. Figure 2 shows the estimated distribution of S for the 128-, 512-, and 4096-taxon trees (solid and dashed lines) compared to the distribution calculated from the true fitness values (vertical shaded bars) for the unimodal data sets. The distribution estimated using the Dirichlet penalty with α = 1.0 on the 128-taxon tree provides an example of applying a strong penalty on limited data, considerably overestimating the proportion of neutral mutations. Congruence with the true distribution improves with the addition of taxa. For the 4096-taxon tree, the estimated distribution is almost identical to the true distribution.
Table 1. Estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations in simulated data sets when the distribution of fitnesses is unimodal for trees with varying number of taxa.
| n taxa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 128 | 512 | 4096 | |||||||
| Penalty | p− | p0 | p+ | p− | p0 | p+ | p− | p0 | p+ |
| True | 0.243 | 0.748 | 0.009 | 0.243 | 0.748 | 0.009 | 0.243 | 0.748 | 0.009 |
| No penalty (λ = 0) | 0.583 | 0.408 | 0.008 | 0.431 | 0.558 | 0.011 | 0.271 | 0.719 | 0.010 |
| Normal | |||||||||
| σ = 1000 | 0.584 | 0.408 | 0.008 | 0.431 | 0.558 | 0.011 | 0.271 | 0.719 | 0.010 |
| σ = 100 | 0.583 | 0.409 | 0.008 | 0.430 | 0.559 | 0.011 | 0.271 | 0.720 | 0.010 |
| σ = 10 | 0.560 | 0.429 | 0.012 | 0.418 | 0.571 | 0.012 | 0.270 | 0.720 | 0.010 |
| Dirichlet | |||||||||
| α = 0.01 | 0.557 | 0.433 | 0.010 | 0.418 | 0.571 | 0.011 | 0.270 | 0.721 | 0.010 |
| α = 0.1 | 0.378 | 0.610 | 0.012 | 0.344 | 0.645 | 0.011 | 0.261 | 0.729 | 0.009 |
| α = 1.0 | 0.088 | 0.910 | 0.002 | 0.143 | 0.852 | 0.004 | 0.216 | 0.776 | 0.008 |
Figure 2.
Estimated and true distribution of S (for nonsynonymous mutations) for simulated data when the fitnesses are sampled from a unimodal distribution. The true distribution is shown as vertical shaded bars. The distributions are calculated using Equation 4 by dividing the range of S from −10 to 10 into equally spaced bins with wI = 0.25. Mutations with S ≤ −10 or those with S ≥ 10 are binned together. We consider mutations to STOP codons to be lethal, and these are included in the calculation of h(−10).
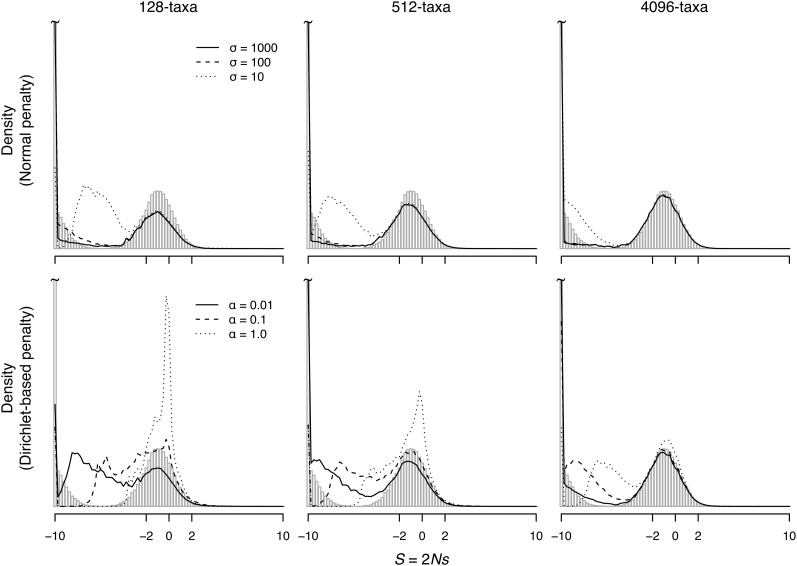
Table 2 and Figure 3 show the corresponding proportions and distribution of S for the simulated data sets where the true fitness distribution is bimodal. Although the estimated proportions approach the true proportions with the addition of taxa, Figure 3 illustrates the difficulty in accurately determining the true shape of the distribution in this case. Contrary to the unimodal data set, where the modes of the distribution were consistent with true distribution, the modes for the estimated distribution for the bimodal data sets are irregular, particularly for the 128-taxon tree under the stronger penalties. The stronger penalties tend to pull the peak of the deleterious mode closer to the neutral mode. The situation improves with the addition of taxa. For the 4096-taxon case, the estimated distribution approaches the true distribution. Furthermore, large values of p− are recovered for the 4096-taxon case even under the stronger penalties (σ = 10 and α = 1).
Table 2. Estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations in simulated data sets when the distribution of fitnesses is bimodal for trees with varying number of taxa.
| n taxa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 128 | 512 | 4096 | |||||||
| Penalty | p− | p0 | p+ | p− | p0 | p+ | p− | p0 | p+ |
| True | 0.598 | 0.397 | 0.004 | 0.598 | 0.397 | 0.004 | 0.598 | 0.397 | 0.004 |
| No penalty (λ = 0) | 0.744 | 0.252 | 0.004 | 0.685 | 0.311 | 0.005 | 0.625 | 0.370 | 0.005 |
| Normal | |||||||||
| σ = 1000 | 0.744 | 0.253 | 0.004 | 0.685 | 0.310 | 0.005 | 0.625 | 0.370 | 0.005 |
| σ = 100 | 0.743 | 0.253 | 0.004 | 0.685 | 0.311 | 0.005 | 0.625 | 0.370 | 0.005 |
| σ = 10 | 0.729 | 0.264 | 0.007 | 0.677 | 0.317 | 0.006 | 0.622 | 0.373 | 0.005 |
| Dirichlet | |||||||||
| α = 0.01 | 0.721 | 0.272 | 0.007 | 0.676 | 0.319 | 0.006 | 0.622 | 0.373 | 0.005 |
| α = 0.1 | 0.545 | 0.443 | 0.012 | 0.598 | 0.393 | 0.009 | 0.607 | 0.388 | 0.005 |
| α = 1.0 | 0.191 | 0.802 | 0.007 | 0.376 | 0.613 | 0.011 | 0.545 | 0.448 | 0.007 |
Figure 3.
Estimated and true distribution of S (for nonsynonymous mutations) for simulated data when the fitnesses are sampled from a bimodal distribution. The true distribution is shown as vertical shaded bars. The distributions are calculated as in Figure 2.
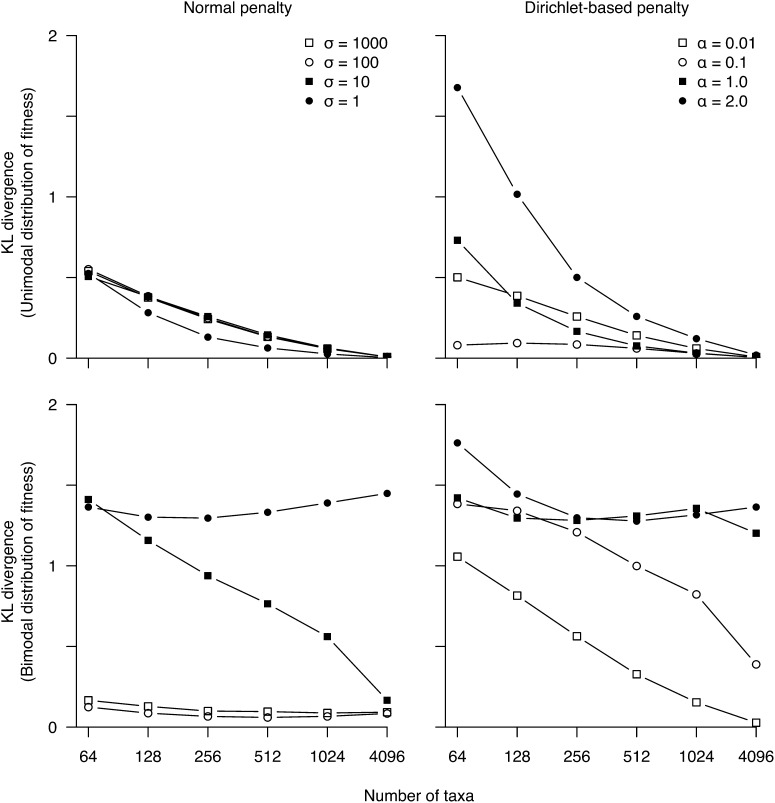
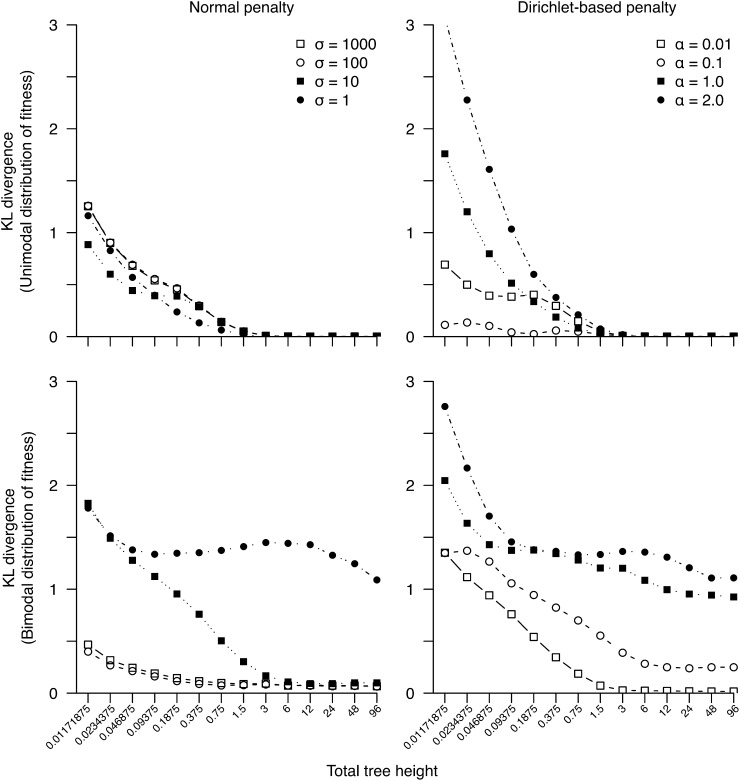
We use the KL divergence to measure the difference between the true and estimated distributions of S for the simulated data sets when the number of taxa is increased (Figure 4). For the unimodal case, the addition of taxa steadily improves the fit of the estimated distribution to the true distribution, until the KL divergence is close to zero (Figure 4, top), i.e., when the estimated distribution is virtually identical to the true distribution. By contrast, the KL divergences for the bimodal case decrease only with the addition of taxa for the weaker penalties, but not for the strong penalties (Figure 4, bottom). Although the shape of the estimated distribution does not always match the true distribution, Table 1 and Table 2 show the relative proportions of mutations do converge to their true values with the addition of taxa in all cases. We note there is a model mismatch between the penalty densities and the data generation density: The penalty densities have a single mode at F = 0, with the probability mass being very concentrated around this mode for the strong penalties (α > 0.1 or σ < 10); while the data generation density has two modes, one at F = 0 and the other at F = −10 × 1. This mismatch between the two densities is expected to be problematic during statistical inference. In other words, the concentrated unimodal penalties indicate a strong prior belief that the distribution of S is unimodal. The use of these strong penalties is thus a stern test on our statistical machinery when the true distribution of S is bimodal. It is remarkable we can still recover a large proportion of deleterious mutations under such strong penalties.
Figure 4.
Kullback–Leibler divergence between the true distribution of S (for nonsynonymous mutations) and its estimate vs. number of taxa for simulated data sets.
Finally, Table 3 and Table 4 list the estimated proportions of nonsynonymous mutations when varying branch lengths on the 4096-taxon tree for unimodal and bimodal distribution of fitnesses, respectively. The method is able to recover increasingly better estimates of proportions as sequence divergence increases. The tree with height 48 recovers almost exactly the true proportions under both weak and strong penalties and for both the unimodal and bimodal distributions of S (Table 3 and Table 4). Although the proportions are recovered with high accuracy, the true shape of the distribution remains elusive as indicated by the KL divergence scores (Figure 5). As with our results from increased taxon sampling, increasing tree height steadily recovers the true shape in the case of unimodal fitness distributions. However, the KL divergence score for the bimodal fitness distributions does not reach zero, except for the weak penalty case, due entirely to the shape of the distribution where S is deleterious (S < −2). In contrast, the neutral mode (−2 < S < 2) is recovered accurately. In other words, although the method recovers the true proportions of S under all penalties, it is unable to determine exactly how the deleterious mutations are distributed. Nevertheless, including more divergent homologous taxa in the data set, in addition to increasing taxon sampling, is a useful method for more accurately estimating the distribution of S.
Table 3. Estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations in simulated data sets when the distribution of fitnesses is unimodal for a 4096-taxon tree with increasing total tree height.
| Tree height | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.01171875 | 0.75 | 48 | |||||||
| Penalty | p− | p0 | p+ | p− | p0 | p+ | p− | p0 | p+ |
| True | 0.243 | 0.748 | 0.009 | 0.243 | 0.748 | 0.009 | 0.243 | 0.748 | 0.009 |
| No penalty (λ = 0) | 0.765 | 0.234 | 0.001 | 0.435 | 0.555 | 0.010 | 0.245 | 0.746 | 0.009 |
| Normal | |||||||||
| σ = 1000 | 0.765 | 0.234 | 0.001 | 0.434 | 0.555 | 0.011 | 0.245 | 0.746 | 0.009 |
| σ = 100 | 0.764 | 0.234 | 0.001 | 0.434 | 0.556 | 0.010 | 0.245 | 0.746 | 0.009 |
| σ = 10 | 0.719 | 0.217 | 0.010 | 0.415 | 0.573 | 0.012 | 0.245 | 0.746 | 0.009 |
| Dirichlet | |||||||||
| α = 0.01 | 0.687 | 0.300 | 0.013 | 0.417 | 0.572 | 0.011 | 0.245 | 0.746 | 0.009 |
| α = 0.1 | 0.286 | 0.705 | 0.008 | 0.327 | 0.663 | 0.011 | 0.245 | 0.746 | 0.009 |
| α = 1.0 | 0.053 | 0.947 | 0.000 | 0.143 | 0.853 | 0.004 | 0.241 | 0.750 | 0.009 |
Table 4. Estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations in simulated data sets when the distribution of fitnesses is bimodal for a 4096-taxon tree with increasing total tree height.
| Tree height | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.01171875 | 0.75 | 48 | |||||||
| Penalty | p− | p0 | p+ | p− | p0 | p+ | p− | p0 | p+ |
| True | 0.598 | 0.397 | 0.004 | 0.598 | 0.397 | 0.004 | 0.598 | 0.397 | 0.004 |
| No penalty (λ = 0) | 0.850 | 0.149 | 0.000 | 0.687 | 0.308 | 0.004 | 0.599 | 0.396 | 0.004 |
| Normal | |||||||||
| σ = 1000 | 0.850 | 0.150 | 0.000 | 0.687 | 0.308 | 0.005 | 0.600 | 0.396 | 0.004 |
| σ = 100 | 0.850 | 0.150 | 0.001 | 0.687 | 0.309 | 0.005 | 0.600 | 0.396 | 0.004 |
| σ = 10 | 0.813 | 0.181 | 0.006 | 0.680 | 0.314 | 0.006 | 0.600 | 0.396 | 0.004 |
| Dirichlet | |||||||||
| α = 0.01 | 0.774 | 0.216 | 0.010 | 0.677 | 0.317 | 0.006 | 0.600 | 0.396 | 0.004 |
| α = 0.1 | 0.390 | 0.599 | 0.011 | 0.581 | 0.410 | 0.008 | 0.599 | 0.396 | 0.004 |
| α = 1.0 | 0.053 | 0.947 | 0.000 | 0.370 | 0.623 | 0.007 | 0.597 | 0.398 | 0.005 |
Figure 5.
Kullback–Leibler divergence between the true distribution of S (for nonsynonymous mutations) and its estimate as a function of total tree height of a 4096-taxon tree.
Analysis of real data sets
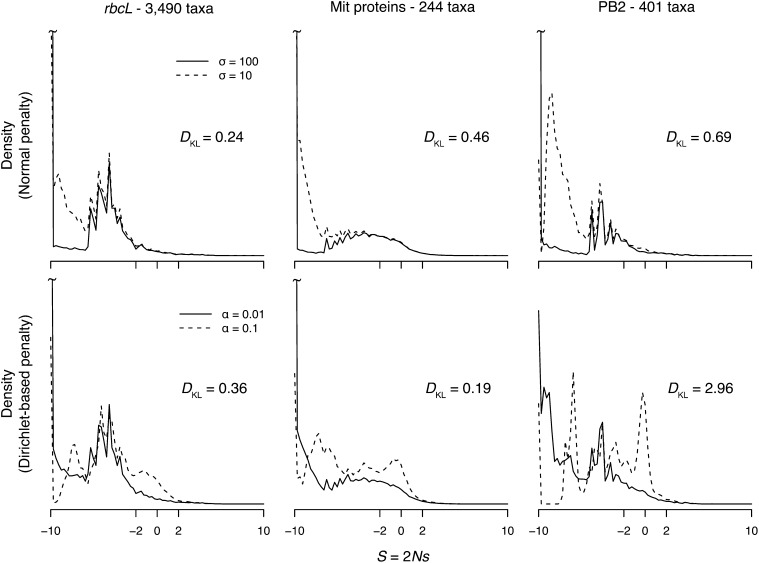
Table 5 lists the estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations for the three real data sets for various penalties. For all three data sets, a large proportion of deleterious mutations is obtained under both weak and strong penalties, with p− ranging from 63.3% (PB2, Dirichlet α = 0.1) up to 95.2% (rbcL, MVN σ = 100), indicating the majority of nonsynonymous mutations in these protein-coding genes are deleterious. Figure 6 shows the estimated distribution of S for each real data set. The shape of the distribution is sensitive to the penalty used. While the weak penalties (MVN σ = 100 and Dirichlet α = 0.01) produce similar distributions for each data set (solid lines in Figure 6), the strong Dirichlet penalty (α = 0.1) has a noticeable mode around S = 0 (dashed lines in Figure 6). The impact of the stronger MVN penalty (σ = 10) on each data set is largely confined to the shape of the distribution over deleterious mutations (S < −2).
Table 5. Estimated proportions of deleterious, neutral, and advantageous nonsynonymous mutations in real data sets.
| Data set | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| rbcL | Mit proteins | PB2 | |||||||
| Penalty | p− | p0 | p+ | p− | p0 | p+ | p− | p0 | p+ |
| No penalty (λ = 0) | 0.952 | 0.042 | 0.006 | 0.893 | 0.103 | 0.005 | 0.948 | 0.047 | 0.005 |
| Normal | |||||||||
| σ = 100 | 0.952 | 0.042 | 0.006 | 0.893 | 0.103 | 0.005 | 0.947 | 0.048 | 0.005 |
| σ = 10 | 0.943 | 0.049 | 0.007 | 0.888 | 0.107 | 0.005 | 0.931 | 0.063 | 0.006 |
| Dirichlet | |||||||||
| α = 0.01 | 0.939 | 0.054 | 0.008 | 0.875 | 0.120 | 0.005 | 0.894 | 0.098 | 0.008 |
| α = 0.1 | 0.805 | 0.184 | 0.010 | 0.747 | 0.245 | 0.008 | 0.633 | 0.356 | 0.011 |
Figure 6.
Estimated distribution of S (for nonsynonymous mutations) for real data sets. The DKL distance is calculated with Equation 12, with h0 being the weaker penalty. The distributions are calculated using Equation 4, setting wI = 0.25 for all I.
For each data set, we calculate the KL divergence between distributions estimated using the strong and weak penalties. This indicates how informative the data sets are about the true distribution of S. The rbcL, mitochondria, and PB2 data sets have KL divergence values of 0.24, 0.46, and 0.69, respectively, using the MVN penalty, and 0.36, 0.19, and 2.96, using the Dirichlet penalty (Figure 6). On average, the long mitochondria and large rbcL alignments are the most informative data sets about the distribution of S. The PB2 alignment is the least informative, with the shape of the distribution being sensitive to the strength of the penalty. How informative a data set is about the distribution of S depends on the length of the alignment, the number of taxa, and the level of divergence among the taxa (Tamuri et al. 2012). For example, a set of 4000 nearly identical sequences is not expected to be informative about the distribution of S.
Discussion
The site-wise mutation–selection model proposed by Halpern and Bruno (1998) has received renewed interest in recent years (Holder et al. 2008; Yang and Nielsen 2008; Rodrigue et al. 2010; Tamuri et al. 2012; Thorne et al. 2012). The model is motivated by a biochemical understanding of protein structure and function, with the site-specific amino acid fitnesses revealing the selective constraints acting on the protein at a given position. For example, buried sites in a protein may accommodate only particular hydrophobic amino acids (Baud and Karlin 1999), or the active site of an enzyme may tolerate only a few amino acids capable of stabilizing a substrate and carrying out the enzymatic reaction (Bartlett et al. 2002). Although the site-wise nature of the model captures the idiosyncratic features of protein sites, its adoption has been considered impractical due to the computational cost of estimating its many parameters in large data sets (Yang and Nielsen 2008; Rodrigue et al. 2010). For example, in their original implementation Halpern and Bruno (1998) could use the model only to estimate the evolutionary distance in pairwise sequence alignments. Holder et al. (2008) made the first implementation where the likelihood of the model could be calculated on a phylogeny to estimate all the parameters. Later Tamuri et al. (2012) provided a full implementation of the model to calculate the distribution of selection coefficients from phylogenetic (species-level) data in real data sets and showed the swMutSel model could recover the large proportion of deleterious mutations expected in real data sets (Akashi 1999; Wloch et al. 2001; Sanjuan 2010; Hietpas et al. 2011) where other phylogenetic models had failed (Nielsen and Yang 2003; Yang and Nielsen 2008; Rodrigue et al. 2010). Furthermore, Tamuri et al. (2012) showed increasing taxon sampling could improve the estimates of fitnesses and the distribution of S, despite the large number of parameters in the model.
Here we extended the work of Tamuri et al. (2012) under a penalized-likelihood framework. The new approach has two main advantages: (1) it regularizes the estimates of fitnesses for small, uninformative data sets, and (2) it can be used to assess whether the estimated distribution of S is robust for a particular data set, by varying the form and strength of the penalty function. Using the new approach, we confirmed the distribution of S among new mutations is indeed bimodal in the mammalian mitochondria and influenza PB2 proteins analyzed by Tamuri et al. (2012) and also for the large subunit of the rubisco protein in plant chloroplasts. Furthermore, our method is useful for estimating the distribution of S in organisms for which mutation–selection experiments cannot be performed, such as mammals or plants.
Rodrigue (2013) criticized the approach of Tamuri et al. (2012) on two counts. First, Rodrigue (2013) suggested the swMutSel model is overparameterized, and simply increasing the number of taxa to improve fitness estimates was unsound. His argument is based on the changing parameterization of the likelihood function as taxa are added: each additional taxon involves a different tree topology and two additional branch lengths, therefore changing the form of the likelihood function. Second, Rodrigue (2013) suggested the large proportion of deleterious mutations estimated by Tamuri et al. (2012) was the result of overfitting by the ML method: unobserved amino acids at particular locations are estimated to have F = −∞, therefore inflating estimates of p−. We deal with these two criticisms here.
Effect of taxon sampling
Site-specific parameters employed in phylogenetic models are themselves often of interest to biologists and a natural way to add more information for analyses is by adding taxa. Despite the changing parametric form of the likelihood function, simulations have consistently demonstrated that estimates improve considerably with the addition of taxa (Pollock and Taylor 1997; Pollock et al. 1999; Zwickl and Hillis 2002; Heath et al. 2008; Tamuri et al. 2012). For example, Pollock and Bruno (2000) found increasing taxon sampling improved phylogenetic inference, more so than increasing sequence length, and reduced the variance of site-specific parameter estimates. Our analysis of simulated data demonstrates the model tends to the true distribution of S given the addition of more taxa and increased evolutionary divergence between taxa. For the unimodal simulated data set, we find the estimated distribution steadily converges to the true distribution even for the strongest penalties. The bimodal simulated data set is not as consistent, due to the penalty imposed on non-unimodal distribution of fitnesses. Only the weaker penalties converge to the true distribution, given a maximum of 4096 taxa analyzed, due to the significant amount of data required to accurately estimate the distribution of deleterious (S < −2) mutations. Importantly, the nearly neutral mode of the distribution and the estimated proportions of S converge readily for all penalties. This may indicate a need for alternative penalty functions. One approach would be to devise penalties more appropriate for bimodal distributions of S. Another interesting way to use prior information about distributions of amino acids at sites is the mixtures of Dirichlet densities proposed by Sjölander et al. (1996). Using the mixture densities in our penalized framework may be useful for dealing with small or skewed samples.
A related question is how one can choose an optimal value of the regularization parameter λ to best balance the trade-off between the fit to the data and the constraint imposed by the penalty function. Clearly, different values of λ can affect fitness estimates using the penalty functions described here, especially for small data sets. The parameter can be user-specified, and several different values can be applied to examine the informativeness of a given data set. The λ value could also be selected by minimizing a cross-validation criterion, as described by Sanderson (2002), or by optimization of a modified Akaike’s information criterion (Harrell 2001; Kim and Pritchard 2007). Here we recommend using α ≤ 0.1 if using the Dirichlet-based penalty or σ ≥ 10 if using the MVN penalty. We find stronger penalties (α > 0.1 or σ < 10) are too informative and may bias the estimated distribution of S.
Effect of unobserved amino acids at a location
There are strong biological reasons indicating the proportion of deleterious mutations is high for most protein-coding genes, and mutation experiments of real protein-coding genes have consistently detected large proportions of deleterious mutants (Wloch et al. 2001; Sanjuan 2010; Hietpas et al. 2011). If a phenotype is lethal, then individuals carrying the phenotype in a population will not be seen. In other words, if an amino acid is lethal at a protein location, it will not be seen at the corresponding location in multiple sequence alignments. Any statistical method devised to estimate the distribution of S from species-level alignments will have to estimate the proportion of highly deleterious mutations based on the amino acids unobserved at particular locations in the alignment. Rodrigue (2013) showed that when removing mutations toward unobserved amino acids in the calculation of mij,k (Equation 3), the peak of deleterious mutations would disappear from the estimated distribution of S. But this is exactly what we expect from population genetics theory! We do not understand why the Bayesian mixture-model approach of Rodrigue et al. (2010) and Rodrigue (2013) failed to produce large estimates of p− in real data. We suspect that if a limited number of site classes is imposed in the model (by the Dirichlet process prior), then the fitness for the site classes will represent averages over the fitnesses at particular locations. We note Rodrigue (2013) tested the performance of his method only for simulated data where the true distribution of S was unimodal, but not for the more biologically realistic case when the distribution is bimodal. A new implementation of the Bayesian mixture model has now become available (Rodrigue and Lartillot 2014), which looks more promising as it produced larger estimates of p− for the PB2 data set.
Challenges and limitations
Tamuri et al. (2012) discussed in detail the limitations and assumptions of the swMutSel model. In particular, they discussed the impact of assuming independently evolving codon sites, that is, assuming free recombination among codon locations and so ignoring linkage, epistasis, and clonal interference. While previous studies (e.g., Lakner et al. 2011; Ashenberg et al. 2013) have suggested the effect on estimates of selective constraints due to epistasis is likely small, a larger effect could be caused by codon locations in a protein being tightly linked. Specifically, the fitness of highly advantageous mutations may be underestimated when linkage is ignored (Bustamante 2005). Some other assumptions are not expected to have much impact on estimates of S. For example, although the mutational component of the model does not vary across sites and lineages, the impact on S is probably small because qij,k is a steeply varying function of Sij,k. Others, such as changes in S over evolutionary time, can be explicitly included in the modeling, using a nonhomogeneous approach (e.g., Tamuri et al. 2012, figure 4). Of particular interest is the effect of uncertainty in branch length estimates on estimates of the distribution of S. We have shown that the height of the phylogeny (Table 3 and Table 4) affects estimates of the distribution of S, so uncertainties in branch length estimates are also expected to have an effect. This is an important issue requiring further investigation.
Our results suggest accurate estimation of the distribution of S using the penalized likelihood method is possible only with a sizable number of sequences, a factor less troublesome due to the rapid increase in the available number of divergent homologous sequences. Highly conserved genes, such as rbcL, will have fewer effective residues per site and, therefore, need many sequences to accurately estimate the distribution of S. If a residue is truly impossible at a given position, overestimation of its fitness by the penalty can be countered by additional informative sequences providing evidence the residue is indeed lethal. Nevertheless, estimating the shape of the left tail of the distribution of S will always be challenging (Eyre-Walker and Keightley 2007), even when analyzing thousands of sequences. For example, imagine a location where the fitness of lysine (K) is 0, the fitness of glutamate (E) is −10, and the fitnesses of all the other amino acids are −∞. The expected frequency of E at equilibrium will be e−10/(1 + e−10) = 4.5 × 10−5 (assuming equal π*). If the fitness of E was instead −7, its equilibrium frequency would be 9.1 × 10−4. In both cases estimating the precise fitness of E is hard because the frequency is very close to zero and we would be unlikely to observe E, even once, in a sequence alignment of 1000 sequences. Nonetheless, examination of hundreds to thousands of sequences is now commonplace in phylogenetic analysis and neither the number of sequences nor the required computational resources are significant obstacles to using our methods to accurately estimate the proportions p−, p0, and p+.
Program availability
The software implementation of the swMutSel model is available to download at https://github.com/tamuri/swmutsel. The program is written in Java and can use multiple and distributed cores, reducing running time considerably. For example, the total running time for each analysis of the 3490-taxa rbcL alignment using several hundred distributed cores ranged from 13 to 20 hr.
Acknowledgments
We thank Alexandros Stamatakis and Guido Grimm for providing the rbcL data set. We thank Ziheng Yang, Adam Siepel, and an anonymous reviewer for valuable discussions and comments. This work was supported by the European Bioinformatics Institute (European Molecular Biology Laboratory–EBI). M.d.R. is supported by a Biotechnology and Biological Sciences Research Council grant awarded to Ziheng Yang.
Appendix
The Jacobian of Equation 10 is the absolute value of the determinant of the Jacobian matrix of the transform, J = |J| = |∂(θ1, … , θ19)/∂(F1, … , F19)|. The elements of matrix J = (Jij) are
| (A1) |
J is of size 19 × 19 because only 19 fitness parameters need to be estimated. Note that J can be written as the product of two matrices
Because the determinant of a product matrix is the product of the determinants, |AB| = |A||B|, the determinant of J can be written as the product of two determinants. The first determinant is the determinant of a diagonal matrix, The determinant of the second matrix is (Grossman 1995, p. 202). Because all θi > 0, the determinant is always positive. Therefore
Footnotes
Communicating editor: J. J. Bull
Literature Cited
- Akashi H., 1999. Within- and between-species DNA sequence variation and the ‘footprint’ of natural selection. Gene 238: 39–51. [DOI] [PubMed] [Google Scholar]
- Ashenberg O., Gong L. I., Bloom J. D., 2013. Mutational effects on stability are largely conserved during protein evolution. Proc. Natl. Acad. Sci. USA 110: 21071–21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett G. J., Porter C. T., Borkakoti N., Thornton J. M., 2002. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 324: 105–121. [DOI] [PubMed] [Google Scholar]
- Baud F., Karlin S., 1999. Measures of residue density in protein structures. Proc. Natl. Acad. Sci. USA 96: 12494–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S., Cusack S., Ruigrok R. W., Hart D. J., 2010. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J. Biol. Chem. 285: 28411–28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C. D., 2005 Population genetics of molecular evolution, pp. 63–99 in Statistical Methods in Molecular Evolution, edited by R. Nielsen. Springer-Verlag, New York. [Google Scholar]
- Cox D. D., O’Sullivan F., 1990. Asymptotic analysis of penalized likelihood and related estimators. Ann. Stat. 18: 1676–1695. [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Grossman, S., 1995 Elementary Linear Algebra. Brooks/Cole Publishing, Belmont, CA. [Google Scholar]
- Halpern A. L., Bruno W. J., 1998. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol. Biol. Evol. 15: 910–917. [DOI] [PubMed] [Google Scholar]
- Harrell, F. E., 2001 Regression Modeling Strategies, Chap. 9. Springer-Verlag, New York. [Google Scholar]
- Hasegawa M., Kishino H., Yano T., 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Heath T. A., Zwickl D. J., Kim J., Hillis D. M., 2008. Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Syst. Biol. 57: 160–166. [DOI] [PubMed] [Google Scholar]
- Hietpas R. T., Jensen J. D., Bolon D. N. A., 2011. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. USA 108: 7896–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder M. T., Zwickl D. J., Dessimoz C., 2008. Evaluating the robustness of phylogenetic methods to among-site variability in substitution processes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 4013–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Pritchard J. K., 2007. Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet. 3: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983 The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge. [Google Scholar]
- Lakner C., Holder M. T., Goldman N., Naylor G. J., 2011. What’s in a likelihood? Simple models of protein evolution and the contribution of structurally viable reconstructions to the likelihood. Syst. Biol. 60: 161–174. [DOI] [PubMed] [Google Scholar]
- Li W. H., 1978. Maintenance of genetic variability under the joint effect of mutation, selection and random drift. Genetics 90: 349–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., 1997. Site-by-site estimation of the rate of substitution and the correlation of rates in mitochondrial DNA. Syst. Biol. 46: 346–353. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Yang Z., 2003. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 20: 1231–1239. [DOI] [PubMed] [Google Scholar]
- Ohta T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23: 263–286. [Google Scholar]
- Pollock D. D., Bruno W. J., 2000. Assessing an unknown evolutionary process: effect of increasing site-specific knowledge through taxon addition. Mol. Biol. Evol. 17: 1854–1858. [DOI] [PubMed] [Google Scholar]
- Pollock D. D., Taylor W. R., 1997. Effectiveness of correlation analysis in identifying protein residues undergoing correlated evolution. Protein Eng. 10: 647–657. [DOI] [PubMed] [Google Scholar]
- Pollock D. D., Taylor W. R., Goldman N., 1999. Coevolving protein residues: maximum likelihood identification and relationship to structure. J. Mol. Biol. 287: 187–198. [DOI] [PubMed] [Google Scholar]
- Rodrigue N., 2013. On the statistical interpretation of site-specific variables in phylogeny-based substitution models. Genetics 193: 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue N., Lartillot N., 2014. Site-heterogeneous mutation-selection models within the PhyloBayes-MPI package. Bioinformatics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue N., Philippe H., Lartillot N., 2010. Mutation-selection models of coding sequence evolution with site-heterogeneous amino acid fitness profiles. Proc. Natl. Acad. Sci. USA 107: 4629–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M. J., 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19: 101–109. [DOI] [PubMed] [Google Scholar]
- Sanjuan R., 2010. Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölander K., Karplus K., Brown M., Hughey R., Krogh A., et al. , 1996. Dirichlet mixtures: a method for improved detection of weak but significant protein sequence homology. Comput. Appl. Biosci. 12: 327–345. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H., 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Göker M., Grimm G. W., 2010. Maximum likelihood analyses of 3,490 rbcL sequences: scalability of comprehensive inference vs. group-specific taxon sampling. Evol. Bioinform. Online 6: 73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamuri A. U., dos Reis M., Hay A. J., Goldstein R. A., 2009. Identifying changes in selective constraints: host shifts in influenza. PLoS Comput. Biol. 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamuri A. U., dos Reis M., Goldstein R. A., 2012. Estimating the distribution of selection coefficients from phylogenetic data using sitewise mutation-selection models. Genetics 190: 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne, J., N. Lartillot, N. Rodrigue, and S. C. Choi, 2012 Codon models as a vehicle for reconciling population genetics with inter-specific sequence data, Codon Evolution: Mechanisms and Models, edited by G. Cannarozzi and A. Schneider. Oxford University Press, New York. [Google Scholar]
- Thorne J. L., Choi S. C., Yu J., Higgs P. G., Kishino H., 2007. Population genetics without intraspecific data. Mol. Biol. Evol. 24: 1667–1677. [DOI] [PubMed] [Google Scholar]
- Wloch D. M., Szafraniec K., Borts R. H., Korona R., 2001. Direct estimate of the mutation rate and the distribution of fitness effects in the yeast Saccharomyces cerevisiae. Genetics 159: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2006. Computational Molecular Evolution. Oxford University Press, Oxford. [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25: 568–579. [DOI] [PubMed] [Google Scholar]
- Yang Z., Rannala B., 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23: 212–226. [DOI] [PubMed] [Google Scholar]
- Zwickl D. J., Hillis D. M., 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 51: 588–598. [DOI] [PubMed] [Google Scholar]