Abstract
The neutral rate of allelic substitution is analyzed for a class-structured population subject to a stationary stochastic demographic process. The substitution rate is shown to be generally equal to the effective mutation rate, and under overlapping generations it can be expressed as the effective mutation rate in newborns when measured in units of average generation time. With uniform mutation rate across classes the substitution rate reduces to the mutation rate.
Keywords: effective mutation rate, generation time, reproductive value, stochastic demography, substitution rate
CONSIDER a haploid population of constant size N without overlapping generations, where neutral mutant alleles are substituted sequentially at a given locus through the constant input of mutations at rate μ per gene. Then, the rate k of allelic substitution is equal to the mutation rate; that is, k = μ (Kimura 1971). Does this simple result extend to more realistic demographic scenarios? In a population with overlapping generations where individuals reproduce at discrete time points and mutations arise only in newborns (the standard assumption), the substitution rate is k = μ0/T, where μ0 is the rate at which a newborn accumulates mutations at the gene locus under focus and T is the generation time (Pollak 1982, equation 11). This is the average age of the parent of a randomly sampled individual among the N0 surviving newborns. When measured in units of T time steps, the substitution rate thus depends only on μ0 (Pollak 1982, equation 15; Charlesworth 1994, p. 94).
There is, however, an even simpler interpretation of k in terms of mutation rate under overlapping generations. This can be reached by observing that the total population size N necessarily satisfies N = TN0, since the generation time is the number of units of time the population takes to produce N newborns when only N0 are produced per unit time (an observation implied by Felsenstein 1971, equation 1). With this, where is the mutation rate at a randomly sampled gene from the population, since mutations arise only in newborns and they form a fraction N0/N of the population.
How do these two different interpretations of k in terms of mutation rate generalize to stochastic demography, where population sizes can fluctuate over time? This article develops a full stochastic demographic model of neutral evolution in a class-structured population (e.g., by sex, age, stage). The model shows that the substitution rate is precisely the effective mutation rate μe, which is the rate at which a gene lineage accumulates mutations (Rousset 2004, p. 158), and can be different from the mutation rate at a randomly sampled gene from a randomly sampled individual in the population. In an age-structured population the effective mutation rate is shown to reduce to the effective mutation rate in newborns when measured in units of average generation time, which can itself be expressed in terms of class reproductive values. The use of such reproductive values turns out to be central in structuring the connections between substitution rate, average mutation rate, effective mutation rate, and generation time.
Model
Biological assumptions
Demography:
We now consider a population structured into a finite number of classes and evolving in discrete time. This class structure could, for instance, result from the presence of males and females, of age classes, of groups of individuals located at different positions in the habitat, or a combination of these or other factors. The number of individuals in class i = 0, 1, 2, … is written Ni and the vector n ≡ (N0, N1, N2, …) denotes a state of the population, which gives the realized number of individuals in each class i at a census point (see Table 1 for a list of symbols).
Table 1. List of symbols.
| Symbol | Definition |
|---|---|
| k | Substitution rate. |
| Ni | No. class i individuals. |
| Average no. gene copies in the population. | |
| Pr(n) | Stationary probability that the population is in state n. |
| Pr(n′|n) | Forward transition probability from state n to state n′. |
| gi | Ploidy of a class i individual. |
| tij | Probability that a gene randomly sampled in a class-i individual was transmitted by a class-j individual. |
| μij | Mutation rate at a gene transmitted by a class-j individual to a class-i individual. |
| Average mutation rate in the stationary demographic regime. | |
| μe | Effective mutation rate. |
| μe,0 | Effective mutation rate in newborns. |
| T | Average generation time (or mean age of the parents of a newborn). |
| T(n) | Average generation time in population state n. |
| wij(n′, n) | Expected no. class-i individuals in a population in state n′ descending from a single class-j individual in population state n. |
| fij(n′, n) | Probability that a gene sampled in a class-i individual when the population is in state n′ is a copy of a gene of a class-j individual given the population was in state n in the parental generation. |
| aij(n′, n) | Probability that a gene sampled in a class-i individual when the population is in state n′ is a copy of a gene of a class-j individual and the population was in state n in the parental generation. |
| πi(n) | Fixation probability of a single mutant residing in a class-i individual. |
| Average fixation probability of a single mutant. |
The change in the demographic state of the population is assumed to follow a discrete-time stochastic process and we denote by Pr(n′|n) the transition probability per unit time that a population in state n will be in state n′ in the next time step. This defines a homogeneous Markov chain, which may be driven by both endogenous and exogenous factors. This demographic process is further assumed to have a finite state space and to be ergodic (irreducible and aperiodic), conditional on the nonextinction of the population. In force of these assumptions, the distribution of the process will approach the stationary probability Pr(n) of being in state n (Karlin and Taylor 1975; Grinstead and Snell 1997), which determines the stationary demographic regime.
Genetics:
Neutral evolution will be investigated at a given locus in the backdrop of the stationary demography. Neutrality means that the number of successful “offspring” (number of descendants over one time step) of all individuals in the same class j are an exchangeable random variable, and this holds for each class. This entails that the forward transition probability Pr(n′|n) and the probability aij(n′, n) that a gene randomly sampled in a class-i individual in population state n′ descends from a class-j individual and from population state n in the previous time step do not depend on the genetic state (allele frequency distribution) of the population
Individuals in different classes, like males and females, may have different ploidies and gi will denote the ploidy of an individual of class i at the locus of interest. Mutation may also differ across classes and μij will denote the mutation rate at a gene transmitted by a class-j individual to a class-i individual. This allows us to evaluate the rate at which a randomly sampled gene in a randomly sampled individual of class i in population state n′ accumulates mutations, which is
| (1) |
This generalizes to any class and to stochastic demography, the standard expression for the rate at which a randomly sampled gene in a newborn accumulates mutations in models of overlapping generations with constant size (Pollak 1982, equations 1 and 2, Charlesworth 1994, equation 2.49b).
From Equation 1, we can define the following average mutation rate,
| (2) |
which gives the ratio of the average number of mutations produced in the population to the average number of genes in the population (average size of the gene pool). The mutation rate gives the rate at which a randomly sampled gene in a randomly sampled individual accumulates mutations, since over replicates of the evolutionary process the probability of sampling an individual in demographic state n is where in which case the probability this is a class-j individual is gjNj/N(n). Hence, the probability of sampling a mutant individual of class j in state n is proportional to the size of its class and the probability of occurrence of that state.
The aim of this note is to express the neutral substitution rate k in terms of a mutation rate averaged over the class specific mutation rates, the μj(n)’s, and we will see that in general this quantity is not equal to
Substitution rate
Forward process:
The substitution rate k is the expected number of mutations that will fix in the population per unit time (Kimura 1971; Pollak 1982). Under the above assumptions, this can be written as
| (3) |
where
| (4) |
is the average fixation probability of a single mutant allele, which depends on the fixation probability πj(n) of a mutant arising as a single copy in a class-j individual in population state n and on the probability that the mutation arises in this state (see Appendix A for a proof). This latter quantity is given in Equation 4 as the ratio of the number of mutations arising in class-j individuals in population state n to the total number of mutations arising per unit time.
Three points are now made about Equation 3. First, this is exactly of the same form as the classical expression for the substitution rate (Kimura 1971, equation 4.2), but where all quantities are averages. Second, if the underlying mutation model at the locus of interest is the infinite-allele model (Kimura and Crow 1964; Kimura 1971), then the average mutation rate must be very small so that novel mutations occur in homoallelic populations; otherwise the population will hardly ever fixate for an allele. By contrast, under the infinite-site mutation model (Kimura 1969, 1971), free recombination among sites and a very small mutation rate per site entail that mutants arise at homoallelic sites, so that several mutations can segregate simultaneously and independently in the population. Then, Equation 3 gives the number of mutants fixing at different sites per unit time and this is the classical situation envisioned for the substitution process (Kimura 1971, p. 183). Third, k does generally not reduce to since depends on the mutation distribution and simplifies to only under special cases. For instance, this is the case when the mutation rate is the same in all classes μij = μ for all i and j, which entails that μj(n) = μ. Then (Lehmann 2012, appendix A), and the substitution rate becomes equal to the mutation rate
| (5) |
Backward process:
To further simplify the expression for k, we now express the fixation probability πi(n) in terms of class reproductive values (e.g., Taylor 1990; Rousset 2004), which allows us to look at neutral evolution in terms of a backward genealogical process, much like coalescent theory does for effective population size (e.g., Wakeley 2008).
Imagine we trace backward in time the ancestral lineage of a randomly sampled gene in the population in the present, at time h = 0. Then, the probability γj, h(n) that this lineage resides in an individual of class j and in population state n at time h in the past satisfies the recursion
| (6) |
Assuming that every gene position (class of individual and population state) can be reached in the long run, the circulation of the ancestral gene lineage among the classes of individuals and population states determined by the aij(n′, n) coefficients defines an ergodic Markov chain that will eventually reach a stationary distribution [given by the left unit eigenvector of the ergodic Markov matrix with transition probabilities aij(n′, n) (Karlin and Taylor 1975; Grinstead and Snell 1997)].
The probability that a gene lineage resides in class i and in population state n under the stationary distribution is denoted γi(n), which is the reproductive value of class (i, n) and is equal to αi(n)Pr(n), where αi(n) is the probability that the ancestral lineage of a randomly sampled gene in the population was in class i, given population state n in the distant past [the reproductive value of class i (Rousset 2004, p. 181)]. With this, we can write the fixation probability of a single mutant allele entering the population in state n as
| (7) |
where 1/[giNi] is the frequency of a single mutant in class i (see Appendix B for a proof).
Substituting Equation 4 and Equation 7 into Equation 3 and rearranging produces
| (8) |
which is a reproductive value weighted average mutation rate.
Substitution rate as effective mutation rate
The effective mutation μe is defined as the average rate at which a gene lineage accumulates mutations (Rousset 2004, equation 9.36). This is precisely an interpretation that can be given to the right-hand side of Equation 8. In effect, γi(n′) can be interpreted as the probability that the ancestral lineage of a randomly sampled gene was in class i in population state n′ in the distant past, aij(n′, n) is the probability that this gene descends from an individual in class j in population state n in the previous time step, and μij is the mutation rate during such a transition. Hence,
| (9) |
which shows that the neutral substitution rate is the effective mutation rate regardless of the genetic and demographic assumptions behind the model. This correspondence is not surprising given the definition of μe, but Equation 8 makes precise that while k has originally been defined in terms of a forward in time looking process (fixation probabilities, see Equation 3), it can be interpreted in terms of a meaningful class-reproductive value weighted average mutation rate, which defines a backward in time looking process (e.g., Equation 6). The substitution rate is thus also the rate at which a randomly sampled gene from a randomly sampled common ancestor of the population accumulates mutations, since with probability γi(n) the common ancestor descends from state (i, n), in which case it carries mutations at rate μi(n).
The substitution rate k will generally depend on population size fluctuations (Balloux and Lehmann 2012) as the reproductive values generally depend on such fluctuations (Whitlock and Barton 1997; Rousset 2004). Further, these are actually very hard to evaluate explicitly and most of the time can be expressed implicitly only as the solution of a very large set of simultaneous equations involving population sizes (e.g., Equation 6). But if
| (10) |
then and from Equation 7 we have
| (11) |
When will the reproductive value of a class be proportional to the number of genes in that class so that this result holds?
If evolution occurs in a patch-structured haploid population of constant size in which migration does not affect group size, migration is said to be conservative and Equation 10 holds (Nagylaki 1998, p. 1600). More generally, however, migration will result in population size fluctuations and density-dependent competition can result in very large populations (or patches) producing only very few individuals. In these cases, the reproductive value of a class is unlikely to be proportional to the number of genes in that class, because the size of a demographic class is not necessarily indicative of its asymptotic contribution to the gene pool. For instance, under good environmental conditions a small group of individuals may contribute a disproportionally larger share to the gene pool than a big group of individuals in poor environmental conditions. Then, μe will differ from if the rate at which mutations accumulate in individuals from different classes differs [the μi(n)’s].
A simple situation where the class reproductive values are not proportional to the number of genes in a class is a population with separate sexes with a constant number Nf of females and Nm of males (constancy of demography entails that we can also drop the dependence on the state n in all quantities). Then, we have γf = αf = tmf/(tmf + tfm), where tij is the probability that a gene randomly sampled in a class-i offspring has been transmitted by a class-j parent (see Appendix D, Equation D17 for a proof; tmf = tfm = 1/2 for diploids, while tmf = 1 and tfm = 1/2 for diploid females and haploid males). With this, the effective mutation rate is
| (12) |
while the average mutation rate is
| (13) |
Thus, if the mutation rate is different in the sexes, then μe and can markedly differ and this stems from the fact that even if the number of females is much larger than the number of males, every offspring has both a father and a mother. Thus, each sex contributes to the gene pool in proportion to its ploidy and not according to the number of its representative individuals (e.g., Taylor 1990).
Overlapping generations and generation time
Haploid reproduction:
We now focus on overlapping generations under haploid reproduction, so that i = 0, 1, 2, … indexes the age classes. An individual will be said of age i if it is between i and i + 1 units of age, where i = 0 stands for newborns. Reproduction is assumed to occur at the end of a time period, that is, when individuals of age i have reached i + 1 units of age.
Following standard formulations (Pollak 1982, equations 1 and 2; Charlesworth 1994, equation 2.49b), we also assume that mutations occur only during gametogenesis and therefore only when newborns are produced. Hence, μij = 0 for i ≠ 0, and we have μ0j ≥ 0 for all j. From Equation 1, the probability that a newborn carries a mutation in demographic state n′ is then given by
| (14) |
whereby the substitution rate (Equation 8) can be written as
| (15) |
We can now define an effective mutation rate μe,0 for newborns, by averaging the mutation rate μ0(n) over the probabilities that the ancestral lineage of a randomly sampled gene will be in a newborn. This yields
| (16) |
whereby where the term in square brackets is related to the average generation time in the next section.
Generation time:
The generation time is now introduced through the quantity
| (17) |
This is the mean age of the parent of an individual of age class zero randomly sampled in population state n′ because a0i(n′, n) is the probability that a newborn sampled in population state n′ descends from a parent of class i living in population state n, in which case the parent has lived i + 1 units of time. For a population with constant size Equation 17 reduces to the equation for the generation time used in population genetics (Felsenstein 1971, p. 583; Pollak 1982, p. 91) and is a direct extension of that case to fluctuating demography.
To evaluate the average generation time, one needs to take into account the probability of sampling a newborn in a given demographic state. Hence, the average generation time is
| (18) |
In Appendix C, it is proved that
| (19) |
which implies that
| (20) |
and shows that the generation time can be expressed in terms of class reproductive values.
Substitution rate in terms of generation time:
From Equations 15, 16, and 20, we can now write
| (21) |
Measured in units of average generation time T, the substitution rate is equal to the effective mutation rate in newborns. If the mutation rate is the same in all individuals, then μ0j = μ in which case μe,0 = μ. In this case, when measured in units of average generation time, the substitution rate is equal to the mutation rate, and this then removes the effect of population size on the substitution rate observed in previous stochastic models of neutral evolution (Balloux and Lehmann 2012), a point that was suggested by Lanfear et al. (2014).
This result (Equation 21) also applies to a population with separate sexes (diploid or haplodiploid), in which case the summations in μe,0 and T must also be taken over the different sexes (Appendix D, Equation D4). For a constant demography, the effective mutation rate in newborns can then be simplified to
| (22) |
which is of the same form as Equation 12 and where μg0 denotes the rate at which a gene randomly sampled in a newborn of sex g ∈ {f, m} accumulates mutations. With this, k (Equation 21) reduces to the standard expression for the neutral substitution rate under overlapping generations for diploid reproduction (Pollak 1982, equation 8) (see Appendix D for the connection). This shows that the average mutation rate in newborns from previous models is the effective mutation in newborns, where the sex-specific mutation rate is weighted by conditional reproductive values, which give the proportion of time a gene lineage will spend in males and females given that it resides in a newborn.
Discussion
The rate k of neutral allelic substitution at a given locus has been analyzed under the assumptions that the evolving population can be structured into classes (e.g., by sex, age, geography, etc.) and that it follows a stationary stochastic demography. Two main results were found. First, the substitution rate is equal to the effective mutation rate: k = μe, which is the rate at which a randomly sampled gene from a randomly sampled common ancestor of the population accumulates mutations. Second, in the presence of overlapping generations, the substitution rate can also be expressed as the product k = μe,0/T, where μe,0 is the effective mutation rate in newborns and T is the average generation time (or the mean age of the parents of a newborn), which itself can be expressed entirely in terms of class reproductive values (Equation 20).
These results entail that the effective mutation rate μe (Equation 8) is not necessarily equal to the rate at which a randomly sampled gene in a randomly sampled individual from the population accumulates mutations (Equation 2). Care must thus be taken in interpreting k as a mutation rate. For instance, it is sometimes said that k is equal to the mutation rate under almost any conceivable complication (Lanfear et al. 2014, box 3). But if this mutation rate is supposed to be that in a randomly sampled gamete, as in the original formulation of the neutral substitution process (Kimura 1971), then the substitution rate can be different, unless mutations occurs at the same rate in every class of individuals (Equation 5). But if mutations occur with different magnitude in different classes of individuals, then even if k is measured in units of generation time, it can be different from the rate at which a randomly sampled gene in a newborn accumulates mutations. This occurs because the effective mutation rate in newborns (μe,0) is a reproductive value weighted average of the rate at which newborns accumulate mutations (Equation 16), which thus weights different classes of individuals according to their asymptotic contribution to the gene pool. Different classes of newborns may be obtained when individuals are born under different types of demographic or environmental conditions (or are of different sexes) and where mutation rates can vary during gametogenesis, owing, for instance, to the fact that irradiation can vary in space and time.
In summary, with uniform mutation across classes, the substitution rate is the mutation rate under stochastic demography and class structure. With nonuniform mutation, how different classes of individuals contribute to the ancestry of the population can matter (e.g., Equations 12 and 13), in which case the substitution rate is the average rate at which the ancestors of the population accumulate mutations. This then justifies the interpretation of the substitution rate as the effective mutation rate.
Acknowledgments
I thank F. Balloux and especially C. Mullon for useful discussions. The two reviewers of this article provided constructive comments that allowed me to improve it; many thanks to them. This work was supported by Swiss National Science Foundation grant PP00P3-123344.
Appendix A
Neutral Substitution Rate
Here, we provide a proof of Equation 3. To that end, we first note that the neutral substitution rate can be expressed as the expectation over all demographic states of the expected number NFix(n) of mutants produced per unit time in a population in state n and that will ultimately fix in the population,
| (A1) |
where
| (A2) |
(Lehmann 2012, equations A1 and A2, assuming no selection). Here, πi(n′) is the fixation probability of a mutation arising as a single copy in a class-i individual in population state n′, wij(n′, n) is the expected number of class-i individuals in population state n′ produced by a single class-j individual in population state n (the fitness of an individual of class j through class-i offspring), gi is the ploidy of a class-i individual, tij is the probability that a gene randomly sampled in a class-i offspring has been transmitted by a class-j individual, and Nj is the number of class-j individuals in population state n. Hence, wij(n′, n)gitijμijNj gives the number of mutations in population state n′ when the population was in state n in the previous time step.
The probability aij(n′, n) that a gene randomly sampled in a class-i individual in population state n′ descends from a class-j individual and in population state n in the previous time step can now be written as
| (A3) |
The second term is the backward transition probability that a population in state n′ derives from a population in state n one time step earlier, while
| (A4) |
is the probability that a class-i individual descends from a class-j individual given state n′ in the offspring generation and state n in the parental generation (Charlesworth 1994, p. 81; Rousset 2004, equation 11.2; Lehmann 2012, equation A15), which is obtained as the ratio of the number of genes in class i descending from class j to the total number of genes in class i.
Substituting Equation A3 into Equation A2 yields
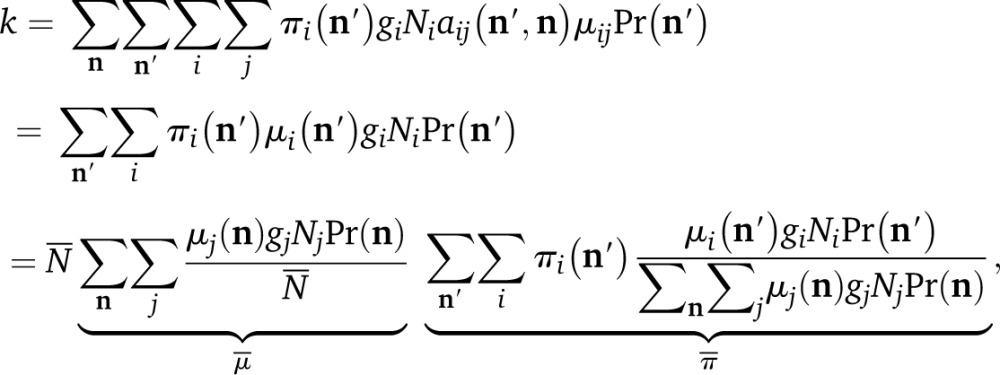 |
(A5) |
which displays k as in Equation 3.
Appendix B
Fixation Probability and Reproductive Value
Here, we provide a proof of Equation 7. To that end, we first write the recursion at steady state for the γ reproductive values, which from Equation 6 reads
| (B1) |
Let us now denote by pi,t(n) the expectation (over replicates of the evolutionary process) of the average frequency of a mutant allele in class i at time t, conditional on the demographic state being n at that time and conditional on some initial mutant frequency distribution at t = 0. This satisfies the recursion
| (B2) |
We can collect the pi,t(n) elements into a vector pt, which satisfies the recursion
| (B3) |
where A is the row stochastic transition matrix collecting the aij(n′, n) elements
Since the fixation probability of a mutant is its asymptotic frequency (e.g., Hill 1972; Emigh and Pollak 1979; Rousset 2004), the fixation probability in a given class i, conditional on the demographic state being n, is limt→∞ pi,t(n). The vector π of fixation probabilities in each class is then given by π = limt→∞ Atp0, where p0 is the initial mutant frequency distribution across classes. Note that each element of the vector π will be the same, as the mutant either fixes in the total population and thus in each class or goes extinct. By standard results for finite ergodic Markov chains, each row of limt→∞ At is equal to the left unit eigenvector of the transition matrix A (Grinstead and Snell 1997), which is precisely the vector satisfying the system of recursions displayed in Equation B1: γ = γA. Hence, using γj(n) = αj(n)Pr(n), the fixation probability of a mutant given initial frequency distribution p0 is
| (B4) |
When a single mutant arises initially in class i and in population state n, this further reduces to αi(n)pi,0(n) (since with probability 1 the state is n), which is precisely Equation 7 since pi,0(n) = 1/[giNi]. This latter equation was also given in Leturque and Rousset (2002, p. 178) for a class-structured population of constant size.
Appendix C
Reproductive Value and Generation Time
Here, we provide a proof of Equation 20. For a haploid age-structured population, with age classes i = 0, 1, 2, … , Equation B1 reduces to
| (C1) |
where the first term in brackets is the contribution to the gene pool of class (j, n) through production of newborns, while the second term is the contribution through survival. For an age-structured population, we also have
| (C2) |
owing to the fact that wj+1j(n′, n) is the survival of an individual of class j given demographic states n′ and n, so that necessarily Nj+1 = wj+1j(n′, n)Nj. This entails that aj+1j(n′, n) = Pr(n′|n)Pr(n)/Pr(n′), whereby Summing both sides of Equation C1 over then yields
| (C3) |
Using the definition of the generation time (Equation 17) and reindexing the last sum, we have
| (C4) |
whereby
| (C5) |
Appendix D
Separate Sexes
Stochastic demography
Here, we extend the result k = μe,0/T found for haploid reproduction (Equation 21) to a population with both males and females, which could be diploid, haplodiploid, or subject to other modes of ploidy. The coefficient a0j(n′, n) for haploids appearing in the previous section is now written as which stems from the probability that a gene randomly sampled in a newborn of sex g ∈ {f, m} in state n′ descends from an individual of sex g′ ∈ {f, m} of age j and that was in state n. Likewise, denotes the mutation rate of a gene in an individual of sex g′ of age j that is transmitted to a newborn of sex g.
With these definitions, the substitution rate (Equation 8) becomes
| (D1) |
where the sex-specific mutation probability in newborns is
| (D2) |
We can define the effective mutation rate among newborns as
| (D3) |
which generalizes Equation 14. Likewise, using the same weights, we can define the average generation time
| (D4) |
which generalizes Equation 18 and where
| (D5) |
Using Equations D1–D5, we can write
| (D6) |
provided that which requires
| (D7) |
We now prove this latter equality by applying the same argument as in the previous section. Namely, the reproductive value γgj(n) satisfies the recursion
| (D8) |
where
| (D9) |
The last equality follows from the fact that necessarily Ngj+1 = wgj+1,gj(n′, n)Ngj and tgg = 1, since during survival no segregation of alleles occurs. If we now take the sum over Equation D8, we obtain
| (D10) |
and by reindexing the last sum, this can be written as
| (D11) |
which shows that Equation D7 holds.
Constant demography
In a population of constant size, the effective mutation rate in newborns and the average generation time can be written
| (D12) |
where
| (D13) |
and
| (D14) |
is the probability that a gene lineage is in a female, given that it has been sampled in a newborn. This probability satisfies the recursion
| (D15) |
since, given that a gene lineage is a newborn female, it descends from a female with probability tff = 1 − tfm and, given that a gene lineage is a newborn male, it descends from a female with probability tmf. At steady state, we have
| (D16) |
Assuming diploid reproduction, tmf = tfm = 1/2, whereby and the expression for the average generation time (Equation D13) can then be seen to be precisely the average of the mean age of the parents of a newborn defined by Pollak (1982, p. 91), since af0,gj (am0,gj) corresponds to () in the notations of Pollak (1982). Likewise, μe,0 in this case is precisely in the notations of Pollak (1982, equations 1 and 2). With all this, we see that in a diploid population of constant size k = μe,0/T reduces precisely to equation 8 of Pollak (1982).
Semelparous populations
Here, we evaluate the class reproductive values and the fixation probability in a population of constant size in the absence of overlapping generations. Since, in this case, wff = 1 and wmf = Nm/Nf, we have from Eqation A4 that fff = tff = 1 − tfm and fmf = tmf. Then from Equations A3 and B1, the reproductive value of the female class satisfies
| (D17) |
whereby
| (D18) |
With this, noting that γi = αi (population size is constant) and using Equation B4, the fixation probability of a single mutant allele that arises in females and males is, respectively, given by
| (D19) |
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Balloux F., Lehmann L., 2012. Substitution rates at neutral genes depend on population size under fluctuating demography and overlapping generations. Evolution 66: 605–611. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1994. Evolution in Age-Structured Populations, Ed. 2 Cambridge University Press, Cambridge, UK. [Google Scholar]
- Emigh T. H., Pollak E., 1979. Fixation probabilities and effective population numbers in diploid populations with overlapping generations. Theor. Popul. Biol. 15: 86–107. [Google Scholar]
- Felsenstein J., 1971. Inbreeding and variance effective numbers in populations with overlapping generations. Genetics 68: 581–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstead C. M., Snell J. L., 1997. Introduction to Probability, Ed. 2 American Mathematical Society, Providence, RI. [Google Scholar]
- Hill W., 1972. Probability of fixation of genes in populations of variable size. Theor. Popul. Biol. 3: 27–40. [DOI] [PubMed] [Google Scholar]
- Karlin S., Taylor H. M., 1975. A First Course in Stochastic Processes. Academic Press, San Diego. [Google Scholar]
- Kimura M., 1969. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1971. Theoretical foundation of population genetics at the molecular level. Theor. Popul. Biol. 2: 174–208. [DOI] [PubMed] [Google Scholar]
- Kimura M., Crow J. F., 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Kokko H., Eyre-Walker A., 2014. Population size and the rate of evolution. Trends Ecol. Evol. 29: 33–40. [DOI] [PubMed] [Google Scholar]
- Lehmann L., 2012. The stationary distribution of a continuously varying strategy in a class-structured population under mutation-selection-drift balance. J. Evol. Biol. 25: 770–787. [DOI] [PubMed] [Google Scholar]
- Leturque H., Rousset F., 2002. Dispersal, kin competition, and the ideal free distribution in a spatially heterogeneous population. Theor. Popul. Biol. 62: 169–180. [DOI] [PubMed] [Google Scholar]
- Nagylaki T., 1998. The expected number of heterozygous sites in a subdivided population. Genetics 149: 1599–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E., 1982. The rate of mutant substitution in populations with overlapping generations. Genet. Res. 40: 89–94. [DOI] [PubMed] [Google Scholar]
- Rousset F., 2004. Genetic Structure and Selection in Subdivided Populations. Princeton University Press, Princeton, NJ. [Google Scholar]
- Taylor P., 1990. Allele-frequency change in a class-structured population. Am. Nat. 135: 95–106. [Google Scholar]
- Wakeley, J., 2008 Coalescent Theory: An Introduction. Roberts and Company, Greenwood Village, CO. [Google Scholar]
- Whitlock M. C., Barton N. H., 1997. The effective size of a subdivided population. Genetics 146: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]


