Abstract
RNA sequencing (RNA-seq) not only measures total gene expression but may also measure allele-specific gene expression in diploid individuals. RNA-seq data collected from F1 reciprocal crosses in mice can powerfully dissect strain and parent-of-origin effects on allelic imbalance of gene expression. In this article, we develop a novel statistical approach to analyze RNA-seq data from F1 and inbred strains. Method development was motivated by a study of F1 reciprocal crosses derived from highly divergent mouse strains, to which we apply the proposed method. Our method jointly models the total number of reads and the number of allele-specific reads of each gene, which significantly boosts power for detecting strain and particularly parent-of-origin effects. The method deals with the overdispersion problem commonly observed in read counts and can flexibly adjust for the effects of covariates such as sex and read depth. The X chromosome in mouse presents particular challenges. As in other mammals, X chromosome inactivation silences one of the two X chromosomes in each female cell, although the choice of which chromosome to be silenced can be highly skewed by alleles at the X-linked X-controlling element (Xce) and stochastic effects. Our model accounts for these chromosome-wide effects on an individual level, allowing proper analysis of chromosome X expression. Furthermore, we propose a genomic control procedure to properly control type I error for RNA-seq studies. A number of these methodological improvements can also be applied to RNA-seq data from other species as well as other types of next-generation sequencing data sets. Finally, we show through simulations that increasing the number of samples is more beneficial than increasing the library size for mapping both the strain and parent-of-origin effects. Unless sample recruiting is too expensive to conduct, we recommend sequencing more samples with lower coverage.
Keywords: allelic imbalance, imprinting, overdispersion, parent-of-origin effect, Xce
HIGH-THROUGHPUT RNA sequencing (RNA-seq) is an increasingly popular technique to measure gene expression abundance (Mortazavi et al. 2008; Wang et al. 2009). RNA-seq offers several advantages over microarrays. For example, RNA-seq data are often less noisy with a larger dynamic range than microarray data. In addition, RNA-seq offers the opportunity to identify new transcripts while the detection capability of microarrays tends to be limited by microarray probes (Wang et al. 2009). Furthermore, RNA-seq is able to measure allele-specific expression (ASE), which requires special methods to attempt using microarrays. The transcript abundance of each allele (i.e., the ASE) allows dissection of cis- and trans-regulation (Doss et al. 2005; Ronald et al. 2005). ASE from reciprocal F1 mouse hybrids (Babak et al. 2008; Wang et al. 2008; Gregg et al. 2010a,b; Deveale et al. 2012; Okae et al. 2012) enables the study of allelic imbalance on gene expression and in particular the imbalance due to parent-of-origin effects.
For RNA-seq data, one analytic strategy to detect differentially expressed genes is to normalize read counts and then to apply linear regression or equivalent approaches commonly used for microarray data (Cloonan et al. 2008; ‘t Hoen et al. 2008; Langmead et al. 2010). However, these approaches do not fully consider the characteristics of read count data and are thus not efficient. More sophisticated approaches are to directly model the count data (Oshlack et al. 2010; Robinson and Oshlack 2010; Skelly et al. 2011; McCarthy et al. 2012), which include generalized regression models and chi-square tests on contingency tables. Count models tend to have higher statistical power for detecting differentially expressed genes than approximate normal models (Robinson and Oshlack 2010). However, overdispersion where the variance of read counts is greater than would be expected from simple Poisson or binomial distribution has been commonly observed in count data, including RNA-seq data (Robinson and Oshlack 2010). To overcome the overdispersion problem of RNA-seq data, several groups have proposed, for example, negative binomial and β-binomial models (Skelly et al. 2011; Zhou et al. 2011; Sun 2012) for detecting differentially expressed genes.
However, these methods are not specifically designed for F1 reciprocals and do not consider the special structure of F1 reciprocal hybrids. They do not specifically model, for example, parent-of-origin effects. The statistical methods used in Wang et al. (2008) and other studies (Babak et al. 2008; Gregg et al. 2010a,b; Deveale et al. 2012; Okae et al. 2012) for reciprocal F1 mouse hybrid data are simply based on binomial distributions. In addition, they test imprinting effects in isolation from strain effects. Joint modeling of strain and parent-of-origin effects is potentially more powerful for detecting imprinting genes. To address these limitations, we extend the eQTL approach of Sun (2012) to F1 reciprocal crosses, simultaneously model the total read counts and allelic-specific counts, and estimate the strain and parent-of-origin effects together. For genes on the X chromosome, we further consider dosage compensation in our model. In mammals, dosage compensation is achieved by inactivating one of the two X chromosomes in female cells. The choice of which X chromosome to be silenced can be nonrandom and has been shown to be biased by alleles at the X-linked X-controlling element (Xce) in mouse. For genes located on the X chromosome, the strain-dependent skewing in X inactivation needs to be modeled to avoid high false positive findings of strain-dependent differentially expressed genes. In addition, for RNA-seq studies with small samples, such as ours, it is critical to check the accuracy of P-values based on asymptotic distributions of test statistics. We use simulations to address this concern and propose a modified procedure to properly control family-wise error or false discovery rate. The rest of the article is arranged as follows. In Methods, we describe the data structure of RNA-seq data and our approach. We then evaluate the method by simulation in the Simulation section. As a case study, we summarize our analysis results on real RNA-seq data derived from brain tissue of reciprocal F1 mouse hybrids and their parental strains. We chose to study three inbred strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ) representing three subspecies of Mus musculus (M. m. castaneus, M. m. musculus, and M. m. domesticus, respectively). These strains were chosen to sample a very high level of genetic diversity and to thoroughly characterize differentially expressed genes among mouse subspecies.
Methods
Throughout this article, we denote each F1 sample by its maternal strain × paternal strain. For example, a CAST × WSB mouse is an offspring of a CAST female that is mated with a WSB male. For simplification, we define the two parental strains as A and B. Suppose there are totals of K1 F1 samples (either A × B or B × A) and K2 inbred samples (either strain A or strain B). For a particular gene of interest, we have the total number of reads from each sample, denoted as ml for l = 1, 2, … , K1 + K2. For each F1 sample, we may have two additional counts, allele-specific reads that are mapped to strain A and strain B, denoted by niA and niB (i = 1, . . . , K1), respectively. Let ni = niA + niB, the total allele-specific read counts. Further, for the ith F1 sample, let xi be the cross indicator such that xi = 1 or −1 if the sample is an A × B or a B × A cross, respectively.
Total read count plus allele specific expression (TReCASE) model
We group genes into two groups, one with both total read count (TReC) and allele specific expression (ASE) and another with only TReC. In this subsection, we describe our TReCASE model for genes in the first group with both TReC and ASE. We further subdivide the genes in the first group into autosomal genes and chromosome X genes since genes on the X chromosome deserve a special treatment.
Autosomal genes:
We assume niB follows a β-binomial distribution that extends a binomial distribution and allows for possible overdispersion,
| (1) |
where πi is the expected proportion of ASE of F1 sample i that are mapped to strain B and φ is the overdispersion parameter. When φ = 0, no overdispersion exists, and niB follows a binomial distribution. To model the sex effect, we create a dummy variable sexi such that sexi = 1 if sample i is a female, otherwise sexi = 0. The following logistic regression is used for linking πi with the strain and parent-of-origin effects plus the sex effect,
| (2) |
where the regression coefficients b0F and b1F correspond to the strain and parent-of-origin effects in females, respectively, and b0M and b1M are the strain and parent-of-origin effects in males, respectively.
The following discussions can help us to understand b0F and b1F (and analogously, b0M and b1M). For a female sample, let define its expected expression of strain B when strain B is its paternal (maternal) allele. Similarly and can be defined. Then from the above logistic regression model, we have
| (3) |
and
| (4) |
For the TReC, ml(l = 1, … , K1 + K2), we assume it follows a negative binomial distribution with mean μl and an overdispersion parameter ϕ. Specifically, we have
| (5) |
where kl = log(library size of sample l), doml = 0 if sample l is an inbred sample, and otherwise doml = 1. The sex effect β2 = . The term ηl is related to the additive allelic effect that we describe below in detail. To facilitate the joint modeling of ASE and TReC, we make the following assumptions for F1 females:
Similar assumptions are made for F1 males. Then for females, the expected TReCs due to the additive allelic effect for the four crosses are
| (6) |
By taking the A × A females as the reference group, we end up withThe joint likelihood of the combined F1 and inbred samples is therefore
where Θ = (b0F, b0M, b1F, b1M, β0, β1, β2, β3, β4, φ, ϕ).
We test the strain and parent-of-origin effects on the following hypotheses,
with likelihood-ratio testing.
In the above model, we assume the strain effects from ASE and TReC are the same for model parsimony. For genes that do not show the consistent strain effects from ASE and TReC, we relax the assumption and replace b0F and b0M in ηl with and , respectively. The hypothesis for the overall strain effect then becomes
We can also test the consistency of the strain effects in ASE and TReC according to
Chromosome X genes:
As mentioned in the Introduction, due to X chromosome inactivation, one of the two X chromosomes in each female cell is silenced but the choice of which chromosome to be silenced can be nonrandom and is biased by the Xce allele. For female F1 sample i, let τi,A and τi,B define the proportions of the cells where the expressed copies of the X chromosome are the A allele and the B allele, respectively. Thus τi,A + τi,B = 1. Further, let and be the expression of the B allele (across a large number of cells) for the ith female sample when the B allele is the paternal or the maternal allele. Similarly, we can define and . For male samples, we define , , , and accordingly.
For ASE, we assume niB follows the β-binomial model (1) but replace πi in (2) with one that satisfies
| (7) |
for F1 females.
For the TReC ml, we again use model (5) but replace ηl in it by
for female samples.
Males only have one X chromosome and it is always maternally inherited. Therefore no parent-of-origin effect and no X inactivation exist, leading us to replace ηl in model (5) by
for male samples.
TReC model
Autosomal genes:
For genes with only TReC, model (5) cannot be directly applied. There is an identifiability problem on the parent-of-origin effect: when no strain effect exists, the parent-of-origin effect in model (5) is unidentifiable. Specifically, when plugging b0F = 0 into the equations of (6), we end up with the same mean expression for all four groups (A × A, A × B, B × A, and B × B), leading to the identifiability problem of b1F. The ASE data help us to avoid the identifiability problem. However, for genes with only TReC, we need an alternative solution, which we propose below by reparameterizing model (5),
| (8) |
where zl = 0 if sample l is an inbred and 1 if it is an A × B sample, and otherwise zl = −1, and
It is easy to check that when b0F = b0M = 0 in model (5), β5 and β6 in (8) become 0. Model (8) avoids the identifiability problem of model (5) but essentially has no power for detecting the imprinting effect in the absence of the strain effect, which is demonstrated in the Simulation section.
Chromosome X genes:
For chromosome X genes with only TReC, we modify model (5) accordingly and consider the following model,
| (9) |
where wl = 0 for all males and also for female inbreds, wl =1 for A × B females, and wl = −1 for B × A females; and
for females. For males,
Note that in the above model, we restrict the parent-of-origin effect, β5 to females. This makes sense since males only have one copy of the X chromosome that is always maternally inherited and gene expression from males does not provide imprinting information for genes on the X chromosome. In models (7) and (9), we need to know τl,A and τl,B, which we propose to estimate globally using all X chromosome genes that have enough allele-specific counts. We may jointly estimate τl,A and τl,B with other parameters, but this can cause model instability for small RNA-seq studies and becomes computationally very intensive as well.
Test statistics inflation adjustment
For each test associated with the models described above, we employ the likelihood-ratio test, which follows a chi-square distribution asymptotically. However, for RNA-seq data with a small number of samples, the asymptotic result may not hold. The P-values based on the chi-square distribution can sometimes be very liberal (see the results in the Simulation section), resulting in a highly inflated type I error or false discovery rate. To overcome this problem, we adopt the genomic control (GC) approach (Devlin and Roeder 1999). The GC approach was originally developed for controlling the inflation of test statistics observed in association studies with population substructures or cryptic relatedness. We follow the same idea of the GC approach. Specifically, we assume that our original test statistics, Tj (j = 1, … , M) ∼ λχ2, where M is the total number of genes tested. When the asymptotic distribution is approximated, λ ≈ 1. However, for studies with limited sample sizes, the asymptotic distribution may not attain, and the inflation factor λ might depart from 1. With the large number of tests performed in RNA-seq studies, we empirically, by following the GC approach, estimate λ as
and rescale the original test statistics Tj to . We then compare with the chi-square distribution for P-value calculation. This procedure should perform well when the number of differentially expressed genes is small. However, if the number of differentially expressed genes is large, λ can be upwardly biased, leading to a severe power loss. For real data where the proportion of differentially expressed genes is high, we alternatively propose the following empirical permutation procedure:
-
1.
For each gene j (j = 1, … , M), permute the sample labels and repeat the data analysis on the permuted data and define the permuted test statistic as .
-
2.
Let .
-
3.
Repeat steps 1 and 2 a large number of times and average the -values.
The final averaged value is set as and used for calculating the ’s.
Simulation
To evaluate the performances of the proposed models, we generated ASE and TReC from model (1) with varying strain and parent-of-origin effects. We also varied the sample size and library size, as well as the proportion of allele-specific reads over TReC, and investigated how each of those factors affects power.
To make fair power comparisons, we first investigated the inflation of the test statistics and evaluated the performance of the proposed GC procedure. Let the numbers of female and male A × B samples be nF,A×B and nM,A×B, the numbers of female and male B × A samples be nF,B×A and nM,B×A, and the numbers of female and male inbred samples be nF,A×A and nM,A×A and nF,B×B and nM,B×B, respectively. In all our simulations, we set nF,A×B = nM,A×B = nF,B×A = nM,B×A = nF,A×A = nF,B×B = n0 and nM,A×A = nM,B×B = n1 with n0 > n1 to mimic the sample size structure of the real mouse data and varied n0 and n1. We set the overdispersion parameter φ of the β-binomial and the overdispersion parameter ϕ of the negative binomial to 1. In addition, we set β0 = log(10−5) = −11.5 and β1 = 1 in all simulations. The parameters were chosen based on the corresponding parameter estimates from the real mouse data. The library size κl of each sample was generated uniformly from [20M, 80M] (l = 1, … , K1 + K2). Conditioning on the sampled library size κl, we simulated TReC for 10,000 genes.
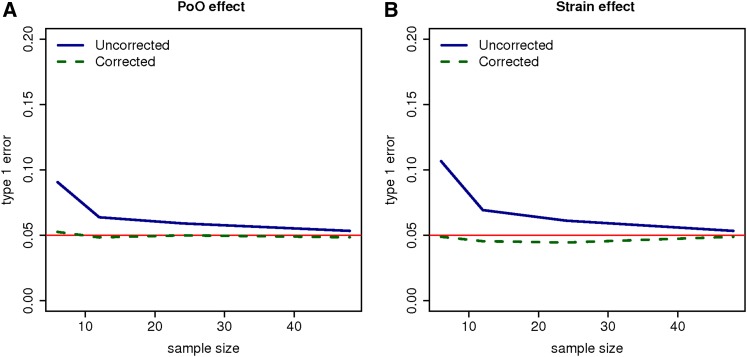
Figure 1 displays the type I error of the TReCASE model before and after the GC correction. Clearly, when the sample size is small, naive P-values from the original uncorrected test statistics are liberal, resulting in highly inflated type I errors. However, as sample size increases, the type I error inflation decreases. The proposed GC correction works well regardless of whether the sample size is small or large and has type I error controlled at the targeted level of 0.05. Similar conclusions were observed for the TReC model. For the remainder of this article, power is calculated based on the GC-corrected test statistics.
Figure 1.
Type I error of the TReCASE model for testing the (A) parent-of-origin (PoO) and (B) strain effects before and after the GC correction. The targeted type I error is 0.05. The red horizontal lines refer the type I error of 0.05.
Our next simulation compared the power of the TReC model with that of the TReCASE model. Table 1 reports the power where the targeted type I error is set to 0.05. In this simulation, the strain and parent-of-origin effects for males and females were set the same and n0 was fixed at 6 and n1 was set to 2. Clearly, the TReCASE model dramatically improves power for detecting both the strain effect and the parent-of-origin effect compared to the TReC model. In addition, the TReCASE model lacks power for testing the parent-of-origin effect in the absence of the strain effect. As expected, the TReC model has almost no power in mapping the parent-of-origin effect. This phenomenon provides strong support for the usage of RNA-seq data over microarray data for studying allelic imbalance on gene expression. For further comparisons, we ignored the overdispersion issue and analyzed the simulated data with simple Poisson and binomial models referred as simple analysis. For testing the strain effect, we combined the test statistics from the Poisson model on TReC and those from the binomial model on ASE. For testing the parent-of-origin effect, we applied the binomial model to ASE. The simple analysis was performed by the R function glm and the results are presented in Table 1. Clearly the simple analysis has a lower power for testing the strain and parent-of-origin effects. Moreover, it is worth mentioning the type I error inflation of the simple analysis in testing the parent-of-origin effect and that the GC-corrected procedure fails the task to control the type I error properly.
Table 1. Power analysis with data from model (1).
| TReC | TReCASE | Simple | |||||
|---|---|---|---|---|---|---|---|
| b0 | b1 | Strain | PoO | Strain | PoO | Strain | PoO |
| 0 | 0 | 0.052 | 0.051 | 0.052 | 0.046 | 0.048 | 0.047 |
| 0 | 0.5 | 0.055 | 0.051 | 0.053 | 0.310 | 0.053 | 0.170 |
| 0 | 1 | 0.051 | 0.049 | 0.049 | 0.841 | 0.049 | 0.525 |
| 0.5 | 0 | 0.106 | 0.049 | 0.288 | 0.050 | 0.201 | 0.103 |
| 0.5 | 0.5 | 0.106 | 0.058 | 0.289 | 0.318 | 0.199 | 0.297 |
| 0.5 | 1 | 0.105 | 0.068 | 0.293 | 0.854 | 0.201 | 0.691 |
| 1 | 0 | 0.321 | 0.051 | 0.866 | 0.054 | 0.589 | 0.210 |
| 1 | 0.5 | 0.331 | 0.062 | 0.878 | 0.337 | 0.591 | 0.458 |
| 1 | 1 | 0.313 | 0.117 | 0.882 | 0.879 | 0.584 | 0.821 |
PoO: parent-of-origin effect.
To evaluate the performances of the proposed models when the model assumptions are violated, we next generated a new set of data, using the Flux Simulator (Griebel et al. 2012), which models RNA-seq experiments in silico. It uses reference genomes according to annotated transcripts to generate sequencing reads. The simulation pipeline adds common sources of systematic bias due to, for example, fragmentation and PCR amplification to the produced reads by in silico library preparation and sequencing. The simulation setups were similar to the previous ones except that we made some minor tweaks to ensure adequate power. That is, we kept the sample size and strain and parent-of-origin effects the same but modified the library sizes. After discarding all poly(A) reads in produced .bed files, we counted the remaining reads gene by gene and sampled a fraction of those reads to produce allele-specific reads. Table 2 summarizes the power where the type I error is set to 0.05. Clearly, the TReCASE model outperforms the simple analysis. Interestingly, the simple analysis has well controlled type I error at 0.05 for testing the strain effect in the previous simulation. However, when data are simulated from the Flux Simulator, the simple analysis has an inflated type I error when testing either the strain or the parent-of-origin effect. The GC-corrected procedure apparently is not powerful enough to deal with the additional noise in the data created by the Flux Simulator. On the other hand, the TReCASE and TReC models are relatively robust to the model misspecification and have the type I error reasonably controlled at 0.05. For genome-wide RNA-seq analysis, it is of great interest to also investigate whether the GC-corrected procedure can control the type I error at lower significance levels. To address this concern, we increased the number of simulations from 10,000 to 2 million and Table 3 summarizes the results under various significance levels. The results confirm that for the TReCASE and TReC models, the GC-corrected procedure works reasonably well even when the significance level is as low as 10−5, no matter whether the data were from model (1) or the Flux Simulator. However, for the simple analysis, the GC-corrected procedure produced poorly controlled type I errors when the data were simulated from the Flux Simulator.
Table 2. Power Analysis with Data from Flux Simulator.
| TReC | TReCASE | Simple | |||||
|---|---|---|---|---|---|---|---|
| b0 | b1 | Strain | PoO | Strain | PoO | Strain | PoO |
| 0 | 0 | 0.061 | 0.049 | 0.054 | 0.053 | 0.164 | 0.162 |
| 0 | 0.5 | 0.055 | 0.046 | 0.052 | 0.233 | 0.147 | 0.245 |
| 0 | 1 | 0.060 | 0.052 | 0.049 | 0.695 | 0.154 | 0.457 |
| 0.5 | 0 | 0.110 | 0.049 | 0.226 | 0.054 | 0.258 | 0.186 |
| 0.5 | 0.5 | 0.106 | 0.056 | 0.226 | 0.252 | 0.230 | 0.268 |
| 0.5 | 1 | 0.120 | 0.067 | 0.204 | 0.689 | 0.201 | 0.436 |
| 1 | 0 | 0.318 | 0.049 | 0.766 | 0.055 | 0.483 | 0.254 |
| 1 | 0.5 | 0.327 | 0.079 | 0.753 | 0.248 | 0.452 | 0.343 |
| 1 | 1 | 0.326 | 0.103 | 0.756 | 0.708 | 0.411 | 0.530 |
PoO: parent-of-origin effect.
Table 3. Type I error analysis.
| TReC | TReCASE | Simple | ||||
|---|---|---|---|---|---|---|
| αa | Strain | PoO | Strain | PoO | Strain | PoO |
| Data generated from model (1) | ||||||
| 5E-02 | 4.95E-02 | 5.06E-02 | 4.94E-02 | 5.07E-02 | 4.99E-02 | 4.89E-02 |
| 1E-02 | 9.74E-03 | 1.01E-02 | 9.74E-03 | 1.01E-02 | 1.01E-02 | 9.55E-03 |
| 1E-03 | 8.88E-04 | 9.76E-04 | 8.92E-04 | 1.02E-03 | 9.88E-04 | 9.40E-04 |
| 1E-04 | 7.90E-05 | 9.40E-05 | 8.25E-05 | 9.75E-05 | 1.02E-04 | 8.25E-05 |
| 1E-05 | 5.50E-06 | 5.50E-06 | 9.00E-06 | 7.50E-06 | 1.40E-05 | 8.50E-06 |
| Data generated from Flux Simulator | ||||||
| 5E-02 | 5.23E-02 | 4.93E-02 | 5.10E-02 | 5.24E-02 | 1.60E-01 | 1.53E-01 |
| 1E-02 | 1.10E-02 | 1.09E-02 | 1.00E-02 | 1.13E-02 | 1.01E-01 | 9.41E-02 |
| 1E-03 | 1.21E-03 | 1.44E-03 | 1.02E-03 | 1.30E-03 | 6.10E-02 | 5.78E-02 |
| 1E-04 | 1.20E-04 | 8.81E-05 | 8.92E-05 | 2.27E-04 | 4.17E-02 | 3.87E-02 |
| 1E-05 | 6.21E-06 | 3.66E-06 | 7.31E-06 | 4.57E-05 | 2.97E-02 | 2.80E-02 |
Targeted type I error.
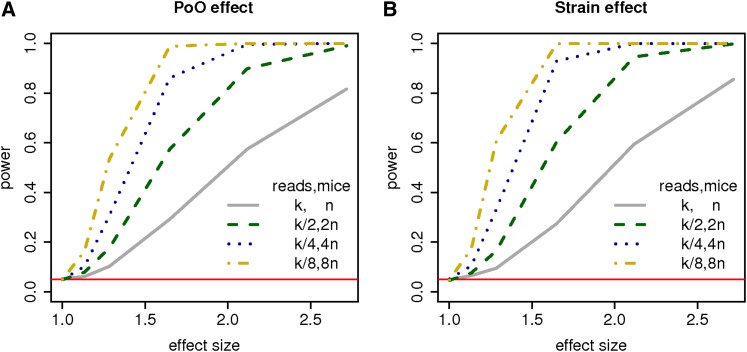
Finally we investigated how each of the following factors—sample size, library size, and the proportion of ASE over TReC—affects power. This addresses an important design question related to RNA-seq studies: With a fixed amount of budget, should we sequence more samples at lower coverage or less samples at higher coverage? To answer this question, we kept the expected total number of reads across all samples constant and varied n0 and n1 (and thus accordingly the library size). The result is presented in Figure 2 and Figure 3. Clearly, increasing the number of samples is more beneficial than increasing the library size for mapping both the strain and parent-of-origin effects. Unless sample recruiting is too expensive to conduct, we recommend sequencing more samples with lower coverage.
Figure 2.
Power of the TReCASE model for the (A) parent-of-origin (PoO) and (B) strain effects with varying sample size and library size. In this simulation, k, the expected TReC, ranges from 201 to 742, and n, the number of samples (mice), is 6. The effect size is calculated as exp(b1) [and exp(b0)] for the PoO effect (and strain effect). The red horizontal lines refer the type I error of 0.05.
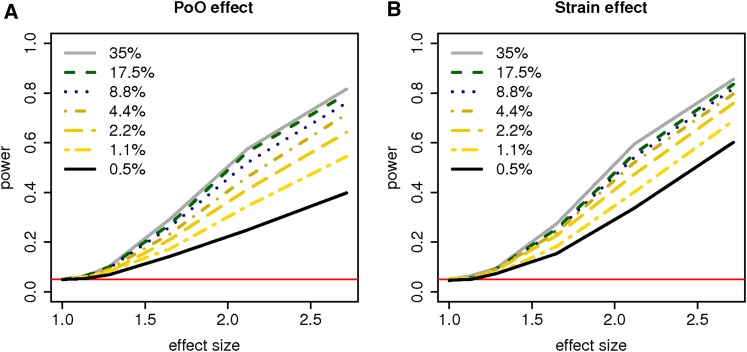
Figure 3.
Power of the TReCASE model for the (A) parent-of-origin (PoO) and (B) strain effects with varying proportion of ASE reads. Each line refers the proportion of ASE simulated out of the TReC. The effect size is defined the same as in Figure 2. The red horizontal lines refer the type I error of 0.05.
We then varied the proportion of ASE to investigate its impact. Figure 3 shows that when the proportion of ASE is low, increasing ASE even by a very small percentage can drastically increase statistical power for testing the strain and parent-of-origin effects. However, when the proportion of ASE is relatively high (e.g., >10%), we gain very little power by further increasing the ASE proportion. Note that the proportion of ASE is determined largely by the DNA similarity of the parental strains, which is out of our control once the parental strains are selected for a given study. We can, however, increase the proportion of ASE by improving the quality of the reference genomes.
Real Data Analysis
This is a small 3 × 3 diallele mouse project conducted for investigating allelic imbalances on gene expression of three wild-derived mouse strains. We focus our analysis on the F1 hybrids from two of the strains, CAST/EiJ and WSB/EiJ. The two strains are incipient species within the M. musculus species group and highly divergent from each other. RNA samples from the whole brains of 12 F1 females (6 of CAST × WSB and 6 of WSB × CAST) and 12 F1 males (again 6 of CAST × WSB and 6 of WSB × CAST) were collected. In addition, RNA samples were also collected from 6 females and 2 males from each of the two inbred strains. The Illumina HiSequation 2000 instrument was used to generate 100-bp paired-end reads (2 × 100) from the 40 samples. The median total number of reads of the 40 samples is ∼28 million after the reads with low-quality score (i.e., phred score <30) were filtered out. Our custom RNA-seq alignment pipeline first aligned reads with high quality from each sample to the pseudogenomes of CAST and WSB, representing each paternal strain genome, using TopHat11 version 1.4. The pseudogenomes are approximations constructed by incorporating all known SNPs and indels of CAST and WSB reported by Wellcome Trust into the mm9 genome. On average, the number of SNPs and/or indels per gene is ∼20 with the standard deviation of 27. We then mapped coordinates from the pseudogenome aligned reads back to mm9 coordinates. Finally, three counts were obtained for each gene in each sample. The first was the total number of (paired-end) reads and the other two were the numbers of allele-specific (paired-end) reads. A paired-end read was allele specific if either end overlapped at least one SNP/indel that was heterozygous between the paternal and maternal strains. If a paired-end read overlapped more than one heterozygous SNP/indel, it was assigned to the allele based on the majority vote of those heterozygous SNPs/indels. We then counted the number of reads mapped to a gene as the number of paired-end reads that overlapped exonic regions of a gene, using the R function isoform/countReads. Exon position information was extracted from the file Mus_musculus.NCBIM37.66.gtf, which was downloaded from Ensembl (http://useast.ensembl.org/info/data/ftp/index.html). Following alignment, we performed a series of quality-control checks, capitalizing on clear expectations for the proportions of reads that should align to each parental strain for the sex, autosomal, and mitochondrial chromosomes. One female CAST sample has nearly 50% of reads mapped to WSB and looks like an F1. We dropped this sample from our analysis.
A gene is defined as expressed if the maximum number of TReCs of the gene across all samples is no less than 50. We restricted our analysis to expressed genes. For each expressed gene, we modeled TReC and ASE jointly unless the maximal ASE of the gene is <5, leaving us to analyze TReC only. The number of significant genes was calculated based on the false discovery rate (FDR) of 0.05 based on the GC-corrected P-values. To further evaluate the GC-corrected P-values, we ran a large number of permutations and pooled all test statistics together, producing in total ∼1 million test statistics that we treated as the null test statistics and used for calculating permutation-based P-values. The GC-corrected procedure allows us to calculate the P-values at finer scales. In contrast, the precision of the permutation P-values is limited by the number of simulations performed. For genes with very large effect sizes, their P-values might be too small to be accurately estimated by the permutation procedure. In Supporting Information, Figure S1 and Figure S2, the GC-corrected and GC-uncorrected P-values for testing the strain effect are plotted against the permutation-based P-values. For genes with corresponding test statistics larger than the maximum of the null test statistics, we arbitrarily set their permutation P-values to 10−6.5. These numbers clearly show that there is a high degree of agreement between the GC-corrected and permutation-based P-values except for the upper right-hand corner ones where the number of permutations is not large enough to allow accurate P-value estimates. However, the GC-uncorrected P-values are consistently larger than the permutation-based ones, again indicating the inflation of the GC-uncorrected P-values. Similar conclusions hold for the P-values testing the parent-of-origin effect (see Figure S3 and Figure S4).
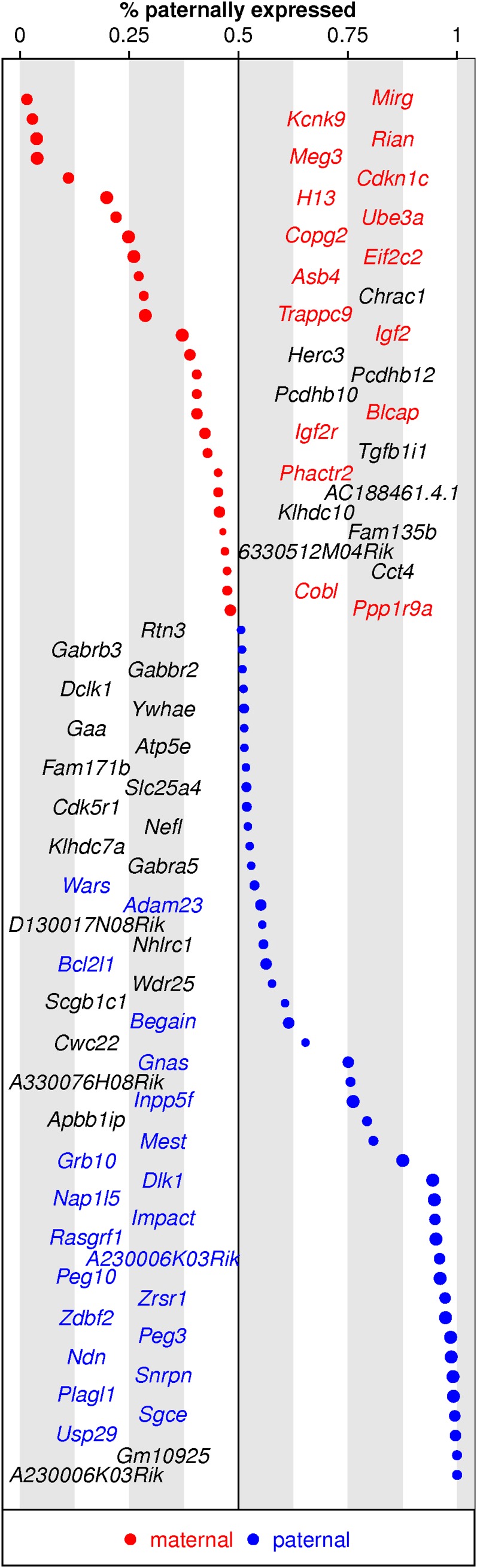
Table 4 summarizes the analysis results. We detected a large number of strain-dependent differentially expressed genes, which we credit to (1) the genetic divergence of CAST and WSB and (2) high-quality RNA-seq data. Figure 4 (left) displays the distribution of the estimated strain effects of the significant genes at FDR = 0.05. We enlarged a small region near the proportion of 0.5 where the strain effects are small (Figure 4, right). The gray and blue curves correspond to the nonsignificant and significant genes, respectively. Clearly, we have declared more nonsignificant genes than significant genes in this small region. However, some genes with small strain effects were detected due to their high number of read counts. The number of significant imprinting genes is smaller. Figure 5 shows that among the 71 identified imprinting genes, 39 of them overlap with the known mouse imprinting genes, the union of imprinting genes collected from the following three sites: http://www.geneimprint.com/site/genes-by-species.Mus+musculus, http://igc.otago.ac.nz, and http://www.mousebook.org/catalog.php?catalog=imprinting. Our estimated imprinting effects are in the same directions as the ones reported. Furthermore, more paternally than maternally expressed genes were detected in our data, which is also consistent with the reported results on the known mouse imprinting genes.
Table 4. Mouse data results.
| No. significant genes | |||||
|---|---|---|---|---|---|
| Chromosome type | No. mapped genes | No. expressed genes | No. expressed genes with ASE | Strain | PoO |
| Autosomes | 30,635 | 14,927 | 11,677 | 8,135 | 71 |
| X chromosome | 1,488 | 522 | 401 | 205 | 0 |
PoO: parent-of-origin effect.
Figure 4.
Histogram of the estimated strain effects of significant genes at FDR = 0.05 (left). The right histogram is an enlarged version of the left histogram around the proportion of 0.5. The x-axis of the right histogram is plotted in the logit scale for easy visualization. The blue and gray curves correspond to the significant and nonsignificant genes, respectively.
Figure 5.
List of the identified imprinting genes. Genes colored red and blue are known maternally and paternally expressed genes, respectively. Genes in black are novel imprinting genes from our analysis. The size of each circle refers to the significance of its corresponding gene (the bigger the circle is, the more significant the gene).
Several studies (Pickrell et al. 2010; Risso et al. 2011) have shown the existence of strong sample-specific GC-content effects on RNA-seq read counts. Our mouse data clearly demonstrate these phenomena (Figure S5). Figure S5 was constructed following exactly the same procedure as that of Pickrell et al. (2010). Although the influence of GC content is clear, the influence is random and nonsystematic with respect to the two parental strains and F1 crosses and thus should have relatively small effects on the differential gene expression analysis. Nevertheless, we included the estimated %GC content as an additional covariate and reanalyzed the data. Figure S6 and Figure S7 are the density scatter plots of the P-values with and without the correction of the %GC content. As expected, the P-values from the two analyses agree reasonably well with each other, especially the P-values for testing the parent-of-origin effect and the ones corresponding to the top ranked genes with strain effects.
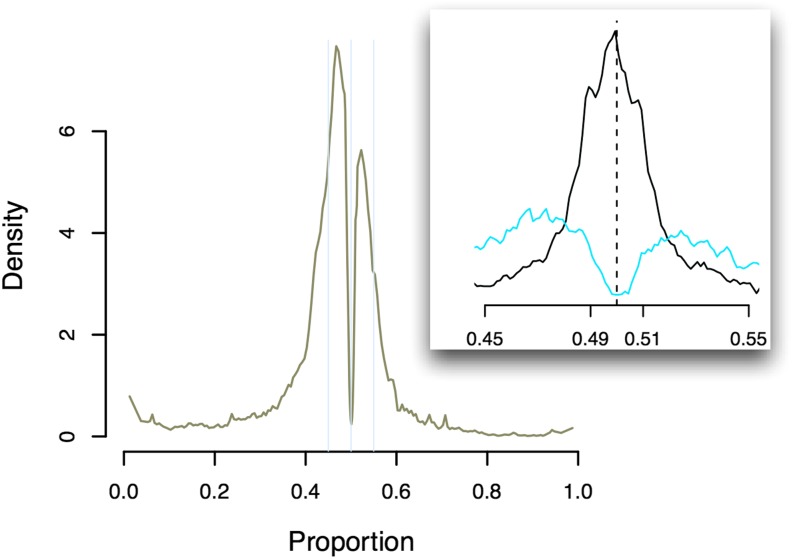
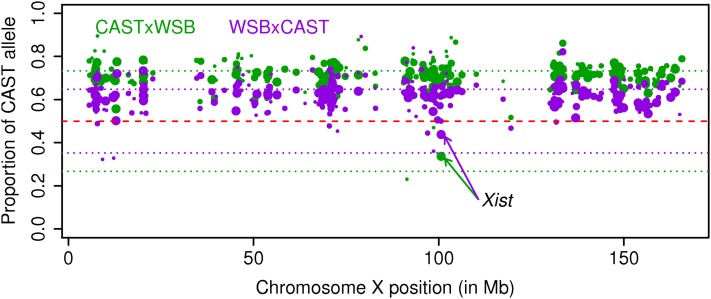
For chromosome X genes, Figure 6 plots the proportion of ASE mapped to the CAST allele of two F1 females (one CAST × WSB and one WSB × CAST). Clearly, for both samples, due to the Xce effect, the CAST allele is overexpressed relative to the WSB allele. The estimated values are 0.73 and 0.63 for the CAST × WSB and WSB × CAST samples, which are far from the 0.5 ratio of autosomal genes. A similar pattern holds for the other F1 samples. Note that one gene, Xist, is known to have a completely opposite inactivation pattern from that of the other genes (Avner and Heard 2001). Our data confirm this. For example, for the same CAST × WSB sample, the proportion of ASE mapped to the CAST allele at gene Xist is ∼0.27 and close to 1 − . Clearly, if the Xce effect were ignored, the majority of the chromosome X genes would be claimed differentially expressed with strain effects. However, after correcting for the Xce effect, our model detected only ∼50% significant genes (Table 4).
Figure 6.
Ratio of the CAST alleles over total ASE for chromosome X genes. The size of each transcript is proportional to the ASE of the gene. The two arrow lines point to gene Xist.
Discussion
In this article, we developed a set of analysis approaches for F1 reciprocal samples coupled with inbred samples. The proposed methods take the special structure of the F1 and inbred samples into consideration and jointly test for strain and parent-of-origin effects. For genes located on chromosome X, our methods adjust the nonrandom X inactivation controlled by the Xce allele, which is important for studying the strain-dependent allelic imbalance on chromosome X. In addition, the methods model both the additive and dominant strain effects and also test the consistency of the strain effects between TReC and ASE. Although the majority of genes show consistent strain effects, we identified some genes with inconsistent strain effects that deserve further investigation. The inconsistency may result from mapping error or other biological reasons.
A particular point of controversy in the mouse community is the number of mouse genes subject to imprinting. Prior to several recent studies, the estimated number of imprinted genes had remained steady at 100–200 for >20 years despite multiple screening efforts. The earliest application of RNA-seq in brain tissue from reciprocal F1 mice yielded a small number of novel imprinted transcripts whereas two more recent studies claimed identification of >1300 imprinted loci (Gregg et al. 2010 a,b). However, a careful reanalysis was unable to replicate these findings and suggested that most of the novel imprinted loci were false due to inaccurate statistical analysis (Deveale et al. 2012; Hayden 2012). As shown by our simulations, for small RNA-seq studies, P-values based on the asymptotic chi-square distribution can be quite liberal, leading to highly inflated type I errors. The studies of Gregg et al. (2010 a,b) are small (with only two F1 samples). One likely reason among many possible reasons for producing such high false positive findings is that their test statistics are highly inflated and the P-values are unadjusted. Our GC procedure greatly reduces the inflation of the type I error and, to our best knowledge, our article is the first to address this important issue.
Due to the nature of this article, we primarily focus on the presentation of the statistical methods and leave the detailed analysis results with more biological insights from the mouse project to another paper (J. J.Crowley, V. Zhabotynsky, W. Sun, S. Huang, I. K. Pakatci, Y. Kim, J. R. Wang, A. P. Morga, J. D. Calaway, D. L. Aylor, Z. Yun, T. A. Bell, R. J. Buus, M. E. Calaway, J. P. Didion, T. J. G. Gooch, S. D. Hansen, N. N. Robinson, G. D. Shaw, J. S. Spence, C. R. Quackenbush, C. J. B. Barrick, Y. Xie, W. Valdar, A. B. Lenarcic, W. Wang, C. E. Welsh, C. P. Fu, Z. Zhang, J. Holt, Z. Guo, D. W. Threadgill, L. M. Tarantino, D. R. Miller, F. Zou, L. McMillan, P. F. Sullivan, F. Pardo-Manuel de Villena, unpublished results). An R package that implements the proposed models can be found online at http://www.bios.unc.edu/~feizou/software/rxSeq.
Supplementary Material
Acknowledgments
The authors are grateful for constructive comments and suggestions from the reviewers and the associate editor. Support was provided in part by National Institute of General Medical Sciences grant R01GM074175 and National Institute of Mental Health/National Human Genome Research Institute Center of Excellence in Genomic Sciences grant P50HG006582.
Footnotes
Communicating editor: C. Kendziorski
Literature Cited
- Avner P., Heard E., 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2: 59–67. [DOI] [PubMed] [Google Scholar]
- Babak T., Deveale B., Armour C., Raymond C., Cleary M. A., et al. , 2008. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 18: 1735–1741. [DOI] [PubMed] [Google Scholar]
- Cloonan N., Forrest A. R. R., Kolle G., Gardiner B. B. A., Faulkner G. J., et al. , 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 5: 613–619. [DOI] [PubMed] [Google Scholar]
- Devlin B., Roeder K., 1999. Genomic control for association studies. Biometrics 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- Doss S., Schadt E., Drake T., Lusis A., 2005. Cis-acting expression quantitative trait loci in mice. Genome Res. 15: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden E. C., 2012. RNA studies under fire. Nature 484: 428. [DOI] [PubMed] [Google Scholar]
- Gregg C., Zhang J., Weissbourd B., Luo S., Schroth G. P., et al. , 2010a High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329: 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C., Zhang J., Butler J. E., Haig D., Dulac C., 2010b Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329: 682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T., Zacher B., Ribeca P., Raineri E., Lacroix V., et al. , 2012. Modelling and simulating generic RNA-Seq experiments with the flux simulator. Nucleic Acids Res. 40: 10073–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Hansen K. D., Leek J. T., 2010. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol. 11: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y., Smyth G. K., 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Okae H., Hiura H., Nishida Y., Funayama R., Tanaka S., et al. , 2012. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 21: 548–558. [DOI] [PubMed] [Google Scholar]
- Oshlack A., Robinson M. D., Young M. D., 2010. From RNA-seq reads to differential expression results. Genome Biol. 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J., Marioni J., Pai A., Degner J., Engelhardt B., et al. , 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Schwartz K., Sherlock G., Dudoit S., 2011. GC-content normalization for RNA-Seq data. BMC Bioinformatics 12: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A., 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J., Brem R., Whittle J., Kruglyak L., 2005. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 1: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly D. A., Johansson M., Madeoy J., Wakefield J., Akey J. M., 2011. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 21: 1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., 2012. A statistical framework for eQTL mapping using RNA-seq data. Biometrics 68: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ‘t Hoen P. A. C., Ariyurek Y., Thygesen H. H., Vreugdenhil E., Vossen R. H. A. M., et al. , 2008. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 36: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sun Q., McGrath S. D., Mardis E. R., Soloway P. D., et al. , 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE 3: e3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M., 2009. Rna-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou Y. H., Xia K., Wright F. A., 2011. A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics 27: 2672–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.