Abstract
Pseudo-homothallism is a reproductive strategy elected by some fungi producing heterokaryotic sexual spores containing genetically different but sexually compatible nuclei. This lifestyle appears as a compromise between true homothallism (self-fertility with predominant inbreeding) and complete heterothallism (with exclusive outcrossing). However, pseudohomothallic species face the problem of maintaining heterokaryotic mycelia to fully benefit from this lifestyle, as homokaryons are self-sterile. Here, we report on the structure of chromosome 1 in mat+ and mat− isolates of strain S of the pseudohomothallic fungus Podospora anserina. Chromosome 1 contains either one of the mat+ and mat− mating types of P. anserina, which is mostly found in nature as a mat+/mat− heterokaryotic mycelium harboring sexually compatible nuclei. We identified a “mat” region ∼0.8 Mb long, devoid of meiotic recombination and containing the mating-type idiomorphs, which is a candidate to be involved in the maintenance of the heterokaryotic state, since the S mat+ and S mat− strains have different physiology that may enable hybrid-vigor-like phenomena in the heterokaryons. The mat region contains 229 coding sequences. A total of 687 polymorphisms were detected between the S mat+ and S mat− chromosomes. Importantly, the mat region is colinear between both chromosomes, which calls for an original mechanism of recombination inhibition. Microarray analyses revealed that 10% of the P. anserina genes have different transcriptional profiles in S mat+ and S mat−, in line with their different phenotypes. Finally, we show that the heterokaryotic state is faithfully maintained during mycelium growth of P. anserina, yet mat+/mat+ and mat−/mat− heterokaryons are as stable as mat+/mat− ones, evidencing a maintenance of heterokaryosis that does not rely on fitness-enhancing complementation between the S mat+ and S mat− strains.
Keywords: mating type, mat region, heterokaryosis, filamentous fungi, Podospora anserina
A dikaryotic stage during a significant portion of the lifecycle is the hallmark of the higher fungi (Ascomycota and Basidiomycota), called for this reason the Dikarya. The dikaryotic part of the life cycle is different in the two groups. In Basidiomycota, mating-competent mycelia fuse and yield the secondary dikaryotic mycelium, upon which basidiospore-bearing dikaryotic fruiting bodies are differentiated. In Ascomycota, fruiting bodies are differentiated around a monokaryotic female gametangium (the ascogonium), which is fertilized by a male gamete (antheridium or spermatium) to yield the dikaryon, which undergoes further development and produces numerous ascospore-containing asci. In Ascomycota, the dikaryotic stage is thus restricted to the sexual lineage inside the fruiting body. There is one exception to this in the Taphrinomycetes, where a dikaryotic mycelium is formed as part of the life cycle (Martin 1940). Ascomycota are nonetheless able to exhibit heterokaryotic mycelia following somatic fusion between genetically different individuals (Buller 1933). In Basidiomycota, a special structure (the clamp) enables the maintenance of the dikaryotic state in the mycelium. No such cellular mechanism faithfully maintaining heterokaryosis in mycelia is known in Ascomycota (while one exists in the sexual lineage). In the absence of nuclear mixing, mathematical modeling showed that heterokaryotic thalli should often break down into homokaryons (Roper et al. 2013).
Pseudohomothallic Ascomycota, such as Podospora anserina and Neurospora tetrasperma, produce mat+/mat− (or mat A/mat a) dikaryotic ascospores, which upon germination yield a self-fertile heterokaryotic mycelium (Raju and Perkins 1994). In addition to being able to engage in self-reproduction, this mycelium is also able to mate with any partner it may encounter. Moreover, in both species, homokaryotic strains can be obtained from ascospores or, in the case of N. tetrasperma from conidia, allowing these organisms to outcross (Dowding 1931; Dodge 1932; Raju 1992). The versatility in breeding behavior probably provides a selective advantage to pseudohomothallism (Billiard et al. 2012). Pseudohomothallic ascomycetes therefore unveil interesting questions regarding maintenance of mycelial heterokaryosis in absence of obvious cellular mechanism of nuclear management. In N. tetrasperma, different populations produce different levels of homokaryotic conidia, arguing for a genetic regulation of heterokaryosis, affecting the possibility of outcrossing (Corcoran et al. 2012). In P. anserina, a coprophilous inhabitant of dung, there are no asexual conidia and reproduction is achieved only by sexual reproduction. Ascospores are produced within a fruiting body and are forcibly expelled at maturity. To germinate they need to pass through the digestive track of an herbivore. Homokaryotic ascospores often remain attached with heterokaryotic ones when they are expelled from the fruiting body (Dowding 1931). Under these conditions, loss of heterokaryosis can be mostly achieved during mycelial growth within dung. It is known from previous studies that phenotypic differences exist between the S mat+ and S mat− mycelia of P. anserina S strain. For example, S mat+ and S mat− grow at the same rate as S mat+/mat−, yet the S mat− strain presents a shorter life span than the S mat+ strain (Marcou 1961), a differential suppression of the su8-1 suppressor tRNA (Silar et al. 2000), triggers the “premature death” syndrome more frequently (Belcour et al. 1991) and is slightly more thermoresistant (Contamine et al. 2004). More recently, genome-wide microarray analysis revealed that many genes are differentially transcribed in S mat+ and S mat− strains (Bidard et al. 2011). Different hypotheses may relate these differences between S mat+ and S mat− strains to the pseudohomothallic lifestyle. It is possible that mat+/mat− heterokaryons may be fitter than either one of the mat+ or mat− homokaryons thanks to some genetic complementation between the mat− and mat+ mating types, and thus promoting inbreeding. On the contrary, mat+/mat− heterokaryons may rapidly break down because of some incompatibility between the mat+ and mat− mating types, resulting in frequent outbreeding. It is also possible that these differences have no relation to the P. anserina lifestyle.
Here, we characterize the mating-type region (mat) of P. anserina strain S, a large polymorphic region that differentiates the S mat+ and S mat− strains. While the mating-type idiomorphs have been well studied (Picard et al. 1991; Debuchy and Coppin 1992; Turgeon and Debuchy 2007), they are embedded in an uncharacterized genomic region devoid of recombination (Marcou et al. 1979). The extent of this “mat region” is not known, nor are the differences between the two mating types apart from those located at the mating-type idiomorphs and at the rmp loci. rmp is a gene that has been shown to be polymorphic between the mat+ and the mat− strains in some P. anserina isolates and its impact on physiology has been characterized (Contamine et al. 2004). By fine genetic mapping and complete sequencing of the P. anserina S mat− strain and comparison with the previously known sequence of the S mat+ strain (Espagne et al. 2008), we define the borders of the mat region and characterize the genetic differences between the two strains. We also identify by microarray analysis the genes differentially expressed between the two strains and whose differential expression does not rely upon the mating-type genes per se, but rather on the polymorphic genes present within the mat region. Finally, we show that mat+/mat− heterokaryotic mycelia are very stable and present evidence that, despite the genetic and phenotypic differences of the S mat+ and S mat− strains, the stability of heterokaryon does not appear to rely on fitness-enhancing complementation.
Materials and Methods
Strains and culture conditions
The strains used in this study derived from the “S” wild-type strain (Rizet and Delannoy 1950). The S mat+ strain was used for resequencing. Its genome sequence and expressed sequenced tag (EST) are available at http://podospora.igmors.u-psud.fr. Standard culture conditions, media, and genetic methods for P. anserina have been described (Rizet and Engelmann 1949; Silar 2013) and the most recent protocols can be accessed at http://podospora.igmors.u-psud.fr/methods.php. Segregation analysis was made with homokaryotic ascospores.
Hygromycin B and phleomycin-resistant strains were obtained by transforming strain S with plasmids derived from pBC-hygro and pBC-phleo (Silar 1995). Primary transformants were crossed with wild type and progeny analysis permitted selection of transgenes segregating independently from the mating type and harboring no phenotypic alteration. In the progeny of these crosses, the same transgenes were associated with either mat+ or mat−.
Construction of the mat− library
DNA from the P. anserina S mat− strain was extracted as described in Cheeseman et al. (2014). The P. anserina S mat− DNA was nebulized and 1.7 µg of fragmented DNA was used for library preparation according to the protocol for multiplexed paired-end sequencing of Illumina (part 1005361 RevB, December 2008). Cloning of an aliquot of the library in the pCR4BluntTOPO– (Invitrogen) and analysis of 20 recombinant plasmids indicated that 15 had an insert. Sequencing of 10 inserts indicated that 6 had the upstream and downstream borders required for Illumina multiplex sequencing.
Sequencing
For the resequencing of S mat+ with the 454 technology, two banks were generated. The first one with 3-kb inserts was sequenced in GsFlex paired reads, generating fivefold coverage. The second bank was used for single Titanium reads generating 17-fold coverage. The combined sequences were assembled with Newbler (software release 2.3-PreRelease-10/19/2009; 454 Life Sciences, Branford, CT). This assembly was colinear with the previous one (Espagne et al. 2008) and was used for gap filling and sequencing errors correction using custom-made programs. Accession number is PRJNA12954. For the Illumina sequencing, this work has benefited from the facilities and expertise of the high-throughput sequencing platform of IMAGIF (Centre de Recherche de Gif, http://www.imagif.cnrs.fr). Custom-made libraries had 300-bp inserts and sequencing was 76-bp paired end. For assembly, this work has benefited from the facilities and expertise of the eBio plateforme (IGM, Orsay). Final assembly of the S mat− genome was completed with the Velvet program and comparison of S mat+ and S mat− sequences (Zerbino and Birney 2008) with the samtools package (Li et al. 2009) and custom-made programs. Accession number for the mat region of the S mat− strain is HG934340.
RNA extraction and microarray analysis
Microarray analysis was conducted as previously described (Bidard et al. 2010, 2011, 2012). Briefly, four biological replicates for each strain (mat−, mat+, fmr1−, and fpr1−) were grown on minimal medium covered with a cellophane sheet at 27° under constant light for 96 hr. This stage corresponds to fertilization competent mycelium (Bidard et al. 2011). Mycelia were harvested by scraping the cellophane surface, frozen in liquid nitrogen and ground in a Mikro-Dismembrator (Sartorius, Göttingen, Germany), and total RNA was purified with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The quality of RNA was controlled on a Bioanalyzed 2100 sytem (Agilent, Santa Clara, CA). One-microgram aliquots of total RNA were amplified and Cy-labeled with Agilent low RNA input fluorescent linear amplification (LRILAK) plus kit and the two-color RNA spike-in kit (Agilent, Santa Clara, CA). The labeling efficiency and the product integrity were checked as described previously (Imbeaud et al. 2005). The microarray consisted of a 4 × 44K platform (Agilent) containing 10,556 probes on each array with each probe present in four replicates (Bidard et al. 2010). The four biological replicates labeled with Cy-3 for each strain were compared with a common reference labeled with Cy-5, in indirect comparisons. The composition of the common reference was as indicated in Bidard et al. (2010). Spot and background intensities were extracted with the Feature Extraction (FE, v. 9.5.3) software (Agilent) using the GE2-v4_95_Feb07 default protocol. FE-software normalized data (Lowess normalized, local background subtracted) were processed with MAnGO (Marisa et al. 2007). A moderated t-test with adjustment of P-value (Benjamini and Hochberg 1995) was computed to measure the significance of each difference of transcript level. Genes were considered as differentially transcribed if P-values were <0.005. Data are accessible through GEO Series accession number GSE27297 and Excel files (Supporting Information, Table S3). Fold-changes for each P. anserina gene in mat+ vs. mat−, fpr1- vs. fmr1−, mat+ vs. fpr1−, and mat− vs. fmr1− comparisons are available from Bidard et al. (2011).
Gene deletions
Centromere-like region (CLR) and Pa_1_18960 were deleted by the split marker techniques as described in Grognet et al. (2012). Pa_1_19950, Pa_1_20280, and Pa_1_18270 were deleted in S mat+ and in S mat− as described in Bidard et al. (2011). Molecular analysis was performed using standard protocols (Ausubel et al. 1987) and as in Grognet et al. (2012).
In silico analysis
Prediction of mitochondrial or extracellular localization was performed with WoLF PSORT (Horton et al. 2007). P-values were computed on http://www.graphpad.com/quickcalcs/contingency1.cfm. The figures used for P-values computing are shown in Table S1.
Heterokaryon generation and analysis
Heterokaryon were generated by mixing 1 mm3 explants of the appropriate genotype in 500 µl sterile water with Fastprep FP 120 (MP Biomedicals, Solon-Ohio, Burlingame, CA) at speed 4 for 20 sec. Inoculations were made with 5 µl of the recovered solution at the edge of the petri plates. Under these conditions, heterokaryons formed spontaneously (Silar 2011). For each heterokaryon tested, the experiments were made at least in triplicate at two different times (e.g., a minimum of six plates were tested).
Results
Structure of the P. anserina mat region
As a prerequisite to obtaining an accurate comparison between the mat+ and mat− strains, we resequenced the S mat+ strain, previously sequenced by the Sanger method (Espagne et al. 2008), using the 454 Roche technology with 22-fold coverage and the GA2x technology from Illumina Technology to an 80-fold coverage. Incorporating these data into the existing assembly enabled us to correct numerous sequencing errors (over 3000), as well as to fill most gaps present in nonrepeated sequences. Finally, the few gaps remaining in nonrepeated sequences were filled manually by sequencing PCR products with extremities unambiguously overlapping gap borders. The S mat+ sequence is thus now composed of seven scaffolds representing the seven chromosomes of the nuclear genome, devoid of gaps in unique sequences, and one for the circular mitochondrial chromosome. Missing sequences remain in repeated regions of the nuclear scaffolds, especially at the telomeres and centromeres. Moreover, orientation for two contigs (one at the centromere of chromosome 1 and one at a telomere of chromosome 7) could not be accurately determined because of lack of meiotic recombination, small sizes, and insufficient paired-end sequence data. These contigs are bordered by large regions containing transposons modified by repeat induced point mutation (RIP; Galagan and Selker 2004) and whose sequences are incomplete.
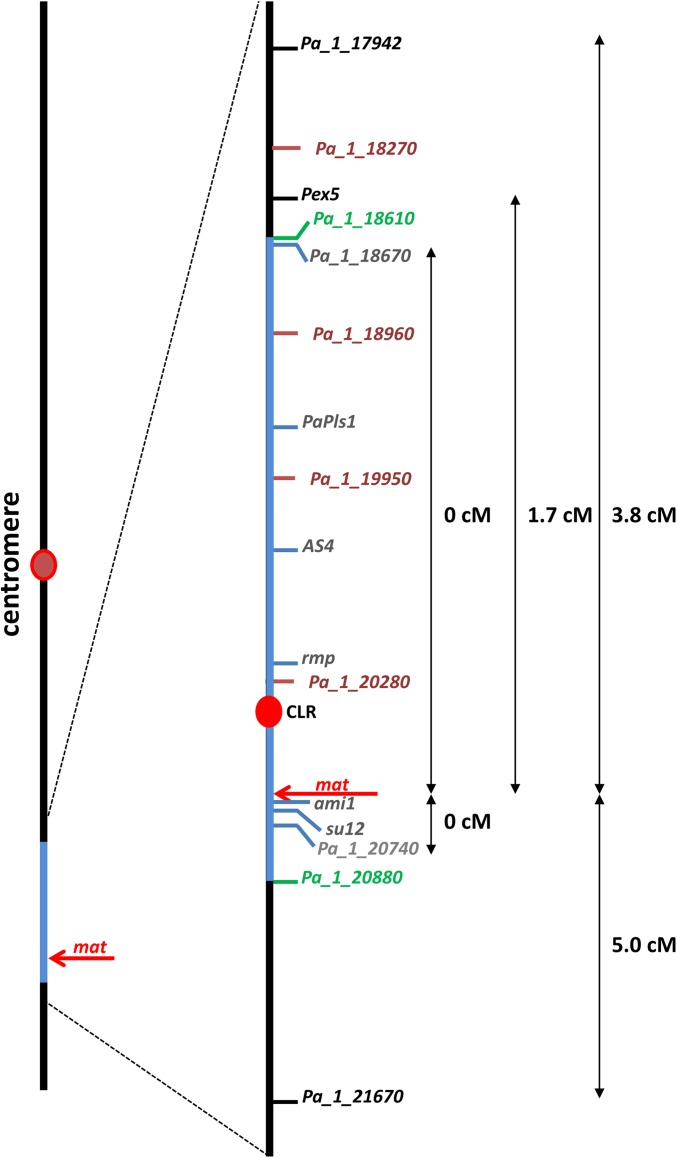
We then sequenced the S mat− strain to 65-fold coverage using the GA2x technology from Illumina Technology. The sequence reads were mapped onto the S mat+ reference sequence and differences were recorded. The two sequences were identical except on chromosome 1 between nucleotides 6634286 and 7471635, where we could detect 687 polymorphisms: 469 SNP and 218 indels (Table 1). The mating type locus is located within the polymorphic region, identifying this 837,349 bp stretch of DNA as the mat region where there is one difference every 1.2 kb in average between the S mat+ and S mat− strains. This situation was expected as the S mat+ and S mat− strains are derived from the same wild-type isolate (S) and have been crossed together yearly for the last 60 years and homokaryotic progeny of each mating type was selected for continuing the line. Differences should thus be present only around the mating-type locus in the region devoid of recombination. To check that we correctly identified the borders of the recombination-less region, we genetically mapped several markers with respect to the mating type (Figure 1). The markers were obtained by gene replacements or classical genetics in diverse projects prior to this study (Silar and Picard 1994; Silar et al. 1997; Graïa et al. 2000; Contamine et al. 2004; Bonnet et al. 2006; Lambou et al. 2008; J. Ait-Benkhali, E. Coppin, and R. Debuchy, unpublished data). Segregation analysis confirmed that no recombination occurred within the mat region, whereas recombination could be observed (Figure 1) between the mating type and Pa_1_17942 (d = 3.8 cM, n = 158), Pex5 (d = 1.7 cM, n = 174), and Pa_1_21670 (d = 5.0 cM, n = 181). Pex5 is located only 50 kb away from the mat region defined by sequencing, suggesting a sharp decline of recombination frequency at the border.
Table 1. Differences in the mat region between the mat+ and mat− strains.
| Transitions | 299 | |
| Transversions | 170 | |
| Indels | 218 | |
| Obs | Exp | |
| Intergenic | 499 | 411 |
| Genic intron | 35 | 19 |
| Genic | 153 | 257 |
| Silent | 75 | 50 |
| Nonsilent | 78 | 103 |
Figure 1.
Scheme of chromosome 1 and expansion of the mat region (in blue). Key features and CDS are mapped on the diagram. CDS labeled in gray type are tightly linked to the mating type (d < 0.5 cM); those in green type are at the borders of the mat region.
The mat− region was assembled with Velvet and remaining gaps were manually filled with targeted PCR sequencing. We did not find any large inversion, deletion, or translocation in the mat region that could account for the lack of recombination, as all differences, except for the mating-type idiomorph per se, affected few contiguous nucleotides. However, in both the mat+ and mat− strains there is a region located between nucleotides 7258560 and 7266980 containing several transposons modified by RIP and whose sequence is incomplete for both S mat+ and S mat−. Because this cluster of ripped transposons is reminiscent of a centromere (Talbert and Henikoff 2010), this region was designated as centromere-like region (CLR). Attempts to fill the gaps in both strains by long-range PCR amplification failed, suggesting that CLR was quite large (>20 kb). To ensure that the structures of CLR in both strains are the ones partially described in the assemblies, we replaced them in vivo with resistance markers (to nourseothricin in S mat− and to hygromycin B in S mat+). We were able to obtain both strains (CLRΔ mat+ and CLRΔ mat−) with the expected replacement (see Figure S1 for Southern blot validation of all replacements made during this study), indicating that the CLR borders defined by sequencing are those actually present in the genome. The mat+ and mat− chromosome 1 of P. anserina are thus totally colinear and have not undergone inversions.
CLR is not involved in repression of recombination in the mat region
Phenotypic analysis of the strains devoid of CLR did not show any obvious phenotypic difference with the wild type. Especially, recombination frequency in the mating-type region remained unchanged, as we could not recombine CLRΔ and mat (d = 0 cM; n = 175). Even in the F5 generation of CLRΔ × CLRΔ crosses, we could not find any recombinant over 180 tested progeny.
Coding differences between the S mat+ and S mat− mat regions
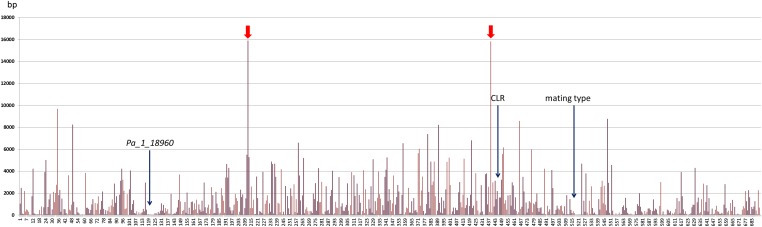
The mat regions of S mat+ and S mat− both contain 229 coding sequences (CDS), as well as the FPR1 gene in the S mat+ strain and the FMR1, SMR1, and SMR2 genes in the S mat− strain. Polymorphisms were preferentially present in intergenic and intronic sequences (Table 1; χ2 test; P < <0.001%) and silent polymorphisms were in much excess to the expected value under a model of random accumulation of mutations (Table 1; χ2 test; P < <0.001%), indicating that nonsilent mutations were counterselected in the CDS of mat region. We then plotted the distance between each consecutive mutation to see if there is accumulation in some regions and scarcity in others (Figure 2). Data show a rather uniform spacing between mutations along the whole region. However, a few large stretches of DNA were devoid of polymorphisms, especially two 15-kb polymorphism-free sequences that split the mat region in three parts of 212, 369, and 246 kb. These two regions show no obvious differences in their coding capacity as compared to the rest of the mat region. Mutations are more closely packed toward the telomere end of the mat region and there appears to be a few hot spots of polymorphisms, including one around the mating type and the other in the Pa_1_18960 CDS. This CDS is highly unusual as it is composed of a ≈3000-amino-acid-long region very rich in threonine/serine, bordered by two domains conserved in other Sordariales, such as Neurospora crassa and Chaetomium globosum (Figure S2). The threonine-/serine-rich domain is similar to those found in yeast agglutinins (Dranginis et al. 2007). The conserved N terminus contains a predicted signal for peptide secretion as well as three carbohydrate binding domains, strongly suggesting that this agglutinin-like protein is inserted into the cell wall for some unknown purpose. We deleted the Pa_1_18960 gene in both S mat+ and S mat− strains and assessed the fertility of the Pa_1_18960Δ deleted strains. We could not see any difference in S mat+ × S mat− and Pa_1_18960Δ mat+ × Pa_1_18960Δ mat− crosses after fertilization (i.e., fruiting bodies and ascospores were in the same amounts); however, the mutant produced three times more spermatia than wild type produced. Finally, like for the deletion of CLR, we could not dissociate the Pa_1_18960Δ deletions from the mating types (n = 61), indicating that this region is not involved in the repression of recombination in the mat region. Intriguingly, three additional CDS-resembling yeast agglutinins are present within the mat region (Pa_1_19950 and Pa_1_20280) or very close (Pa_1_18270). None are polymorphic between S mat+ and S mat−. Inactivation of Pa_1_19950 and Pa_1_18270 did not result in any obvious phenotype, while inactivation of Pa_1_20280 resulted in mature perithecia that contained ascospores, yet none were discharged as occurred in the wild type (Figure S3).
Figure 2.
Plot of the distances between each consecutive mutation. The y-axis gives the size of the region up to the next mutation for all 687 mutations ordered on the x-axis according to their succession on the chromosome starting from the centromere toward the telomere. Red arrows point to two 15-kb regions devoid of polymorphism.
Few additional genes contained polymorphisms that would present obvious phenotypic outcome (Table S2). The rmp1 gene (Pa_1_20180) has G/E and STOP/Q polymorphisms (Contamine et al. 2004; Table S2) and has previously been shown to account for differences in the timing of the premature death syndrome and thermosensitivity of S mat+ and S mat−. Large indels are found in Pa_1_19010, a CDS of unknown function specific to P. anserina, and Pa_1_20400, a CDS conserved in some Ascomycota and carrying an EOS1 domain involved in sensitivity to high-sucrose stress (Nakamura et al. 2007). A polymorphism removes in S mat− the predicted start codon of Pa_1_20750. However, this CDS is small and specific to P. anserina; it may be miscalled a gene. A few other CDS carry polymorphisms in excess of what is expected, indicative of possible positive selection: Pa_1_19460 (six polymorphisms, three nonsilent) coding a glycoside hydrolase of CAZy family 76, Pa_1_20560 (three polymorphisms, two nonsilent) coding a putative subunit of the anaphase-promoting complex, Pa_1_20650 (three nonsilent polymorphisms) coding a putative DNA polymerase zeta subunit, as well as Pa_1_19040 (five polymorphisms, three nonsilent), Pa_1_19560 (three nonsilent polymorphisms), and Pa_1_20140 (four polymorphisms, three nonsilent) coding proteins of unknown function conserved in Pezizomycotina. Thirty-nine additional CDS carry polymorphisms with changed coding capacity resulting in polymorphic proteins that can possibly account for phenotypic differences between the S mat+ and S mat− strains.
Microarray analysis evidences differences in transcript accumulation between S mat+ and S mat−
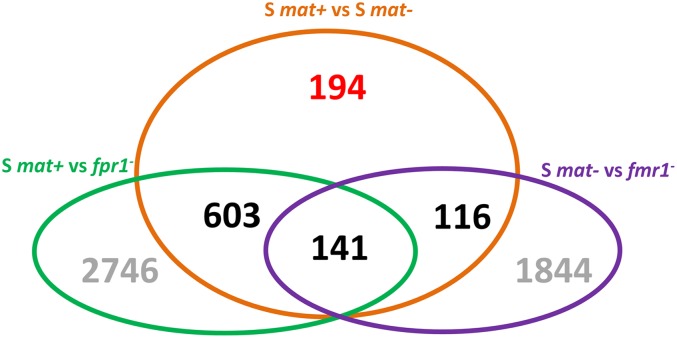
To evaluate potential effects of the mat region vs. the mating type on the physiological differences between the S mat+ and S mat− strains, we identified the genes differentially expressed at the level of transcription between the two strains. Genes specifically regulated by the mating-type genes were previously investigated (Bidard et al. 2011).The analysis uncovered 683 genes expressed higher in mat− strain and 371 genes expressed higher in mat+ strains under crossing conditions. These 1054 differentially transcribed genes mapped on all chromosomes, including the mitochondrial one, and there appears to be no preferential clustering in a particular chromosome (χ2 test; P-value > 0.1). The comparison of wild type with fpr1− and fmr1− strains, which have loss-of-function mutations in the mating-type idiomorphs (El-Khoury et al. 2008), allowed us to assess the role of the mating-type genes in differential expression and to dissociate their effect from that of the mat region (mating-type genes not included). Among the 1054 genes, the vast majority (860 genes) was under the control of the mat idiomorphs and only 194 genes were differentially transcribed due to differences between mat+ and mat− strains excluding the mat idiomorphs (Figure 3). Among the 194 genes, 131 genes had higher expression in the mat− strain, while 63 genes had higher expression in the mat+ strain (Table S3). Analysis of the functions of the 194 genes revealed consistent differences between S mat+ and S mat−, indicative of metabolic differences between the two strains (Table S4). A total of 21 glycoside hydrolase genes had higher expression in the S mat− strain, as well as various other genes involved in carbon metabolism [e.g. Pa_6_11500 and Pa_5_1860 (Pec_lyase_C) and Pa_7_1080 (O-FucT)]. The enrichment in glycoside hydrolases among the 194 genes was highly significant (P-value < 0.0001). Strikingly, the set of genes with higher expression in S mat+ did not contain any gene belonging to this family. As most glycoside hydrolases were expected to be secreted, the S mat− highly expressed category was enriched in putative extracellular proteins (P-value = 0.0003). The nad2 and nad3 mitochondrial genes had high expression in the mat− strain, but the genes highly expressed in S mat+ showed a strong enrichment in nuclear genes encoding proteins with known or predicted mitochondrial localization (P-value = 0.0005). A total of 13 nuclear genes encode well-known mitochondrial proteins among the genes with high expression in the mat+ strain, including three proteins related to the iron–sulfur cluster binding domain. Most of the 194 genes had less than twofold differences between S mat+ and S mat−. In a second analysis, we investigated in more detail 46 genes that were strongly regulated by the mat region (i.e., they had a statistically significant twofold or more differences in their expression between S mat+ and S mat−), but that would present little control by the mating type, which had less than twofold differences when S mat+ (resp. mat−) was compared to the fpr1− (resp. fmr1−) mutant (Table S5). Only 19 of these genes belonged to the 194 genes not regulated at all by the mating type. None of the 46 selected genes had a fold change above five; most had a two- to threefold change. Intriguingly, among the genes identified in the analysis, 21 have no obvious function that can be deduced from their sequence, 3 encode putative plasma membrane transporters, and 20 encode enzymes involved in various metabolisms, including respiration. One encodes an enzyme involved in the post-translation modification of proteins (Pa_7_9690) and one (Pa_1_30) is similar to Aspergillus nidulans TmpA, a gene encoding an oxidoreductase involved in the production of a metabolite controlling the sexual and asexual cycles (Soid-Raggi et al. 2006). Three of the 46 genes are in the mat region and two (Pa_1_19170 and Pa_1_20140) are polymorphic between S mat+ and S mat−. Both encode proteins of unknown function. Still, Pa_1_20140 contains more than expected polymorphisms (four polymorphisms, three nonsilent). The fact that half of the 46 genes were involved in metabolic processes confirms that the physiology of the S mat+ and S mat− strains may be slightly different.
Figure 3.
Venn diagram of genes differentially regulated (P-value < 0.005) between the S mat+ and S mat− strains, and S mat+ and S mat− mating-type mutants, under crossing conditions. Mating-type mutants have loss-of-function mutations in FPR1 (mat+) and FMR1 (mat−) (El-Khoury et al. 2008). Orange set: genes differentially expressed in the S mat+ vs. S mat− comparison. Green set: genes differentially expressed in the mat+ vs. fpr1− comparison. Violet set: genes differentially expressed in the mat− vs. fmr1− comparison. A total of 194 genes are differentially expressed between mat+ and mat− strains and not controlled by mating-type genes (see Table S3). A total of 141 genes are differentially expressed between mat+ and mat− strains and controlled by FMR1 and FPR1. A total of 603 and 116 genes are differentially expressed between mat+ and mat− strains and controlled by FPR1 and FMR1, respectively.
mat+/mat− heterokaryons are stable
To check if S mat+/mat− heterokaryons had some advantage over S mat+ and S mat− homokaryons, we measured apical growth and monitor mycelium morphology under various conditions. Growth rate of the heterokaryotic mat+/mat− cultures was not different from that of homokaryotic mat+ or mat− cultures on M2 and M0 (M2 lacking dextrin) covered with a layer of cellophane as carbon source, at 18°, 23°, 27°, and 30°; 37° was not assayed as at this temperature S mat− and S mat+ grew differently (Contamine et al. 2004). Moreover, we did not see any obvious morphological differences aside from the presence of perithecia in heterokaryotic cultures. Stability of these S mat+/S mat− heterokaryons was tested by inoculating heterokaryotic mycelia on various media. On sterile dung, M2 minimal medium and M0 medium supplemented with wood shavings as carbon sources (a medium with an heterogeneity similar to that of P. anserina natural substrate, but with a composition better controlled), heterokaryons were stable as the culture retained both mating types, as seen by the ability of the cultures to differentiate fertilized fruiting bodies, up to senescence (Figure 4).
Figure 4.
Stability of S mat+/mat− heterokaryons. S mat+/S mat− heterokaryons were inoculated at the positions indicated by the red stars and the plates were incubated for 10 days before the pictures were taken. Fruiting bodies appear as small black dots (green arrows on the M0 + wood shavings plate) all over the plates. As perithecia are not readily visible on dung, the cover of the plates where ascospores have been expelled is shown, indicative that perithecia are present all over dung. On M0 + wood shavings, perithecia are differentiated only on the shavings. On M2, perithecia are mostly differentiated in a 1-cm-thick ring located 1 cm away from the inoculation, but additional perithecia are differentiated further away. Only at the end of the plate perithecia are missing. However, at this location senescence has occurred, preventing fruiting.
We then assayed if mat+/mat+ or mat−/mat− heterokaryons were less stable than mat+/mat− ones. To this end, we used strains marked with either hygromycin B or phleomycin-resistance markers and used the M0 medium supplemented with wood shavings as a carbon source. We tested two independent couples of hygromycin B and phleomycin-resistant transgenes as the insertion point may affect the stability of heterokaryons. The same transgene was associated with each mating type, by genetic crosses. Phenotypic analysis of all transgenic strains, either mat+ or mat−, did not show any phenotypic differences with wild type: the homokaryotic strains grew at the same rate as wild type on M0 with wood shavings, indicating that each nuclear genotype has the same rate of division. To measure stability, hygromycin B- and phleomycin-resistant heterokaryons in all mat combinations (i.e., mat+/mat+, mat−/mat−, mat+/mat−, and mat−/mat+) were inoculated in three replica plates containing M0 with wood shavings and lacking both antifungal compounds. Mycelia were allowed to grow for 6.5 cm, at which point five 0.5 × 0.5 × 0.5 mm explants were taken at the growing edge of the thalli. The 15 explants were then tested for their resistance to hygromycin B and phleomycin. Data are reported in Table 2. For transgenic couple 1, all explants showed resistance to both hygromycin B and phleomycin, in all mat combinations, while for transgenic couple 2, phleomycin resistance was lost from all explants originating from hygR mat+/phleoR mat− and hygR mat−/phleoR mat− heterokaryon plates. This showed (1) that mat+/mat+ and mat−/mat− heterokaryons appear as stable as mat+/mat− ones, at least in the conditions investigated here, and (2) that one of the phleomycin-resistant transgene promoted a disadvantage in heterokaryon only when associated with the mat− mating type, although we could not detect any noticeable phenotypic difference in all used homokaryotic strains.
Table 2. Stability of mat+/mat− vs. mat+/mat+ and mat−/mat− heterokaryons.
| Transgene couple | Hyg mat + /phleo mat+ | Hyg mat−/phleo mat− | Hyg mat−/phleo mat+ | Hyg mat+/phleo mat− |
|---|---|---|---|---|
| 1 | 15 HR PR | 15 HR PR | 15 HR PR | 15 HR PR |
| 2 | 15 HR PR | 15 HR PS | 15 HR PR | 15 HR PS |
Discussion
Like many eukaryotic organisms, heterothallic fungi need to find a suitable partner to engage sexual reproduction. Given the numerous sexual lifestyles found in fungi, solutions to this problem are very diverse in these organisms (Billiard et al. 2012). Like homothallism, pseudohomothallism, a sexual lifestyle chosen by several fungi, enables both outcrossing and inbreeding. However, unlike homothallic species, pseudohomothallic species are faced with the problem of regulating heterokaryosis. In the basidiomycetes, the problem of maintaining heterokaryons is solved by the presence of a dedicated cellular structure, since dikaryons are a normal part of the life cycle in most species of Basidiomycota and, following plasmogamy, dikaryosis is faithfully maintained by clamp connections. In pseudohomothallic ascomycetes, whether selfing-competent dikaryons are maintained or counterselected by a dedicated mechanism is unknown. To better understand this important feature of the fungal life cycle, we have first characterized the mat region of P. anserina, i.e., the chromosomic region encompassing the mating-type idiomorphs and in which recombination is severely inhibited. We found it to be a ≈800-kb region, containing > 200 genes, yet with little coding differences and lacking rearrangements between the two mating-partner nuclei.
Inhibition of recombination around the sex-determining mating type is a common phenomenon in fungi (Heitman et al. 2013). It has been well documented for two pseudohomothallic ascomycetes, N. tetrasperma and P. anserina, and for the heterothallic basidiomycetes Cryptococcus spp. and Microbotrym lychnidis-dioicae (Marcou et al. 1979; Merino et al. 1996; Gallegos et al. 2000; Fraser et al. 2004; Jacobson 2005; Hsueh et al. 2006; Metin et al. 2010; Ellison et al. 2011; Hood et al. 2013). In N. tetrasperma, complete sequencing of the genomes of mat A and mat a strains has revealed several inversions on a ≈7.8-Mb region of the ≈9.4-Mb chromosome 1 carrying the mating type (Ellison et al. 2011). In Cryptococcus ssp., different genes are present in the mating-type regions, which are ≈100 kb large, preventing meiotic pairing (Fraser et al. 2004; Hsueh et al. 2006; Metin et al. 2010). In M. lychnidis-dioicae, optical mapping has shown extensive divergence over 90% of the sex chromosomes (Hood et al. 2013). In all these fungi, improper pairing can thus account for the lack of recombination. The data reported here indicate that the mechanism for recombination inhibition is likely to be very different in P. anserina. The mat region devoid of recombination is only ≈0.8 Mb of the ≈8.8-Mb chromosome 1 that carries the mating type. Regions from both mating types are perfectly colinear in both S mat+ and S mat−, except at the mating-type idiomorphs, which call for a recombination inhibition that does not rely on inversion complexes or sequence divergence. This is in line with previous studies showing that, although severely repressed, recombination can occur in the mat region in P. anserina (Contamine et al. 1996). Such a low level of recombination could be sufficient to maintain the colinearity of both mat regions. In C. neoformans, meiotic recombination hotspots flank the unpaired region (Hsueh et al. 2006) and conversion occurs within the mat locus (Sun et al. 2012), showing that patterns of recombination in this fungus may be more complex than expected. A similar phenomenon may occur in P. anserina, as we identified two potential converted regions of 15 kb.
A candidate cis-element for shaping the recombination pattern of the mat region is CLR as it bears some resemblance to a centromere with its accumulation of transposons mutated by RIP. Data from several organisms show that crossing-over is inhibited around centromeres by as-yet unclear mechanism(s) possibly involving repeat-promoted heterochromatin and/or centromere-specific epigenetic marks (Talbert and Henikoff 2010). Deletion of CLR did not result in increased recombination in the mat region, even after five generations that should have enabled the removal of an epigenetic mark. Although we cannot rule out a mark stable enough to pass through several rounds of meiosis, we speculate that, like for centromere, another cis-acting element may be involved (Talbert and Henikoff 2010). Unfortunately, close inspection of the mat region does not reveal any obvious candidate. We speculate that structuration by such elements results in a special kind of chromatin domain over the mat region endowed with normal transcriptional activity, i.e., not akin to heterochromatin. Indeed, several highly transcribed genes, such as AS4 encoding translation elongation factor eEF1A and su12 encoding a ribosomal protein, are present within the mat region. Another possibility is that this element structures heterochromatin specifically during meiosis or even only prophase 1 of meiosis, allowing expression of the mat region during the other stages of the life cycle.
The N. tetrasperma genome sequences uncovered 190,728 nucleotide differences between the 7.8-Mb nonrecombining regions (Ellison et al. 2011) resulting in degeneration in codon usage for both mat A and mat a (Whittle et al. 2011). This amounts to one difference on average every 40 nucleotides, a rate 30 times higher than the differences observed between the P. anserina mat regions and that extends in a stretch of DNA 10 times larger. Two hypotheses may account for the limited differences between the mat regions in P. anserina. First, they may have a more recent origin. However, sequencing the genome of strain T showed that strain T genome has an average 1–2% divergence with strain S genome (P. Silar, unpublished data), a value 10 times higher than the differences in the mat region of strain S. Yet, this strain, like all the known strains of P. anserina and its sibling species Podospora comata, is also pseudohomothallic. It can be crossed with strain S and the progeny of such mating are also pseudohomothallic and have normal mating ability. Therefore, pseudohomothallism appears to be an ancestral condition in P. anserina and P. comata, arguing against a recent origin of the mating-type region. Alternatively, conversions may homogenize the mat region. The fact that the sequences directly bordering the mating type contain a high number of polymorphisms and the presence of several large regions devoid of polymorphisms that could be traces of conversion support this hypothesis. Moreover, conversion has been detected in the mat region (Contamine et al. 1996) and has also been evidenced in the mating-type region of N. tetrasperma (Menkis et al. 2010). If recombination inhibition proceeds as in centromeres, conversion may nonetheless occur, because it has been observed in centromeres (Talbert and Henikoff 2010).
Regarding the differences in coding capacity, a few polymorphic genes are present in the mat region in addition to the previously characterized mating idiomorphs (Picard et al. 1991; Debuchy and Coppin 1992) and the rmp1 gene (Contamine et al. 2004), although nonsilent polymorphisms are strongly counterselected. The role(s) of the polymorphisms in most of these genes is presently unknown, but none seems involved in the definition of the mating identity, which relies exclusively on the mat idiomorphs (Coppin et al. 1993). Moreover, our microarray analysis uncovered dissimilarities in transcript accumulation of 1054 genes. This gene number is much higher than the 196 genes that exhibited a mating-type biased expression in N. tetrasperma (Samils et al. 2013). As proposed by the authors, it is possible that they underestimated the differences between N. tetrasperma mating types because they used microarrays designed for N. crassa. N. tetrasperma displayed an excess of mat A (MAT1-1 in the standard nomenclature; Turgeon and Yoder 2000) upregulated genes on crossing medium (Samils et al. 2013). This feature is conserved in P. anserina, which also displayed a large excess of genes highly expressed in mat− (MAT1-1 in the standard nomenclature) in crossing conditions (683 highly expressed genes in mat−, 371 highly expressed genes in mat+). However, there is no evidence in P. anserina to support the idea that sex-specific selection resulted in the feminization of the mat− chromosome and masculinization of the mat+ chromosome, as proposed in N. tetrasperma. In fact, the conclusion in N. tetrasperma was based on quantitative analyses of microarray, while any valid conclusion in this matter will require functional analyses of the genes differentially expressed in strains of opposite mating types. Moreover, the phenotype of homokaryotic N. tetrasperma strains seems to contradict the proposal of Samils et al. (2013), as the mat A strain, which was proposed to contain a feminized mat region, produces in fact fewer protoperithecia (Howe 1964) than the supposedly masculinized mat a strain. Size differences were also reported for protoperithecia in mat a and mat A strains (Howe 1964). This observation suggests that some metabolic differences may be related to quality differences in protoperithecial production. It is likely that protoperithecial quality differences correlate with ascospore production per perithecium, which could be a more pertinent indicator of feminization than protoperithecium number. Further analyses will be required to determine whether ascospore number per perithecium is different in mat a and mat A strain.
Analysis of the linkage group distribution of the 1054 genes showed no overrepresentation of genes, especially in the mat region, whereas the recombination-suppressed regions of N. tetrasperma are enriched in highly expressed genes. Samils et al. (2013) observed that the mating-type bias in gene expression accumulates as a consequence of sequence divergence. It is likely that the number of polymorphisms is too small in the P. anserina mat region to induce a bias in the number of differentially expressed genes on chromosome 1. The 194 investigated genes regulated specifically by the mat region and the 46 genes with the highest differences are mostly genes coding enzymes from various metabolisms, suggesting that S mat+ and S mat− present metabolic differences. While the triggering of premature death and thermosensitivity are controlled by rmp1, previous data suggest that rmp1 does not control the life span difference between S mat+ and S mat− (Contamine et al. 1996). Longevity in P. anserina appears to be strongly connected to metabolism (Rossignol and Silar 1996; Silar et al. 2001), especially respiration (Dufour et al. 2000). The potential metabolic differences evidenced here, especially the differences in the level of the mitochondria-encoded nad1, nad2, and nad3 transcripts, may thus account for the different life span of the S mat+ and S mat− strains. The last detected difference between S mat+ and S mat− is in the efficiency of the su8-1 suppression (Silar et al. 2000). The su8-1 suppressor tRNA acts by pairing with UGA stop codon and allowing translational readthrough (Debuchy and Brygoo 1985). In the list of mat+/mat− polymorphic genes (Table S2), there is no obvious candidate gene whose product would interact with tRNAs or the translation machinery. Action on translation could result from an indirect effect. For example, the S mat+ vs. S mat− differentially regulated Pa_7_9690, participating in the post-translational modification of proteins, may be involved and could act on a factor that directly interacts with su8-1, such as eEF1A or the ribosome. Alternatively, differences in the expression of ribosomal proteins may be involved since Rp115, Rp127, Rp1P0, and Rp17 are differentially regulated in S mat+ and S mat− (Table S3). It is currently not possible to determine whether all these differences between mat+ and mat− strains are random, or selected to promote complementation between S mat+ and S mat− nuclei (i.e., inbreeding) or heterokaryons breakthrough (i.e., outcrossing).
Inoculation of P. anserina on its natural substrate or on artificial substrates shows that heterokaryotic mycelia retain both mat+ and mat− nuclei. This suggests that in nature P. anserina mat+/mat− heterokaryotic mycelia are very stable and that P. anserina, or at least the strains that behave like S, are unlikely to lose one of the mating types by mitotic segregation during growth on its restricted biotope. It thus appears that this species prefers inbreeding to outcrossing. However, we cannot rule out that under natural conditions, when other fungi compete for dung exploitation or when environmental conditions are more variable, heterokaryosis is lost more frequently than anticipated from our controlled lab experiments. Indeed, although we did not detect differences between heterokaryotic and homokaryotic strains in our controlled experiments, we know that at 37° S mat+ and S mat− behave clearly differently (Contamine et al. 2004), arguing that under more stressful conditions, having both mat+ and mat− nuclei may be advantageous. Data regarding the choice of inbreeding or outcrossing in N. tetrasperma are inconclusive. On one hand, homokaryotic conidia may frequently be recovered (Metin et al. 2010; Corcoran et al. 2012), and on the other, outcrossing is associated with sexual dysfunctions (Jacobson 1995). In both species, the versatility of pseudohomothallism may be advantageous in selecting for inbreeding as the preferred mode of reproduction, yet sometimes forcing outcrossing when heterokaryons break down.
A surprising result from this study is the fact that despite physiological differences in the S mat+ and S mat− strains, this does not seem to affect the stability of heterokaryons in either way: more or less stability. Indeed, we found that P. anserina undergoes senescence before losing nuclei in the absence of selective pressure. That genetic factors may control heterokaryon stability in P. anserina is demonstrated by transgene couple 2 (Table 2). While we did not detect any obvious difference in growth of all homokaryotic strains for all transgenic strains, the phleoR mat− nuclei of couple 2 are systematically lost when associated with either the hygR mat+ or hygR mat− nuclei, while the phleoR mat+ nuclei are retained. Yet, in both cases, the resistance to phleomycin is promoted by the same transgene. This does not happen with couple 1, although the same plasmid was used to create the phleomycin-resistant strain, showing that this effect is actually due to the insertion of the transgene in couple 2.
Intriguingly, in A. bisporus, a pseudohomothallic basidiomycete, clamp connections are missing and hyphae are multinucleated. Like in the case of P. anserina, it is not clear how heterokaryosis could be maintained in this fungus (Raper et al. 1972; Raper and Kaye 1978). A first candidate is anastomosis. However, leading hyphae growing at the edge of the colony, which are those most likely to generate homokaryotic sectors by loss of one of the two kinds of nuclei, do not undergo anastomosis (Buller 1933). A second possibility could be the nuclear mixing recently discovered in N. crassa (Roper et al. 2013). This mechanism is not involved in the maintenance of sexually compatible nuclei, but likely enables mycelium fitness in variable environments. In this instance, stability of heterokaryons in P. anserina may not have required the selection of a special mechanism to maintain sexual heterokaryosis. Nuclear mixing can indeed explain the stability of mat+/mat− heterokaryons in P. anserina. It may be a general phenomenon in fungi and may also account for the stability of heterokaryons in A. bisporus, but also in Heterobasidion parvisporum, in which nuclear ratios are imbalanced, genetically determined, and stable over time (James et al. 2008).
Supplementary Material
Acknowledgments
We thank Sylvie François for expert technical assistance and Ibtissen Grissa who participated in the early stages of this work. P.G. is a recipient of a “nouveau contrat doctoral” fellowship from Université Paris Diderot, Paris 7, Sorbonne Paris cité.
Footnotes
Communicating editor: E. U. Selker
Literature Cited
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 1987 Current Protocoles in Molecular Biology. Wiley Interscience, New York. [Google Scholar]
- Belcour L., Begel O., Picard M., 1991. A site-specific deletion in mitochondrial DNA of Podospora is under the control of nuclear genes. Proc. Natl. Acad. Sci. USA 88: 3579–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bidard F., Imbeaud S., Reymond N., Lespinet O., Silar P., et al. , 2010. A general framework for optimization of probes for gene expression microarray and its application to the fungus Podospora anserina. BMC Res. Notes 3: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard F., Ait Benkhali J., Coppin E., Imbeaud S., Grognet P., et al. , 2011. Genome-wide gene expression profiling of fertilization competent mycelium in opposite mating types in the heterothallic fungus Podospora anserina. PLoS ONE 6: e21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard F., Coppin E., Silar P., 2012. The transcriptional response to the inactivation of the PaMpk1 and PaMpk2 MAP kinase pathways in Podospora anserina. Fungal Genet. Biol. 49: 643–652. [DOI] [PubMed] [Google Scholar]
- Billiard S., Lopez-Villavicencio M., Hood M. E., Giraud T., 2012. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J. Evol. Biol. 25: 1020–1038. [DOI] [PubMed] [Google Scholar]
- Bonnet C., Espagne E., Zickler D., Boisnard S., Bourdais A., et al. , 2006. The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol. Microbiol. 62: 157–169. [DOI] [PubMed] [Google Scholar]
- Buller, A. H. R., 1933 The formation of hyphal fusions in the mycelium of the higher fungi, pp. 1–74 in Researches on Fungi. Longmans Green & Co., London. [Google Scholar]
- Cheeseman K., Ropars J., Renault P., Dupont J., Gouzy J., et al. , 2014. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat Commun 5: 2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine V., Lecellier G., Belcour L., Picard M., 1996. Premature death in Podospora anserina: sporadic accumulation of the deleted mitochondrial genome, translational parameters and innocuity of the mating types. Genetics 144: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine V., Zickler D., Picard M., 2004. The Podospora rmp1 gene implicated in nucleus-mitochondria cross-talk encodes an essential protein whose subcellular location is developmentally regulated. Genetics 166: 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin E., Arnaise S., Contamine V., Picard M., 1993. Deletion of the mating type sequences in Podospora anserina abolishes mating without affecting vegetative functions and sexual differentiation. Mol. Gen. Genet. 241: 409–414. [DOI] [PubMed] [Google Scholar]
- Corcoran P., Jacobson D. J., Bidartondo M. I., Hickey P. C., Kerekes J. F., et al. , 2012. Quantifying functional heterothallism in the pseudohomothallic ascomycete Neurospora tetrasperma. Fungal Biol 116: 962–975. [DOI] [PubMed] [Google Scholar]
- Debuchy R., Brygoo Y., 1985. Cloning of opal suppressor tRNA genes of a filamentous fungus reveals two tRNASerUGA genes with unexpected structural differences. EMBO J. 4: 3553–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Coppin E., 1992. The mating types of Podospora anserina: functional analysis and sequence of the fertilization domains. Mol. Gen. Genet. 233: 113–121. [DOI] [PubMed] [Google Scholar]
- Dodge B. O., 1932. Crossing hermaphroditic races of Neurospora. Mycologia 24: 7–13. [Google Scholar]
- Dowding E. S., 1931. The sexuality of the normal, giant and dwarf spores of Pleurage anserina. (Ces). Kuntze. Ann. Bot. 45: 1–14. [Google Scholar]
- Dranginis A. M., Rauceo J. M., Coronado J. E., Lipke P. N., 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour E., Boulay J., Rincheval V., Sainsard-Chanet A., 2000. A causal link between respiration and senescence in Podospora anserina. Proc. Natl. Acad. Sci. USA 97: 4138–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury R., Sellem C. H., Coppin E., Boivin A., Maas M. F., et al. , 2008. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53: 249–258. [DOI] [PubMed] [Google Scholar]
- Ellison C. E., Stajich J. E., Jacobson D. J., Natvig D. O., Lapidus A., et al. , 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E., Lespinet O., Malagnac F., Da Silva C., Jaillon O., et al. , 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Diezmann S., Subaran R. L., Allen A., Lengeler K. B., et al. , 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Selker E. U., 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20: 417–423. [DOI] [PubMed] [Google Scholar]
- Gallegos A., Jacobson D. J., Raju N. B., Skupski M. P., Natvig D. O., 2000. Suppressed recombination and a pairing anomaly on the mating type chromosome of Neurospora tetrasperma. Genetics 154: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graïa F., Berteaux-Lecellier V., Zickler D., Picard M., 2000. ami1, an orthologue of the Aspergillus nidulans apsA gene, is involved in nuclear migration events throughout the life cycle of Podospora anserina. Genetics 155: 633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grognet P., Lalucque H., Silar P., 2012. The PaAlr1 magnesium transporter is required for ascospore development in Podospora anserina. Fungal Biol. 116: 1111–1118. [DOI] [PubMed] [Google Scholar]
- Heitman J., Sun S., James T. Y., 2013. Evolution of fungal sexual reproduction. Mycologia 105: 1–27. [DOI] [PubMed] [Google Scholar]
- Hood M. E., Petit E., Giraud T., 2013. Extensive divergence between mating type chromosomes of the anther-smut fungus. Genetics 193: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., et al. , 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35: W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe H. B., Jr, 1964. Vegetative traits associated with mating type in Neurospora tetrasperma. Mycologia 56: 519–525. [Google Scholar]
- Hsueh Y.-P., Idnurm A., Heitman J., 2006. Recombination hotspots flank the Cryptococcus mating type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet. 2: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeaud S., Graudens E., Boulanger V., Barlet X., Zaborski P., et al. , 2005. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 33: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson D. J., 1995. Sexual dysfunction associated with outcrossing in Neurospora tetrasperma, a pseudohomothallic ascomycete. Mycologia 87: 604–617. [Google Scholar]
- Jacobson D. J., 2005. Blocked recombination along the mating type chromosomes of Neurospora tetrasperma involves both structural heterozygosity and autosomal genes. Genetics 171: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. Y., Stenlid J., Olson A., Johannesson H., 2008. Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution 62: 2279–2296. [DOI] [PubMed] [Google Scholar]
- Lambou K., Malagnac F., Barbisan C., Tharreau D., Lebrun M. H., et al. , 2008. The crucial role during ascospore germination of the Pls1 tetraspanin in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot. Cell 7: 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcou D., 1961. Notion de longévité et nature cytoplasmique du déterminant de la sénescence chez quelques champignons. Ann. Sci. Natur. Bot. Paris Sér. 12 2: 653–764. [Google Scholar]
- Marcou D., Masson A., Simonet J. M., Piquepaille G., 1979. Evidence for non-random spatial distribution of meiotic exchanges in Podospora anserina: comparison between linkage groups 1 and 6. Mol. Gen. Genet. 176: 67–79. [DOI] [PubMed] [Google Scholar]
- Marisa L., Ichante J. L., Reymond N., Aggerbeck L., Delacroix H., et al. , 2007. MAnGO: an interactive R-based tool for two-colour microarray analysis. Bioinformatics 23: 2339–2341. [DOI] [PubMed] [Google Scholar]
- Martin E. M., 1940. The morphology and cytology of Taphrina deformans. Am. J. Bot. 27: 743–751. [Google Scholar]
- Menkis A., Whittle C. A., Johannesson H., 2010. Gene genealogies indicates abundant gene conversions and independent evolutionary histories of the mating type chromosomes in the evolutionary history of Neurospora tetrasperma. BMC Evol. Biol. 10: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino S. T., Nelson M. A., Jacobson D. J., Natvig D. O., 1996. Pseudohomothallism and evolution of the mating type chromosome in Neurospora tetrasperma. Genetics 143: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metin B., Findley K., Heitman J., 2010. The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet. 6: e1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Ando A., Takagi H., Shima J., 2007. EOS1, whose deletion confers sensitivity to oxidative stress, is involved in N-glycosylation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 353: 293–298. [DOI] [PubMed] [Google Scholar]
- Picard M., Debuchy R., Coppin E., 1991. Cloning the mating types of the heterothallic fungus Podospora anserina: developmental features of haploid transformants carrying both mating types. Genetics 128: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju N. B., 1992. Functional heterothallism resulting from homokaryotic conidia and ascospores in Neurospora tetrasperma. Mycol. Res. 96: 103–116. [Google Scholar]
- Raju N. B., Perkins D. D., 1994. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora, and Podospora. Dev. Genet. 15: 104–118. [DOI] [PubMed] [Google Scholar]
- Raper C. A., Kaye G., 1978. Sexual and other relationships in the genus Agaricus. J. Gen. Mic. 105: 135–151. [Google Scholar]
- Raper C. A., Raper J. R., Miller R. E., 1972. Genetic analysis of the life cycle of Agaricus bisporus. Mycologia 64: 1088–1117. [Google Scholar]
- Rizet G., Delannoy G., 1950. Sur la production par des hétérozygotes monofactoriels de Podospora anserina de gamétophytes phénotypiquement différents des gamétophytes parentaux. C. R. Acad. Sci. Paris 231: 588–590. [Google Scholar]
- Rizet G., Engelmann C., 1949. Contribution à l’étude génétique d’un Ascomycète tétrasporé: Podospora anserina (Ces.). Rehm. Rev. Cytol. Biol. Veg. 11: 201–304. [Google Scholar]
- Roper M., Simonin A., Hickey P. C., Leeder A., Glass N. L., 2013. Nuclear dynamics in a fungal chimera. Proc. Natl. Acad. Sci. USA 110: 12875–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol M., Silar P., 1996. Genes that control longevity in Podospora anserina. Mech. Ageing Dev. 90: 183–193. [DOI] [PubMed] [Google Scholar]
- Samils N., Gioti A., Karlsson M., Sun Y., Kasuga T., et al. , 2013. Sex-linked transcriptional divergence in the hermaphrodite fungus Neurospora tetrasperma. Proc. Biol. Sci. 280: 20130862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silar P., 1995. Two new easy-to-use vectors for transformations. Fungal Genet. Newsl. 42: 73. [Google Scholar]
- Silar P., 2011. Grafting as a method for studying development in the filamentous fungus Podospora anserina. Fungal Biol 115: 793–802. [DOI] [PubMed] [Google Scholar]
- Silar P., 2013. Podospora anserina: from laboratory to biotechnology, pp. 283–309 in Genomics of Soil- and Plant-Associated Fungi, edited by P. K. M. Benjamin, A. Horwitz, M. Mukherjee, and C. P. Kubicek, Springer, Heidelberg/New York/Dordrecht/London. [Google Scholar]
- Silar P., Picard M., 1994. Increased longevity of EF-1 alpha high-fidelity mutants in Podospora anserina. J. Mol. Biol. 235: 231–236. [DOI] [PubMed] [Google Scholar]
- Silar P., Koll F., Rossignol M., 1997. Cytosolic ribosomal mutations that abolish accumulation of circular intron in the mitochondria without preventing senescence of Podospora anserina. Genetics 145: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silar P., Rossignol M., Haedens V., Derhy Z., Mazabraud A., 2000. Deletion and dosage modulation of the eEF1A gene in Podospora anserina: effect on the life cycle. Biogerontology 1: 47–54. [DOI] [PubMed] [Google Scholar]
- Silar P., Lalucque H., Vierny C., 2001. Cell degeneration in the model system Podospora anserina. Biogerontology 2: 1–17. [DOI] [PubMed] [Google Scholar]
- Soid-Raggi G., Sanchez O., Aguirre J., 2006. TmpA, a member of a novel family of putative membrane flavoproteins, regulates asexual development in Aspergillus nidulans. Mol. Microbiol. 59: 854–869. [DOI] [PubMed] [Google Scholar]
- Sun S., Hsueh Y. P., Heitman J., 2012. Gene conversion occurs within the mating type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 8: e1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S., 2010. Centromeres convert but don’t cross. PLoS Biol. 8: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon B. G., Debuchy R., 2007. Cochliobolus and Podospora: Mechanisms of Sex Determination and the Evolution of Reproductive Lifestyle in Sex in Fungi: Molecular Determination and Evolutionary Implications, edited by J. Heitman, J. W. Kronstad, J. W. Taylor and L. A. Casselton ASM Press, Washington, DC. [Google Scholar]
- Turgeon B. G., Yoder O. C., 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31: 1–5. [DOI] [PubMed] [Google Scholar]
- Whittle C. A., Sun Y., Johannesson H., 2011. Degeneration in codon usage within the region of suppressed recombination in the mating type chromosomes of Neurospora tetrasperma. Eukaryot. Cell 10: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E., 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.