Abstract
Studies in tunicates such as Ciona have revealed new insights into the evolutionary origins of chordate development. Ciona populations are characterized by high levels of natural genetic variation, between 1 and 5%. This variation has provided abundant material for forward genetic studies. In the current study, we make use of deep sequencing and homozygosity mapping to map spontaneous mutations in outbred populations. With this method we have mapped two spontaneous developmental mutants. In Ciona intestinalis we mapped a short-tail mutation with strong phenotypic similarity to a previously identified mutant in the related species Ciona savignyi. Our bioinformatic approach mapped the mutation to a narrow interval containing a single mutated gene, α-laminin3,4,5, which is the gene previously implicated in C. savignyi. In addition, we mapped a novel genetic mutation disrupting neural tube closure in C. savignyi to a T-type Ca2+ channel gene. The high efficiency and unprecedented mapping resolution of our study is a powerful advantage for developmental genetics in Ciona, and may find application in other outbred species.
Keywords: mutant, Ciona, whole-genome sequencing, genetic variation
A valuable attribute of many model organisms is the ability to conduct forward and reverse genetics. The availability of sequenced genomes and transcriptomes have streamlined reverse genetic approaches, but forward genetic approaches remain time consuming and cumbersome. Even for organisms with well-developed mutation mapping strategies and resources, classical linkage analysis can be slow and subject to chance. Genome-wide association studies now provide an alternative approach, but are severely limited by the need for high-frequency alleles and very large samples (Marchini et al. 2007; Cheng et al. 2010). A need remains for additional phenotype-to-genotype strategies in, for example, the investigation of quantitative traits, natural variation, and disease loci (Hillier et al. 2008; Jelier et al. 2011; Liti and Louis 2012; Lehner 2013). In recent years, new and inexpensive deep sequencing technologies have created opportunities for forward genetic approaches (Hobert 2010). By taking a snapshot of variation across the genome of an outbred population, a researcher can now quickly identify a region of homozygosity unique to mutant individuals. Variations of this method then use a fine-mapping parameter to define a high-confidence mapping interval and to retrieve a list of variable sites in the interval as a list of possible causal mutations. Modeling shows that the mapping power using whole-genome sequencing (WGS) is a function of how many genomes are sampled from mutant individuals, the recombination rate, and genome coverage (Leshchiner et al. 2012; Obholzer et al. 2012). This approach has worked efficiently and accurately for the well-assembled, annotated, and inbred genomes of model organisms such as Drosophila melanogaster, Caenorhabditis elegans, and Mus musculus (Blumenstiel et al. 2009; Andersen et al. 2012; Leshchiner et al. 2012).
Tunicates, such as the ascidian Ciona intestinalis, are classic model organisms for developmental biology, and as the closest living relatives of the vertebrates they are a key group for understanding chordate development and evolution (Satoh 1994; Delsuc et al. 2006). As larvae, ascidians exhibit a conserved chordate body plan that includes a notochord and a dorsal hollow central nervous system (Passamaneck and Di Gregorio 2005). However, the embryos and larvae of ascidians are much simpler than those of vertebrates (e.g., a C. intestinalis larva consists of ∼2600 cells) and develop according to a fixed cell lineage (Satoh 1994). Reference genome sequences are now available for both C. intestinalis and the closely related species C. savignyi. Both Ciona species have compact genomes (∼160 Mb) with relatively few protein-encoding genes (∼16,000 genes) (Shoguchi et al. 2006; Small et al. 2007b). In addition, both Ciona species display very high levels of natural genomic variation; the two haplotypes in the reference genomes of C. intestinalis and C. savignyi are ∼1 and ∼4% different, respectively (Dehal et al. 2002; Small et al. 2007a; Satou et al. 2012). Our laboratory has found that a significant fraction of wild-caught individuals from both species carry recessive mutations that can be uncovered by self-fertilization (Veeman et al. 2011). Because establishing inbred lines of Ciona has proven to be very difficult, we maintain mutant and transgenic lines by outbreeding them to wild-caught stocks (Veeman et al. 2011). Despite this limitation, we have been able to isolate, maintain, and map naturally occurring mutations in C. intestinalis and C. savignyi that disrupt processes such as notochord morphogenesis and neural plate development (Jiang et al. 2005; Veeman et al. 2008; Tresser et al. 2010; Hackley et al. 2013). The high level of genetic variation between individuals has actually aided mapping via bulk segregant analysis by providing an abundance of single-nucleotide variation (SNV) markers, although our current mapping procedures are very time consuming.
The WGS mapping method presents a tremendous opportunity to investigate mutations and genetic variation in wild (i.e., outbred) animals. The relatively small size, high recombination rate (∼25–200 kb/cM) (Kano et al. 2006; Small et al. 2007a), and relatively complete reference genomes make the two Ciona species ideal models for applying the WGS mapping method for highly polymorphic wild models. Two wild-isolated recessive developmental mutants, one in C. intestinalis and one in C. savignyi, were used to test the WGS mutation-mapping strategy. We report that via homozygosity mapping we are able to quickly, and with high resolution, identify mutant loci in both species. The smaller genomes and higher polymorphism rate of the Ciona species allowed us to define narrower candidate regions than has been reported for vertebrate models.
Materials and Methods
Genomic DNA isolation
Larvae were incubating in 0.5M EDTA with gentle pipetting for 30 min to remove maternally derived follicle cells before being homogenized for gDNA isolations as described previously (Silva and Smith 2008). Upward of 300 larvae were used for sufficient genomic DNA collection.
Ciona mutation mapping strategy
The C. savignyi and C. intestinalis unmasked reference genomes were downloaded from ftp://ftp.ensembl.org/pub/release-73/fasta/ciona_savignyi/dna/ and http://ghost.zool.kyoto-u.ac.jp/datas/JoinedScaffold.zip, respectively. The Illumina sequencing reads were aligned to their corresponding reference genomes using the Burrows Wheeler Aligner program (v. 0.5.9) (Li and Durbin 2009). Default parameters for the ‘bwa aln’ command were used for all alignments. Following alignment, Samtools (v. 0.1.18) were used to generate pileup files from reads of mapping quality ≥15 and base quality ≥15 (Li et al. 2009). The pileup file was used to compute homozygosity for genome-wide mapping using custom scripts in Python and R (Python v. 2.7 and http://r-project.org). For the homozygosity calculation, the program first discerned whether a genomic base position was covered by >10 reads; if so, it then proceeded to count the percentage of aligned bases at that position that matched each other. If 85% or more of the aligned bases at the position were in agreement then that site was considered homozygous. The program summed the amount of homozygous sites within a designated genomic window size (5 kb for C. savignyi and 10 or 20 kb for C. intestinalis) and divided by the number of informative sites (>10× coverage sites) in that window. This yielded a proportion or percentage of homozygosity for each genomic window across the entire genome. These values were transferred to R and graphically depicted. The peak homozygosity values are also called from the output file and left to the user to visually verify in the R plot.
Once the region of interest (ROI) is identified from the second step, the corresponding pileup file is used for the fine-mapping program. In step 2 of the program, the major allele frequencies for each variant site (with coverage greater than five reads) is calculated. The fine-mapping program used only SNV allele frequencies between 40 and 85%. We used this allele-frequency filter to exclude the high amount of population-specific variation in the >85% allele-frequency range and the inherently large number of homozygous SNVs as we approached the mutant locus. The <40% filter was used to exclude minor-allele frequencies and potential sequencing errors and focuses on the major allele of each SNV. These 40–85% allele frequencies were binned in 100-bp windows and listed in the output file, which is used for plotting in R. The program outputs the position of the largest stretch of windows with zero heterozygosity and the user can verify if the bin output is unique to the ROI, within a trough trending to zero values (as this was not the case for the C. savignyi bug fine mapping). All LOESS lines for the C. savignyi data were computed in R by calling the LOESS function and using a span of 0.2 (R project, Manuals).
Downstream gene model alignment was carried out using the Integrative Genomics Viewer software from the Broad Institute (Robinson et al. 2011; Thorvaldsdottir et al. 2013).
qRT–PCR
RNA was extracted from mutant and wild-type larvae using Trizol (Invitrogen) after follicle cells were removed, as above. A total of 200 ng RNA was used for cDNA synthesis using Superscript III First-Strand Synthesis kit [Oligo(dT) primer, Invitrogen]. One microliter of this cDNA reaction was used for each qRT–PCR reaction with Fast Sybr Green 2X MasterMix (ABI). Primers are listed in Supporting Information, Table S2. Genes used for the first ΔCT normalization calculation were C. intestinalis actin and C. savignyi RPS27A (Olinski et al. 2006). Wild-type values were used for the second ΔCT calculation. qRT–PCR reactions were run in triplicate and each experiment consisted of three biological repeats.
Results
Ciona mutant lines
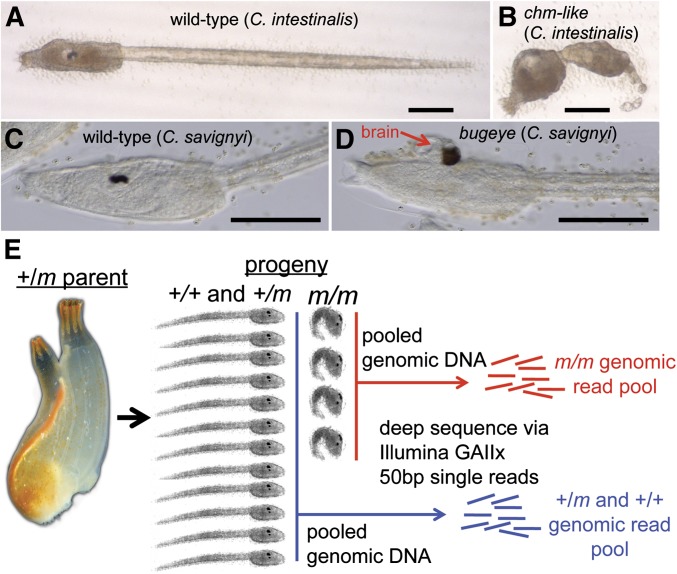
The bottleneck for large-scale mutation screening and characterization has been the time-consuming process of mapping mutant loci. We asked whether homozygosity mapping via WGS, as used in previous inbred model organisms (Blumenstiel et al. 2009; Leshchiner et al. 2012; Obholzer et al. 2012), would accurately and efficiently allow mutation identification in Ciona. To test this, we developed a bioinformatics strategy and applied it to two novel mutant lines, one in C. intestinalis and the other in C. savignyi. The C. intestinalis mutant had a short-tail phenotype (Figure 1, A and B) closely resembling the chongmague (chm) mutation previously isolated from the related species C. savignyi (Veeman et al. 2008) and was named chm-like. A cross-species fertilization test indicated that the C. intestinalis mutant was in the same complementation group as the C. savignyi chm mutant (data not shown). In a previous report we showed that the C. savignyi chm phenotype is due to a profound disruption in notochord morphogenesis caused by a null mutation in the α-laminin3,4,5 gene (Veeman et al. 2008). The second mutant, in C. savignyi, was identified in a self-fertilization screen and has the defining characteristic of an open anterior neural plate, with a protruding and exposed brain, and was named bugeye (bug) (Veeman et al. 2011, Figure 1, C and D). Both Ciona mutants were scored as phenotypically recessive.
Figure 1.
Ciona mutation mapping. (A) Wild-type C. intestinalis larvae. (B) C. intestinalis larvae homozygous for the chongmague-like (chm-like) mutation. (C) Wild-type C. savignyi larvae. (D) C. savignyi bugeye (bug) mutant. The brain protruding from the bug mutant is indicated by a red arrow. (E) Spawning and self-/cross-fertilizing adult Ciona heterozygous for the recessive mutant m allele. DNA was isolated from the hundreds of mutant progeny (m/m), and when included, their corresponding wild-type (WT) siblings (+/+ and +/m). Bars in A–D, ∼100 μm.
The first step of the strategy consisted of spawning heterozygous parents (+/m) to generate F1 progeny (Figure 1E). An adult Ciona will typically spawn several hundred eggs (and countless sperm). For mapping of the C. savignyi mutant we tested samples of both self-fertilized gametes from a single hermaphroditic parent (SPP samples, Table 1) and of crossed gametes from several +/m parents spawned together (MPP samples, Table 1). For the C. intestinalis mutant only a crossed-gamete sample was tested. Samples consisted of the pooled genomic DNA from 600–800 homozygous mutant (m/m) progeny, and for the C. savignyi mutant a separate pool +/+ and +/m siblings, called the WT sample, was prepared (Table 1). The genomic DNA pools were used to create libraries for 50 cycle/single-end Illumina sequencing. The resulting reads (∼150 million) were then used as the input for the next step of the mapping program.
Table 1. Ciona deep sequencing statistics.
| Sample | Total reads | %Reads ≥ MapQ 15 | Avg. coverage | % of sites ≥ 10× coverage | Avg. homozygosity(%) |
|---|---|---|---|---|---|
| C. intestinalis chm | 159,931,687 | 68.5 | 34.22 | 95.42 | 98.23 |
| MPP C. savignyi bugeye | 116,448,947 | 37.4 | 13.62 | 67 | 96.97 |
| MPP C. savignyi WT-sib | 181,200,319 | 40.3 | 22.81 | 76.64 | 96.45 |
| SPP C. savignyi bugeye | 175,967,795 | 41.1 | 22.60 | 77.97 | 96.71 |
| SPP C. savignyi WT-sib | 166,735,236 | 39.6 | 20.63 | 74.42 | 96.78 |
Homozygosity maps
Construction of genome-wide homozygosity maps was the next step in the program. Loci linked to the causative mutation should be evident by a cluster of windows with higher than average homozygosity—approaching complete homozygosity (i.e., 100% homozygous values). For bug we generated both a cross-fertilized data set, in which 12 heterozygous bug adults were spawned together, and a self-fertilized data set in which all embryos contributing to the genomic DNA pool derived from a single heterozygous bug adult (Table 1). For chm-like only a single cross-fertilized data set was generated. For the initial round of mapping we used the two cross-fertilized data sets. Mapping with self vs. cross-fertilized data sets will be treated separately, below.
To map the homozygosity, the Illumina sequences were first aligned to their respective reference genomes and filtered for mapping and sequence base quality (Dehal et al. 2002; Small et al. 2007b). Using a cutoff of MapQ ≥15 (Table 1 and Figure S1), 68% of the C. intestinalis reads mapped to the genome, vs. ∼40% of the C. savignyi reads (Table 1). The overall coverage from the C. savignyi data sets was highly variable, due to poor mapping quality (Figure S1). The C. savignyi reference genome consists of 374 reftigs with an N50 of 1.8 Mb (Vinson et al. 2005; Small et al. 2007b). Two hundred and twenty-seven of the largest reftigs were roughly mapped into 14 linkage groups, corresponding to the 14 chromosomes (Hill et al. 2008). The difficulty in aligning the C. savignyi reads to the reference genome is primarily a reflection of the highly polymorphic nature of the wild C. savignyi population, with an ∼3–5% genome-wide variation between haplotype isolates (Small et al. 2007a). Thus a lower percentage of the C. savignyi reads were aligned to the reference genome; there were more mismatches per 50-bp read in this species, and thus more alignment penalties and lower mapping quality (Figure S1 and Figure S2). This biased coverage in C. savignyi to areas of lower heterozygosity, such as exons (Table S1). Nevertheless, we were able to obtain 13–22× coverage of the C. savignyi reference genome with the MapQ ≥ 15 reads, for the four data sets. The difference in the mapping quality of the two species resulting from the higher variation of C. savignyi can be seen in comparisons of the edit distances (i.e., the minimum number of changes required to transform one sequence into the other) of two randomly chosen sets of 5 million reads from the two genomes (Figure S2). Thus many reads fail to map to the reference genome due to natural variations causing multiple mismatches that exceed the cutoff. The higher sequence variation of C. savignyi also likely resulted in a high fraction of reads scoring as “repeated” vs. those from C. intestinalis (Figure S2). We did not investigate whether reducing the mapping stringency could lead to a higher percentage of the reads mapping, as this also has the potential to result in reads being incorrectly mapped on the genome. Moreover, as we describe below, we were able to successfully map the C. savignyi mutation despite the low percentage of reads that mapped.
The post-alignment, quality-filtered, sequence reads were used for mapping analysis. A relative homozygosity value was calculated for nonoverlapping windows of 5 kb for C. savignyi, and 10 kb for C. intestinalis data sets. Homozygosity is calculated by determining the percentage of base positions, within the windows, in which ≥85% of the sequence reads at each position are in agreement. This calculation simplifies the analysis and reduces computing time by focusing on regions with high homozygosity. With expected sequencing error rate of ≈1–5%, we would not expect estimates of homozygosity to reach 100% for all sites within any window (Luo et al. 2012). We set our homozygosity cutoff well below this error rate (i.e., 85%). Therefore a 100% homozygosity window in our analysis would be defined as one in which all 5000 or 10,000 base positions are at least 85% in agreement. Other cutoff values were tested, and values >85% yielded minor differences in window values, but did not change the identity of the windows with peak homozygosity. Within each window, we considered only bases with ≥10-fold read coverage. The relative homozygosity measure also adjusted for areas of variable coverage by adjusting window sizes to the number of usable sites in a window (e.g., if only 9680 bases of 10,000 in a window had coverage >10, then homozygosity was calculated as a percentage of the 9680 bases in that window). On average, the C. savignyi data sets had ∼74% usable sites per window. In contrast, the C. intestinalis data set yielded an average of 95% usable sites per window (Table 1). Because of the lower overall coverage for C. savignyi, we added a minimal requirement that at least 40% of the bases within a window have ≥10-fold coverage to be used in the analysis.
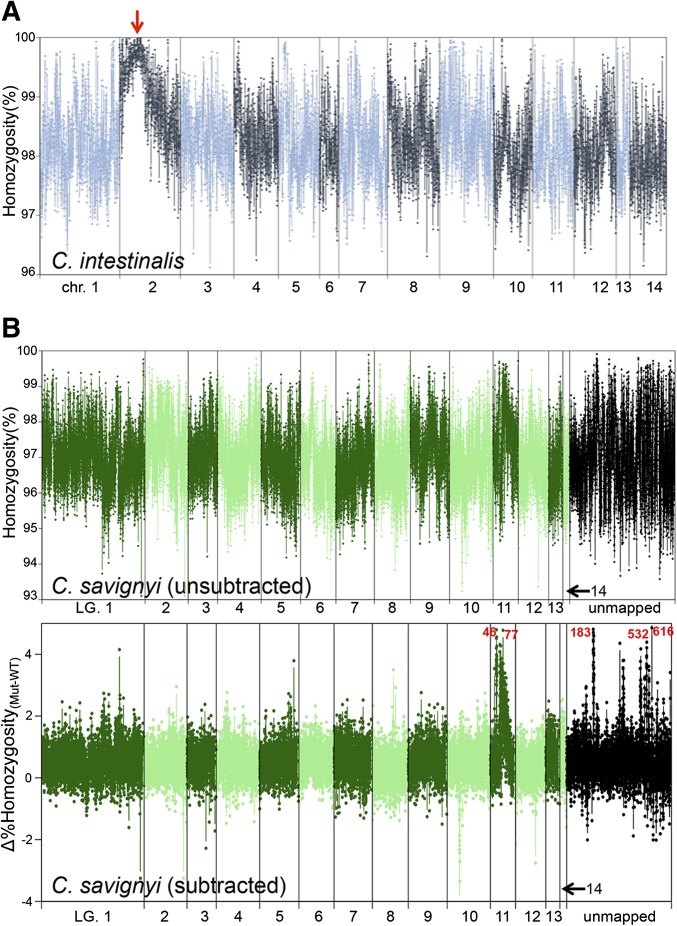
For the C. intestinalis data set, the average homozygosity of all windows was 98.3% (i.e., 1.7% variable); while for the C. savignyi data sets the average homozygosity was ∼96.7% (i.e., ∼3.3% genomic variability; Table 1). These values are in agreement with previous estimates of genomic variation in C. intestinalis and C. savignyi (Small et al. 2007a; Satou et al. 2012). When the distribution of percentage homozygosity values across the genome for the C. intestinalis chm-like was examined, it was apparent that chromosome 2 contained a uniquely homozygous region with a single peak (100% value) at 2.76 Mb (red arrow, Figure 2A).
Figure 2.
Genome-wide homozygosity maps. (A) Percentage homozygosity for nonoverlapping 10-kb windows from a pooled sample of embryos homozygous for the C. intestinalis chm-like mutation. The highest homozygosity value was at ∼2.76 MB in chromosome 2 (red arrow). Chromosomes are depicted in alternating dark and light colored lines. (B) Two homozygosity plots are shown for the C. savignyi bugeye (bug) mutation data set. The top plot (unsubtracted) shows the percentage homozygocity values for 5-kb windows of aligned sequence reads from homozygous bug mutants (m/m) across the 14 linkage groups, as well as unmapped reftigs. Alternating light and dark lines delineate the linkage groups. Unmapped reftigs are place at the end (black points). In the bottom plot (subtracted) the percentage homozyogosity values of aligned genomic reads from the wild-type siblings were first subtracted from the m/m values, generating Δhomozygosity values. In the lower plot, several regions of high relative homozygosity were observed. Reftigs 183, 616, 532, 46, and 77 had the highest values as marked in red text. Lower peaks in homozygosity were on reftig 370, 494, and 556. LG, linkage group.
For the C. savignyi data set, a simple plot of percentage homozygosity vs. the assembled linkage groups (LG) produced a noisier plot than that for C. intestinalis, in which the peak homozygosity was less apparent (top plot, Figure 2B). Complicating the situation, a large number of small reftigs in the C. savignyi reference genome have not mapped to linkage groups (Hill et al. 2008). These are shown clustered at the right end of the plot. To better identify the genomic region of peak sequence homozygosity in the homozygous mutant genomes we subtracted the percentage homozygosity values of the WT sample from the m/m sample to generate Δhomozygosity values. The subtraction potentially controls for the variation in homozygosity across the genome of natural populations introduced by mutation rate variation, genetic drift, and natural selection.
From the “subtracted” plot in Figure 2B a ROI can readily be identified on linkage group 11, with reftigs 46 and 77 showing high homozygosity values. We also observed the high homozygosity values for several of the unmapped reftigs, which are shown grouped together at the right side of the plot (reftigs 183, 532, and 616). The highest cluster of genome-wide Δhomozygosity values centered around the 60 kb position of reftig 183. These highly homozygous reftigs correspond to highly linked loci, and consequently we can roughly place the unmapped contigs in the vicinity of linkage group 11. Because most of the reftigs had been mapped using only single markers for linkage analysis, their relative orientations are not known (Hill et al. 2008). Contiguity of the roughly mapped reftigs remains largely unknown, unlike in C. intestinalis whose physical map was made by a combination of BAC and FISH data (Shoguchi et al. 2006). The quantitative linkage data provided by the C. savignyi data sets allowed us to estimate the relative positions of previously unmapped reftigs 183, 616, and 532 relative to reftigs 46 and 77. A large gene model is split between the 3′-end of reftig 532 and the 3′-end of reftig 183, indicating that reftig 532 sequence is continuous with and in reverse orientation relative to reftig 183. There are also large gaps (stretches of N’s) near the high homozygosity values found on reftig 616 and 183. Our discontinuous high homozygosity values on each of these reftigs, near large gaps, may indicate misassembly at these specific reftig locations. Our results suggest that reftigs 183, 532, and 616 likely belong on linkage group 11 between reftig 77 and 46. Several other potentially linked (but unmapped) reftigs have slightly lower homozygosity values (e.g., reftig 370, 494, and 556; Figure 2B) and likely belong on linkage group 11, but no placement or orientation information could be confidently inferred from our linkage analysis.
The incomplete nature of the C. savignyi reference genome and the poor mapping of the reads (Table S1 and Figure S2) resulted in multiple reftig peaks, and interspersed coverage within the reftigs, so we analyzed the distribution of Δhomozygosity values with 1-kb intervals for each of the individual candidate reftigs separately (Figure S4A). Overall, reftig 183 had the highest cluster of Δhomozygosity values and the highest median (max = Δ 4.8%, median = Δ 3.46%, Figure S4A). Based on these data we chose the 340-kb reftig 183 as our ROI.
To assess the utility of calculating Δhomozygosity values, we examined the genome-wide homozygosity using only the m/m data set (i.e., without subtraction of WT data set homozygosity values). Although the plot was much noisier, reftig 183 could still be discerned as having the highest homozygosity values (Figure 2B and Figure S4). The five highest values of homozygosity in the m/m data set were found on reftig 183 (three values), reftig 117 (one value), and reftig 209 (one value). Reftigs 117 and 209 were also found to have the same high homozygosity values in the matching wild-type data. In the m/m-only plot, linkage group 11 also had a unique clustering of windows with high homozygosity values, although it was much less apparent (compare subtracted vs. unsubtracted plots, Figure 2B). For the m/m-only analysis, the peak of homozygosity on reftig 183 was calculated to be at ∼217 kb, while for the Δhomozygosity analysis, the peak on reftig 183 was determined to be at ∼57 kb (Figure 4A). These results demonstrated that the wild-type data set eliminated many of the unlinked, inherently high homozygosity values found in the C. savignyi data set.
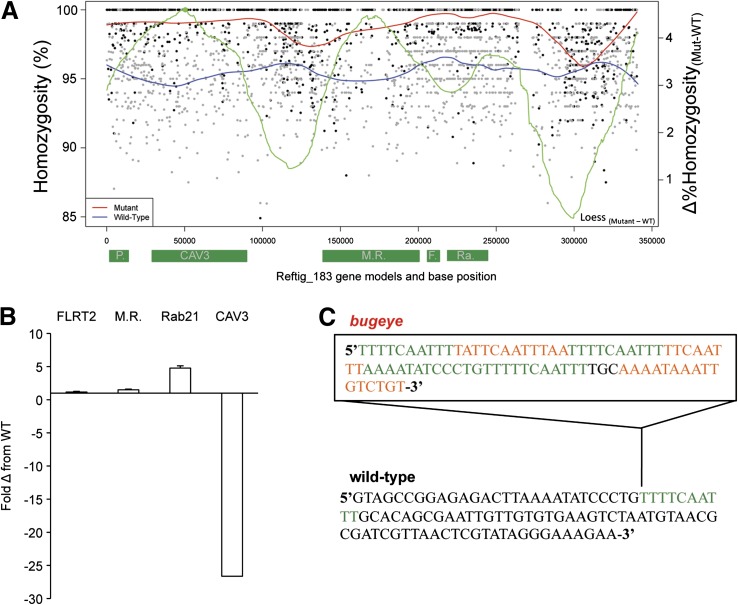
Figure 4.
Homozygosity mapping on the bugeye ROI, reftig 183. (A) Values were calculated for windows of 100 bp. Black points depict mutant values and gray points depict WT sibling homozygosity values. The differential value for the best fit lines of bug/bug (red) minus WT sibling (blue) homozygosity is shown in green; the peak of this differential line is shown as a solid green point. Gene models and their approximate positions along reftig 183 are depicted along the x-axis as green rectangles. P, phosholipid scramblase; CAV3, CAV3; M.R., cation-independent mannose receptor; F., fibronectin leucine-rich transmembrane receptor 2; Ra, ras-related Rab21. (B) qRT–PCR of candidate genes on reftig 183. The plot shows the relative expression in bugeye compared to WT samples for the indicated genes. Expression levels were normalized to a housekeeping gene (RPS27A) first, and bugeye values were then compared to WT values in parallel reactions. Bar indicates average of three samples; error bars were calculated on the basis of standard deviation in three biological replicates. (C) Eighty-one base pair insertion (box) found in the putative cis-regulatory region of the mutant CAV3 gene. A repeating 10-bp element found in the insertion is shown in green.
Having identified ROIs for both the chm-like and bug mutants we proceeded to higher resolution mapping to identify the causative mutations for the two lines. Because of inherent differences in the degree of polymorphism of the two genomes, and differences in reference genome quality, we found that we had to use different fine-mapping approaches for the two mutants, and thus they are treated separately, below.
Fine mapping C. intestinalis chm-like
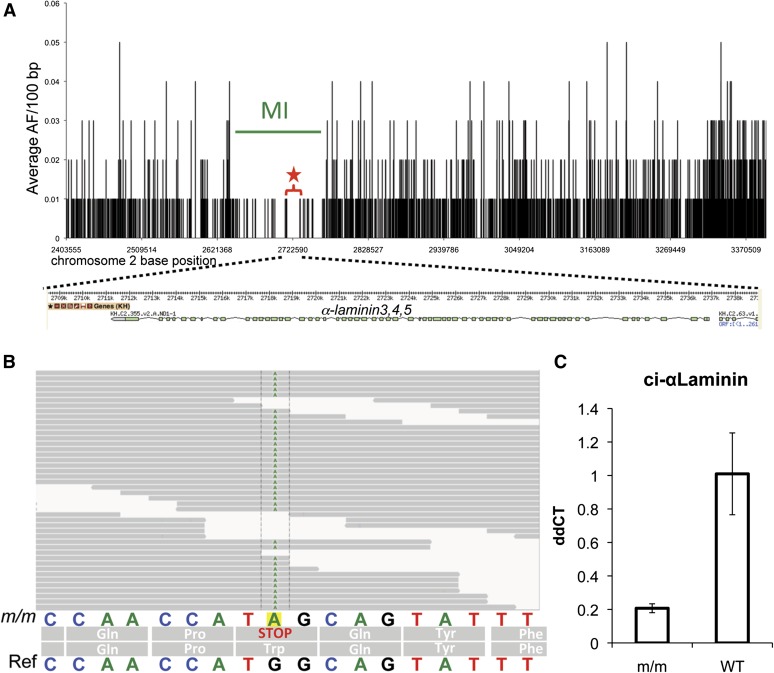
For fine mapping of chm-like, a 1-Mb ROI centered on the peak of the genome-wide map (chromosome 2:2.4–3.4 Mb) was used (Figure 3A). Unlike for genome-wide mapping, in which only the proportion of invariable sites was used to measure homozygosity (equivalent to Watterson’s theta, Watterson 1975), for fine mapping in C. intestinalis we computed the frequency of the major allele at each SNV, using only SNVs covered by five or more reads. One hundred base pair bins of the average major allele frequencies were then calculated across the ROI. We lowered the fold-coverage threshold in this analysis to exclude fewer base positions in the interval and achieve the most complete picture of linkage. By definition, the ROI is characterized by the presence of high-frequency alleles, and for fine mapping we want to visualize the disappearance of lower-frequency alleles as we approach the causative mutation. Our analysis included only bins where the nonreference allele frequency was in the 40–85% range, thereby excluding both the lowest (and possibly erroneous) allele frequencies and the highest (possibly completely homozygous) alleles. These bins were plotted against genomic position in the ROI. A distinct trough ∼100 kb in heterozygosity is seen centered around position 2.73 Mb and was defined as the mapping interval (MI; Figure 3A). The trough contained five predicted genes, including α-laminin3,4,5, which was located in the longest segment with zero heterozygosity (star, Figure 3A).
Figure 3.
Fine-mapping of the chm-like mutation. (A) The average allele frequency (AF) for 100-bp windows is plotted against nucleotide position for a 1-Mb region of interest (ROI) from chromosome 2 (2.4–3.4 Mb). The likely causative mutation was found in a 17-kb segment from 2.714 to 2.731 Mb. Interestingly, this region was the longest stretch with a complete absence of heterozygous allele frequencies (red star and bracket). A schematic of the α-laminin3,4,5 gene model is shown below the graph. (B) The nonsense mutation (G to A)in the α-laminin3,4,5 gene was found in all the of the aligned m/m reads. The sequence of the reference genome (ref) is shown at the bottom. (C) qRT–PCR of α-laminin3,4,5 in homozygous mutant (m/m) and WT cDNA samples. Bars indicate average of three samples and error bars calculated on the basis of standard deviation in the three replicates. ddCT values were calculated by normalizing to actin cDNA levels within each sample and then comparing mutant and wild-type levels.
Although the window of peak homozygosity (Figure 2A) did not include the α-laminin3,4,5 gene (it was located two windows away), we found that if the window size of genome-wide homozygosity analysis was changed from 10 to 20 kb, the peak homozygosity window landed directly on top of the α-laminin3,4,5 gene (Figure S3). Increasing the window size appeared to eliminate minor differences in the number of measurable sites between windows (e.g., a 100% homozygous window compromising only 9984 measurable sites vs. a window with 9999 homozygous sites of 10,000 measurable sites).
Examination of the aligned read sequences from the homozygous mutants identified a nonsense mutation in the third exon of α-laminin3,4,5 gene at position 2,734,938 of chromosome 2 (Figure 3B). This mutation was found in all 36 reads that covered this position. As this nonsense mutation lies at the beginning of the protein-coding region of the gene, it would almost certainly create a null allele. In addition, the α-laminin3,4,5 transcript of the C. intestinalis mutant was reduced fivefold, as measured by qRT–PCR (Figure 3C). A similar reduction in transcript level has been seen for other Ciona mutant genes, including in C. savignyi chm (Veeman et al. 2008). Based on the similar phenotype, complementation results, and nature of the mutation we concluded that this short-tail mutation is a C. intestinalis ortholog of the C. savignyi chm mutation.
Mapping a neural tube closure mutation in C. savignyi
Because of the low and uneven coverage for the C. savignyi sample, we found that analyzing windows of average allele frequency, as was done above for the C. intestinalis chm mutation, yielded inconclusive results, with several regions showing apparent drops in heterozygosity (Figure S2 and data not shown). As an alternative approach, we tested homozygosity mapping with smaller windows in the ROI. In addition to calculating homozygosity, we computed a local polynomial regression line of best fit (LOESS) based on the available homozygosity values of the ROI and to make the best estimate of peak homozygosity given incomplete coverage and information (Figure 4A). In the LOESS curve a gradual increase in homozygosity is seen in the 50- to 90-kb region of the ROI, with a corresponding peak over a predicted T-type calcium channel gene (CAV3, Okamura et al. 2005; Figure 4A). qRT–PCR analysis revealed that the expression of CAV3 is reduced ∼25-fold in bug/bug embryos relative to WT embryos (Figure 4B). None of the genes flanking CAV3 in the ROI appeared to be strong candidates for the bug mutation. Neighboring genes included a cation-independent mannose receptor (MR) and a fibronection leucine rich transmembrane receptor 2 (FLRT2). Only synonymous changes were found in the mutant FLRT2 sequence and a few nonsynonymous changes in the mutant MR sequence (data not shown). Moreover, the expression level of these genes was similar between mutant and wild-type animals (Figure 4B). Other genes in the ROI, a phospholipid scramblase, a ribosomal protein, and Ras-related Rab21 (Figure 4B), were excluded as bug candidate loci on the basis of the ubiquitous nature of their gene function and lack of nonsynonymous changes. The sequence for CAV3 from the homozygous bug mutants revealed a number of nucleotide changes resulting in nonconservative amino acid substitutions (Table 2). In addition to the amino acid substitutions, we identified an 81-bp insertion upstream from the predicted start methionine, in the putative cis-regulatory region. This insertion was bound at each end by a direct repeat of a 10-bp element found as a single copy in the WT assembly (Figure 4C). This 10-bp element was repeated an additional three times within the insertion. This insertion has the characteristics of a footprint from an excised transposable element (Scott et al. 1996; Kawakami et al. 2004). This insertion in the putative cis-regulatory region may account for the reduced expression of CAV3 observed in the mutant.
Table 2. Bugeye sequence changes for CAV3 gene.
| Genomic position | Mutation |
|---|---|
| Promoter/5′-UTR region | Large insertion unique to mutant |
| 45,007 | D > N |
| 45,011 | I > T |
| 46,850 | T > R |
| 47,973 | M > T |
| 50,330 | Q > K |
| 54,194 | P > Q |
| 55,701 | E > K |
| 66,750 | S > C |
| 66,926 | A > S |
| 68,702 | T > A |
| 69,589 | E > D |
| 90,325 | C > S |
| 90,811 | 3′-UTR A > G |
| 90,836 | 3′-UTR C > A |
| 90,858 | 3′-UTR G > C |
Given the mapping predictions for bug and the reduced expression of the CAV3 allele in bug, we hypothesize that disruption of CAV3 underlies the bug phenotype and the large insertion in the putative CAV3 promoter to be the likely causal mutation.
Self vs. cross-fertilization mapping
To further demonstrate the utility of the mapping strategy with wild isolated mutants in Ciona, we also sequenced the self-fertilized progeny (homozygous mutant and WT progeny, ∼600 each) from a single heterozygous bug adult. We used a gravid adult, heterozygous for bug, different from those used in the above analysis, and spawned it multiple times to collect enough progeny for sequencing. The mean genome-wide homozygosity values for both mutant and WT samples were 96.7% (only 0.2% higher than cross-fertilized bugeye data set; Table 1). The genome-wide homozygosity mapping from the single parent identified the same reftig peaks as were found in the data set from multiple parents (Figure 5). Although the Δhomozygosity values were slightly lower for the self-data set, the overall noise was reduced. The ROI, reftig 183, and the predicted genetic mutation in the two data sets were also found to be similar. These results demonstrate the feasibility of mapping directly from the mutant progeny of a single wild C. savignyi founder adult, which will considerably accelerate the screening and mapping of mutants.
Figure 5.
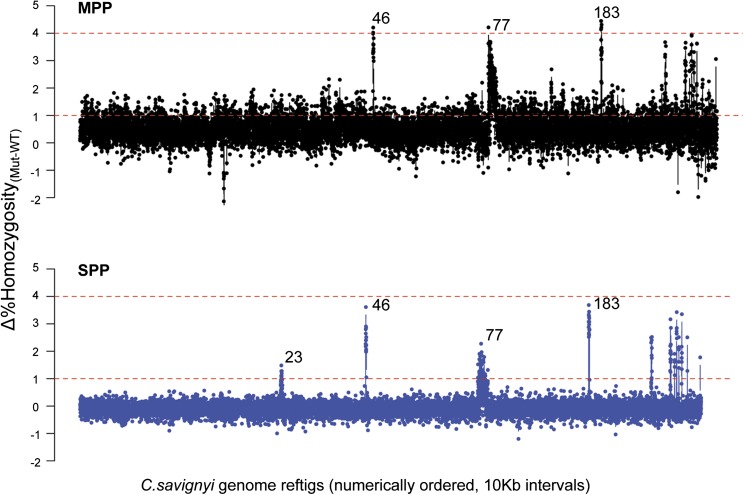
Comparison of outcrossed vs. self-fertilized genomic data sets for mapping the C. savignyi bugeye mutation. MPP, multiple parents used for cross-fertilization and progeny generation; data are shown in black. SPP, self-fertilized parent used for progeny generation, data are shown in blue. Δ% homozygosity values were computed for 10-kb windows across C. savignyi genomic reftigs. For the genome-wide plot, reftigs are presented in arbitrary order (according to assigned number) rather than as linkage groups. Reftigs <10 kb were not included. Dashed lines represent reference lines at Δ1% and Δ4% homozygosity values for comparing the two data sets. The three largest peaks fall on reftig 46, reftig 77, and reftig 183 in order. The extra peak seen in the SPP sample but not the MPP sample is reftig 23 (also known to belong to linkage group 11, Hill et al. 2008).
Discussion
Our results demonstrate the successful application of a whole-genome mapping strategy in two outbred Ciona mutant lines. WGS mutation mapping strategies have been reported for several genomically well-defined models organisms, including D. melanogaster, C., Danio rerio, and M. musculus (Blumenstiel et al. 2009; Leshchiner et al. 2012; Obholzer et al. 2012). The theoretical linkage analysis behind published WGS mapping methods and our method is very much the same. The differences lie in how each method computes linkage for the different genomic backgrounds and subtleties of each model organism. Published methods have made use of databases of SNP markers and reference or parental genetic background information in computing homozygosity (Doitsidou et al. 2010; Miller et al. 2013). By contrast, our homozygosity analysis makes no assumption and uses no information about parental or background genetic markers. Our unique approach simply asks for the greatest preservation of homozygous sequence (as opposed to individual SNP markers) as determined by the aligned and variable genomic sequencing reads. This works for Ciona and should work for other highly polymorphic/wild genomes where both genetic marker information may be unavailable and polymorphisms disrupt sequence homogeneity frequently. This approach may not be as effective for inbred genomes, which by nature contain large intervals of high homozygosity, or for species that are inherently highly homozygous. However, given a highly polymorphic genome, our approach simplifies the computation for homozygosity analysis by only calculating a simple binary problem across the whole genome and not deriving allele frequencies for millions of SNPs. Unlike the results of other methods, this simple computation may be sufficient to identify the candidate gene in genomes like Ciona as revealed by our narrow and extremely close peak calls at the whole-genome scale. Previous methods supplemented homozygosity analysis with fine-scale allele frequency mapping and still yielded, at best, 800-kb mapping intervals (Leshchiner et al. 2012).
Ciona presents a unique system for identifying spontaneous mutants. In temperate regions of the world Ciona species are present in enormous numbers in places such as harbors, where they are considered a nuisance. The facts that both C. savignyi and C. intestinalis are hermaphrodites with a capacity to self-fertilize and that the wild population of both species harbor lethal recessive mutations at high frequency make them ideal models (Veeman et al. 2011). The WGS mapping strategy presented here will greatly accelerate gene discovery in Ciona. The two species used, C. intestinalis and C. savignyi, differ significantly in their natural levels of heterozygosity (Dehal et al. 2002; Small et al. 2007a), which necessitated different approaches for fine mapping. The more polymorphic species, C. savignyi, presented several challenges due to its high sequence variation. The C. savignyi data set was generated from the progeny of 12 heterozygous parents, giving the potential for high genetic variation at all loci. This was evident in the lower percentage of sequence reads mapping to the reference genome when compared to C. intestinalis. Relaxing the stringency of read mapping might allow sequences with a greater degree of variation from the reference genome to be successfully aligned, but this will also increase misalignment and add noise. Mapping using longer reads or only transcriptome or coding regions might be another feasible approach for C. savignyi (Hill et al. 2013).
The C. intestinalis chm-like mutation proved to be an ideal test for the WGS strategy. The similar phenotype and complementation data meant that we were starting with a strong candidate. Although we strongly suspected that the C. intestinalis chm-like mutation would map to α-laminin3,4,5, the availability of orthologous mutations in the two Ciona species will provide a valuable research tool. Previous studies of the C. savignyi chm mutation show that a loss-of-function allele severely disrupts convergent extension in the notochord (Veeman et al. 2008). Our initial genome-wide homozygosity mapping with 10-kb windows identified a peak-homozygosity window 25 kb from the causative mutation. In retrospective analysis, we found homozygosity mapping with 20-kb windows would have mapped peak homozygosity directly on the α-3,4,5-laminin gene. The mapping of other C. intestinalis mutations in the future will indicate whether mutant genes can be identified from plots of homozygosity, without the need for fine mapping, as is now carried out in other species (Leshchiner et al. 2012; Obholzer et al. 2012). We have reason to be confident that the WGS strategy in Ciona will consistently give higher-resolution mapping than, for example, in zebrafish, in which mapping intervals have been reported to be in the range of 800 kb (Leshchiner et al. 2012). Modeling with zebrafish data indicated that much smaller mapping intervals could be achieved with greater sampling of individuals (Leshchiner et al. 2012). Because of the relatively small size of the Ciona genome compared to the zebrafish genome (160 Mb vs. 1.9 Gb), and our large input pools (>400 individuals), we are able to sample many more individuals for an equivalently sized data set. The highest average coverage we achieved was 34×. However, the sampling from the input pool of individuals within a window is much higher than 34×. For example, a 1-kb window with 34× coverage would contain an average of 680 50-bp reads. Each of these reads would be randomly derived from the genomes of the input pool. Thus, the ability to detect rare recombination events in genomic regions immediately flanking the causative mutation is very high. The mapping interval we defined for the C. intestinalis data set was ∼100 kb. However the causative mutation was found in a 17-kb interval corresponding to the longest stretch with 100% homozygosity, suggesting that the mapping ability may be much higher and smaller than the average per-site recombination rate (25–49 kb/cM) (Kano et al. 2006). Should the same be found for mutations mapped in the future, this would indicate a mapping precision higher than 2 cM. Furthermore, the narrowest mapping intervals depend on the availability of informative SNVs across a genome. The more sites in a genomic region, the more potential there is to detect a recombination event. Ciona has the highest known abundance of SNVs of any sequenced animal: In a 10-kb region, Ciona has on average ∼170 potential sites useful for detecting rare recombination events that can be used for linkage analysis. By contrast, zebrafish would, on average and depending on the strain, have only 20 of these potentially informative sites within the same window (Guryev et al. 2006). One can imagine how combining this increased site frequency with increased sampling would yield narrow mapping intervals for linkage analysis.
For mapping C. savignyi bug we included a WT data set that would potentially control for unlinked variation in homozygosity. Subtracting the WT homozygosity values reduced the noise and appears to have allowed us to map closer to the candidate mutated gene in the genome-wide mapping (comparing the 217-kb peak prediction of m/m only with the 57-kb peak prediction of m/m–WT on reftig 183; Figure 3D). Additonally, the fact that a large number of reftigs remain unmapped in C. savignyi gives extra value to the WT data set in providing confidence that unmapped reftigs are truly linked. Moreover the WT sample contained the progeny from 12 outbred parents and thus may provide a reference for population-wide genomic regions of low heterozygosity that can be used with future C. savignyi m/m data sets.
The bug mutation severely disrupts anterior neural tube closure. There are several lines of evidence supporting CAV3 as being the causative gene for the bug mutation. Aside from the genetic linkage, the transcript of this gene is downregulated 25-fold. We have previously shown that depletion of Ca2+ during C. intestinalis neurulation causes profound defects to anterior neural tube development, including an open neural plate phenotype in ∼40% of embryos, which is consistent with our findings here for CAV3 (Hackley et al. 2013). Although T-type calcium channels are known to have early embryonic expression in the developing vertebrate anterior neural tube (Perez-Reyes 2003; Lewis et al. 2009), to our knowledge this is the first implication of their involvement in neurulation and potentially neural tube closure. One of the advantages of Ciona compared to their vertebrate cousins is their lower genetic redundancy. The Ciona genomes appear to encode a single T-type Ca2+ channel (Cav3), which is the ortholog of the vertebrate Cav3.1, Cav3.2, and Cav3.3 genes (Okamura et al. 2005). Further characterization of CAV3 expression and the bug mutant will reveal new insights into how Ca2+ channels may contribute to the proper development of chordate CNS. A full description of the phenotype, and its causative link to CAV3 disruption, will follow in a separate publication.
Thus the small genome size, high heterozygosity (i.e., high density of genetic markers), and fecundity of Ciona all favor the whole-genome approach to mutation mapping. Finally, we show that it is possible to map a mutation using self-fertilized progeny from a single founder animal. Taken together, this mapping strategy removes the single largest bottleneck in the characterization of spontaneous mutants in these key chordate organisms.
Supplementary Material
Acknowledgments
S.A.W. thanks Daniel Bean and Jonathon Hill for their advice on the computational work of this study. We acknowledge the use of the UCSB CNSI computing facility supported by NSF grants MRSEC DMR-1121053 and CNS-0960316. Funding for this work was also provided by NIH Grants HD038701 to WCS, and GM098614 to TLT.
Footnotes
Communicating editor: B. Sullivan
Literature Cited
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Lim J. E., Samocha K. E., Sokoloff G., Abney M., et al. , 2010. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185: 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P., Satou Y., Campbell R. K., Chapman J., Degnan B., et al. , 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298: 2157–2167. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H., 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439: 965–968. [DOI] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V., Koudijs M. J., Berezikov E., Johnson S. L., Plasterk R. H., et al. , 2006. Genetic variation in the zebrafish. Genome Res. 16: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackley C., Mulholland E., Kim G. J., Newman-Smith E., Smith W. C., 2013. A transiently expressed connexin is essential for anterior neural plate development in Ciona intestinalis. Development 140: 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. T., Demarest B. L., Bisgrove B. W., Gorsi B., Su Y. C., et al. , 2013. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 23: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M. M., Broman K. W., Stupka E., Smith W. C., Jiang D., et al. , 2008. The C. savignyi genetic map and its integration with the reference sequence facilitates insights into chordate genome evolution. Genome Res. 18: 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Marth G. T., Quinlan A. R., Dooling D., Fewell G., et al. , 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5: 183–188. [DOI] [PubMed] [Google Scholar]
- Hobert O., 2010. The impact of whole genome sequencing on model system genetics: get ready for the ride. Genetics 184: 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelier R., Semple J. I., Garcia-Verdugo R., Lehner B., 2011. Predicting phenotypic variation in yeast from individual genome sequences. Nat. Genet. 43: 1270–1274. [DOI] [PubMed] [Google Scholar]
- Jiang D., Munro E. M., Smith W. C., 2005. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr. Biol. 15: 79–85. [DOI] [PubMed] [Google Scholar]
- Kano S., Satoh N., Sordino P., 2006. Primary genetic linkage maps of the ascidian, Ciona intestinalis. Zool. Sci. 23: 31–39. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Imanaka K., Itoh M., Taira M., 2004. Excision of the Tol2 transposable element of the medaka fish Oryzias latipes in Xenopus laevis and Xenopus tropicalis. Gene 338: 93–98. [DOI] [PubMed] [Google Scholar]
- Lehner B., 2013. Genotype to phenotype: lessons from model organisms for human genetics. Nat. Rev. Genet. 14: 168–178. [DOI] [PubMed] [Google Scholar]
- Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C. A., et al. , 2012. Mutation mapping and identification by whole-genome sequencing. Genome Res. 22: 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. B., Wester M. R., Miller L. E., Nagarkar M. D., Johnson M. B., et al. , 2009. Cloning and characterization of voltage-gated calcium channel alpha1 subunits in Xenopus laevis during development. Dev. Dynamics 238: 2891–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2012. Advances in quantitative trait analysis in yeast. PLoS Genet. 8: e1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Tsementzi D., Kyrpides N., Read T., Konstantinidis K. T., 2012. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS ONE 7: e30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Howie B., Myers S., McVean G., Donnelly P., 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Miller A. C., Obholzer N. D., Shah A. N., Megason S. G., Moens C. B., 2013. RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res. 23: 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obholzer N., Swinburne I. A., Schwab E., Nechiporuk A. V., Nicolson T., et al. , 2012. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 139: 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y., Nishino A., Murata Y., Nakajo K., Iwasaki H., et al. , 2005. Comprehensive analysis of the ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol. Genomics 22: 269–282. [DOI] [PubMed] [Google Scholar]
- Olinski R. P., Dahlberg C., Thorndyke M., Hallbook F., 2006. Three insulin-relaxin-like genes in Ciona intestinalis. Peptides 27: 2535–2546. [DOI] [PubMed] [Google Scholar]
- Passamaneck Y. J., Di Gregorio A., 2005. Ciona intestinalis: chordate development made simple. Dev. Dynamics 233: 1–19. [DOI] [PubMed] [Google Scholar]
- Perez-Reyes E., 2003. Molecular physiology of low-voltage-activated T-type calcium channels. Physiol. Rev. 83: 117–161. [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N., 1994. Developmental Biology of Ascidians. Cambridge Univ. Press, Cambridge, UK/New York. [Google Scholar]
- Satou Y., Shin-i T., Kohara Y., Satoh N., Chiba S., 2012. A genomic overview of short genetic variations in a basal chordate, Ciona intestinalis. BMC Genomics 13: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L., LaFoe D., Weil C. F., 1996. Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E., Kawashima T., Satou Y., Hamaguchi M., Sin I. T., et al. , 2006. Chromosomal mapping of 170 BAC clones in the ascidian Ciona intestinalis. Genome Res. 16: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N., Smith W. C., 2008. Inverse correlation of population similarity and introduction date for invasive ascidians. PLoS ONE 3: e2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K. S., Brudno M., Hill M. M., Sidow A., 2007a Extreme genomic variation in a natural population. Proc. Natl. Acad. Sci. USA 104: 5698–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K. S., Brudno M., Hill M. M., Sidow A., 2007b A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 8: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresser J., Chiba S., Veeman M., El-Nachef D., Newman-Smith E., et al. , 2010. doublesex/mab3 related-1 (dmrt1) is essential for development of anterior neural plate derivatives in Ciona. Development 137: 2197–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T., Nakatani Y., Hendrickson C., Ericson V., Lin C., et al. , 2008. Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development 135: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T., Chiba S., Smith W. C., 2011. Ciona genetics. Methods Mol. Biol. 770: 401–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson J. P., Jaffe D. B., O’Neill K., Karlsson E. K., Stange-Thomann N., et al. , 2005. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 15: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.