Abstract
Platynereis dumerilii is a marine polychaete and an established model system for studies of evolution and development. Platynereis is also a re-emerging model for studying the molecular basis of circalunar reproductive timing: a biological phenomenon observed in many marine species. While gene expression studies have provided new insight into patterns of gene regulation, a lack of reverse genetic tools has so far limited the depth of functional analyses in this species. To address this need, we established customized transcriptional activator-like effector nucleases (TALENs) as a tool to engineer targeted modifications in Platynereis genes. By adapting a workflow of TALEN construction protocols and mutation screening approaches for use in Platynereis, we engineered frameshift mutations in three endogenous Platynereis genes. We confirmed that such mutations are heritable, demonstrating that TALENs can be used to generate homozygous knockout lines in P. dumerilii. This is the first use of TALENs for generating genetic knockout mutations in an annelid model. These tools not only open the door for detailed in vivo functional analyses, but also can facilitate further technical development, such as targeted genome editing.
Keywords: marine, invertebrate, genome editing, evolution, chronobiology
MANY fascinating biological phenomena, of which we have little to no molecular understanding, are observed in organisms outside of those that constitute conventional molecular model systems. An interesting system that has re-emerged in recent years as model for chronobiology and evolutionary studies is the marine polychaete worm Platynereis dumerilii (Annelida, Lophotrochozoa). However, in Platynereis and other emerging model systems alike, dissecting gene function in vivo remains challenging.
Platynereis is slowly evolving, compared with other more conventional molecular model protostomes such as Caenorhabditis elegans and Drosophila melanogaster (Raible et al. 2005). Evidence from comparative morphology and development suggest that nereidid annelids like P. dumerilii possess a body plan that is likely ancestral for Bilateria (Dohrn 1875; Arendt and Nübler-Jung 1994, 1997; Tessmar-Raible and Arendt 2003). This, together with its phylogenetic position in the Lophotrochozoa, makes Platynereis an ideal model for understanding how developmental gene regulation might have evolved from that present in the last common ancestor of all bilaterians. Additionally, Platynereis is also studied to understand principles of animal development (reviewed in Fischer et al. 2010), the hormonal regulation of regeneration vs. maturation (Hauenschild 1974; Hofmann 1976), and chronobiology (Hauenschild 1960; Zantke et al. 2013).
Descriptive studies have contributed significantly to the understanding Platynereis biology. These have been facilitated by reliable techniques such as in situ hybridization (Tessmar-Raible et al. 2005), quantitative PCR (Dray et al. 2010; Zantke et al. 2013), and image registration (Tomer et al. 2010). Established techniques for transgenesis also provide the opportunity to label specific cell types (Backfisch et al. 2013). Transgenic reporter lines in particular facilitate in-depth analyzes of spatio-temporal regulation of gene expression across the entire life cycle. This approach has already contributed to the discovery of r-opsin1+ peripheral photoreceptor cells (Backfisch et al. 2013). Coupling transgenesis with chemical-mediated cell ablation can be further used to identify the functional importance of specific cells that express a gene of interest (Veedin Rajan et al. 2013). While highly useful, this approach can unravel only the function of the ablated cell types, although the function of a specific gene of interest, and the protein(s) that it encodes, may well deviate from this. In addition, generating cell-type-specific transgenic lines is not trivial, considering that only few specific enhancers have been characterized for Platynereis.
Analysis of protein functions is also possible using methods that include the incubation of Platynereis larvae with small neuropeptides for studying phenotypic or behavioral phenotypes (Conzelmann et al. 2011), and treatment with small-molecule activators and inhibitors, to directly modulate protein functions in vitro (estrogen receptor) (Keay and Thornton 2009) and in vivo (Denes et al. 2007; Schneider and Bowerman 2007; Dray et al. 2010; Demilly et al. 2013; Lidke et al. 2014). While useful for gaining spatio-temporal insight into protein activity, the penetration (i.e., effective dosage), affinity, and specificity of small-molecule inhibitors (often developed for use in other systems) may limit the pathways and targets that can be investigated in vivo in Platynereis.
Currently, the only technique available to interfere with the function of any given gene of interest in Platynereis is injection of morpholino antisense oligonucleotides into early stage embryos (Conzelmann et al. 2013). Morpholinos mediate efficient knockdown of target gene expression in vivo and are particularly useful for analyzing genetic phenotypes during embryogenesis and early larval development (Nasevicius and Ekker 2000). However, due to their transient action and difficulties regarding delivery into adult tissues, morpholinos are not suitable for analyzing gene function at later stages of the life cycle. Hence, to enable functional genetics studies across the entire life cycle of Platynereis, new tools for engineering stable and heritable gene knockouts are needed.
Transcriptional activator-like effector nucleases (TALENs) have rapidly become a technique of choice for precision genome engineering. TALENs are custom-designed nucleases that consist of a modular DNA-binding domain fused to a monomeric, C-terminal FokI nuclease domain (Christian et al. 2010). TALENs work in pairs and are designed to recognize and bind to tandem-oriented sequences in genomic DNA, separated by a short spacer (15–30 bp). TALEN binding causes dimerization and activation of the FokI nuclease domains, which results in cleavage of the DNA within the spacer region. Small insertions or deletions (indels) are frequently introduced at this site, as the result of errors made during DNA repair by nonhomologous end-joining (NHEJ). These indels can be up to several hundred base pairs in length and result in frameshift mutations that lead to the production of truncated or nonfunctional proteins (Sander et al. 2011; Lei et al. 2012; Ansai et al. 2013).
Successful use of TALENs for inducing targeted mutations has been reported in many conventional models, for example: mice (Davies et al. 2013; Qiu et al. 2013; Wang et al. 2013), teleost fish [zebrafish (Danio rerio)] (Huang et al. 2011; Bedell et al. 2012; Zu et al. 2013), medaka (Oryzias latipes) (Ansai et al. 2013, 2014), Xenopus (Ishibashi et al. 2012; Lei et al. 2012; Suzuki et al. 2013), and D. melanogaster (Liu et al. 2012). TALENs are also reported to be functional in a variety of other invertebrate arthropods, including mosquitos (Aedes aegypti) (Aryan et al. 2013) and Anopheles gambiae (Smidler et al. 2013), silkworm (Bombyx mori) (Ma et al. 2012; Sajwan et al. 2013; Takasu et al. 2013), and cricket (Gryllus bimaculatus) (Watanabe et al. 2012). The efficacy of TALENs across a wide variety of species, together with the availability of open-source target prediction tools, construction protocols, and reagents (available via Addgene: http://www.addgene.org/TALEN/), make this technology ideally suited for precision genome engineering in nonconventional model organisms. We therefore reasoned that TALEN technology could be used for genetic manipulation in P. dumerilii.
Here we report the establishment of P. dumerilii strains carrying TALEN-induced targeted mutations. We outline the workflow for TALEN design, construction, and in vitro validation and describe methods to facilitate rapid in vivo mutation screening of Platynereis larvae and adult worms, all of which can easily be adapted to other nonconventional model species. Using this workflow, we detected a variety of TALEN-induced mutations in three endogenous Platynereis genes. Mutation efficiencies were variable in animals raised from TALEN-injected zygotes, including evidence of biallelic mutation rates. Detection of TALEN-induced mutations in G1 offspring demonstrated that such mutations are heritable. The use of TALEN-mediated genome modification provides a new opportunity to move from inferring gene functions from gene expression profiles to assigning genetic causality to biological phenotypes.
Materials and Methods
Animal culture, breeding, and sampling procedures
Platynereis cultures were maintained at 18° according to standard protocols as described previously (Hauenschild and Fischer 1969). Animals used for genotyping and TALEN injection were from either PIN or VIO inbred strains (as described in Zantke et al. 2014).
Injected larvae (G0, i.e., raised from TALEN-injected zygotes, two- or four-cell-stage embryos) were sampled for mutation screening at 24 hr post fertilization (hpf) in pools of four to five individuals or as single larvae. G0 adult worms (raised from TALEN-injected embryos and sibling controls) were sampled at 1–3 months of age for genomic DNA (gDNA) extraction. Worms were anesthetized for at least 5 min in a 1:1 mixture of artificial sea water (ASW):7.5% magnesium chloride (hexahydrate, AppliChem), and a small piece of the tail (∼10 segments) was removed into gDNA lysis buffer (see below for extraction). After sampling, the animals were placed into individual wells of eight-well plastic boxes containing fresh ASW and grown to maturity under standard conditions. For G1 sampling, at least 20 individual larvae (in pools of four) were screened from each batch, and remaining offspring were set out into culture boxes to be raised.
Genomic DNA extraction from adult worm samples
Genomic DNA was extracted from whole mature, spawned worms or tail samples from adult worms using the NucleoSpin Tissue kit (#740952, Machery-Nagel). The protocol for extraction from human or animal tissue and cultured cells was followed as described except that tail samples were extracted using half-volumes of buffers T1, proteinase K, buffer B3, and 100% ethanol, and DNA was finally eluted using 40 μl of elution buffer (gDNA from whole mature worms was eluted in 100 μl of elution buffer). Samples were stored at −20°.
Platynereis genes and genomic sequences used in this study
Coding sequences were previously identified for l-cry (GU322429: Zantke et al. 2013), vtn (EF544399: Tessmar-Raible et al. 2007), and er (EU482033: Keay and Thornton 2009). These sequences were used for designing primers for genotyping and screening PCR. Reference sequences for er, l-cry, and vtn genomic loci were obtained from assembly of shotgun sequence reads from BAC clones identified to include the coding sequences of each gene (lcry: CH305_21G18; er: CH305_148E21; vtn: CH305_55J15 and CH305_88I16). All BAC clones are available from the CHORI-305 BAC library, BAC PAC Resources, Children’s Hospital Oakland Research Institute, Oakland, California. BAC screening was performed as described previously (Raible et al. 2005). PCR-verified genomic reference sequences for er, lcry, and vtn loci are available from GenBank.
Genotyping of Platynereis strains to identify target gene alleles
Primers were designed to amplify genomic regions for each target gene. The primer combinations used were vtnF1/R1, erF1/R1, and l-cryF1/R1 (see Supporting Information File S1). Genomic regions were amplified from gDNA samples prepared from 8–10 individual, mature worms from different inbred lab strains (VIO and PIN: for PCR reaction mixes see File S1). Bands corresponding to the size expected for the BAC reference allele as well as secondary bands representative of different alleles with putative size polymorphisms were subcloned. To identify single nucleotide polymorphisms (SNPs), plasmid DNA for multiple clones for each product was double-digested with BglII to excise the cloned insert, and with HinfI, a frequent cutter, to generate a restriction fragment profile for each clone. Subclones representative of different restriction fragment profiles were sequenced, and sequences were compared to the BAC reference sequence to map the position and identity of the SNPs present.
TALEN design and construction
All TALENs were designed using the TALEN Targeter prediction tool (TAL Effector nucleotide targeter v1.0 and 2.0; https://tale-nt.cac.cornell.edu/) (Doyle et al. 2012). Exon sequences were used as input. The design criteria applied were the following: equal length of left and right recognition sites [i.e., equal repeat variable diresidue (RVD) lengths], spacer length of 15–20 bp, NN for G recognition, and the presence of a unique restriction enzyme site in the spacer. To minimize the chance of translational bias between left and right TALENs, we aimed to standardize the size of TALEN recognition sites to 15 bp each (i.e., 14.5 RVDs). TALENs (Table S1) were constructed with the GoldenGate plasmid kit (v1.0, v2.0 Addgene: 1000000024) using the reaction mixture recipes generated by inputting RVD sequences into the Golden Gate TAL Assembly form (Excel file available from https://tale-nt.cac.cornell.edu/protocols). Assembly steps were performed according to the published protocol (Cermak et al. 2011).
To enable in vitro validation of TALEN messenger RNA (mRNA) using the Sp6 mMESSAGE mMACHINE system, we constructed a new GoldenGate compatible expression vector with a pCS2+ backbone: pCS2+_TAL3-WT (Figure S1). The pCS2+_TAL3-WT vector features the long N287/C230 N- and C- terminal architecture and was constructed by inserting the TALEN cloning site from pTAL3 (GoldenGate TALEN Construction kit v1.0) (Cermak et al. 2011; see Figure S1 and File S1 for construction details). TALENs were also (re)constructed using the heterodimeric FokI expression plasmids pCS2TAL3-DD and pCS2TAL3-RR (Dahlem et al. 2012), obtained from Addgene (plasmids #37375 and #37376). All final TALEN expression vectors were sequence-verified using seqTAL_1-5 forward and TAL_R2 primers (Cermak et al. 2011).
Transcription/translation in vitro cleavage assay
TALEN efficacy was evaluated using an in vitro DNA cleavage assay [referred to here as transcription/translation (TnT) assay] similar to that previously described (Mussolino et al. 2011). Reactions were assembled using 40 μl of Sp6 Quick-coupled TnT reaction master mix (#L2080, Promega), 1 μg of each TALEN plasmid, 225 ng of specific target DNA, and 225 ng of nontarget DNA (egfp-coding sequence). Negative control reactions contained only left TALEN plasmid DNA. Reactions were incubated for 3 hr at 30°C. DNA was purified by phenol/chloroform extraction. Extracted DNA was treated with RNaseA and analyzed on 1.5% TAE agarose gels.
mRNA transcription
Plasmids encoding full-length TALENs were linearized by NotI digestion, gel-purified (Gel Extraction kit, Qiagen), and used as templates for in vitro transcription using mMESSAGE mMACHINE Sp6 Kit (AM1340, Life Technologies). The majority of injections were performed using RNA that was not subject to further cleanup steps.
Embryo microinjection
Fertilized embryos were injected with TALEN mRNA at the one- to four-cell stage. Preparation of embryos for microinjection and microinjection techniques were performed as described previously (Backfisch et al. 2013). Injection solutions were prepared in a final volume of 10 μl, with 1 μl of 3% tetramethylrhodamine isothiocyanate (TRITC)–dextrane in 0.2 M KCl (Invitrogen) and equal concentrations of each TALEN mRNA (between 40 ng/µl/TALEN and 300 ng/µl/TALEN). Injection solutions were filtered using 0.45 μm PVDF centrifugal filters (Ultrafree-MC-HV, Millipore) and centrifugation at 12,000 × g for 1–3 min. Average injection volumes were estimated to range from 25 to 60 pl per embryo.
Rapid digestion of single or pooled larvae for mutation screening
Individual or pools of larvae (n = 4–5) were harvested at, or after, 24 hpf and digested in Proteinase K/1× PCR digestion buffer (10× Qiagen PCR buffer with 1 µl Proteinase K solution per 10 μl of solution) (NucleoSpin Tissue, Machery-Nagel) at 56° for 2–3 hr. The reaction was terminated by incubation at 95° for 10 min, and samples were stored either at 4° or −20° or used directly as PCR template (1–2 μl/reaction).
PCR and restriction digest screening assays
PCR reaction mixes contained DNA polymerase [either HotStar Taq Plus (Qiagen) or Phusion Polymerase (Fermentas)], 1.5–3 mM MgCl2, 400 μM dNTPs, 200 μM of each primer, 1× reaction buffer (according to the enzyme used), 1–2 µl of DNA template in final volume of 25 or 50 µl. Cycling conditions included an initial hot start at 95°–98° for 30 sec to 2 min, 35–40 cycles of 95°–98°/30 sec; 60°–68°/1 min; 72°/1–2.5 min, plus a final 72° extension for 5–10 min.
Unless otherwise stated, restriction enzymes were from New England Biolabs. In general, restriction digest reactions to screen for mutations, were incubated for 1–3 hr at 37° and heat-inactivated if necessary according to the manufacturer’s instructions. See Table S2 for primer and enzyme combinations used for individual screening assays.
Calculation of mutation rates using relative band intensity analysis
We defined mutation frequency as “the percentage of samples showing evidence of TALEN-induced mutations” (as uncut PCR products from restriction digest or with extra bands representing larger insertions or deletions). We defined the mutation rate as “the percentage of mutant genome copies per sample.” These values were inferred from the percentage of the total PCR product represented by mutant bands (i.e., uncut PCR bands or extra insertion/deletion bands).
The relative percentages of mutant bands were calculated from gel images (TIFF format) using gel analysis tools in ImageJ (Schneider et al. 2012: http://rsbweb.nih.gov/ij/). The intensity of each band per lane was measured, and intensities were normalized to band size by dividing the band intensity value by the size of the band. The intensities of all bands per lane were summed to give an intensity value for the total PCR product from which the percentage of uncut, or deletion band, was calculated: i.e., [i(uncut band)/i(total)] × 100 = % uncut band. These values are reported in Table 1, Table 2, and Figure S5.
Table 1. TALEN mutation frequencies in larvae.
| Gene/TALEN pairs(s) | [TALEN mRNA]a (ng/µl) | Total no. of screened larvae | No. of larval samples screened (n = no. of pooled/sample) | Mutation frequency (% mutation-positive samples) | % mutant genome copies/sample (range)b | % mutant genome copies/sample (average ± SD) |
|---|---|---|---|---|---|---|
| erEx3_L2/_R2 | 100 | 144 | 36 (n = 4) | 0.0 | 0 | 0 |
| erEx3_L2/_R2 | 180 | 20 | 5 (n = 4) | 20.0 | Not measured | NA |
| erEx3_L2/_R2 | 200 | 48 | 12 (n = 4) | 50.0 | Not measured | NA |
| erEx3_L2/_R2 | 267 | 68 | 17 (n = 4) | 70.6 | 2.94 - 6.08 | 4.98 ± 1.44 |
| erEx3_L2/R2 | 300 | 92 | 23 (n = 4) | 39.1 | 5.7–13.1 | 9.02 ± 3.2 |
| erEx2_L/_R + erEx3_L2/_R2 | 300 | 44 | 11 (n = 4) | 18.2 (deletion) | 22.3–27.3 (large deletion) | 24.8 ± 3.5 (large deletion) |
| l-cryEx2_L/_R + l-cryEx3_L/_R | 300 | 186 | 54 (n = 4 × 44; n = 1 × 10) | 18.5 (uncut bands); 16.7 (large deletions) | 2.21–8.84 (uncut bands); 12.7–56.2 (large deletions) | 5.33 ± 2.50 (uncut bands); 35.7 ± 18.9 (large deletions) |
| vtnEx2_L/R | 40 | 18 | 18 (n = 1) | 5.6 | Not measured | NA |
| vtnEx2_L/R | 100 | 32 | 8 (n = 4) | 75.0 | Not measured | NA |
| vtnEx2_L/R | 200 | 44 | 11 (n = 4) | 27.3 | 0.9–21.8 | 7.45 ± 9.00 |
Final concentration of mRNA in a 10-μl injection solution per TALEN.
Calculated from intensity of uncut or deletion band for samples of pooled larvae (n = 4–5). See also Figure S5, A and B.
Table 2. Mutation frequencies in adult worms and germline carriers in adult worms.
| Gene/TALEN pair(s) | No. of worms raised | No. of mature adults screened | No. of mutation-positive adults | Mutation frequency (% mutation-positive adults) | % mutant genome copiesa | No. of germline carriers (carrier ID) |
|---|---|---|---|---|---|---|
| erEx2_L/R + erEx3_L2/R2 | 60 | 60 | 6 | 5% (3/60 indels); 3% (2/60: large deletion); 1% (1/60: inversion) | 10–23% (indels) 6–12% (large deletions) <1% (inversion) | 2 (er+32; er+59: large deletions) |
| erEx3_L2/R2 | 6 | 6 | 0 | 0% | NA | NA |
| l-cryEx2_L/R + l-cryEx3_L/R | >80 | 62 | 27 | 43% (27/60: indels) 9% (6/60: large deletions and indels) | 5–∼100% (indels) Not measured | 1 (l-cry+36: indels and large deletion) |
| vtnEx2_L/R | 13 | 13 | 3 | 20% (indels) | Not measured | 0 |
Calculated from intensity of uncut or deletion band. Indicates individual mutation rate. See also Figure S5C.
Results
To assess whether TALENs generate potential loss-of-function mutations in P. dumerilii, we chose to target three previously described endogenous genes: light-receptive cryptochrome (l-cry), a putative circadian light receptor (Zantke et al. 2013); estrogen receptor (er), a verified ortholog of vertebrate ligand-dependent estrogen receptors (Keay and Thornton 2009); and vasotocin-neurophysin (vtn), an ortholog of oxytocin/vasopressin-neurophysin prepro-hormones (Tessmar-Raible et al. 2007).
Genotyping to identify appropriate TALEN target sites in endogenous Platynereis genes
TALEN efficiency is highly dependent upon specific recognition of target sequences (Cade et al. 2012). Hence, it is important to ensure the fidelity of reference sequences used for TALEN design and avoid targeting regions that contain polymorphisms. Genome sequencing analyses of marine invertebrates such as Capitella teleta, Lottia gigantea, sea urchin (Strongylocentrus purpuratus), and sponge (Amphimedon queenslandica) report high rates of polymorphisms (∼3–5%) (Sodergren et al. 2006; Srivastava et al. 2010; Simakov et al. 2013b). We and others have also identified frequent SNPs present in both intronic and coding sequences of various genes in Platynereis laboratory strains (Simakov et al. 2013a; C. Lohs and F. Raible, unpublished results). Therefore, to design effective TALENs against endogenous genes, we first needed to identify the most frequent alleles present in our worm strains and determine appropriate reference sequences for TALEN design.
To identify allelic variations for l-cry, er, and vtn, including intron size and SNPs, each locus was analyzed by PCR-based genotyping of two related and commonly used laboratory strains of P. dumerilii (VIO and PIN). As frameshift mutations introduced at the 5′ end of coding sequences have a higher chance of producing truncated, nonfunctional proteins, we aimed to target the 5′ coding exons of each gene. PCR amplicons for genotyping were restricted to the first three exons of each gene, and 8–10 individuals were screened per strain. PCR products were then analyzed for size polymorphisms by restriction digest and gel electrophoresis. Clones with different restriction fragment profiles, indicative of SNPs, were sequenced, and allelic variations were classified by aligning cloned sequences to BAC reference sequences.
We detected an ∼2.5-kb polymorphism in intron 2 of the vtn locus in PIN strain worms (data not shown). The majority of SNPs identified for each gene were intronic and primarily present in PIN strain subclones (Figure S2 and Table S3). Only one exonic SNP was found at the 3′ end of er exon 2 (Table S3). In VIO worms, the majority of sequences obtained were identical to the BAC reference sequences. Hence, we concluded that the exonic BAC reference sequences were sufficient for designing TALENs to target exons of l-cry, vtn, and er loci.
TALEN design and construction
Potential TALEN target sites were predicted for the 5′ exons of each target gene. We selected final TALEN pairs for construction based on several criteria: (1) target sites did not overlap with known SNPs or exon–intron boundaries; (2) target sites were upstream of, or within critical protein functional domains; (3) a “T” residue preceded the first nucleotide in each TALEN-binding site; and (4) spacer regions contained a unique restriction site to facilitate mutation screening by restriction digest.
Using these criteria, we selected TALEN pairs for each gene (Table S1): one pair targeting the region encoding the mature Vasotocin peptide (yellow in Figure 1A); three pairs for er targeting exons 2 and 3, encoding part of the N-terminal and DNA binding domains, respectively (yellow in Figure 2A); and two pairs targeting exons 2 and 3 of l-cry, which encode the N-terminal half of the photolyase domain (yellow in Figure 3A).
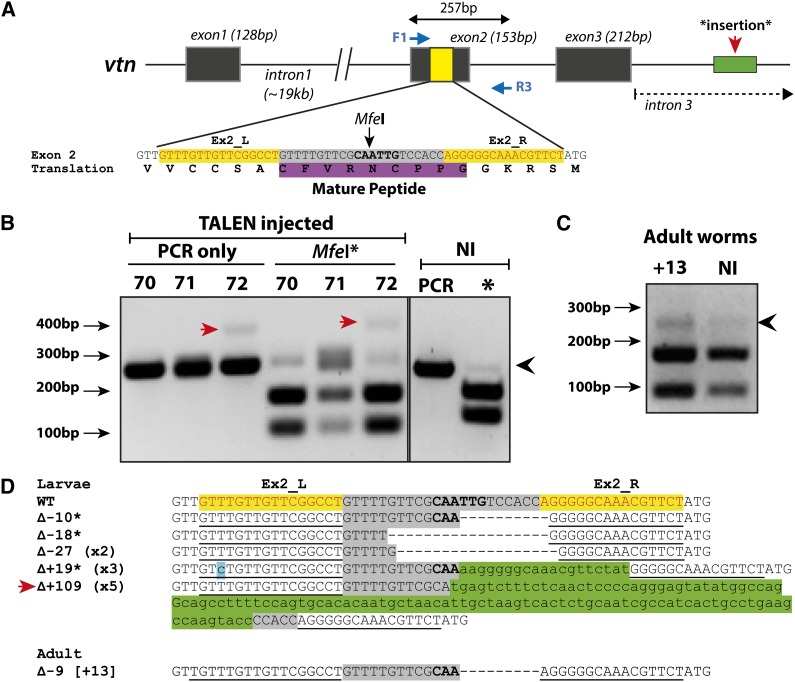
Figure 1.
Evidence for TALEN-induced mutation of Platynereis vasotocin-neurophysin. (A) Schematic of the vtn locus showing TALEN target site (yellow) in exon 2 (L, left TALEN; R, right TALEN). Blue arrows (F1/R3) indicate primers used for mutation screening PCR [size of amplicon (bp) indicated by double-ended arrows]. Green block indicates position of sequence from intron 3 detected as an insertion (sample 72). The mature peptide (highlighted in purple) is included by the majority of the TALEN spacer and includes the MfeI-screening site. (B) Restriction digest screening of 24-hpf larvae injected with vtnEx2_L and vtnEx2_R TALEN mRNA at concentration of 200 ng/µl/TALEN. Samples contain pools of four larvae. Black arrow indicates size of uncut PCR product; red arrow indicates 109-bp insertion detected in sample 72. (C) Results of MfeI digest of PCR products from adult worms: uncut PCR product (black arrowhead) cloned from adult injected worm vtn+13, representing 9-bp deletion. (B and C) PCR, un-digested PCR product; NI, non-injected; asterisk (*) indicates samples digested with MfeI. (D) Results of sequence analysis of uncut and insertion bands from samples in B and C. Length of mutations indicated by ∆ symbol with “−” indicating deletions and “+” indicating insertions. Restriction site is shown in boldface type; asterisks indicate frameshift mutations. Shading key: yellow, TALEN-binding sites; gray, spacer; blue, nucleotides differing from wild type; green, inserted nucleotides.
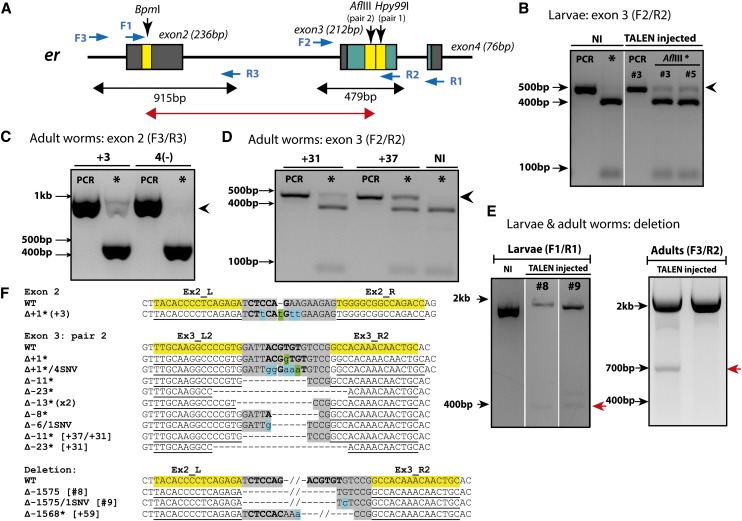
Figure 2.
TALEN-induced mutations in the Platynereis estrogen receptor. (A) Schematic of the er locus showing target exons 2 and 3 with TALEN target sites (yellow). Blue arrows, primer positions; red double-ended arrow, region of sequence deleted in E; green, DNA-binding domain. Primer combinations used for screening are shown above in B–E. (B–E) PCR, undigested PCR product; NI, non-injected. (B) Restriction digest screening of larvae injected with erEx3_L2/R2 TALENs (mRNA concentration: 267 ng/µl/TALEN mRNA). Arrowhead indicates uncut PCR product following AflIII digestion (asterisk). (C) Mutation evidence at exon 2 site: uncut band adult worm +3 vs. fully digested product from mutation-negative (−) TALEN-injected worm. (D) Adult worms er+31 and er+37 with mutations at exon 3 site. (E) Deletions (red arrow) detected in larvae and adult worms resulting from simultaneous cleavage at exons 2 and 3 using 300 ng/µl/TALEN mRNA: deletion positive (+); deletion negative (−). Please note different primer pairs used for larval vs. adult samples. (F) Mutant sequences obtained from digest screening for exons 1, 2, and long deletions. Numbers in brackets indicate the sample or worm from which the sequence was obtained; all other sequences are from injected larvae shown in B. Length of mutations are indicated by ∆ with “−” indicating deletions and “+” indicating insertions. Restriction site is shown in boldface type; asterisks indicate frameshift mutations. Shading key: yellow, TALEN binding sites; gray, spacer; blue, nucleotides differing from wild type; green, inserted nucleotides.
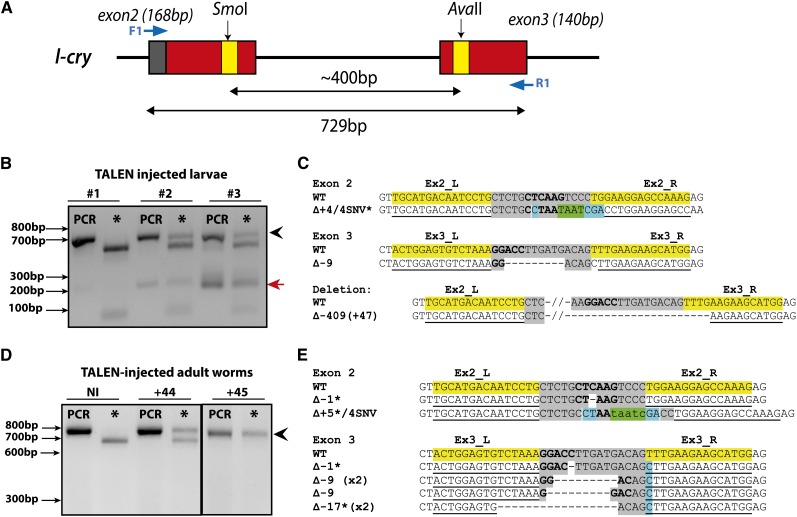
Figure 3.
Evidence of TALEN-induced mutations at l-cry locus. (A) Schematic of the l-cry genomic locus. Gray, exons; red, photolyase domain; yellow, TALEN sites; blue arrows, position of primers. Size of PCR amplicons and distance between TALEN sites indicated by double-ended arrows. Mutation screening results from larvae (B) and adult worms (D) injected with TALENn mRNA (l-cryEx2_L/R and l-cryEx3_L/R: 300 ng/µl/TALEN), showing uncut PCR bands following AvaII digestion (black arrow) and/or long deletions (red arrow). (D) Worm l-cry+45 shows completely undigested band suggesting biallelic mutation. Mutant sequences obtained from injected larvae (C) and adult worms (E) following AvaII digestion (asterisks). Length of mutations indicated by ∆ with “−” indicating deletions and “+” indicating insertions. Restriction site is shown in boldface type; asterisks indicate frameshift mutations. Shading key: yellow, TALEN0binding sites; gray, spacer; blue, nucleotides differing from wild type; green, inserted nucleotides. (B and D) PCR, undigested PCR product; NI, non-injected.
We assembled TALENs featuring two alternative backbone architectures [either with long N- and C-terminal domains (N287/C230) and the wild-type FokI domain from the pTAL3 vector (Cermak et al. 2011) or with the shorter backbone architecture (N136/C63) with heterodimeric DD/RR FokI domains (Dahlem et al. 2012). TALENs with the latter architecture are reported to mediate better mutation efficiencies and reduced capacity for nonspecific genome cleavage (Miller et al. 2011; Cade et al. 2012; Dahlem et al. 2012; Ansai et al. 2013).
Validation of TALEN activity for target-specific cleavage in vitro
To test whether the TALENs were able to recognize and cleave the specific target sequence, we performed an in vitro cleavage assay (TnT assay), whereby TALEN expression plasmids were incubated with target DNA and nontarget DNA in a transcription translation reaction mixture. TALEN activity was assessed by the ratio of cut vs. uncut target DNA present following incubation. Irrespective of the assembly method, all TALEN pairs tested were found to cleave the specific target DNA with efficiencies ranging from 40 to 97% (Figure S3). Nontarget DNA was not cleaved, consistent with the observation that TALEN activity is specific for the intended target. We further compared the activity of TALENs constructed using the alternative N- and C-terminal architectures N287/C230 and N136/C63. The same TALEN pairs targeting vtn (vtnEx2_L/R) and er (erEx3_L1/R1) with the shorter N136/C63 architecture were more efficient by 30 and 7%, respectively, compared to those with the longer N287/C230 architecture (Figure S3D). Based on these findings, we proceeded to validate the activity of TALENs made using the N136/C63 architecture in vivo.
Validation of TALEN activity and efficiency in vivo
To test whether the constructed TALENs were active in vivo, embryos were injected with various concentrations of mRNA encoding either a single TALEN pair per gene (vtn and er) or two TALEN pairs targeting different exons of the same gene (er and l-cry). To screen for TALEN-induced indel mutations in the endogenous target genes, we established a method for digesting single or small pools of larvae and using this lysate as template for PCR (see Materials and Methods). This facilitated mutation screening of TALEN-injected animals (injected at zygote/two- or four-cell stage) at larval stages of development (as early as 24 hpf), thereby enabling rapid validation of TALEN efficacy and activity in vivo.
If mutations are induced that disrupt the restriction site, the PCR product should be either completely or partially resistant to digestion. PCR without digestion was also used to screen for larger indel mutations and long-range deletions resulting from simultaneous cleavage by two TALEN pairs.
Predicted loss-of-function mutations detected in TALEN-injected larvae and adult worms
Evidence of mutations in TALEN-injected larvae was detected for all three genes (Figure 1, Figure 2, Figure 3). Sequencing of subcloned, digest-resistant (here referred to as “uncut”) PCR products revealed a variety of small deletions ranging in size from 1 to 23 bp and insertions of 1–19 bp (Figure 1, B and D; Figure 2, B and F; Figure 3, B and C). In one case, we detected a 109-bp insertion at the vtn target site (Figure 1, red arrow in B and green sequence in D). Blastn analysis revealed exact homology of this inserted sequence with part of intron 3 of the vtn locus (Figure 1A).
Subsequently, adult worms were raised from embryos injected with TALENs against each of the three target genes. Sequencing of uncut PCR products from these worms recovered similar indel mutations as those detected in larval samples (Figure 1, C and D; Figure 2, C–F; Figure 3, C–E).
Of the mutant sequences obtained, ∼70% represented frameshift mutations resulting in premature stop codons. These results validate that TALENs induce targeted, predicted loss-of-function mutations in endogenous genomic loci in vivo.
Simultaneous cleavage by two TALEN pairs induces long-range deletions
Simultaneous cleavage by two TALEN pairs targeting different sites of the same locus or chromosome was shown to induce long-range deletions in different species (Gupta et al. 2013). We therefore investigated whether co-injection of two TALEN pairs targeting different exons of the same gene induce large genomic deletions in Platynereis. We screened for deletions using PCR assays that amplify a region of the locus containing both TALEN recognition sites in the er and l-cry genes, respectively (Figure 2A and Figure 3A).
In addition to wild-type PCR products, smaller bands corresponding to the size expected for long-range deletions were detected in animals co-injected with TALENs targeting different exons of er and l-cry (Figure 2, E and F; Figure 3, B and C). Sequencing of subcloned small bands confirmed the presence of deletions with breakpoints that correspond to the spacer regions of the two respective TALEN target sites.
At the er locus, deletions of 1568 and 1575 bp were recovered in both larvae and individual adult worms (Figure 2E, red arrows; Figure 2F, bottom). These deletions remove the majority of the DNA-binding domain. In addition, we detected one inversion event at the er locus (er+7: 1/60 animals, Figure S4). At the l-cry locus, a 409-bp long-range deletion was sequence-confirmed, which resulted in a frameshift and thus a premature stop codon (Figure 3B, red arrow; Figure 3C). Together, these results show that co-injection of two TALEN pairs targeting the same locus can induce long-range deletions in P. dumerilii.
Individual mutation rates in TALEN-injected worms
To estimate the efficiency of individual TALEN pairs, or TALEN pair combinations, we analyzed mutation frequencies as the percentage of larval samples or individual worms for which evidence of mutant PCR products could be detected. Uncut PCR products representative of indel mutations were detected in 5–75% of TALEN-injected larval samples (Table 1) and in 5–43% of adult TALEN-injected worms (Table 2). Long-range deletions resulting from cleavage with two TALEN pairs were detected in larval samples at frequencies of 16–18% (Table 1) and in 3–9% of individual adult worms (Table 2).
To estimate the percentage of the genome modified in individual animals or larval samples (i.e., mutation rate), we calculated the percentage of mutant PCR products (uncut, or short deletion bands) relative to the total PCR product. TALEN efficiencies in pooled larvae ranged between ∼1 and 21% and between 6 and 52% in adult worms (Table 1; Table 2; Figure S5). The highest efficiency was detected for the l-cry exon 3 TALENs, with completely uncut PCR products representing likely biallelic mutations detected in 2 of 62 injected worms (Figure 3D: l-cry+45, Table 2). The high frequency of TALEN-induced mutations detectable in individual worms, combined with individual mutation rates of up to 100%, demonstrate that TALENs are highly efficient tools for engineering targeted mutations in endogenous Platynereis genes.
Germline transmission of TALEN-induced mutations
In a variety of animal models, TALEN-induced mutations have been shown to occur in the germline, thus facilitating the establishment of mutant lines (Cade et al. 2012; Aryan et al. 2013; Qiu et al. 2013; Sajwan et al. 2013). To validate that TALENs can also mediate germline mutations in Platynereis, we analyzed the offspring of mutation-positive adult worms for evidence of transmitted mutations. We screened small pools of G1 larvae from mutation-positive mature worms. Germline transmission rates were estimated based on the number of heterozygous mutant G1 larvae detected.
We identified three germline mutation carriers (Table 2): one transmitting both long-range deletions and frameshift mutations at the l-cry exon 3 site (l-cry+36: Figure 4, A and B) and two transmitting long-range deletions of the er locus (er+32 and er+59: Figure 4, C and D).
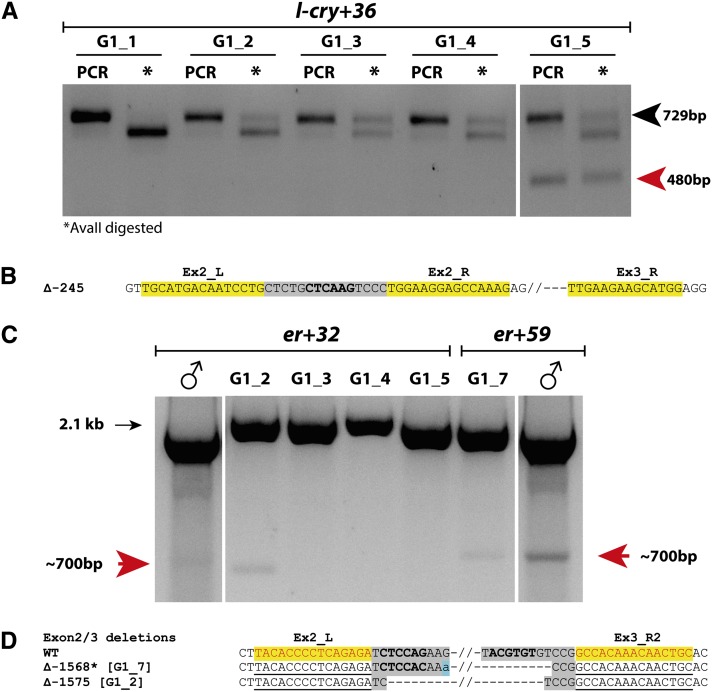
Figure 4.
Evidence for germline transmission of TALEN-mediated mutations. (A) Mutation screening results: G1 from male worm (l-cry+36) injected with l-cry TALENs. Pools of four G1 larvae were digested using proteinase K buffer and analyzed by PCR and AvaII digestion. Aliquots of undigested PCR products (PCR) are run next to a digested aliquot (*) for each sample. Sample G1_5 shows both uncut (black arrowhead) and deletion (red arrowhead) bands. (B) Sequence of uncut band in sample G1_5 in A. (C and D) Evidence of long-range deletions (red arrows) in pooled G1 larvae from er TALEN-injected worms er+32 and er+59 (male symbols indicate PCR products from these male worms). (D) Sequenced deletions from G1 samples: G1_2 and G1_7 shown in C. Asterisks indicate frameshift mutations.
Mutations were detected in ∼3% of the G1 screened from er carriers and 40% of the G1 screened from the l-cry carrier (Table 2). Sequencing revealed that the same mutations were present in the G1 and the respective parent. These results demonstrate that TALEN-induced mutations are both stable and heritable in Platynereis.
Survival rates of TALEN-injected worms
To assess the possible impact of the TALEN injection procedure on the normal development of Platynereis embryos, we analyzed larval survival data. In general, rates of normal development were highly variable among different batches (Figure S6A), a phenomenon frequently observed when crossing animals to maintain the laboratory culture. Neither higher concentrations of TALEN mRNA nor co-injection of two TALEN pairs vs. a single pair appeared to have a negative impact upon survival at 24 hpf (Figure S6A and Table S4). This is evidenced by the fact that we did not observe any obvious differences in the proportions of normal vs. mis-developed embryos in TRITC+ vs. TRITC− embryos (Figure S6B).
Of the embryos injected and set out to be raised, between 77 and 100% survived to adulthood (Table S4). Spawning data were also recorded for vtn and er TALEN-injected worms. The majority of worms spawned during new moon phases (Figure S7), in line with normal spawning rhythms observed under our laboratory conditions (Zantke et al. 2013). These collective observations suggest that delivery of TALEN mRNA into Platynereis is generally nontoxic.
Taken together with the results of the mutation analyses and germline transmission evidence, we conclude that TALENs represent a highly suitable approach for generating stable and heritable genomic modifications in P. dumerilii.
Discussion
We have adapted TALENs to engineer heritable, predicted loss-of-function mutations in three endogenous genes of the marine annelid P. dumerilii. Detectable mutations included short indels that localize to spacer regions of TALEN recognition sites, as well as long-range deletions when co-injecting two TALEN pairs targeting the same locus. The detection of potential biallelic mutation of l-cry in injected worms exemplifies that TALENs are highly efficient tools for genome modification in Platynereis that can be used to establish genetic mutant lines.
TALENs cause a variety of mutation types at variable frequencies in Platynereis
Injection of TALEN mRNA into Platynereis zygotes induced a range of mutations at target sites. Indel mutations varied in size from 1 to 110 bp while long-range deletions of up to 1.5 kb were generated by co-injection of two TALEN pairs. This variety of mutations is similar to what has been shown for TALEN-mediated in vivo genome modification studies in other model systems (Ma et al. 2012; Qiu et al. 2013; Xiao et al. 2013). The use of PCR and restriction digest screening, as we demonstrate here, is both rapid and effective for detecting mutations. However, this approach may not detect mutations at single TALEN sites where the restriction site is not disrupted; hence, our results may under-represent the actual mutation rates present in individual animals.
Compared with long-range deletions, inversion events are rare and have only been reported in vivo at rates of 0.2–1% and 4% in zebrafish (Gupta et al. 2013; Xiao et al. 2013) and pigs (Carlson et al. 2012), respectively. In agreement with this, we detected long-range deletions in a number of animals at mutation rates of up to 9%, while only one inversion event was observed in a single animal, for which the rate is likely <1% (Figure S4).
Our collective data show that TALEN efficiencies are highly variable in Platynereis. We could detect mutations in 3–43% of adult TALEN-injected worms, and mutation rates detected in individual worms ranged from 6% up to potential biallelic (∼100%) modification. Mutation rates calculated for individual worms were also variable, both between different individuals injected with the same TALENs and between the different TALEN pairs tested (Figure S5). Why different TALEN pairs elicit such variable mutation efficiencies in vivo is generally not well understood. Factors that likely relate to the sequence of the TALENs themselves or accessibility of the target sites could explain at least part of the observed variation.
Interestingly, a large insertion that we detected at the vtn TALEN site was found to be exactly homologous to part of intron 3. Gene insertion can occur following TALEN-induced cleavage, via homologous recombination, or NHEJ mechanisms (reviewed in Gaj et al. 2013 and Joung and Sander 2013). We noted that a 3-bp GCA motif was shared between the 5′ end of this insertion and the 5′ breakpoint in the TALEN spacer. Aside from this, however, we could not find any contiguous sequence homology between intron 3 in the region flanking the inserted sequence and the TALEN target site. While this 3-bp homology might argue for NEHJ-mediated insertion, we cannot explain exactly how or why this particular sequence was inserted. Regardless of the underlying mechanism, however, this integration event already provides evidence that TALEN-induced cleavage can mediate insertion of alternative sequences at TALEN sites in vivo.
Workflow and recommendations for TALEN use in emerging molecular model organisms
In the process of establishing TALENs as tools for genome modification in Platynereis, we streamlined several protocols together into a single workflow (for a summarizing workflow scheme, see also Zantke et al. 2014). Our workflow encompasses everything from screening for allelic variation at the target genomic locus to TALEN design and construction, in vitro validation, and in vivo mutation screening approaches, all of which can be applied to facilitate establishment of TALENs for genome modification in other emerging molecular model species. In addition to adapting and combining existing methods described for TALEN design (Miller et al. 2011), construction (Cermak et al. 2011), in vitro validation, and mutation screening via restriction digestion (Mussolino et al. 2011), we included two additional steps: (1) establishing quality reference sequences through initial genotyping and analysis of allelic frequencies and (2) developing methods to screen for in vivo TALEN activity in small pools of larvae shortly after injection.
The initial genotyping to identify SNPs and validate appropriate genomic reference sequences was crucial for TALEN design. TALEN efficiency in vivo is reported to be significantly decreased where mismatches are present in one or both of the TALEN recognition sequences (Cade et al. 2012; Dahlem et al. 2012). Thus, for emerging model systems with possible high rates of polymorphisms we recommend to first genotype and define appropriate reference genomic sequences for the target strain to maximize potential TALEN efficiencies.
The second adaptation of our workflow was the optimization of a protocol to prepare crude gDNA templates from small pools of larvae. This was crucial for rapid validation of TALEN activity in vivo by PCR. By generating crude gDNA lysates from injected larvae, we could analyze different batches of injected animals separately, rather than pool animals together from several injections, which would have been required for gDNA extraction using kit-based methods. This enabled both a rapid assessment of TALEN activity in vivo and a more accurate estimation of mutation frequencies. In other species, phenotype scoring has been used to demonstrate proof of TALEN activity in vivo (Dahlem et al. 2012; Liu et al. 2012; Ma et al. 2012; Aryan et al. 2013; Suzuki et al. 2013; Wang et al. 2013). However, this is difficult to apply when evaluating TALEN efficiency for genes with unpredictable phenotypes and may be impractical for rapidly assessing TALEN functionality in vivo against genes where phenotypes are not observable until adult stages of the life cycle.
The genotyping and rapid larval digestion protocols that we introduced here can also be equally applied to other genome-editing systems, such as the recently described Cas9/CRISPR system (Cong et al. 2013; Mali et al. 2013). Cas9/CRISPR technology involves the use of short guide RNAs (sgRNAs) that direct a bacterially derived endonuclease (Cas9) to recognize specific sites in the genome and induce DNA double-strand breaks (Jinek et al. 2012). The initial version of the system caused high numbers of off-target mutations (Fu et al. 2013), which was improved by the simultaneous use of two sgRNAs and a mutant nickase version of the Cas9 protein (Ran et al. 2013) and most recently by shortening the length of the sgRNA (Fu et al. 2014). While the production of Cas9/CRISPR reagents is more straightforward than constructing TALENs and thus potentially useful for multiplex functional screens, possible target sites are more limited than in the case of TALENs (reviewed in Blackburn et al. 2013). Furthermore, as both systems use very different initial mechanisms of DNA sequence recognition, it is possible that in some cases where one system fails or is inefficient, the other might work better (Hwang et al. 2013; Auer et al. 2014). In the future, it will be useful to explore the efficiency of TALEN and Cas9/CRISPR systems in parallel to further enhance the capacity for targeted genome modification in P. dumerilii.
In conclusion, we have demonstrated that TALENs mediate targeted and heritable mutation of endogenous genes in the marine annelid P. dumerilii and provide a streamlined workflow that can serve as a template for the establishment of TALEN technology in other nonconventional and emerging model organisms.
Supplementary Material
Acknowledgments
We thank all three anonymous reviewers for their positive comments and constructive feedback on our manuscript and Poonam Sharma, Andrij Belokurov, and Verena Hoellbacher for feeding and maintaining the Platynereis cultures and assistance with animal care for TALEN-injected worms. This work was supported by funds of the Max F. Perutz Laboratories (http://www.mfpl.ac.at/home.html), the research platform “Marine rhythms of Life” of the University of Vienna (to K.T.-R. and F.R.); START award (#AY0041321) from the Fond zur Förderung der wissenschaftlichen Forschung (www.fwf.ac.at); and a Human Frontier Science Program (http://www.hfsp.org/) research grant (#RGY0082/2010) (to K.T.-R.). The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement 260304 (to F.R.) and (FP7/2007-2013)/ERC Grant Agreement 337011 (to K.T.-R.). S.B. is funded by the Max F. Perutz Laboratories Vienna International PostDoctoral Program for Molecular Life Sciences (funded by Austrian Ministry of Science and Research and City of Vienna, Cultural Department—Science and Research). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Note added in proof: See Zantke et al. 2014 (pp. 19–31) in this issue for a related work.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Ansai S., Sakuma T., Yamamoto T., Ariga H., Uemura N., et al. , 2013. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 193: 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansai S., Inohaya K., Yoshiura Y., Schartl M., Uemura N., et al. , 2014. Design, evaluation, and screening methods for efficient targeted mutagenesis with transcription activator-like effector nucleases in medaka. Dev. Growth Differ. 56: 98–107. [DOI] [PubMed] [Google Scholar]
- Arendt D., Nübler-Jung K., 1994. Inversion of dorsoventral axis? Nature 371: 26. [DOI] [PubMed] [Google Scholar]
- Arendt D., Nübler-Jung K., 1997. Dorsal or ventral: similarities in fate maps and gastrulation patterns in annelids, arthropods and chordates. Mech. Dev. 61: 7–21. [DOI] [PubMed] [Google Scholar]
- Aryan A., Anderson M. A., Myles K. M., Adelman Z. N., 2013. TALEN-based gene disruption in the dengue vector Aedes aegypti. PLoS ONE 8: e60082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer T. O., Duroure K., De Cian A., Concordet J. P., Del Bene F., 2014. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backfisch B., Veedin Rajan V. B., Fischer R. M., Lohs C., Arboleda E., et al. , 2013. Stable transgenesis in the marine annelid Platynereis dumerilii sheds new light on photoreceptor evolution. Proc. Natl. Acad. Sci. USA 110: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., et al. , 2012. In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn P. R., Campbell J. M., Clark K. J., Ekker S. C., 2013. The CRISPR system: keeping zebrafish gene targeting fresh. Zebrafish 10: 116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade L., Reyon D., Hwang W. Y., Tsai S. Q., Patel S., et al. , 2012. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 40: 8001–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson D. F., Tan W., Lillico S. G., Stverakova D., Proudfoot C., et al. , 2012. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA 109: 17382–17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., et al. , 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann M., Offenburger S. L., Asadulina A., Keller T., Munch T. A., et al. , 2011. Neuropeptides regulate swimming depth of Platynereis larvae. Proc. Natl. Acad. Sci. USA 108: E1174–E1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann M., Williams E. A., Tunaru S., Randel N., Shahidi R., et al. , 2013. Conserved MIP receptor-ligand pair regulates Platynereis larval settlement. Proc. Natl. Acad. Sci. USA 110: 8224–8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., et al. , 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8: e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B., Davies G., Preece C., Puliyadi R., Szumska D., et al. , 2013. Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS ONE 8: e60216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demilly A., Steinmetz P., Gazave E., Marchand L., Vervoort M., 2013. Involvement of the Wnt/beta-catenin pathway in neurectoderm architecture in Platynereis dumerilii. Nat. Commun. 4: 1915. [DOI] [PubMed] [Google Scholar]
- Denes A. S., Jekely G., Steinmetz P. R., Raible F., Snyman H., et al. , 2007. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129: 277–288. [DOI] [PubMed] [Google Scholar]
- Dohrn A., 1875. The Origin of Vertebrates and the Principle of Functional Change (in German). Verlag von Wilhelm Engelmann, Leipzig, Germany. [Google Scholar]
- Doyle, E. L., N. J. Booher, D. S. Standage, D. F. Voytas, V. P. Brendel et al., 2012 TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40(Web Server issue): W117–W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray N., Tessmar-Raible K., Le Gouar M., Vibert L., Christodoulou F., et al. , 2010. Hedgehog signaling regulates segment formation in the annelid Platynereis. Science 329: 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A. H., Henrich T., Arendt D., 2010. The normal development of Platynereis dumerilii (Nereididae, Annelida). Front. Zool. 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., et al. , 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K., 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., III, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Hall V. L., Kok F. O., Shin M., McNulty J. C., et al. , 2013. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 23: 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenschild C., 1960. Lunar periodicity. Cold Spring Harb. Symp. Quant. Biol. 25: 491–497. [DOI] [PubMed] [Google Scholar]
- Hauenschild C., 1974. Endocrine control of sexual development in some polychaetes (in German). Fortschr. Zool. 22: 75–92. [PubMed] [Google Scholar]
- Hauenschild C., Fischer A., 1969. pp. 1–55 in Platynereis dumerilii. Mikroskopische Anatomie, Fortpflanzung, Entwicklung [Platynereis dumerilii. Microscopical Anatomy, Reproduction and Development]. Groses Zoologisches Praktikum Heft, Stuttgart, Germany. [Google Scholar]
- Hofmann D. K., 1976. Regeneration and endocrinology in the polychaete Platynereis dumerilii: an experimental and structural study. Dev. Genes Evol. 180: 47–71. [DOI] [PubMed] [Google Scholar]
- Huang P., Xiao A., Zhou M., Zhu Z., Lin S., et al. , 2011. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29: 699–700. [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S., Cliffe R., Amaya E., 2012. Highly efficient bi-allelic mutation rates using TALENs in Xenopus tropicalis. Biol. Open 1: 1273–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J. K., Sander J. D., 2013. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keay J., Thornton J. W., 2009. Hormone-activated estrogen receptors in annelid invertebrates: implications for evolution and endocrine disruption. Endocrinology 150: 1731–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Guo X., Liu Y., Cao Y., Deng Y., et al. , 2012. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. USA 109: 17484–17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke A. K., Bannister S., Lower A. M., Apel D. M., Podleschny M., et al. , 2014. 17beta-Estradiol induces supernumerary primordial germ cells in embryos of the polychaete Platynereis dumerilii. Gen. Comp. Endocrinol. 196: 52–61. [DOI] [PubMed] [Google Scholar]
- Liu J., Li C., Yu Z., Huang P., Wu H., et al. , 2012. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics 39: 209–215. [DOI] [PubMed] [Google Scholar]
- Ma S., Zhang S., Wang F., Liu Y., Xu H., et al. , 2012. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS ONE 7: e45035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., et al. , 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Mussolino C., Morbitzer R., Lutge F., Dannemann N., Lahaye T., et al. , 2011. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39: 9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S. C., 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26: 216–220. [DOI] [PubMed] [Google Scholar]
- Qiu Z., Liu M., Chen Z., Shao Y., Pan H., et al. , 2013. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res. 41: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F., Tessmar-Raible K., Osoegawa K., Wincker P., Jubin C., et al. , 2005. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science 310: 1325–1326. [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C. Y., Gootenberg J. S., Konermann S., et al. , 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajwan S., Takasu Y., Tamura T., Uchino K., Sezutsu H., et al. , 2013. Efficient disruption of endogenous Bombyx gene by TAL effector nucleases. Insect Biochem. Mol. Biol. 43: 17–23. [DOI] [PubMed] [Google Scholar]
- Sander J. D., Cade L., Khayter C., Reyon D., Peterson R. T., et al. , 2011. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29: 697–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S. Q., Bowerman B., 2007. beta-Catenin asymmetries after all animal/vegetal-oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev. Cell 13: 73–86. [DOI] [PubMed] [Google Scholar]
- Simakov O., Larsson T. A., Arendt D., 2013a Linking micro- and macro-evolution at the cell type level: a view from the lophotrochozoan Platynereis dumerilii. Brief. Funct. Genomics 12: 430–439. [DOI] [PubMed] [Google Scholar]
- Simakov O., Marletaz F., Cho S. J., Edsinger-Gonzales E., Havlak P., et al. , 2013b Insights into bilaterian evolution from three spiralian genomes. Nature 493: 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidler A. L., Terenzi O., Soichot J., Levashina E. A., Marois E., 2013. Targeted mutagenesis in the malaria mosquito using TALE nucleases. PLoS ONE 8: e74511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E., Weinstock G. M., Davidson E. H., Cameron R. A., Gibbs R. A., et al. , 2006. The genome of the sea urchin Strongylocentrotus purpuratus. Science 314: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E., et al. , 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. T., Isoyama Y., Kashiwagi K., Sakuma T., Ochiai H., et al. , 2013. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol. Open 2: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu Y., Sajwan S., Daimon T., Osanai-Futahashi M., Uchino K., et al. , 2013. Efficient TALEN construction for Bombyx mori gene targeting. PLoS ONE 8: e73458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmar-Raible K., Arendt D., 2003. Emerging systems: between vertebrates and arthropods, the Lophotrochozoa. Curr. Opin. Genet. Dev. 13: 331–340. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K., Steinmetz P. R., Snyman H., Hassel M., Arendt D., 2005. Fluorescent two-color whole mount in situ hybridization in Platynereis dumerilii (Polychaeta, Annelida), an emerging marine molecular model for evolution and development. Biotechniques 39: 460–, 462, 464.. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K., Raible F., Christodoulou F., Guy K., Rembold M., et al. , 2007. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell 129: 1389–1400. [DOI] [PubMed] [Google Scholar]
- Tomer R., Denes A. S., Tessmar-Raible K., Arendt D., 2010. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142: 800–809. [DOI] [PubMed] [Google Scholar]
- Veedin Rajan V. B., Fischer R. M., Raible F., Tessmar-Raible K., 2013. Conditional and specific cell ablation in the marine annelid Platynereis dumerilii. PLoS ONE 8: e75811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu Y. C., Markoulaki S., Welstead G. G., Cheng A. W., et al. , 2013. TALEN-mediated editing of the mouse Y chromosome. Nat. Biotechnol. 31: 530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Ochiai H., Sakuma T., Horch H. W., Hamaguchi N., et al. , 2012. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat. Commun. 3: 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wang Z., Hu Y., Wu Y., Luo Z., et al. , 2013. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zantke J., Ishikawa-Fujiwara T., Arboleda E., Lohs C., Schipany K., et al. , 2013. Circadian and circalunar clock interactions in a marine annelid. Cell Rep. 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zantke, J., S. Bannister, V. B. Veedin Rajan, F. Raible, and K. Tessmar-Raible, 2014 Genetic and genomic tools for the marine annelid Platynereis dumerilii Genetics 197: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Tong X., Wang Z., Liu D., Pan R., et al. , 2013. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10: 329–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.