Abstract
Researchers have proposed a genetic differential sensitivity to social environmental (GDSE) model positing that individuals with certain genetic makeups are more sensitive to favorable and unfavorable environmental influences than those without these genetic makeups. We discuss several issues facing researchers who want to use GDSE to examine health: (1) the need for greater theorizing about the social environment to properly understand the size and direction of environmental influences; (2) the potential for combining multiple genetic markers to measure an individual’s genetic sensitivity to environmental influence; (3) how this model and exogenous shocks deal with gene–environment correlations; (4) implications of this model for public health and prevention; and (5) how life course and developmental theories may be used to inform GDSE research.
Studies of human molecular genetics and social environment interactions have increased dramatically during the past decade.1 Although the majority of these studies rely on a classic diathesis-stress model that emphasizes genetic variations and social environments that are unfavorable or risky, researchers have proposed a differential susceptibility or biological sensitivity model, which postulates that individuals vary in their sensitivity to environments, with those having more genetic sensitivity experiencing more negative outcomes in unfavorable environments and more positive outcomes in favorable environments compared with those having less genetic sensitivity.1-4 A third model referred to as the social distinction model mirrors the diathesis-stress model by arguing that in the harshest environments outcomes are confined to a narrow range because the environment overwhelms any specific biological pathway. However, given a more supportive environment, the variance of outcomes is larger and that variance is partly caused by genetic factors. Interestingly, little research examines this model, even less than is found for differential susceptibility or biological sensitivity.5,6Early research primarily focused on behavioral indicators of sensitivity (negative emotional reactivity to psychosocial challenges or what was sometimes termed difficult temperament) and physiological responses to such challenges (autonomic, immune, and cortisol changes).7 Interest expanded from considering susceptibility to negative environments to a consideration of sensitivity to positive environments as well. With advances in genotyping, researchers are now using measured gene variations as markers of individuals’ biological sensitivity, creating what we term the genetic differential sensitivity to social environment model (GDSE). In addition to hypothesizing a crossover effect, GDSE models imply that heterogeneity of response to environments (including social interventions, treatments, and policies) may be partly genetic. For this reason, GDSE models can be an important tool not only for understanding population health but also for designing public health policies and programs that are more targeted and more effective.
Our goal for this article is to discuss some of the broader theoretical and methodological issues confronting researchers who are using or plan to use GDSE models to enrich their understanding of population health. Others have more formally documented the process of detecting a genuine GDSE model; therefore, we do not review that process here.3,8 Our first section is a brief review of the 2 theoretical approaches to GDSE.1–4,7,9–11 In subsequent sections, we discuss the importance of developing positive measures of both environments and health; the advantages and disadvantages of using multiple indicators of the same genetic construct; how the GDSE model and exogenous environmental events can help social scientists to deal with causality; and how GDSE approaches may be useful for public health and prevention. We close with a discussion of the importance of integrating GDSE models with developmental and life course frameworks to further research and prevention efforts.
MODELS OF GENETIC DIFFERENTIAL SENSITIVITY TO ENVIRONMENT
In the late 1990s, Boyce et al.11 and Belsky7 independently proposed models for explaining individual variability to environmental conditions or events. Both researchers were struck by the fact that not all individuals (both focused on children and youths) exposed to negative events had compromised health or development nor did all individuals exposed to positive environments have enhanced health or development. Both researchers posited that certain individuals were more sensitive to their environments, for better and for worse. These insights were based in part on work with children who had difficult temperaments or who exhibited strong behavioral reactivity to the environment. Belsky7 used the term differential susceptibility theory, whereas Boyce et al.11 used the term biological sensitivity to context theory. The former emphasizes genetic sensitivity as possibly responsible for individual characteristics, such as temperament; the latter emphasizes the physiological features of behavioral reactivity in various biological systems. Each researcher argued that such individual differences in sensitivity had evolutionary advantages, in part, because humans can exist in a wide range of environments (or what is sometimes called multiniche environments), and therefore, biological variation is beneficial because, at a population level, such genes hedge bets in terms of which characteristics are most adaptive in more and less difficult circumstances and which environments humans are likely to experience in the future.7,12 Both approaches argue that bet-hedging helps explain within-family (across-sibling) variability in phenotypes and the phenomena of relatively large nonshared environmental effects in behavioral genetics.13
These models were derived, in large part, from developmental theory focusing on the intersection of person, process, and context as outlined by Bronfenbrenner.14 Less emphasis was placed on changes over the life course (individuals) or over cohorts (history). The models also differed with respect to their emphasis on the possibility that early experiences would influence biological sensitivity itself, with Boyce et al.2 highlighting the fact that biological reactivity to the environment could be influenced by previous experience (i.e., individuals who experience very negative or positive environments are likely to become more biologically reactive over time). Although most genetic approaches to differential sensitivity do not have such bidirectional effects built into their models, the epigenetic processes (e.g., methylation, histone modification, etc.) could have the same effect (i.e., very positive or negative experiences being more likely to reset or dial up or down the expression of a particular gene variant).15,16 Also, neither model considers variability across genes or biological systems, instead focusing to a large extent on behavioral reactivity and difficult temperament and their underlying biological processes (neurotransmitters and certain brain systems, such as the amygdala and prefrontal cortex).
To date, of the many studies searching for gene by environment interactions (G×E), probably less than 5%, use GDSE models. Nevertheless, 2 recent reviews devoted to GDSE models signal an emerging literature.1,3 At the time of this writing, the majority of GDSE work is primarily with children, in part because of the historical focus on difficult temperament and the interest in behavioral reactivity. However, more work is being done on adolescents and adults, as well as intergenerational approaches where GDSE is being examined in parents.17–19
Our own use of the GDSE model borrows from these 2 traditions, with a particular focus on genetic variation and regulation. As such, it includes an examination of genetic and epigenetic processes (here we focus on genetic variation more than epigenetic regulation of expression, although the latter may be critical for understanding how early experiences may become instantiated in an individual’s biology and have long-term behavioral effects).
GDSE AND MEASUREMENT OF THE ENVIRONMENT
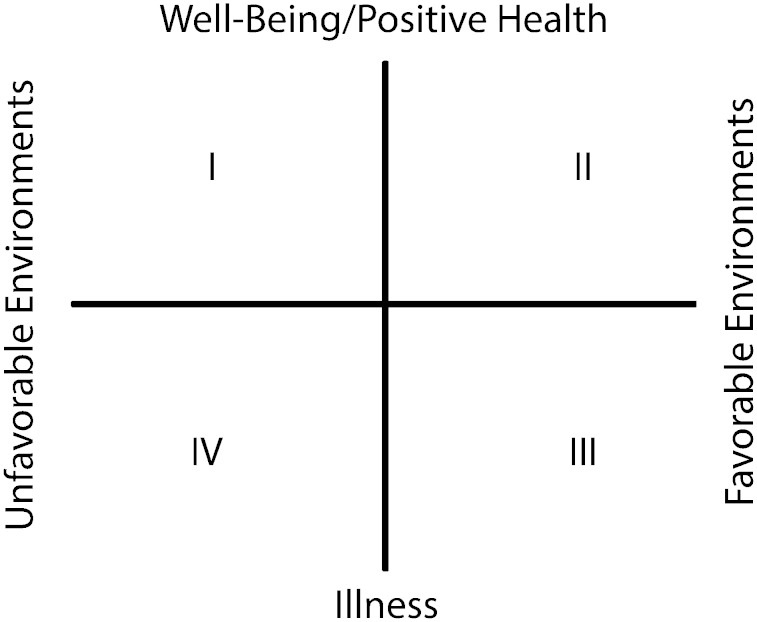
A key contrast between the GDSE model and the diathesis-stress model is that the latter predicts a unidirectional influence of G×E, whereas GDSE predicts a bidirectional influence. Interestingly, there are a few G×E studies where, depending on the beneficial environment under study, researchers may not be searching for a crossover effect, but for a sensitivity effect on the positive or bright side of environments.20-22 Figure 1 illustrates the 4 possible quadrants in which G×E interactions might be observed. The top 2 quadrants (I and II) represent positive outcomes or health, whereas the bottom 2 quadrants (III and IV) represent negative outcomes or illness. Quadrants I and IV represent unfavorable environments, whereas quadrants II and III represent favorable environments. In Figure 1, the diathesis-stress model focuses almost exclusively on interactions in quadrant IV (the association between unfavorable environments and illness). In contrast, the GDSE model pushes researchers to conceptualize and measure both favorable and unfavorable environments (quadrants III and IV).3 Environmental influences can be thought of as a continuum where one end is expected to result in a positive outcome, the other end is expected to result in a negative outcome effect, with the middle showing no effect. Alternatively, they can be thought of as discrete events or changes with opposite outcomes. Good examples of continuous measures include family income or parental education, whereas good examples of discrete events include events such as union formation or dissolution or residential moves. In both cases, the goal is to construct a measure of the environment that allows testing for the effect of both favorable and unfavorable environments on outcomes within the same domain.3,4 In the absence of a favorable environmental dimension, results from the GDSE and diathesis-stress models should appear to be very similar. By illustration, we present data from the Fragile Family and Child Wellbeing Study, in which we examine the association between socioeconomic status (SES; which was measured in terms of family income and maternal education) and maternal depression 1 year after birth as a function of genotype based on 2 genetic markers (5-HTTLPR and STin2) on the 5-HTT gene.17 In Figure 2, we classified mothers based on their number of sensitive alleles (given the 2 markers, the theoretical range is 0–4). If only the negative effect of parents’ low education is examined, the 3–4 alleles group would appear to be the risky genetic variant. However, if the positive effects of parents’ higher education are studied, the 0–1 allele group would be the risky version. Looking at both environments simultaneously provides a more nuanced understanding of the G×E influence on maternal depression, suggesting that the risky variant is more accurately described as a reactive or sensitive variant.
FIGURE 1—
Conceptual research opportunities of gene–environment research on health.
Note. Quadrant IV, which examines genetic differences by negative environments on illness, has received the most attention and is the main location of diathesis-stress research. Genetic differential sensitivity to social environmental research has been conducted primarily in quadrants III and IV.
FIGURE 2—
Probability of postpartum depression across socioeconomic status (SES) by the number of reactive 5-HTT alleles.
Note. The G×E model does not constrain SES to be related across allele types. We combined 0 and 1 alleles into 1 group and 3 and 4 alleles into a second group, with those with 2 alleles in a third group (excluded from the graph). Those with 0, 1, or 2 alleles do not have a significant effect of SES on postpartum depression (PPD). Those with 3 or 4 alleles have a significant 18% decrease in the odds of PPD for every year of education (b = −0.199; SE = 0.089; Z = −2.24, 0.025).
Source. Reprinted from Mitchell et al.17 with permission.
The GDSE model also encourages scholars to think carefully about the theoretical and substantive meaning of having favorable and unfavorable environmental influences. For example, it is not always clear which environmental effects are opposite. By illustration, what is the favorable equivalent of a high conflict home—remembering that the absence of an unfavorable environmental influence is not the same as the presence of a favorable environment influence? Similarly, even when clear opposites are found, environments may not always be symmetric in their effects. For example, research shows that decreases in resources are much more powerful than equally sized increases in resources.23 In our work, we found that negative responses to income losses are stronger than positive responses to income gains. Similarly, we found that union dissolutions are more negative than the positive responses to union formations.24 An important concern is that measures of unfavorable environments are much more common than measures of favorable environments in most health studies.
Finally, in addition to pushing social scientists to think harder about how to measure favorable and unfavorable environments, the GDSE model also encourages researchers to consider both positive and negative health outcomes. Although the majority of GDSE work has focused on negative outcomes, such as poor mental health, child behavioral problems, etc., GDSE has the potential to provide important insights into positive health as well.24 That is, GDSE encourages researchers to focus on quadrants I and II in Figure 1 just as much as quadrants III and IV. Interestingly, the diathesis-stress model could also apply to positive outcomes (quadrant I), but very little research has been directed there. Again, however, the field is limited, in that measures of positive health are less common than measures of poor health and illness. Possible candidates for measuring positive health or wellbeing outcomes include happiness (or positive affect), self-efficacy, conscientiousness, empathy or concern for others, openness to new experiences, positive relationships or interactions, faster than expected recovery times, and high age-specific health on any outcome. Just as favorable environments are more than an absence of unfavorable environmental influences, positive health and wellbeing is not just an absence of illness.
GDSE AND MEASUREMENT OF GENES
Another issue for social scientists interested in GDSE is how to parsimoniously identify and measure genes that indicate differential sensitivity. At least 2 issues are relevant, the first having to do with whether sensitivity should be conceptualized as a continuum or not and the second having to do with the specificity of genetic effects. With regard to the first issue, we agree with others that sensitivity ought to be seen as a continuum, rather than defining individuals as sensitive or not sensitive.7 Past research has tended to define groups, such as individuals with difficult temperaments or with 1 sensitive gene variant. However, a more profitable and probably realistic approach is to look at distributions of sensitivity. To do so, multiple gene variants need to be measured, rather than a single genetic marker (as has been the case for virtually all of the G×E research to date). Research is just beginning to take this approach, typically summing gene variants within a specific neurotransmitter system (i.e., either the serotonergic or dopaminergic system). Focusing on a single gene marker within a complex system and then categorizing an individual as sensitive or not ignores the fact of variation across gene markers within an individual. It also has probably led in part to the sometimes divergent findings of G×E where genes appear to have different directions of effects in some environments compared with others.
With respect to the second issue, following standard procedures for social sciences, most of the genes used by researchers are based on some theoretically plausible role and a previously discovered interaction. For example, most of the GDSE literature on social emotional wellbeing has relied on the serotonergic and dopaminergic systems.25,26 Serotonin and dopamine are neurotransmitters, chemicals that transmit signals in between the nerve cells (neurons) of the brain. These systems appear to regulate thought, movement, mood, attention, motivation, and learning.25,27–34 The idea is to gather multiple markers for these systems in hopes of producing a more accurate measure of an individual’s latent differential sensitivity. In a recent article on maternal depression in the first year after birth, we constructed a measure based on a count of the number of sensitive alleles from 2 polymorphisms of the prominent 5-HTT gene. We found that using a measure based on multiple indicators of the same gene yields a more robust interaction between 5-HTT and the social environment (SES in this case) on depression than using a single marker.17 For some genes, there may be dozens, if not hundreds, of possible genetic markers (although many of these will be highly correlated or in high linkage disequilibrium), so choosing the appropriate marker takes some knowledge of the literature and sample characteristics.35 Utilizing multiple genetic markers of either the same gene or even of a system may provide a better representation of a G×E interaction, whereas most G×E research typically uses 1 genetic marker of many possible markers of that gene.
After choosing the genetic markers of interest, researchers will need to determine the best procedure for combining the genetic information, if at all. Because of the novelty of GDSE, there is little guidance in how to determine the reactivity or sensitivity of a genetic variant or polymorphism. To date, most studies have taken markers classified as risky and reclassified them as sensitive.1,3 Often these risky (now sensitive) alleles are often the variants associated with lower transcriptional efficiency.36,37 Many GDSE studies assume a recessive genetic effect, requiring the gene to have sensitive alleles from both the mother and the father for it to be considered a sensitive genotype.1,3,38 Multiple genes are combined with an equal weighting strategy, creating a count of the number of homozygote sensitive genes for the individual. Essentially, this approach constitutes what some may label a genetic risk score, but under the GDSE framework it becomes a genetic sensitivity score. Although smaller in number, other GDSE studies use an additive genetic approach so that each person can have 0, 1, or 2 sensitive alleles for each genetic marker, which are then summed across multiple genetic markers.17 Again, multiple genes are combined by equally weighting each allele for each gene. Both of these methods tend to result in peaked distributions that are usually strongly positively skewed because the homozygote sensitive genetic markers are often the minor alleles.
As larger amounts of genetic information become available, researchers may want to incorporate larger numbers of genetic markers into their measure of biological sensitivity. At that point, weighting methods and other data reduction strategies may be more useful in creating this biological sensitivity measure. For example, research from molecular biology may suggest that some genetic effects may be additive, whereas others are recessive or dominant. It may be possible that genes at different points in the pathway should be weighted differently because of their importance in determining system function. Similarly, certain genetic markers of the same gene (but at different loci) may be more influential for gene function, thus deserving a higher weight. Further, with molecular biology increasing understanding of epistasis and gene networks, researchers may also want to incorporate this information into their measure of genetic sensitivity, possibly in ways similar to methods now measuring social networks.
As the detail and complexity of genetic factors increase, however, so do the statistical power requirements for obtaining reliable estimates of direct and interaction effects.39 As with most studies of interactions, the power of the test is a product of the distribution of the 2 independent variables (in this case the gene and the environment), the dependent variable, the sample size, and the expected effect size. Few data sets have sufficient sample sizes to power studies that properly take account of the multiple comparisons that arise from testing several different genes (or environments) and G×E interactions. By using theory and previous empirical research to construct scales that combine multiple genetic markers, less exploratory examination is needed. The cost, however, is that the inclusion of markers with no effect may offset the influence of those with strong effects (i.e., measurement error). In addition, the multiple indicator approach does not allow researchers to test for specific genetic mechanisms.
High-throughput sequencing and genotyping has led to a dramatic explosion of genetic data available for G×E research. At the time of this writing, most GDSE work comes from data with relatively few genetic markers (i.e., candidate gene approaches). At this point it is unknown to what extent GDSE genetic markers are prevalent in the genome. However, as mentioned previously, dealing with the multiple comparison concerns is paramount. One approach to handle the possible millions of genetic markers (and hundreds of environmental ones), is to be completely atheoretical as to the environment and the genetic markers. A way to do this is to test variance equality for a specific outcome across the genotypes, so that in the case of the 5-HTTLPR marker, we would expect significantly higher variance in the SS genotype compared with the LL genotype.40 After finding the reactive variants, however, the search for a possible environment(s) inducing the variation begins—with its own multiple comparison issues. Under GDSE, the main effect of a genetic marker will be a result of the distribution of the environment in the sample, and conditional on the environment, the genetic marker would typically not have a main effect.3,8Therefore, methods that use only genetic markers with large main effects to test for GDSE would be expected to miss the markers most likely to convey differential sensitivity.
A second approach requires more knowledge of the environment, but still tests all the genetic markers, by specifying the expected direction of the interaction; for example, understanding that low education has worse outcomes and high education confers positive outcomes, compared with normal levels of education.41 A third approach follows from the preceding discussion and limits testable markers to those in specific systems or even genes of interest. This then would require knowledge of the biological and social underpinnings of the health outcome, but require a less stringent P value because of the reduction in multiple comparisons.
Even among genome-wide association studies, the genetic markers often identified are relatively common, often occurring in at least 5% of the population. Considering the more recent expansion of genetic work on rare variants, this is an area GDSE has not examined. However, because GDSE is based on an evolutionary model of hedging bets, rare variants—especially those that are a result of very recent mutations—may be less likely to be markers that convey differential sensitivity.7,42 Further, because allele frequency tends to be inversely correlated with penetrance, we may expect much smaller environmental interactions with those markers.43 However, at this point, this is speculation because we are unaware of any GDSE models using rare variants. A major hurdle of rare variant research in GDSE will be having sufficient samples in multiple environmental contexts to properly test a GDSE.
One of the major challenges all G×E studies, not just GDSE, face is to make progress in understanding the pathways and mechanisms involved in linking the external experience to molecular genetic responses. Starting at the molecular level, epigenetic and gene expression studies can prove informative and potentially supportive, such as the linking of 5HTTLPR to differential processing of serotonin.15 One of the nicest examples of exploring the molecular basis of a G×E pathway is for the interleukin (IL)-6 gene.44 This work provides some evidence that depression in later adulthood operates through the sympathetic nervous system to increase norepinephrine levels, which lead to greater GATA1 activity, but that GATA1 only binds as a transcription factor to the IL-6 gene when the rs1800795 genotype is GG. This, in turn, means that IL-6 gene production of C-reactive protein is much greater for those with the GG marker. Empirical findings show a strong G×E interaction with greatly increased mortality from inflammation-related causes of death for the depressed who have the GG marker on rs1800795 on the IL-6 gene, and that this is mediated by C-reactive protein levels.
Recent discussions of the issues involved in teasing out these epigenetic and gene expression pathways and mechanisms linking the social environment to genes and physical health outcomes provide a roadmap for incorporating epigenetic and expression data in G×E work.45–47 However, further discussion of these broad issues in the context of GDSE is beyond the scope of this article. Careful research is required to explore how far GDSE pathways involve epigenetic or transcription binding elements that link to altered gene expression, and beyond that, to link to plausible or evidence-based pathways whereby the social environment enters the body and gets transmitted down to the molecular level. Equally, there is a need for much research that links the molecular biology upward to health outcomes (largely still bodily, whether physical or mental health) and eventually to behavioral outcomes.
For the reasons just listed, some social science studies, including the Fragile Families and Child Wellbeing Study, are beginning to collect more variable genetic markers such as DNA methylation and (RNA) gene expression. Interestingly, this effort will make measures of genetic characteristics more similar to standard social science and health measures that require multiple time points and proper temporal ordering to appropriately analyze possible causal pathways. More importantly, the idea that the social environment may influence these variable genetic markers provides a fundamental paradigm shift in understanding the interplay of genes and the social environment. To what extent the social environment can influence more changes in variable genetic traits may indicate how the social environment not only gets under the skin but onto the genes. This is a very exciting direction of G×E research, and is one of our main purposes for attempting to integrate genetics and the social sciences to study health.
GDSE AND GENE–ENVIRONMENT CORRELATION
Long-standing challenges for all social scientists include (1) addressing how unobserved variables may lead to biased estimates of true environment effects through spurious relationships and selection on unobservable characteristics, and (2) certifying that the outcomes are not causing the environment (i.e., reverse causation). G×E work is no different, although in addition to concerns about environmental exogeneity, there is also concern about the possible role of genetic heritability in explaining apparent G×E through gene–environment correlation (rGE).39,48 There are 3 types of rGEs: passive, evocative, and active. Passive rGE exists when children receive genotypes already correlated with their environments from their parents, thus causing a spurious relationship. Evocative rGE occurs when the environment reacts to the child’s genetic makeup; this is reverse causation. Active rGE occurs when individuals, based on their genetic makeup, select or modify their environment to be more inline with their genes, resulting in a selection on unobservable characteristics. Understanding the extent to which rGE influences an apparent G×E goes directly to the underlying mechanism of causation, resulting in distinctly different recommendations for interventions, policies, and treatments. Note that all 3 types of rGEs predict a unidirectional effect of genes and environment. The effect may be positive or negative, but not both. In other words, when we find that a more sensitive child responds more negatively to a harsh environment, we can easily explain it as a rGE. In contrast, the GDSE model provides a more powerful tool for identifying true G×E interactions because the previously described arguments cannot easily explain how the same child would then be more likely to respond more positively to a favorable environment and more negatively to an unfavorable environment.
In our study of child externalizing behavior, we found that a biological father’s exit from the household (separation or divorce) was associated with poorer outcomes among children with more sensitive genes, whereas a biological father’s entrance into the household was associated with better outcomes for children with these same genes.38 If we had focused exclusively on negative environments and negative outcomes, an alternative explanation of our findings would have been passive rGE. Difficult parents create difficult environments and have difficult children.49 This argument, however, cannot explain why children with the same genotypes experience a greater improvement in outcomes if their biological father moves into the household than children with fewer sensitive genes. In short, although passive rGE may easily explain 1 reaction, it is more difficult to explain both reactions at the same time. Similarly, there is some evidence that temperamentally difficult children evoke less paternal involvement and negatively influence parental relationship quality, which may result in union dissolution.50 This suggests evocative rGE, but this evidence also fails to explain how children with the same previously described genotype would encourage biological parents to form a residential relationship.8 Depending on the design of the study, 1 or more of the rGEs may be ruled out. In keeping with the preceding example, we argue that children—especially those in early childhood—could not actively seek out an environment of parental stability or instability. Other studies, such as adoption studies, could exclude passive rGE and may be used to help estimate the extent to which evocative rGE is evident.48 Further, having parental genes, or even measures expected to be associated with those genes, can be useful in examining 1 or more of types of rGEs. Controlling for both parent’s DNA can statistically exclude all rGE.41 However, doing so would reduce the analysis sample, and thus the power, by eliminating all cases where both parents are homozygote for the same allele (resulting in a linear combination with the child). This is not unlike sibling studies, where if both experience the same event (i.e. Head Start or no Head Start), the analysis sample is limited to siblings experiencing different environments. The less common the homozygosity in both alleles, and the lower the assortative mating on that genetic marker, the fewer the number of excluded cases.
Another method for verifying a G×E is to utilize an exogenous environment such as a natural or laboratory experiment.48 In our own research, we observed that mothers’ parenting differences associated with the recent Great Recession were moderated by the dopamine receptor gene (DRD2) responsible for coding a protein that ultimately aides in signal transduction in the neurons, and is associated with motivation, attention, and several mental health outcomes.51 Mothers with the more sensitive genetic marker exhibited more hash parenting practices at the beginning of the recession than mothers with the less sensitive variant of the marker. Because genes are not expected to affect macroeconomic changes, and because the recession was so dramatic, we are fairly confident that we observed a true G×E. However, because the changes associated with the Great Recession only went in 1 direction (negative for the majority of people), the GDSE model could not be fully tested. Another study, using experimental procedures more common to psychology, found evidence that toddlers with a sensitive dopamine receptor D4 gene (DRD4) genotype were more affected by experimentally induced changes in (positive) parenting than children without the DRD4 variant.20 Much like most observational social science studies, well-designed experimental or quasi experimental studies provide an important additional test of GDSE, although they are often less generalizable to normal life.
GDSE AND PREVENTION
Formally testing for a GDSE would be difficult within prevention research because that would require both a positive and negative treatment in addition to a control. However, 3 studies found a differential response to (positive) preventative treatments. The first randomly assigned a small sample (< 200) of toddlers to a parenting intervention involving video feedback. The purpose was to reduce young children’s externalizing behavior. Children with the DRD4 7-repeat polymorphism, which is considered the sensitive variant, were influenced by the parenting treatment, whereas children with other DRD4 repeat variants were not influenced.21 It was thought that the children with the sensitive variant responded more to their mother’s positive discipline (the behavior which the intervention was targeting) than the other children. The second intervention that focused on changing child behavior directly rather than indirectly through the parent was implemented with preschool children.21 Children were randomly assigned to receive positive feedback or not during a literacy program provided via computer. Here, increases in literacy skills were seen in those children with the DRD4 7-repeat who received positive feedback but not in those without the feedback.
In these examples, children with the more sensitive DRD4 genetic variant were the ones who benefited from the positive experiences offered by the intervention. Another recent prevention trial, which compared orphans in foster care or institutional care in Romania, found positive effects for children with the sensitive variant of the 5-HTTLPR of the serotonin transporter (the short allele), suggesting that such effects are not limited to the dopamine system.22 Clearly, more prevention work looking at gene by treatment interactions would be welcome. The emerging work does suggest genetic differential sensitivity to positive environments, however, while controlling via randomization for possible rGE issues. Combining this with other, typically negative, exogenous environmental interactions discussed previously provides greater support for the GDSE. From a public health standpoint, the fact that individuals’ responses to treatment may vary as a function of genetic sensitivity suggests that the efficacy of certain interventions is sometimes underestimated because average effects may mask that some groups show very large responses. It is also likely that ways will be found to target interventions by genes. Targeting by genes may or may not be good public health policy. Research on the benefits and costs of such targeting will be useful in ascertaining its worth.
INTEGRATING GDSE RESEARCH AND LIFE COURSE AND DEVELOPMENTAL THEORIES
Social science theory can enrich the GDSE model by highlighting the importance of considering life course and development frameworks in examining human behavior. These frameworks imply that G×E interactions may not be constant across an individual’s life, may vary as a function of the context in which individuals operate, and may not be constant over cohorts or historical periods. With the risk of oversimplification, we argue that developmental theory has as its primary focus the interaction of person, process, and context,14,52 as studied with regard to age and age-graded transitions in processes and relationships.53 The life course theories focus on context as well, but often with a broader sweep (cohort, period, and historical contexts).54 Little GDSE work has used this broader historical context, and its full discussion is beyond the scope of this article. However interested researchers might begin with a broad description of the paradigm and methods55 and an excellent example with behavioral genetic research that used cohort and periods effects to examine the change in heritability of smoking.56 Specific transitions that are fairly universal within a particular society or time period are often the focus (childbearing, mate selection, work, retirement, school entry, and completion). Both theories consider the (individual, societal, and historical) timing of transitions and the adaptation to various transitions (and possible influences upon adaptation to later transitions or turning points).57,58
Developmental theories have focused more on personal characteristics (for example, temperament and behavioral regulation, as discussed previously) and on processes underlying development (cognitive, biological, and emotional changes). Contexts such as the family (parents, siblings, larger family systems), the peer group, and school and health settings are most often considered within developmental theory, whereas contexts such as the neighborhood, the community, and society have been the province of life course theories, although these distinctions are blurring (i.e., developmental theorists have incorporated larger contexts, and life course theorists have incorporated more proximal contexts into their work).
In general, individuals tend to increase control over their environments as they age59; they are also more likely to actively seek out environments that match their personal characteristic and interests. Moreover, their position within their society (education, income, neighborhood residence) and the organization of society are likely to impact how individuals cope with environmental change.60,61 For all these reasons, the impact of life events will depend on the developmental stage, social context, and historical period within which they occur.54
Given that GDSE work has just begun, most of these insights are not reflected in the existent research. However, they provide a powerful lens in understanding the interplay between individual biological and psychological change and the timing, duration, and sequencing of environmental influences.62,63 Specifically, the 2 perspectives guide researchers to consider not only the age of the individual, but the timing, stability, sequencing, and accumulation of environmental events under study.64
In addition, experiences at certain ages may have larger or more lasting effects than similar experiences at other ages. Biological systems may also be especially open to environmental inputs at certain time points (such as the prenatal and the infancy-toddler periods). Finally, environments are expected to be more exogenous in the first years of life than later (given opportunities for choice) so that G×E interactions seen earlier are more likely to be a true interaction rather than a correlation. In our own research, we found that changes in family structure during early childhood (aged ≤ 3 years) showed stronger G×E interactions than changes in middle childhood (aged 5–9 years).38 Although these age-linked differences were not always statistically significant in our analyses, they were large enough to suggest that more attention should be paid to the role of age of occurrence and timing of events in examining GDSE.
Not only are there individual differences in the adaptation to environmental events, but adaptation to an event often occurs, suggesting that longitudinal follow-up is critical. Perhaps the GDSE model will yield even more powerful findings when considering which individuals recover from a negative event or series of events, not just which individuals are affected in the short term. At the same time, effects of earlier events may fade with time, because the individual is influenced by what is happening (given GDSE theory that certain individuals are more sensitive to their current environment).62 Specifically, more recent events may yield larger G×E interactions than earlier events for individuals with the most sensitive genotypes.38 This finding is consistent with the idea that if people are truly more sensitive to their environments, recent events should be more predictive of current outcomes than earlier events. These 2 insights—the importance of timing and the importance of recency—highlight an important tension; namely, although early life course events may be more important than later events for human development in general, for some groups, more recent events may show larger effects. Little GDSE research has provided longitudinal research to address this tension.
GDSE research also needs to consider the sequencing of events. For instance, although not connected to GDSE work yet, other work in the Fragile Families and Child Wellbeing Study finds that having a child before marriage—even among those who eventually marry—has important consequences for family formation and child wellbeing.63 In addition, the accumulation of events, specifically the number of partner transitions that a mother experiences in the child’s first 5 years of life, is associated with lower child wellbeing.65 Even if each environmental event has a small influence on outcomes, research suggests that the cumulative effect of many events over time may have a large effect. Some of the first G×E work utilized measures of childhood stress that included counts of several harsh events. However, 1 difficulty of these measures is consistently defining what events matter, how many times they can be counted, and over what period of time they are counted.25
Finally, the life course development framework pushes researchers to consider the implications of a changing versus a stable environment. Although a consistently poor environment is most likely worse than a consistently nurturing environment, it is less clear where a highly volatile environment ranks. Previous research suggests individuals may actually do worse in volatile environments, where access to resources are less certain.66 In our own work, we found that change itself appears to have a negative effect on outcomes. For example, although poorer neighborhoods are unfavorable for many child outcomes, experiencing decreasing poverty in initially high poverty neighborhoods does not always result in more favorable outcomes.67 This may help explain some of the asymmetry in our GDSE work. Although having a father move into the home (presumably a favorable change) is associated with positive outcomes, the effect is not as large as the decrease if the father moves out (a typically unfavorable change). The smaller positive effect may be a result of the net effect of the favorable move in and the unfavorable effect of a major disruption, whereas the stronger negative effect is the net effect of the unfavorable exit of the father and the unfavorable effect of any change.
At the time of this writing, GDSE research has not explicitly tested the life course developmental framework because most of the research is cross sectional. However, to be fair, it is also clear that many GDSE researchers have relied on the developmental and life course theories to narrow the possible time periods in which to look for GDSE. One reason for the lack of integration of developmental or life course frameworks into GDSE is because incorporating multiple waves of longitudinal experiences is a more difficult test for researchers who have been more focused on demonstrating the existence of GDSE.
CONCLUSIONS
We argue that the use of the GDSE model provides both opportunities and challenges for researches who hope to use this approach to study health. The GDSE model pushes researchers to consider the theoretical and substantive meaning of both favorable and unfavorable environmental influences. GDSE affords important tools for addressing traditional concerns about rGE. Issues of endogeneity and measurement error are still important, but can be dealt with using methods familiar to many health researchers. It may also provide novel insights when considering timing of an environmental influence, which can lead to more robust measures of G×E effects. In reality, the GDSE model suggests that integration of genetic and social science data may not be as impossible as it sometimes appears. Biologists and social scientists will still need to collaborate for this area to progress, but hopefully, the previous discussion provides greater evidence that these collaborations could be incredibly fruitful.
For GDSE to become a more central framework some important questions need to be addressed. First, how pervasive is this sensitivity? Does it apply to all environmental influences or is it just limited to certain types of influences? Second, does this biological sensitivity simply heighten sensitive periods or is it a long-term sensitivity? How accurately can we measure GDSE? Is GDSE a broad neurologic issue, or is it system (e.g., dopaminergic, serotonergic) specific? Also, can we find (or create) equally favorable environmental measures to contrast with our measures of unfavorable environments? Once GDSE is better understood, researchers could more easily integrate these measures into their studies without extensive training. Essentially, we are in scale formation stage, and we may soon learn to what extent there are multiple scales, how they should be weighted, how reliable they are, and when they can be best applied.
The purpose of this article has been to demonstrate the use of one type of G×E model, both its drawbacks, strengths, and possible future integration in social science research on health. More specifically, we have utilized the Fragile Families data, which focused on child development, and women in child-bearing years. The GDSE framework, however, should not be thought of just related to these ages, but could be examined across the entire life course. Further, although we have emphasized the family environment as our measure of the social environment, there are many other types of environments, such as schools, neighborhoods, cities, regions, and other macro environments.14 In addition, although GDSE (and more broadly G×E) work is broadly international, little work has been done in countries that are not members of Organisation for Economic Co-operation and Development (e.g., most countries located in Latin America, Africa, and Asia). As this research begins, attention will need to be paid to population stratification because the distribution of gene variants differs by ancestry populations; however, methods for addressing population stratification are well known in the genetic literature.68 Nevertheless, in addition to the important genetic differences that may exist in these areas, there are important cultural, social, and health differences that could help tease out environmental specificities and mechanisms. That is, because of the unique cultural situations, norms and values may influence what is considered a favorable environment in some places yet may be considered unfavorable in others, with the obvious exceptions of severe environments such as abuse, extreme poverty, war, or forced migration. In sum, the growing interest in G×E models and epigenetics has the potential not only to bridge the gap between the social and biological sciences but also to reach across cultures and geographical boundaries.
Acknowledgments
This study was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development grants (R01HD36916, R01HD39135, R01HD40421), as well as by a consortium of private foundations that supported the Fragile Families and Child Wellbeing Study.
A previous version of this article was presented at the Eunice Kennedy Shriver National Institute Child Health and Human Development Nature and Nurture Conference on March 20-21, 2013, in Washington, DC.
Human Participant Protection
The University of Michigan, Princeton University, Columbia University, and Penn State Hershey Medical Center human investigation review boards approved the study.
References
- 1.Belsky J, Pluess M. Beyond diathesis stress: differential susceptibility to environmental influences. Psychol Bull. 2009;135(6):885–908. doi: 10.1037/a0017376. [DOI] [PubMed] [Google Scholar]
- 2.Boyce WT, Ellis BJ. Biological sensitivity to context: an evolutionary-developmental theory of the origins and functions of stress reactivity. Dev Psychopathol. 2005;17(2):271–301. doi: 10.1017/s0954579405050145. [DOI] [PubMed] [Google Scholar]
- 3.Belsky J, Bakermans-Kranenburg MJ, van IJzendoorn MH. For better and for worse: differential susceptibility to environmental influences. Curr Dir Psychol Sci. 2007;16(6):300–304. [Google Scholar]
- 4.Ellis BJ, Boyce WT. Biological sensitivity to context. Curr Dir Psychol Sci. 2008;17(3):183–187. [Google Scholar]
- 5.Raine A. Biosocial studies of antisocial and violent behavior in children and adults: a review. J Abnorm Child Psychol. 2002;30(4):311–326. doi: 10.1023/a:1015754122318. [DOI] [PubMed] [Google Scholar]
- 6.Boardman JD, Domingue BW, Fletcher J. How social and genetic factors predict friendship networks. Proc Natl Acad Sci USA. 2012;109(43):17377–17381. doi: 10.1073/pnas.1208975109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belsky J. Variation in susceptibility to rearing influences: an evolutionary argument. Psychol Inq. 1997;8(3):182–186. [Google Scholar]
- 8.Seltzer MM, Barker ET, Greenberg JS, Hong J, Coe C, Almeida D. Differential sensitivity to life stress in FMR1 premutation carrier mothers of children with fragile X syndrome. Health Psychol. 2012;31(5):612–622. doi: 10.1037/a0026528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakermans-Kranenburg MJ, van Ijzendoorn MH. Genetic vulnerability or differential susceptibility in child development: the case of attachment (research review) J Child Psychol Psychiatry. 2007;48(12):1160–1173. doi: 10.1111/j.1469-7610.2007.01801.x. [DOI] [PubMed] [Google Scholar]
- 10.Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes of plasticity genes? Mol Psychiatry. 2009;14(8):746–754. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyce WT, Chesney M, Alkon A et al. Psychobiological reactivity to stress and childhood respiratory illness: results of two prospective studies. Psychosom Med. 1995;57(5):411–422. doi: 10.1097/00006842-199509000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Shonkoff JP, Boyce WT, McEwen BS. Neuroscience, molecular biology, and the childhood roots of health disparities: building a new framework for health promotion and disease prevention. JAMA. 2009;301(21):2252–2259. doi: 10.1001/jama.2009.754. [DOI] [PubMed] [Google Scholar]
- 13.Belsky J. Differential susceptibility to rearing influences: an evolutionary hypothesis and some evidence. In: Ellis B, Bjorklund D, editors. Origins of the Social Mind: Evolutionary Psychology and Child Development. New York, NY: Guildford; 2005. pp. 139–163. [Google Scholar]
- 14.Bronfenbrenner U. The Ecology of Human Development: Experiments by Nature and Design. Cambridge, MA: Harvard University Press; 1979. [Google Scholar]
- 15.Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet Part B. 2007;144B(1):101–105. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 16.Rutter M. Achievements and challenges in the biology of environmental effects. Proc Natl Acad Sci USA. 2012;109(suppl 2):17149–17153. doi: 10.1073/pnas.1121258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell C, Notterman D, Brooks-Gunn J et al. The role of mother’s genes and environment on postpartum depression. Proc Natl Acad Sci U S A. 2011;108(20):8189–8193. doi: 10.1073/pnas.1014129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick DM, Meyers JL, Latendresse SJ et al. CHRM2, parental monitoring, and adolescent externalizing behavior: evidence for gene-environment interaction. Psychol Sci. 2011;22(4):481–489. doi: 10.1177/0956797611403318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van IJzendoorn MH, Bakermans-Kranenburg MJ, Mesman J. Dopamine system genes associated with parenting in the context of daily hassles. Genes Brain Behav. 2008;7(4):403–410. doi: 10.1111/j.1601-183X.2007.00362.x. [DOI] [PubMed] [Google Scholar]
- 20.Bakermans-Kranenburg MJ, van IJzendoorn MH, Pijlman FT, Mesman J, Juffer F. Experimental evidence for differential susceptibility: dopamine D4 receptor polymorphism (DRD4 VNTR) moderates intervention effects on toddlers’ externalizing behavior in a randomized controlled trial. Dev Psychol. 2008;44(1):293–300. doi: 10.1037/0012-1649.44.1.293. [DOI] [PubMed] [Google Scholar]
- 21.Kegel CAT, Bus AG, van IJzendoorn MH. Differential susceptibility in early literacy instruction through computer games: the role of the dopamine D4 receptor gene (DRD4) Mind Brain Educ. 2011;5(2):71–79. [Google Scholar]
- 22.Drury SS, Gleason MM, Smyke AT et al. Genetic sensitivity to the caregiving context: the influence of 5httlpr and BDNF val66met on indiscriminate social behavior. Physiol Behav. 2012;106(5):728–735. doi: 10.1016/j.physbeh.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahneman D, Krueger A, Schkade D, Schwarz N, Stone A. Would you be happier if you were richer? A focusing illusion. Science. 2006;312(5782):1908–1910. doi: 10.1126/science.1129688. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell C, Brooks-Gunn J, Hobcraft J, Garfinkel I, Notterman D, McLanahan S. The influence and interplay of family instability and genes on children’s prosocial behavior. Paper presented at: Annual Meeting of the Population Association of America; May 3–5, 2012; San Francisco, CA.
- 25.Caspi A, Hariri AR, Holmes A, Uher R, Moffitt T. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167(5):509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakermans-Kranenburg MJ, van IJzendoorn MH. Differential susceptibility to rearing environment depending on dopamine-related genes: new evidence and a meta-analysis. Dev Psychopathol. 2011;23(1):39–52. doi: 10.1017/S0954579410000635. [DOI] [PubMed] [Google Scholar]
- 27.Fan JB, Sklar P. Meta-analysis reveals association between serotonin transporter gene STin2 VNTR polymorphism and schizophrenia. Mol Psychiatry. 2005;10(10):928–938. doi: 10.1038/sj.mp.4001690. [DOI] [PubMed] [Google Scholar]
- 28.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Young SE, Smolen A, Corley RP et al. Dopamine transporter polymorphism associated with externalizing behavior problems in children. Am J Med Genet. 2002;114(2):144–149. doi: 10.1002/ajmg.10155. [DOI] [PubMed] [Google Scholar]
- 30.Williams RB, Marchuk DA, Gadde KM et al. Serotonin-related gene polymorphisms and central nervous system serotonin function. Neuropsychopharmacology. 2003;28(3):533–541. doi: 10.1038/sj.npp.1300054. [DOI] [PubMed] [Google Scholar]
- 31.Ungless MA, Magill PJ, Bolam JP. Uniform inhibition of dopamine neurons in the ventral tegmental area by aversive stimuli. Science. 2004;303(5666):2040–2042. doi: 10.1126/science.1093360. [DOI] [PubMed] [Google Scholar]
- 32.Propper C, Willoughby M, Halpern CT, Carbone MA, Cox M. Parenting quality, DRD4, and the prediction of externalizing and internalizing behaviors in early childhood. Dev Psychobiol. 2007;49(6):619–632. doi: 10.1002/dev.20249. [DOI] [PubMed] [Google Scholar]
- 33.Zald DH, Cowan RL, Riccardi P et al. Midbrain dopamine receptor availability is inversely associated with novelty-seeking traits in humans. J Neurosci. 2008;28(53):14372–14378. doi: 10.1523/JNEUROSCI.2423-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brischoux F, Chakraborty S, Brierley DI, Ungless MA. Phasic excitation of dopamine neurons in ventral VTA by noxious stimuli. Proc Natl Acad Sci USA. 2009;106(12):4894–4899. doi: 10.1073/pnas.0811507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaykin DV, Pudovkin A, Weir BS. Correlation-based inference for linkage disequilibrium with multiple alleles. Genetics. 2008;180(1):533–545. doi: 10.1534/genetics.108.089409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heils A, Teufel A, Petri S et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 37.Hranilovic D, Stefulj J, Schwab S et al. Serotonin transporter promoter and intron 2 polymorphisms: relationship between allelic variants and gene expression. Biol Psychiatry. 2004;55(11):1090–1094. doi: 10.1016/j.biopsych.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell C, Brooks-Gunn J, Hobcraft J, Garfinkel I, Notterman D, McLanahan S. Genes, mother’s relationship instability, and children’s socioemotional wellbeing. Paper presented at: Annual Meeting of the Population Association of America; March 31–April 2, 2011; Washington, DC.
- 39.Freese J, Shostak S. Genetics and social inquiry. Annu Rev Sociol. 2009;35:107–128. [Google Scholar]
- 40.Paré G, Cook NR, Ridker PM, Chasman DI. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the Women’s Genome Health Study. PLoS Genet. 2010;6(6):e1000981. doi: 10.1371/journal.pgen.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boardman J. Gene-environment interactions in a genome-wide association framework: the example of college education. Paper presented at: New Approaches to Child Development and Risky Behavior Research: Gene-environment Interaction and Epigenetics; January 30, 2012; Columbia Population Research Center, Columbia University, New York, NY.
- 42.Manolio TA, Collins FS, Cox NJ et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11(20):2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 44.Cole SW, Arevalo JMG, Takahashi R et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci USA. 2010;107(12):5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller G, Chen E, Cole SW. Health psychology: developing biologically plausible models linking the social world and physical health. Annu Rev Psychol. 2009;60:501–524. doi: 10.1146/annurev.psych.60.110707.163551. [DOI] [PubMed] [Google Scholar]
- 46.Stein RA. Epigenetics and environmental exposures. J Epidemiol Community Health. 2012;66(1):8–13. doi: 10.1136/jech.2010.130690. [DOI] [PubMed] [Google Scholar]
- 47.Slavich GM, Cole SW. 2013. The emerging field of human social genomics. Clin Psychol Sci. Epub ahead of print. March 5, 2013 doi: 10.1177/2167702613478594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fletcher J, Conley D. The challenge of causal inference in gene-environment interaction research: leveraging research designs from the social sciences. Paper presented at: New Approaches to Child Development and Risky Behavior Research: Gene-environment Interaction and Epigenetics; January 30, 2012; Columbia Population Research Center, Columbia University, New York, NY.
- 49.Meadows SO, McLanahan S, Brooks-Gunn J. Family structure changes and maternal health trajectories. Am Sociol Rev. 2008;73(2):314–334. doi: 10.1177/000312240807300207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewin-Bizan S. Identifying the associations between child temperament and father involvement: theoretical considerations and empirical evidence. Princeton, NJ: Center for Research on Child Wellbeing; 2006. Working paper 2006-24-FF.
- 51.Lee D, McLanahan S, Brooks-Gunn J, Notterman D, Garfinkel I. The effect of the Great Recession and dopamine receptor gene DRD2 on maternal harsh parenting. Paper presented at: Annual Meeting of the American Sociological Association; August 20–23, 2011; Las Vegas, NV.
- 52.Overton WF. Life-span development: concepts and issues. In: Overton WF, editor. Cognition, Biology, and Methods across the Lifespan. Volume 1. Handbook of Life-Span Development. Hoboken, NJ: Wiley; 2010. pp. 1–29. [Google Scholar]
- 53.Baltes PB, Lindenberger U, Staudinger UM. Life span theory in developmental psychology. In: Damon W, Lerner RM, editors. Handbook of Child Psychology Theoretical Models of Human Development. 6th ed. Vol 1. New York, NY: Wiley; 2006. pp. 569–664. [Google Scholar]
- 54.Elder GH. The life course as developmental theory. Child Dev. 1998;69(1):1–12. [PubMed] [Google Scholar]
- 55.Mayer KU. New directions in life course research. Annu Rev Sociol. 2009;35:413–433. [Google Scholar]
- 56.Boardman JD, Blalock CL, Pampel FC. Trends in the genetic influences on smoking. J Health Soc Behav. 2010;51(1):108–123. doi: 10.1177/0022146509361195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graber JA, Brooks-Gunn J. Transitions and turning points: navigating the passage from childhood through adolescence. Dev Psychol. 1996;32(4):768–776. [Google Scholar]
- 58.Rutter M. Stress, coping, and development: some issues and some questions. In: Garmezy N, Rutter M, editors. Stress, Coping, and Development in Children. New York, NY: McGraw Hill; 1983. pp. 1–42. [Google Scholar]
- 59.Burt SA. The importance of the phenotype in explorations of gene-environment interplay. In: Booth A, McHale S, Landale N, editors. Biosocial Foundations of Family Processes. Series: National Symposium on Family Issues. New York, NY: Springer; 2010. pp. 85–94. [Google Scholar]
- 60.Rutter M. Genes and Behavior. London, UK: Blackwell; 2006. [Google Scholar]
- 61.Shanahan M, Hofer SM. Molecular genetics, aging, and well-being: sensitive period, accumulation, and pathway models. In: Binstock RH, George LK, editors. Handbook of Aging and the Social Sciences. New York, NY: Elsevier; 2011. pp. 135–147. [Google Scholar]
- 62.Zeidner M, Endler NS. Handbook of Coping: Theory, Research, Applications. New York, NY: John Wiley and Sons; 1996. [Google Scholar]
- 63.Carlson MJ, McLanahan S, England P. Union formation and stability in fragile families. Demography. 2004;41(2):237–261. doi: 10.1353/dem.2004.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Addison JT. Urie Bronfenbrenner. Hum Ecol. 1992;20(2):16–20. [Google Scholar]
- 65.Waldfogel J, Craigie T-A, Brooks-Gunn J. Fragile families and child wellbeing. Future Child. 2010;20(2):87–112. doi: 10.1353/foc.2010.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenblum LA, Paully GS. The effects of varying environmental demands on maternal and infant behavior. Child Dev. 1984;55(1):305–314. [PubMed] [Google Scholar]
- 67.Leventhal T, Brooks-Gunn J. Changes in neighborhood poverty from 1990 to 2000 and youth’s problem behaviors. Dev Psychol. 2011;47(6):1680–1698. doi: 10.1037/a0025314. [DOI] [PubMed] [Google Scholar]
- 68.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]