Abstract
Cytochrome c oxidase generates a proton motive force by two separate mechanisms. The first mechanism is similar to that postulated by Peter Mitchell, and is based on electrons and protons used to generate water coming from opposite sides of the membrane. The second mechanism was not initially anticipated, but is now firmly established as a proton pump. A brief review of the current state of our understanding of the proton pump of cytochrome oxidase is presented. We have come a long way since the initial observation of the pump by Mårten Wikström in 1977, but a number of essential questions remain to be answered.
Keywords: Oxidase, Cytochrome, Proton, Redox, Energy coupling, Pump, Heme, Copper
Introduction
The genius of Peter Mitchell established the central importance of charge separation across the biological membrane in driving oxidative phosphorylation as well as many other biological functions, including active transportof solutes and ions. The molecular mechanisms of how charge separation is generated, on the one hand, and how it is utilized, on the other hand, have been the central theme of bioenergetics research for the past three decades. During this time it has become evident that numerous molecular mechanisms have evolved both to generate and to utilize the proton motive force (now extended to include the sodium motive force). The universality of the principle of using a proton (or sodium) motive force (pmf or smf) as a means to couple energy-generating and energy-utilizing processes is impressive, including eukaryotes, prokaryotes, aerobic respiration, anaerobic respiration and photosynthesis. This modular character of the design, being able to “plug in” different means of generating transmembrane charge separation and couple them to different pmf/smf-driven processes, makes clear the evolutionary advantage of the overall architecture. Bioenergetics can evolve in a modular fashion without redesigning the entire system.
Figure 1 schematically illustrates several ways to couple redox reactions to the generation of a proton motive force: (A) A redox “loop” in which a neutral carrier (e.g., quinone) transports both electrons and protons across the membrane from a substrate oxidation site on the inside (negative) of the membrane to a site on the opposite side of the membrane where the quinol is oxidized. Electrons are transferred across the membrane to a second substrate on the inside, generating a voltage (positive out). The Q-cycle is a variation of this scheme.; (B) Same as shown in scheme a, except in this case the second substrate is reduced on the outside of the membrane, utilizing protons from the inside, again generating a voltage (positive out). (C) A substrate is oxidized on the outside of the membrane and electrons are transferred to a site on the opposite side of the membrane where a second substrate is reduced, generating a voltage (positive out); (D) A schematic for the arrangement of the redox sites of cytochrome oxidase. Cytochrome c is oxidized at a site on the outside (four consecutive one-electron reactions), and electrons are transferred to the oxygen redox site located in the middle of the membrane, utilizing protons and electrons from opposite sides of the membrane; (E) Same as in part d, with the addition of an indirectly coupled proton pump, indicated by the thick red arrow. The existence of the proton pump in cytochrome oxidase was established by Wikström (1977) in 1977 and has intrigued investigators since that time. The last mechanism (E) can be adapted, in principle, to the active transport or pumping of any ion (or solute).
Fig. 1.
Several schemes by which redox reactions are coupled to the generation of a transmembrane potential and proton motive force. In each case, the upper side of the membrane is the positive side (P-side or outside), and corresponds to the bacterial periplasm or mitochondrial intermembrane space
Examples of each of these mechanisms have been demonstrated in different respiratory enzymes, as well as other ways of generating a voltage across the membrane. Cytochrome oxidase is represented by scheme E. Mutants that are decoupled from the proton pump (see detailed discussion below) still operate according the scheme D.
The heme–copper superfamily
Initially, all the experimental work on cytochrome oxidase was on the mitochondrial enzyme, primarily from bovine heart since it is a convenient source. We now know that the mitochondrial cytochrome c oxidase is a member of a large superfamily called the heme–copper superfamily (Pereira et al. 2001). These enzymes include both O2 reductases (respiratory oxidases) and NO reductases from prokaryotes. The superfamily is defined by amino acid sequence homology of a core subunit, which corresponds to subunit I of the mitochondrial oxidase. Much progress over the past 20 years has benefited from the application of molecular genetics techniques to prokaryotic versions of cytochrome oxidase.
Redox centers
All of the heme–copper respiratory oxidases have in common (1) a low-spin heme ligated to two histidines; (2) a heme–copper binuclear center consisting of a high-spin heme (one histidine ligand) and a nearby copper, CuB, (three histidine ligands); (3) a tyrosine at the active site which is covalently linked to one of the histidine ligands to CuB (Hemp et al. 2006). The modified tyrosine may be redox-active, transiently forming a neutral radical during the catalytic cycle of the enzymes (Proshlyakov et al. 2000). The hemes occupying the low-spin and high-spin sites are variations of one of three chemical types: heme B, heme O and heme A (Ferguson-Miller and Babcock 1996). There is no functional correlation with the type of heme located within the enzyme. Heme B is identical to the heme in myoglobin and hemoglobin, but heme O and heme A are found only in the heme–copper enzymes. For historical reasons, the oxygen-binding heme, which is in the heme–copper binuclear center, is designated with a subscript “3”. The mitochondrial oxidase contains heme A in each the low-spin and the high-spin site and is, thus, referred to as an “aa3-type” oxidase, or cytochrome aa3.
X-ray structures
The first X-ray structures of the bovine heart oxidase (Tsukihara et al. 1995, 1996; Yoshikawa et al. 2000; Shinzawa-Itoh et al. 2007) and of the aa3-type oxidase from Paracoccus denitrificans (Iwata et al. 1995; Ostermeier et al. 1997) were reported in 1995. Structures are also available now for the aa3-type oxidase from Rhodobacter sphaeroides (Svensson-Ek et al. 2002; Qin et al. 2006, 2007), the bo3-type oxidase from E. coli (Abramson et al. 2000) and the ba3-type oxidase from Thermus thermophilus (Soulimane et al. 2000; Luna et al. 2008). These structures all show the same general architecture of the transmembrane helices of subunit I and the same arrangement of the hemes and copper ligated within subunit I.
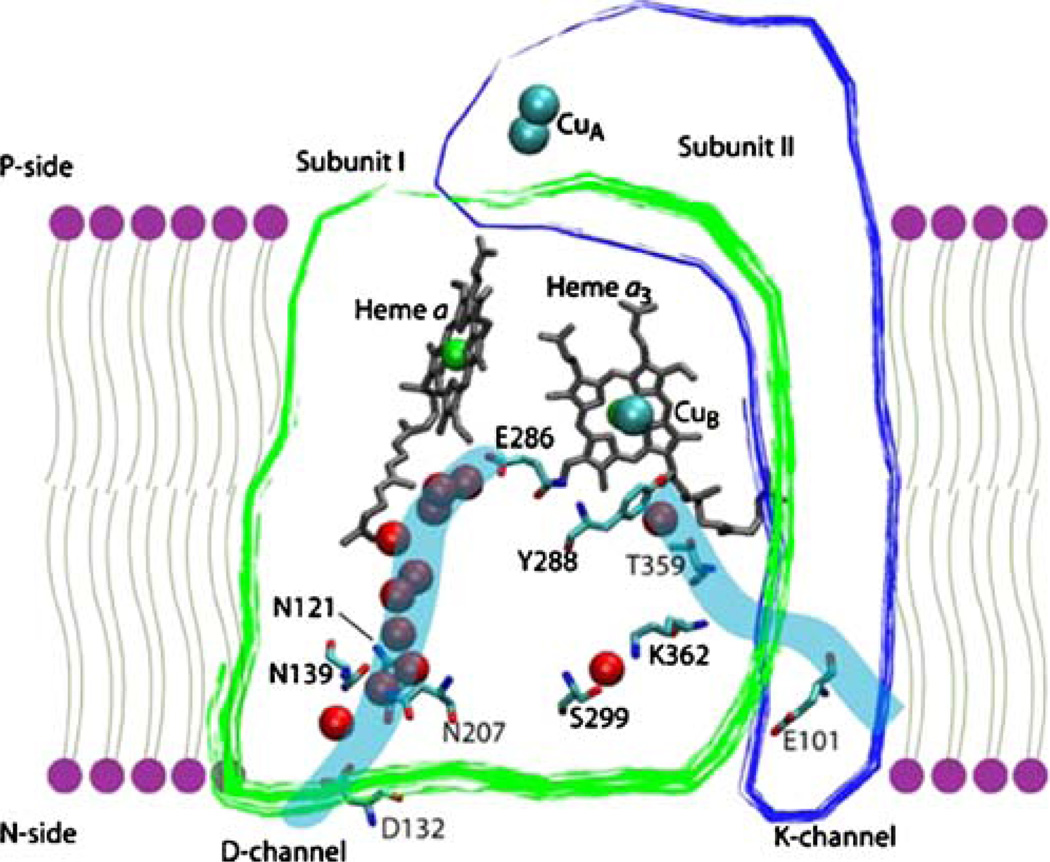
The structures largely confirmed models based on spectroscopic and molecular genetics evidence, showing the two hemes at about the same depth in the membrane. The hemes are nearly within van der Waals contact at the edges, which may be important for rapid electron transfer. Figure 2 shows the structure of the enzyme from R. sphaeroides illustrating the redox centers. Figure 3 shows an early model of the oxidase from 1978 (Artzatbanov et al. 1978) based on biochemical experiments and the knowledge that the enzyme was a true proton pump.
Fig. 2.
Schematic of subunits I and II of the cytochrome c oxidase from R. sphaeroides, showing the metal centers and two proton-input channels
Fig. 3.
Model of cytochrome oxidase from 1978 illustrating the early recognition of the need for proton channels and the positioning of the two hemes. From (Artzatbanov et al. 1978)
Two mechanisms of charge separation
The reaction catalyzed by cytochrome oxidase is the four-electron reduction of O2 to water.
| (1) |
The site where reduced cytochrome c is oxidized is located on the “outside” or P-side of the membrane (bacterial periplasm or mitochondrial intermembrane space) and the protons come from the “inside” or N-side (bacterial cytoplasm or mitochondrial matrix). The heme–copper-active site of the enzyme is buried within the protein at a depth corresponding to about one third of the membrane dielectric from the outside. Oxygen reaches the active site through a channel that opens to the hydrophobic membrane interior, where the equilibrium concentration of oxygen would be expected to be higher than in the surrounding solution. The chemistry in Eq. 1 will result in the separation of charge, one charge per electron consumed in the chemistry because the protons and electrons originate from opposite sides of the membrane, as in scheme (D) in Fig. 1.
Proton pumping
Experimentally, proton ejection from the P-side of the membrane can be measured and with the mitochondrial enzyme the stoichiometry is 1 proton per electron. Therefore, the net reaction catalyzed by the enzyme needs to be modified from Eq. 1 to the following.
| (2) |
In other words, in addition to coupling by scheme D (Fig. 1), the oxidase also utilizes a true proton pump (scheme E, Fig. 1) to separate charges across the membrane. For each turnover 8 charges cross the membrane (per O2) and one proton is pumped for each electron used in the chemistry. By far the major component of the proton motive force in the mitochondrion is the voltage (ΔΨ) rather than ΔpH.
Proton pumping has also been determined for several of the prokaryotic heme–copper oxidases, and it appears that the enzymes from P. denitrificans and from R. sphaeroides also pump with a stoichiometry of 1 proton per electron. However, the ba3-type oxidase from T. thermophilus appears to pump only half as many protons per catalytic cycle (Kannt et al. 1998; Siletsky et al. 2007). There are mutants of both the oxidases from P. denitrificans (Pfitzner et al. 2000) and from R. sphaeroides (Pawate et al. 2002; Namslauer et al. 2003a; Siletsky et al. 2004; Brändén et al. 2006; Han et al. 2006; Lepp et al. 2008a) which are fully active but which do not pump protons at all or have a low stoichiometry of protons/electron (discussed in “Separating chemical and pumped protons by using uncoupled mutants”). These prove that the pumping mechanism is separated from the oxygen chemistry (which is not altered) and also shows that the stoichiometry is not locked-in by any mechanistic considerations. There is no reason why all the heme–copper oxidases must have the same stoichiometry of pumping as the mitochondrial oxidase. There is also no mechanistic constraint that the proton pumping needs to have an integer value per electron (i.e., 1:1) or that the stoichiometry would be the same in each of the four electron-transfer steps of the oxidase. On the other hand, values lower than one proton/electron could result from numerous experimental artifacts and must be regarded in this context.
Oxygen chemistry and reaction intermediates
A series of beautiful time-resolved spectroscopic methods have been used to define the sequence of events during the catalytic cycle and chemically define the nature of the intermediate states of the enzyme (Verkhovsky et al. 1992, 1994; Babcock and Varotsis 1993; Ogura et al. 1993; Varotsis et al. 1993; Hallén et al. 1994; Kitagawa and Ogura 1997; Han et al. 2000). Generally, the electron distribution is known with greater certainty than the proton distribution because the optical spectra are more sensitive to where the electrons reside. As a result, states of the enzyme with different proton distributions and different biochemical properties may appear identical spectroscopically. At the current time, the following is a reasonable summary of the sequence of events. We will start with the oxidized enzyme encountered during turnover. It has been demonstrated that just after formation, the oxidized enzyme is in an “activated” state in which CuB has a very high electron affinity (Bloch et al. 2004; Belevich et al. 2007). This is the OH state. The overall sequence is
| (3) |
Below, the redox states of heme a3, CuB and the active-site tyrosine are indicated next to the letter designation of each state. There is a fair degree of speculation in this, especially as regards hydroxides, water and the amino acid radical.
: One electron is taken up along with 2 protons from the inside. One proton is ejected to the outside (pumped). The electron ends up on CuB. This reaction only works if the enzyme is reduced immediately after it has been oxidized. The product state, EH, is not well defined, but is a presumed “activated” one-electron reduced form of the enzyme. The non-activated forms of the OH and EH states are designated the O and E states. Pumping has been demonstrated for this step.
: A second electron is added to the binuclear center. This is the least understood step and is putatively as written. Again, two protons are taken up and presumably one is pumped.
: Oxygen transiently binds to the doubly-reduced binuclear center, forming a ferrous heme a3/O2 called compound A (not shown; Verkhovsky et al. 1994). The ensuing reaction forms an oxygenated intermediate, the PM state, in which the O–O bond is broken and the active site tyrosine is thought to form a neutral radical (Proshlyakov et al. 2000). Some data suggest that a nearby tryptophan may be the source of the “fourth” electron required to break the O–O bond (de Vries 2008). This step is not associated with proton pumping or charge separation.
: A third electron reduces the radical back to tyrosine, a proton is taken up by the active site and another proton is pumped. Pumping has been demonstrated for this step.
: This is perhaps the most studied step in the catalytic cycle and has been directly shown to pump one proton. The product takes us back to the beginning.
Energetics
The free energy available from reaction (1) depends on the steady state concentration of O2 and on the extent of reduction of the electron donor (cytochrome c). Hence, this may vary considerably depending on the physiological state, particularly for prokaryotes which may be in oxygen-depleted environments. For the mitochondrial enzyme, the free energy available per electron is about 500 mV. As described above, two charges cross the membrane from the N-side to the P-side for each electron. If the membrane potential is 220 mV (positive out), the work required to move two charges across the membrane is equivalent to 440 mV. Thus, cytochrome oxidase is very efficient in converting chemical energy into the proton motive force. The postulated activated forms of the enzyme, OH and EH, are required if electron transfer to each state is associated with proton pumping against a voltage of 220 mV. Otherwise, the redox chemistry of the “as isolated”, unactivated forms of the enzyme does not provide sufficient free energy to drive the pump.
Principle of electroneutrality
The heme–copper binuclear center is buried within the protein, but is accessible to electrons (reduction via heme a), protons (via proton-conducting channels, described below) and to small anions such as chloride (presumably also via the proton channels). Experimentally, it has been shown, largely by the work of Rich and colleagues (Rich 1995; Rich et al. 1996, 1997), that every negative charge added to the binuclear center is always accompanied by a proton due to a need to maintain electroneutrality and charge balance. During catalytic turnover, this means that each electron transferred to the binuclear center will create a large electrostatic drive for protonation of a group at or near the binuclear center in order to keep charge balance. This electrostatic coupling is at the heart of most current models of how the proton works.
Proton-conducting channels
There must be pathways for protons to reach the active site and for pumped protons to traverse the protein from the N-side to the P-side. This was evident 30 years ago before anything was known about the protein structure (e.g., see Fig. 3). Site-directed mutagenesis on the prokaryotic oxidases (Konstantinov et al. 1997), followed by the X-ray structures have defined two proton-input channels, called the D-channel and the K-channel (Fig. 2). The D-channel contains about ten water molecules that are observed in the X-ray structures and which provide a continuous sequence of hydrogen bonds facilitating proton diffusion by the Grotthus mechanism (Nagle and Tristam-Nagle 1983). Whereas electrons can tunnel rapidly (τ<1 ms) between redox centers that are separated by 10 to 15 Å, protons in biological systems diffuse by being transferred from a hydrogen bond donor to a hydrogen bond acceptor, which can then become the hydrogen bond donor to the next element along the pathway.
Unexpectedly, in prokaryotes the two proton-input channels do not have the anticipated roles of providing chemical protons versus pumped protons (Konstantinov et al. 1997). The K-channel provides two chemical protons accompanying two of the electron transfer steps to the active site, whereas the D channel provides the remaining two chemical protons plus all four of the pumped protons (per O2). The functional advantage of this system is not clear.
A third channel, the H channel has been defined for the mammalian cytochrome oxidases (Muramoto et al. 2007; Shimokata et al. 2007). It is postulated that the H channel is used for all pumped protons and that, in mammals only, the role of the D and K channels is to provide chemical protons (two each). The H channel does not exist in the prokaryotic oxidases (Lee et al. 2000; Pfitzner et al. 2000) and, though supported by experimental evidence, remains controversial.
While the free energy generated by the redox chemistry catalyzed by cytochrome oxidase must be coupled to drive the proton pump, there must be strict barriers to separate the pumped protons from those required for the chemistry. The use of the D channel for the input of both chemical and pumped protons would appear to violate this restriction. However, there is a separation of the flow of chemical and pumped protons after a branch point, which is a glutamic acid (Glu286 in the R. sphaeroides oxidase, Fig. 2). Water is a critical component of each of the proton pathways leading from Glu286 to the proton acceptor in the pump pathway and to the enzyme active site (chemical protons), and it is postulated that either the presence/absence of water or the orientation of the water molecules determines which pathway is “open” and which is “closed” (Riistama et al. 1997; Wikström et al. 2003; Zheng et al. 2003). It appears that the pathways for pumped protons and chemical protons alternate in being open and closed, providing a clear temporal separation for the flow of pumped protons and chemical protons.
Requirements for the proton pump
Among the mitochondrial respiratory enzymes, only cytochrome oxidase qualifies as a true proton pump. Protons that are pumped by a “true pump” are distinguished by not being involved directly in the chemistry. It seems likely that Complex I may also utilize a pumping mechanism, however the mechanism is not yet known. The ATP synthase, when operating in the reverse direction of using ATP hydrolysis to generate a proton motive force, is also a true proton pump.
In this section, we will define the minimal functional and structural components required for the proton pump of cytochrome oxidase. Following this, experimental results will be summarized.
The proton pump must have the following components.
The active site: The reduction of O2 to water requires four electrons, which are delivered to the heme–copper site in a sequence of four one-electron transfers from heme a. It appears that each of the one-electron transfer steps results in pumping one proton, and translocating two charges across the membrane. The driving force is provided by the high proton affinity of the intermediates at the active site during each step of the reduction, and by the electrostatic drive to maintain electroneutrality.
The proton loading site or pump site: There should be at least one site which alternates proton affinity depending on the redox state of the active site. In most current models, the coupling between the events at the active site and the proton affinity of the pump site is purely electrostatic, summarized by the electroneutrality principle (see “Principle of electroneutrality”). However, in principle, a conformational change of the protein could also be responsible for the coupling. The identity of the pump site is not known, but prime candidates are the A-propionate of heme a3 and one of the histidine ligands to CuB (His334 in R. sphaeroides; Popovic and Stuchebrukhov 2004; Belevich et al. 2007; Sugitani et al. 2008).
Proton-conducting pathways: The input pathways are well defined as the D and K pathways for the prokaryotic oxidases. The strongest evidence that the D pathway is used by all the pumped protons is the existence of point mutations within the D channel which decouple the proton pump from the oxidase chemistry (see “Separating chemical and pumped protons by using uncoupled mutants”). Little is known about the exit pathway (Popovic and Stuchebrukhov 2005), and there may be multiple routes out beyond the proton loading site.
- Gating or valve mechanisms: It is essential for the proton pump that the input and output of protons occur through the correct channels. This means that there are kinetic barriers that are increased and decreased for proton movement which are directed by the charge distribution within the enzyme during the different steps in catalysis (Siegbahn and Blomberg 2007). In terms of the current models (Wikstrom and Verkhovsky 2007), this means the following.
- Protons are rapidly delivered to the pump site from the N-side of the membrane to stabilize the electron delivered to the active site. Both, the rate of proton delivery to the active site (chemical proton) from the N-side, and the rate of proton delivery to the pump site from the P-side of the membrane, must be at least 100-fold slower than the proton delivery rate to the pump site from the N-side of the membrane.
- Protons are delivered to the active site from the N-side of the membrane and not from the proton already residing at the pump site, which would represent a short circuit of the process. Once there, the presence of the proton at the active site electrostatically repels the proton at the pump site.
- Proton ejection from the pump site occurs to the P-side of the membrane and not back through the input channel to the N-side of the membrane. It has been proposed, for example, that the conformation of the side chain of Glu286 will serve as a valve, preventing backflow of the proton from the pump site to the N-side of the membrane through the D channel (Kaila et al. 2008a, b). Rapid reprotonation of Glu286 may also be sufficient to prevent backflow from the pump site.
Separating proton and electron transfer by the flow-flash technique
As outlined above, four electrons are required to reduce O2 to H2O. During cytochrome oxidase turnover, these electrons are transferred from cytochrome c, through CuA and heme a to the catalytic site, one at a time. This means that four different states with one to four electrons at heme a3-CuB site are formed during turnover (as outlined in “Oxygen chemistry and reaction intermediates”), where each of the transitions between these states is linked to proton and electron transfer to the catalytic site and proton pumping, presumably utilizing the same mechanism. However, the chemistry catalyzed at each of the fours steps is different, and the detailed structures of the heme-copper site are also different at each step. Consequently, identifying the detailed structures of the catalytic intermediates of cytochrome oxidase and understanding the pumping mechanism represents two separate problems and in the latter case we need to focus on the intramolecular electron and proton transfer reactions in cytochrome oxidase. A complication when approaching the nature of these reactions is that their time constants are typically in the microsecond time range, which makes it difficult to study them.
One technique that has been used frequently to overcome this problem is the so-called flow-flash method where the oxidase is first fully reduced, i.e. with four electrons, and a blocking CO ligand is bound to heme a3, i.e. at the same site where O2 would normally bind at the catalytic site. Next, the anaerobic oxidase-CO solution is mixed with an O2-containing solution, after which the CO ligand is removed by a short (typically a few nanoseconds) laser flash. Now, the reduced enzyme binds O2 and reactions linked to its step-wise reduction to water can be followed in time using various spectroscopic techniques. Even though this approach allows only studies of one half of the reaction cycle, i.e. oxidation of the reduced enzyme , the results from experiments using this technique have presented us with a wealth of information, providing mechanistic insights into the function of the oxidase. In addition, in this experiment all four electrons needed to reduce dioxygen to water are present within the oxidase upon initiation of the reaction with O2. Hence, in the initial part of the reaction electron transfer is not rate-limited by transfer from an external donor, allowing internal electron transfer to be observed separated in time from proton transfer.
When heme a3 and CuB become reduced during turnover (i.e. two electrons are transferred to the catalytic site), O2 binds and the PM state is formed as in reaction (3). Because the next (third) electron is transferred all the way from an external electron donor to the catalytic site, the overall rate of the PM→F transition is determined by the rate of electron transfer, and the electron and proton transfers occur simultaneously:
| (4) |
In the flow-flash reaction described above, two electrons are initially present at the catalytic site and another electron is found at heme a. Consequently, upon binding of O2 to heme a3 (τ≅10 µs at 1 mM O2) the third electron can now be transferred rapidly into the catalytic site. The time constant of this electron transfer has been found to be in the range 30–50 µs, which is faster than that of the proton transfer to the catalytic site (τ≅100 µs at pH 7). Therefore, in this case a state is formed at the catalytic site which presumably has the same chemical structure as PM, but with one additional electron, which is transferred to the active-site tyrosine radical (see R2→PM described for “Oxygen chemistry and reaction intermediates”). This state is called PR and it decays spontaneously into F, accompanied by proton transfer from solution with a time constant of ~100 µs. Starting with the four-electron reduced enzyme (R4) the PM intermediate is not observed. If present at all, it decays rapidly to the observed PR state, as shown below.
| (5) |
In other words, in studies of the reaction of the fully reduced oxidase with O2 using the flow-flash approach, the electron and proton-transfer reactions can be separated in time and studied independently.
The proton is transferred through the D pathway, via Glu286. Early studies of the flow-flash reaction with this pathway, blocked near the protein N-side surface (by replacement of the Asp132 residue by, e.g. Asn; Fetter et al. 1995) showed that the PR→ F transition still can take place with the same rate as in the wild type oxidase, which implies that the proton that is necessary for F formation can be taken internally from the D pathway (Smirnova et al. 1999). The proton donor was suggested be the Glu286 residue, which was recently confirmed from FTIR studies (Gorbikova et al. 2007). These results show that even though the Glu286 residue is located in the membrane-spanning part of the protein, it can be transiently deprotonated. Furthermore, these results imply that the pKa of Glu286 is dramatically elevated as compared to the solution value, which was also implied from independent FTIR studies (Hellwig et al. 1998; Nyquist et al. 2001).
The pH dependence of the PR→F transition rate, i.e. proton transfer through the D pathway, displays a simple Henderson–Hasselbalch titration with a pKa of 9.4 where the rate is ~1 × 104 s−1 at low pH, and it decreases with increasing pH (Namslauer et al. 2003b). Knowing that Glu286 is an internal proton donor/acceptor, the reaction can be modeled in terms of a rapid (>104 s−1) proton equilibrium between solution and the Glu286, and proton transfer from Glu286 to the catalytic site with a time constant of 104 s−1. In other words, the protonation state of the Glu determines the overall proton-transfer rate, kPF, from solution to the catalytic site.
| (6) |
Expressing this mathematically, the measured rate constant for the PR→F transition can be written as follows.
| (7) |
where αEH is the fraction of protonated Glu286, pKE286 is its pKa and kH is the proton-transfer rate from Glu286 to the catalytic site.
Understanding the proton-pumping mechanism of the oxidase is complicated by the fact that protons are both substrate for the O2-reduction reaction and for the pumping machinery, and both “types” of protons are taken up through the same (D) pathway. In the above-described studies of the PR→F transition, proton transfer from solution to the catalytic site is modeled using Eq. 7 and assumes transfer of only one proton. However, the reaction is also linked to proton pumping (Verkhovsky et al. 1997; Jasaitis et al. 1999; Faxén et al. 2005), which implies that two protons are taken up through the D pathway over the time scale of the PR→F transition. Nevertheless, it appears that the relative rates of the proton transfers are such that the Eq. 7 can correctly describe the process, but nevertheless we have to consider the pumped proton as well.
Separating chemical and pumped protons by using uncoupled mutants
When addressing this problem of the pumping mechanism, a particularly important class of structural variants of the oxidase includes mutants which are able to reduce oxygen to water, but are unable to pump protons (so-called uncoupled mutants). Remarkably, such uncoupling can be achieved through mutation of single amino acid residues. Understanding the nature of uncoupling at a molecular level is expected to contribute towards unraveling the molecular design of the proton-pumping machinery.
There are two classes of uncoupled mutant oxidases, one in which the turnover activity is dramatically decreased due to slowed proton uptake and one in which the turnover activity is similar to that of the wild type oxidase. One example of the first class is the Asp132Asn mutant (Fetter et al. 1995) in which, during oxidase turnover, proton uptake to the D pathway is slowed by two orders of magnitude due to removal of the acidic residue at the orifice of the pathway (Smirnova et al. 1999). Since both the pumped and chemical proton utilize the same input channel, the rate of uptake of both protons during each step of the catalytic cycle must be slowed down to an equal extent. Hence, the relative rates of input of the two protons are not the critical issue. The data can be interpreted to indicate that the value of the rate constant for delivery of the pumped proton must be equal to or faster than some threshold value. There is a limited time-window during which the pumped proton must be available to protonate the pump site. The length of this time-window must be a function of the charge redistribution at the enzyme active site once the electron arrives at heme a and the kinetic barriers for proton flux through the channels become altered.
When attempting to understand the pumping mechanism, the second class of uncoupled mutants (Pfitzner et al. 2000; Namslauer et al. 2003a; Han et al. 2006; Lepp et al. 2008b) is particularly exciting because in this case the proton-transfer rate into and through the D pathway is similar to that of the wild-type oxidase. One member of this class of mutant oxidases is that in which Asn139, ~7 Å from Asp132, is replaced by an Asp residue. The Asn139Asp mutant is able to reduce O2 to water at a rate that is about a factor of two higher than that of the wild-type CytcO and internal electron transfer and proton transfer through the D pathway are not perturbed. The only apparent alteration in the Asn139Asp mutant oxidase is that the apparent pKa (of Glu286) in the pH dependence of the PR→F transition is increased by about ~2 units to ≥11. Furthermore, this mutant oxidase can be re-coupled by introduction of a second mutation, replacing Asp132 by an Asn (Brändén et al. 2006). At the same time, in this double-mutant oxidase, the pKa of Glu286 drops to a value similar to that of the wild type enzyme, which suggests that the Glu286 pKa is critical for maintaining a tightly coupled oxidase. These structural manipulations involve the introduction and removal of charged residues, which readily explain the observed changes in pKa. However, the pump can also be uncoupled without altering any charges (Lepp et al. 2008a). For example, in the uncoupled Asn139Thr mutant oxidase the Glu286 pKa drops by about two units to 7.6 (Lepp et al. 2008b). At present, the explanation for the observed pKa changes is not known, but one possibility is that the water structure in the D pathway changes as a result of the mutations, propagating the effect to Glu286 (Vakkasoglu et al. 2006). Nevertheless, the above-described results indicate that the dynamics of the Glu286 side chain is critical for maintaining a tight coupling (Kaila et al. 2008a, b), where changes in the Glu286 environment are reflected in changes of its pKa value.
Separating chemical protons from pumped protons by isotope effects
Rapid kinetics procedures, including the flow-flash method described in “Separating proton and electron transfer by the flow flash technique” have been used to monitor (1) electron transfer events using optical spectroscopy; (2) proton uptake and release by the enzyme, either incorporated in phospholipid vesicles or in detergent solution, using pH-sensitive dyes; and (3) voltage changes across a membrane into which the enzyme is incorporated, primarily due to protons moving perpendicular to the plane of the membrane. During each of the four electron transfer steps (“Oxygen chemistry and reaction intermediates”) the transfer of the electron transfer triggers a sequence of events resulting in the uptake of two protons and release of the pumped proton. The rates of some of the steps making up the entire process have been shown to the very sensitive to whether the reaction is carried out in H2O or D2O. The primary reason for this is that many proton binding sites within the enzyme are able to exchange protons or deuterons from solution, and then the internal transfer of a deuteron (D+) can be substantially slower than the transfer of a proton (H+). Some processes will be much more sensitive to H/D exchange than others, depending on the details of the process and, of course, whether the H/D transfer is rate limiting. The rates of protein conformational changes, which may depend on the making and breaking of hydrogen (or deuterium) bonds, can also display significant H/D isotope effects.
When the flow-flash reaction is performed in H2O buffer, the rates of proton uptake and release during the PR→F transition are essentially the same, and they appear coincident in time (Salomonsson et al. 2005). This is observed in phospholipid vesicles. When D2O solvent is used in place of H2O, the rate of proton release is slowed down about sevenfold, whereas the rate of proton uptake is slower by only a factor of about 1.5 (Salomonsson et al. 2005). This allows one to clearly observe that the uptake of both the pumped and chemical protons during the PR→F transition can occur prior to proton release. Recall from “Requirements for the proton pump” that the PR→F transition involves no electron transfer, but is due to proton transfer from Glu286 to the heme–copper center. The following step in the flow-flash sequence is the F→OH transition, which is initiated by electron transfer from heme a to the heme–copper center. In this case, the H/D effect is also large (about 7) but proton uptake and release are both slowed down equally. The simplest interpretation is that when the step is initiated by electron transfer, as it would be during normal steady state turnover of the enzyme, the entire process is rate-limited by the release of the pumped proton from the pump site. Why is proton release so sensitive to the H/D kinetic isotope effect? This is not clear, but could be due to a required conformational change of the protein or due to proton transfer occurring through a highly structured series of hydrogen bonds.
H/D kinetic isotope effects have also been useful to separate voltage changes due to the uptake of the pumped proton and chemical proton during the F→OH transition (Siletsky et al. 2004). In this experiment, the protein was initially in the F state, mimicked by a reaction with H2O2, and one electron was rapidly “injected” into the enzyme using a photoreductant. Accompanying electron transfer to the heme–copper center, a voltage is generated across the membrane into which the enzyme is incorporated. Presumably, this voltage is due to uptake of the chemical and pumped protons. There are two phases with similar rates, and these two phases can be distinguished by different H/D effects by performing the experiment in D2O. By comparison of the wild type enzyme with a decoupled mutant that does not pump protons, it was determined that the first phase of proton uptake must be due to the pumped proton.
These studies, together with others, support a general sequence of events:
(1) Electron transfer→(2) uptake of a proton to the pump site from the N-side of the membrane through Glu286→(3) uptake of a proton to the active site from the N-side through Glu286→(4) release of the pumped proton from the pump site to the P-side of the membrane.
Future prospects
Not surprisingly, we know vastly more about cytochrome oxidase today compared with 30 years ago. The X-ray structures have added enormously but by their static nature, have not answered the principle questions of how the rates of proton and electron transfer reactions are regulated at a molecular level so that the enzyme does not waste energy. The fact that a single point mutation at a sight near the surface of this large protein (e.g., Asn139Asp) can completely eliminate proton pumping speaks to degree of fine tuning required for this wonderful molecular machine to operate properly. What are the kinetic barriers that increase and decrease in precise sequence to be sure protons enter and leave in ways that are productive? What is the proton pump site (or sites)? What is the advantage of having two proton-input channels? What is the nature of the so-called “activated” states, OH and EH? These are all questions that can and will be answered. Molecular modelling and the application of computational chemistry is starting to have an impact (Olsson and Warshel 2006; Siegbahn and Blomberg 2007; Xu et al. 2007; Fadda et al. 2008; Kaila et al. 2008a, b; Pisliakov et al. 2008; Sugitani et al. 2008), and this will doubtless increase as the amount of experimental facts provide a solid framework of constraints. Comparisons with prokaryotic heme–copper oxidases that are phylogenetically distant but which still pump protons despite substantial structural differences will be very useful to tease out the essential common features (Pereira et al. 2008). New time-resolved spectroscopic methods appear promising, as is the prospect of trapping intermediates in crystals and determining their structure.
The study of cytochrome oxidase brings together state-of-the-art techniques of modern enzymology with questions of membrane biology. How does nature convert chemical energy into a transmembrane potential? Even though we know much more than 30 years ago, many essential questions remain unanswered. The current state of knowledge is far from the point of diminishing returns where only details remain to be clarified.
Acknowledgements
This work was supported by grants from the National Institutes of Health (HL16101 to RBG), the Swedish Research Council (to PB), the Knut and Alice Wallenberg Foundation (to PB) and the Wenner-Gren Foundations (PB and RBG).
Contributor Information
Peter Brzezinski, Department of Biochemistry and Biophysics, The Arrhenius Laboratories for Natural Sciences, Stockholm University, 106 91 Stockholm, Sweden.
Robert B. Gennis, Email: r-gennis@uiuc.edu, Department of Biochemistry, University of Illinois, 600 South Goodwin Avenue, A320 CLSL, Urbana, IL 61801, USA
References
- Abramson J, Riistama S, et al. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nature Struct Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- Artzatbanov VY, Konstantinov AA, et al. Involvement of intramitochondrial protons in redox reactions of cytochrome a. FEBS Lett. 1978;87:180–185. doi: 10.1016/0014-5793(78)80327-5. [DOI] [PubMed] [Google Scholar]
- Babcock GT, Varotsis C. Discrete steps in dioxygen activation—the cytochrome oxidase/O2 reaction. J Bioenerg Biomemb. 1993;25(2):71–80. doi: 10.1007/BF00762849. [DOI] [PubMed] [Google Scholar]
- Belevich I, Bloch DA, et al. Exploring the proton pump mechanism of cytochrome c oxidase in real time. Proc Natl Acad Sci U S A. 2007;104(8):2685–2690. doi: 10.1073/pnas.0608794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Belevich I, et al. The catalytic cycle of cytochrome c oxidase is not the sum of its two halves. PNAS. 2004;101(2):529–533. doi: 10.1073/pnas.0306036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brändén G, Pawate AS, et al. Controlled uncoupling and recoupling of proton pumping in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103(2):317–322. doi: 10.1073/pnas.0507734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S. The role of the conserved tryptophan272 of the Paracoccus denitrificans cytochrome c oxidase in proton pumping. Biochim Biophys Acta. 2008;1777(7–8):925–928. doi: 10.1016/j.bbabio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Fadda E, Yu CH, et al. Electrostatic control of proton pumping in cytochrome c oxidase. Biochim Biophys Acta. 2008;1777(3):277–284. doi: 10.1016/j.bbabio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Faxén K, Gilderson G, et al. A mechanistic principle for proton pumping by cytochrome c oxidase. Nature. 2005;437:286. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- Ferguson-Miller S, Babcock GT. Heme/copper terminal oxidases. Chem Rev. 1996;7(96):2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- Fetter JR, Qian J, et al. Possible proton relay pathways in cytochrome c oxidase. Proc Natl Acad Sci U S A. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbikova EA, Belevich NP, et al. Time-resolved ATR-FTIR spectroscopy of the oxygen reaction in the D124N mutant of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2007;46(45):13141–13148. doi: 10.1021/bi701614w. [DOI] [PubMed] [Google Scholar]
- Hallén S, Brzezinski P, et al. Internal electron transfer in cytochrome c oxidase is coupled to the protonation of a group close to the bimetallic site. Biochemistry. 1994;33:1467–1472. doi: 10.1021/bi00172a024. [DOI] [PubMed] [Google Scholar]
- Han S, Takahashi S, et al. Time dependence of the catalytic intermediates in cytochrome c oxidase. J Biol Chem. 2000;275(3):1910–1919. doi: 10.1074/jbc.275.3.1910. [DOI] [PubMed] [Google Scholar]
- Han D, Namslauer A, et al. Replacing Asn207 by aspartate at the neck of the D channel in the aa3-type cytochrome c oxidase from Rhodobacter sphaeroides results in decoupling the proton pump. Biochemistry. 2006;45(47):14064–14074. doi: 10.1021/bi061465q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig P, Behr J, et al. Involvement of glutamic acid 278 in the redox reaction of the cytochrome c oxidase from Paracoccus denitrificans investigated by FT-IR spectroscopy. Biochemistry. 1998;37:7390–7399. doi: 10.1021/bi9725576. [DOI] [PubMed] [Google Scholar]
- Hemp J, Robinson DE, et al. Evolutionary migration of a posttranslationally modified active-site residue in the proton-pumping heme–copper oxygen reductases. Biochemistry. 2006;45(51):15405–15410. doi: 10.1021/bi062026u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S, Ostermeier C, et al. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- Jasaitis A, Verkhovsky MI, et al. Assignment and charge translocation stoichiometries of the major electrogenic phases in the reaction of cytochrome c oxidase with dioxygen. Biochemistry. 1999;38:2697–2706. doi: 10.1021/bi982275l. [DOI] [PubMed] [Google Scholar]
- Kaila VR, Verkhovsky M, et al. Prevention of leak in the proton pump of cytochrome c oxidase. Biochim Biophys Acta. 2008a;1777(7–8):890–892. doi: 10.1016/j.bbabio.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Kaila VR, Verkhovsky MI, et al. Glutamic acid 242 is a valve in the proton pump of cytochrome c oxidase. Proc Natl Acad Sci U S A. 2008b;105(17):6255–6259. doi: 10.1073/pnas.0800770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannt A, Soulimane T, et al. Electrical current generation and proton pumping catalyzed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS. 1998;434:17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- Kitagawa T, Ogura T. Oxygen activation mechanism at the binuclear site of heme–copper oxidase superfamily as revealed by time-resolved resonance Raman spectroscopy. Prog Inorg Chem. 1997;45:431–479. [Google Scholar]
- Konstantinov AA, Siletsky S, et al. The roles of the two proton input channels in cytochrome c oxidase from Rhodobacter sphaeroides probed by the effects of site-directed mutations on time-resolved electrogenic intraprotein proton transfer. Proc Natl Acad Sci U S A. 1997;94:9085–9090. doi: 10.1073/pnas.94.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-m, Das TK, et al. Mutations in the putative H-channel in the cytochrome c oxidase from Rhodobacter sphaeroides show that this channel is not important for proton conduction but reveal modulation of the properties of heme a. Biochemistry. 2000;39:2989–2996. doi: 10.1021/bi9924821. [DOI] [PubMed] [Google Scholar]
- Lepp H, Salomonsson L, et al. Impaired proton pumping in cytochrome c oxidase upon structural alteration of the D pathway. Biochim Biophys Acta. 2008a;1777(7–8):897–903. doi: 10.1016/j.bbabio.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepp H, Svahn E, et al. Charge transfer in the K proton pathway linked to electron transfer to the catalytic site in cytochrome c oxidase. Biochemistry. 2008b;47(17):4929–4935. doi: 10.1021/bi7024707. [DOI] [PubMed] [Google Scholar]
- Luna VM, Chen Y, et al. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: structural analysis and role of oxygen transport channels in the heme–Cu oxidases. Biochemistry. 2008;47(16):4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- Muramoto K, Hirata K, et al. A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc Natl Acad Sci U S A. 2007;104(19):7881–7886. doi: 10.1073/pnas.0610031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle JF, Tristam-Nagle S. Hydrogen bonded chain mechanisms for proton conduction and proton pumping. J Membr Biol. 1983;74:1–14. doi: 10.1007/BF01870590. [DOI] [PubMed] [Google Scholar]
- Namslauer A, Aagaard A, et al. Intramolecular proton-transfer reactions in a membrane-bound proton pump: the effect of pH on the peroxy to ferryl transition in cytochrome c oxidase. Bichemistry. 2003a;42:1488–1498. doi: 10.1021/bi026524o. [DOI] [PubMed] [Google Scholar]
- Namslauer A, Pawate A, et al. Redox-coupled proton translocation in biological systems: proton shuttling in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2003b;100(26):15543–15547. doi: 10.1073/pnas.2432106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyquist RM, Heitbrink D, et al. Perfusion-induced redox differences in cytochrome c oxidase: ATR/FT-IR spectroscopy. FEBS Letters. 2001;505:63–67. doi: 10.1016/s0014-5793(01)02769-7. [DOI] [PubMed] [Google Scholar]
- Ogura T, Takahashi S, et al. Time-resolved resonance Raman elucidation of the pathway for dioxygen reduction by cytochrome c oxidase`. J Am Chem Soc. 1993;115:8527–8536. [Google Scholar]
- Olsson MH, Warshel A. Monte Carlo simulations of proton pumps: on the working principles of the biological valve that controls proton pumping in cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103(17):6500–6505. doi: 10.1073/pnas.0510860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier C, Harrenga A, et al. Structure at 2.7 Å resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody Fv fragment. Proc Natl Acad Sci U S A. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawate AS, et al. A mutation in subunit I of cytochrome oxidase from Rhodobacter sphaeroides results in an increase in steady-state activity but completely eliminates proton pumping. Biochemistry. 2002;41:13417–13423. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Santana M, et al. A novel scenario for the evaluation of haem-copper oxygen reductases. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Sousa FL, et al. Looking for the minimum common denominator in haem-copper oxygen reductases: towards a unified catalytic mechanism. Biochim Biophys Acta. 2008;1777(7–8):929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- Pfitzner U, Hoffmeier K, et al. Tracing the D-pathway in reconstituted site-directed mutants of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39(23):6756–6762. doi: 10.1021/bi992235x. [DOI] [PubMed] [Google Scholar]
- Pisliakov AV, Sharma PK, et al. Electrostatic basis for the unidirectionality of the primary proton transfer in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2008;105(22):7726–7731. doi: 10.1073/pnas.0800580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic DM, Stuchebrukhov AA. Proton pumping mechanism and catalytic cycle of cytochrome c oxidase: coulomb pump model with kinetic gating. FEBS Lett. 2004;566:126–130. doi: 10.1016/j.febslet.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Popovic DM, Stuchebrukhov AA. Proton exit channels in bovine cytochrome c oxidase. J Phys Chem B. 2005;109:1999–2006. doi: 10.1021/jp0464371. [DOI] [PubMed] [Google Scholar]
- Proshlyakov DA, Pressler MA, et al. Oxygen activation and reduction in respiration: involvement of redox-active tyrosine 244. Science. 2000;290:1588–1591. doi: 10.1126/science.290.5496.1588. [DOI] [PubMed] [Google Scholar]
- Qin L, Hiser C, et al. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103(44):16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Mills DA, et al. Crystallographic location and mutational analysis of zn and cd inhibitory sites and role of lipidic carboxylates in rescuing proton path mutants in cytochrome C oxidase. Biochemistry. 2007;46(21):6239–6248. doi: 10.1021/bi700173w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich PR. Towards an understanding of the chemistry of oxygen reduction and proton translocation in the iron–copper respiratory oxidases. Aust J Plant Physiol. 1995;22:479–486. [Google Scholar]
- Rich PR, Meunier B, et al. Coupling of charge and proton movement in cytochrome c oxidase. Biochim Biophys Acta. 1996;1275:91–95. [Google Scholar]
- Rich PR, Jünemann S, et al. Protonmotive mechanism of haemcopper oxidases. J Bioenerg Biomembr. 1997;30(1):131–137. doi: 10.1023/a:1020524014920. [DOI] [PubMed] [Google Scholar]
- Riistama S, Hummer G, et al. Bound water in the proton translocation mechanism of the heme–copper oxidases. FEBS Lett. 1997;414(2):275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- Salomonsson L, Faxen K, et al. The timing of proton migration in membrane-reconstituted cytochrome c oxidase. Proc Natl Acad Sci U S A. 2005;102(49):17624–17629. doi: 10.1073/pnas.0505431102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokata K, Katayama Y, et al. The proton pumping pathway of bovine heart cytochrome c oxidase. Proc Natl Acad Sci U S A. 2007;104(10):4200–4205. doi: 10.1073/pnas.0611627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26(6):1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegbahn PE, Blomberg MR. Energy diagrams and mechanism for proton pumping in cytochrome c oxidase. Biochim Biophys Acta. 2007;1767(9):1143–1156. doi: 10.1016/j.bbabio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Siletsky SA, Pawate AS, et al. Transmembrane charge separation during the Ferryl-oxo ∅ oxidized transition in a nonpumping mutant of cytochrome c oxidase. J Biol Chem. 2004;279(50):52558–52565. doi: 10.1074/jbc.M407549200. [DOI] [PubMed] [Google Scholar]
- Siletsky SA, Belevich I, et al. Time-resolved single-turnover of ba3 oxidase from Thermus thermophilus. Biochim Biophys Acta. 2007;1767(12):1383–1392. doi: 10.1016/j.bbabio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Smirnova IA, Ädelroth P, et al. Aspartate-132 in cytochrome c oxidase from Rhodobacter sphaeroides is involved in a two-step proton transfer during Oxo-Ferryl formation. Biochemistry. 1999;38:6826–6833. doi: 10.1021/bi982865j. [DOI] [PubMed] [Google Scholar]
- Soulimane T, Buse G, et al. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19(8):1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani R, Medvedev ES, et al. Theoretical and computational analysis of the membrane potential generated by cytochrome c oxidase upon single electron injection into the enzyme. Biochim Biophys Acta. 2008;1777:1129–1139. doi: 10.1016/j.bbabio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson-Ek M, Abramson J, et al. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Vakkasoglu AS, Morgan JE, et al. Mutations which decouple the proton pump of the cytochrome c oxidase from Rhodobacter sphaeroides perturb the environment of glutamate 286. FEBS Lett. 2006;580(19):4613–4617. doi: 10.1016/j.febslet.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Varotsis C, Zhang Y, et al. Resolution of the reaction sequence during the reduction of O2 by cytochrome oxidase. Proc Natl Acad Sci U S A. 1993;90:237–241. doi: 10.1073/pnas.90.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky MI, Morgan JE, et al. Intramolecular electron transfer in cytochrome c oxidase: a cascade of equilibria. Biochemistry. 1992;31:11860–11863. doi: 10.1021/bi00162a026. [DOI] [PubMed] [Google Scholar]
- Verkhovsky MI, Morgan JE, et al. Oxygen binding and activation: early steps in the reaction of oxygen with cytochrome c oxidase. Biochemistry. 1994;33:3079–3086. doi: 10.1021/bi00176a042. [DOI] [PubMed] [Google Scholar]
- Verkhovsky MI, Morgan JE, et al. Translocation of electrical charge during a single turnover of cytochrome-c oxidase. Biochim Biophys Acta. 1997;1318:6–10. [Google Scholar]
- Wikström M. Proton pump coupled to cytochrome c oxidase in mitochondria. Nature. 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- Wikstrom M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme–copper oxidases. Biochim Biophys Acta. 2007;1767(10):1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Wikström M, Verkhovsky MI, et al. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim Biophys Acta. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- Xu J, Sharpe MA, et al. Storage of an excess proton in the hydrogen-bonded network of the d-pathway of cytochrome C oxidase: identification of a protonated water cluster. J Am Chem Soc. 2007;129(10):2910–2913. doi: 10.1021/ja067360s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S, Shinzawa-Itoh K, et al. X-ray structure and the reaction mechanism of bovine heart cytochrome c oxidase. J Inorg Biochem. 2000;82:1–7. doi: 10.1016/s0162-0134(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Zheng X, Medvedev DM, et al. Computer simulation of water in cytochrome c oxidase. Biochim Biophys Acta. 2003;1557:99–107. doi: 10.1016/s0005-2728(03)00002-1. [DOI] [PubMed] [Google Scholar]