Transcription factor-like proteins without a DNA binding domain, are involved in a potentially ubiquitous layer of transcriptional regulation.
Abstract
Truncated transcription factor-like proteins called microProteins (miPs) can modulate transcription factor activities, thereby increasing transcriptional regulatory complexity. To understand their prevalence, evolution, and function, we predicted over 400 genes that encode putative miPs from Arabidopsis (Arabidopsis thaliana) using a bioinformatics pipeline and validated two novel miPs involved in flowering time and response to abiotic and biotic stress. We provide an evolutionary perspective for a class of miPs targeting homeodomain transcription factors in plants and metazoans. We identify domain loss as one mechanism of miP evolution and suggest the possible roles of miPs on the evolution of their target transcription factors. Overall, we reveal a prominent layer of transcriptional regulation by miPs, show pervasiveness of such proteins both within and across genomes, and provide a framework for studying their function and evolution.
Organismal complexity may arise from increasingly elaborate regulation of gene expression by transcriptional regulatory complexes arising from gene duplication (Levine and Tjian, 2003). In addition, individual protein complexity can evolve by protein domain accretion (Koonin et al., 2000). Are there other ways of increasing regulatory complexity? For example, can domain loss contribute to evolving regulatory complexity? Transcription factor (TF)-like proteins carrying a protein-protein interaction domain (PPID) but lacking a DNA-binding domain (DBD) can regulate TF complexes (Van Doren et al., 1991; Alifragis et al., 1997; Ulmasov et al., 1997; Campuzano, 2001; Kim et al., 2002; Haller et al., 2004; Arata et al., 2006; Noro et al., 2006; Roig-Villanova et al., 2007; Dubos et al., 2008; Hu et al., 2008; Galstyan et al., 2011; Staudt and Wenkel, 2011). Recently coined as microProteins (miPs; Staudt and Wenkel, 2011), they can either prevent or contribute to the formation of active transcription complexes by interacting directly or indirectly with their target TFs (TFs that share sequence similarity to the miPs). For instance, Arabidopsis (Arabidopsis thaliana) LITTLE ZIPPER (ZPR) miPs prevent their target homeodomain (HD) TFs from dimerizing by protein sequestration (Wenkel et al., 2007; Kim et al., 2008). By contrast, Drosophila melanogaster HD-less Homothorax (Hth) forms active complexes with its target TFs (Noro et al., 2006). miPs and their target TFs may not physically interact but share a common third interaction partner. For example, Arabidopsis KNATM miP and KNOTTED1-like (KNOX) TFs compete to interact with BEL1-like (BELL) TFs (Kimura et al., 2008; Magnani and Hake, 2008). To date, miPs have been shown to regulate developmental pathways, hormone signaling, and the circadian clock (Staudt and Wenkel, 2011). Notwithstanding a few examples in the literature, little is known about the origin and prevalence of miP-dependent transcriptional regulation.
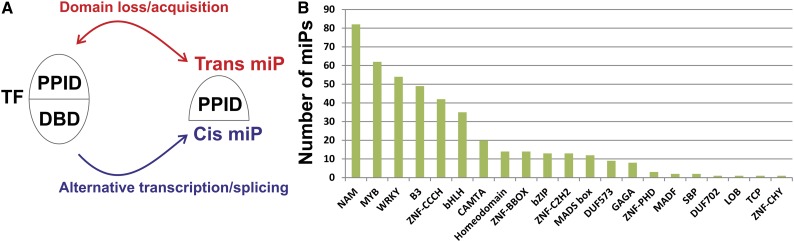
How did miPs arise? miPs might have predated the evolution of their target TFs or evolved from them. In addition, miP/TF couples could have evolved through domain loss or acquisition, or by alternative transcription or splicing. Alternatively, miPs could have arisen by convergent evolution. We define miPs that might have arisen by domain loss/acquisition as trans-miPs because they lie in independent loci from their target TFs, and miPs that are generated by alternative transcription or splicing are defined as cis-miPs (Fig. 1A).
Figure 1.
Distribution of predicted miPs across Arabidopsis TF families. A, Models for miP evolution from TFs. B, Distribution of predicted Arabidopsis miPs, expressed in absolute numbers, across target TF families.
We combined bioinformatics, phylogenetics, molecular genetics, biochemistry, and physiology to understand the prevalence, evolution, and function of miPs.
RESULTS
Prevalence of miPs in Arabidopsis
To study the function and evolution of miPs systematically, we sought all putative miPs in Arabidopsis by searching for proteins whose sequences were similar to those of TFs but were missing DBDs, with additional filters to reduce potential false positives (Supplemental Fig. S1; “Materials and Methods”). A total of 438 putative miPs were identified, of which 12 were cis-miPs (Supplemental Table S1), including all Arabidopsis miPs that have been experimentally characterized to date (Ulmasov et al., 1997; Kim et al., 2002, 2008; Hyun and Lee, 2006; Roig-Villanova et al., 2007; Wenkel et al., 2007; Dubos et al., 2008; Hu et al., 2008; Kimura et al., 2008; Magnani and Hake, 2008; Wang et al., 2009; Zhang et al., 2009; Galstyan et al., 2011). Over 85% (375/439) of putative miP gene models have transcript support by complementary DNA (cDNA) or EST clones (The Arabidopsis Information Resource, http://www.arabidopsis.org). We found putative miPs for 97 of the 1,687 Arabidopsis TFs belonging to 21 of the 62 TF families. Four TF families (NO APICAL MERISTEM, MYB, WRKY, and B3) were targeted by more than 50% of the putative miPs (Fig. 1B).
We asked whether putative miPs tended to interact physically with their target TFs in protein interaction databases (AIMC, 2011; Kerrien et al., 2012; Chatr-Aryamontri et al., 2013). From 18,255 physical interactions among 6,829 Arabidopsis proteins, 38 miPs interacted with 32 predicted target TFs (out of 118 putative miPs with 1,041 physical interactions and 212 predicted target TFs with 1,438 interactions). The frequency of miP/TF interactions in these databases is significantly higher than expected by chance (P = 2.15 × 10–25, binomial distribution). We further tested for physical interactions between 10 predicted miPs and their target TFs in the HD TF family using yeast (Saccharomyces cerevisiae) two-hybrid assays. Six miPs interacted with their predicted HD TF targets (Supplemental Fig. S2). Putative miPs that did not interact with their predicted target TFs might still be valid candidates because they could interact with a partner of their target TFs, similar to KNATM (Kimura et al., 2008; Magnani and Hake, 2008). In line with this hypothesis, eight of the 10 predicted miPs carry a known conserved domain in common with their putative target TFs and might therefore act as agonists or antagonists (Supplemental Fig. S2).
Discovering Functions of Novel miPs
To investigate the range of processes that could be regulated by miPs, we performed a Gene Ontology (GO) enrichment analysis of the predicted target TFs in Arabidopsis. We detected significant enrichments in GO annotations in development, stress response, and metabolism (Supplemental Table S2). To date, most of the miPs that have been described in the literature affect development (Van Doren et al., 1991, 1992; Campuzano, 2001; Haller et al., 2004; Arata et al., 2006; Noro et al., 2006; Hu et al., 2008; Staudt and Wenkel, 2011). To test our predictions and assess the functional roles of novel miPs, we characterized the in vivo activity of two novel miPs, an uncharacterized protein we call LITTLE SIPPER (SPPR; AT4G26920; Zalewski et al., 2013), which we predicted to affect flowering time, and RESPONSE TO ABSCISIC ACID AND SALT1 (RAS1; AT1G09950), a previously characterized negative regulator of salt tolerance (Ren et al., 2010), which we predicted to also regulate immunity against bacterial pathogens.
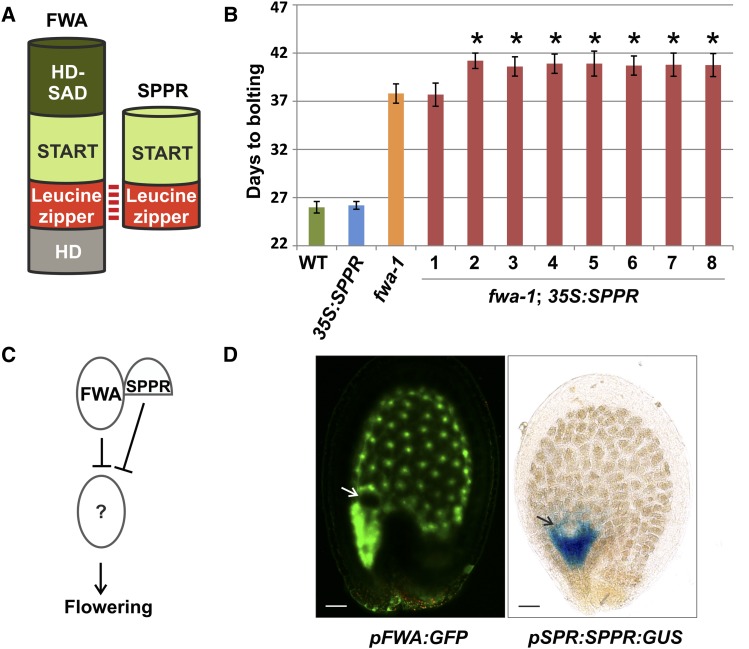
First, we predicted SPPR to regulate class IV HD-Leu zipper (HD-Zip IV) TFs, including FLOWERING WAGENINGEN (FWA). Compared with HD-Zip IV TFs, SPPR carries only the steroidogenic acute regulatory protein-related lipid-transfer (START) and Leu zipper (for protein-protein interaction) domains and lacks the HD (for DNA binding) and HD-START-associated domain (function unknown; Fig. 2A). As predicted, SPPR physically interacted with FWA in yeast two-hybrid assays (Supplemental Fig. S2). Plants carrying the fwa-1 mutation show a late flowering phenotype due to FWA ectopic expression because of hypomethylation in the 5′ region of the gene (Soppe et al., 2000). To test the effect of SPPR on FWA, we overexpressed SPPR using the strong, constitutive 35S promoter in wild-type or fwa-1 backgrounds. Wild-type plants overexpressing SPPR did not show any detectable difference in flowering time (Fig. 2B). By contrast, seven of eight independent transgenic lines overexpressing SPPR in the fwa-1 background flowered significantly later than fwa-1 plants (P value < 0.05, Student’s t test), showing a synergistic interaction between SPPR and FWA (Fig. 2B). Therefore, SPPR might act redundantly with FWA in repressing flowering (Fig. 2C).
Figure 2.
A novel miP regulates flowering time. A, Scheme of FWA and SPPR interacting through the Leu zipper domain. HD-SAD, HD-START-associated domain. The N-terminal region of FWA and SPPR lies at the bottom of the protein schemes. B, Number of days to bolting of wild-type (WT), 35S:SPPR, fwa-1, and eight independent fwa-1;35S:SPPR plants. Error bars indicate 95% confidence interval. Asterisks indicate fwa-1;35S:SPPR transgenic lines that are statistically different from fwa-1 (P < 0.05, Student’s t test). C, Model for SPPR molecular function. D, Arabidopsis seeds at globular embryo stage expressing GFP with the FWA promoter (pFWA:GFP) or GUS as a translational fusion to the SPPR promoter and genomic region (pSPPR:SPPR:GUS). Arrows indicate embryos. Bars = 20 μm.
Reported SPPR and FWA mRNA expression patterns support a potential synergistic relationship. Laser dissection transcriptomics analysis indicated that SPPR is specifically expressed in the seed micropylar endosperm region at the embryo globular stage (Le et al., 2010). We confirmed these results by expressing a translational fusion of the SPPR promoter and genomic region with the GUS reporter gene (pSPPR:SPPR:GUS) in planta. SPPR-GUS was expressed in a subdomain of the FWA expression pattern (pFWA:GFP) that marks all endosperm tissue (Fig. 2D; Kinoshita et al., 2004). The spatial restriction of SPPR-GUS expression relative to that of FWA promoter activity suggests that SPPR may fine-tune FWA’s action in the endosperm.
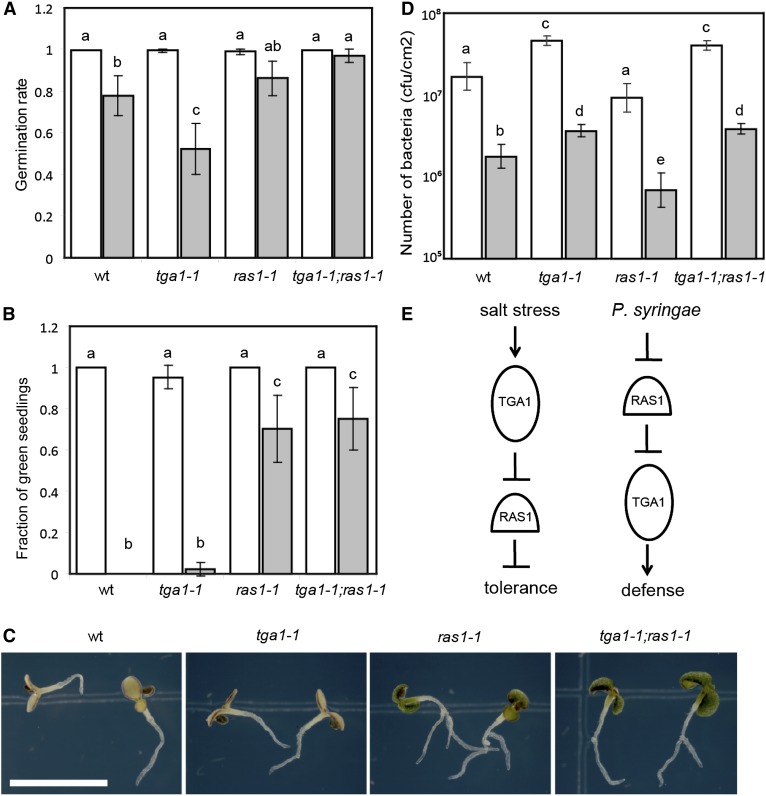
Second, we identified several miPs that were predicted to regulate TFs involved in stress responses, including RAS1 (Supplemental Table S2). RAS1 was originally identified as a novel protein of unknown function that negatively regulates salt tolerance during early seedling development (Ren et al., 2010). RAS1 belongs to a family of small proteins containing the DELAYED IN GERMINATION1 (DOG1) domain (Bentsink et al., 2006; Supplemental Fig. S3). In addition to the RAS1/DOG1 family, the only other proteins that contain the DOG1 domain are TGACG SEQUENCE-SPECIFIC BINDING PROTEIN (TGA) TFs (Gatz, 2013). The presence of DOG1 domain in TGA family was the basis for inferring RAS1/DOG1 family as miPs for TGA TF family in our prediction scheme. Most members of TGA TFs are involved in defense against bacterial pathogens (Gatz, 2013). Therefore, we sought to find the role of TGA TFs in salt tolerance and the role of RAS1 in defense against bacterial pathogens. We first tested the effect of 150 mm NaCl on germination using knockout mutants of TGA genes previously implicated in immunity (tga1-1, tga4-1, tga1-1;tga4-1 double mutant, and tga2-1;tga3-1;tga5-1;tga6-1 quadruple mutant; Kesarwani et al., 2007). Only tga1-1 mutants were significantly more sensitive to salt during germination than the wild type (two-way ANOVA and Tukey’s multiple comparison test, P < 0.05, Fig. 3A; data not shown for tga4-1, tga1-1;tga4-1, and tga2-1;tga3-1;tga5-1;tga6-1 mutants). By contrast, mutants carrying the ras1-1 allele were similar to the wild type in germination and insensitive to salt during seedling greening (Fig. 3, A–C). Interestingly, ras1-1;tga1-1 double mutant lines showed phenotypes similar to ras1-1 (Fig. 3, A–C), indicating that RAS1 may act downstream of TGA1 to achieve salt tolerance in seedlings (Fig. 3E).
Figure 3.
A novel miP-TF pair regulates salt tolerance and SAR. A, Seed germination determined on MS (white bars) or MS + 150 mm NaCl (gray bars) medium. Bars summarize at least nine experiments. B, Green seedling frequency of 16-d-old plants grown on MS (white bars) or MS + 150 mm NaCl (gray bars) medium. Bars summarize nine experiments. C, Sixteen-day-old seedlings on MS + 150 mm NaCl medium. Bar = 5 mm. D, P. syringae pv maculicola ES4326 growth in Arabidopsis leaves primed with 1 mm MgCl2 (white bars) or P. syringae pv tomato DC3000 (avrRpt2; gray bars). Data points represent mean cfu cm–2 of six plants/genotype. A representative result from three experiments is shown. E, Genetic models of TGA1 and RAS1 action. Different letters above bars indicate statistically significant differences between samples (Tukey’s multiple comparison test, P < 0.05). Error bars represent 95% confidence interval.
We then asked whether RAS1 can work with TGA1 in regulating defense against bacterial pathogens. We tested ras1-1, tga1-1, and ras1-1;tga1-1 double mutant lines for systematic acquired resistance (SAR) against Pseudomonas syringae. We found that ras1-1 plants exhibited significantly more SAR than the wild type, whereas tga1-1 plants had less SAR (two-way ANOVA and Tukey’s multiple comparison test, P < 0.05, Fig. 3D). Contrary to salt stress, ras1-1;tga1-1 double mutants showed SAR phenotypes similar to tga1-1 single mutants (Fig. 3D), suggesting RAS1 acts upstream of TGA1 during defense (Fig. 3E). We did not detect physical interaction between RAS1 and TGA1 in yeast or Nicotiana benthamiana (data not shown), suggesting that RAS1 may interact with a partner of TGA1. These results indicate that RAS1 and TGA1 work together to regulate salt tolerance and immunity, although the mechanism of their action appears to be different for these two responses. To date, RAS1 is the first example of an miP regulating stress response in any organism.
Evolution of miPs from TFs
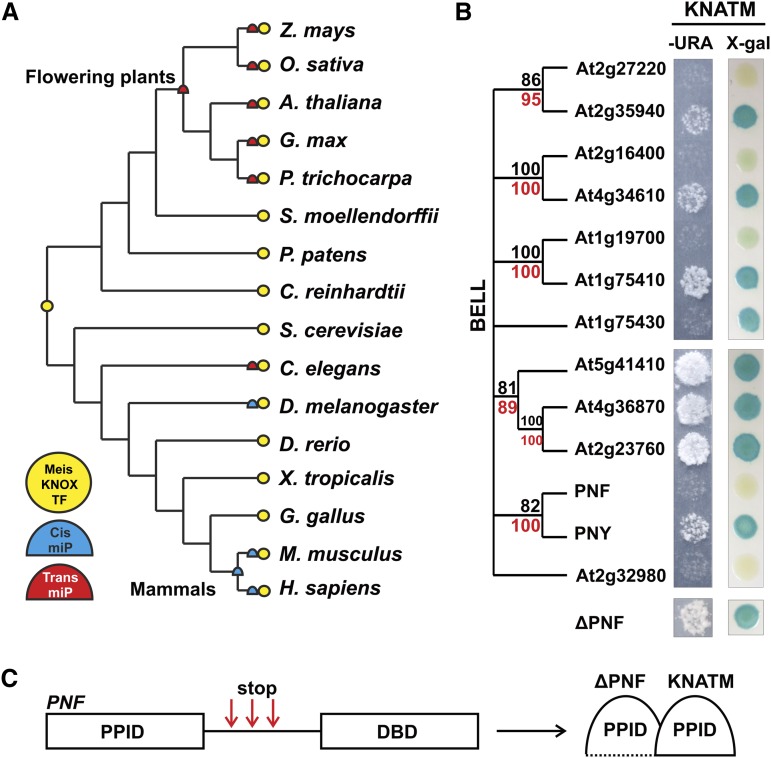
The potential prevalence of miP-mediated transcriptional regulation led us to ask how miPs might have evolved. We focused on the three-amino acid loop extension (TALE) family of HD proteins because it is the only TF family homologous among plants, fungi, and metazoans (Bürglin, 1997) known to be regulated by miPs (Yang et al., 2000; Arata et al., 2006; Noro et al., 2006; Kimura et al., 2008; Magnani and Hake, 2008), which allows tracing of their evolutionary history to the divergence of plants from metazoans. If miPs played a role in increasing biological complexity, they would have evolved after their target TFs. Alternatively, miPs might be ancestral forms that led to the evolution of TFs after the acquisition of a DBD. We predicted miPs and their target TALE TFs from 16 organisms belonging to taxa relevant for studying the evolution of plants and metazoans (Supplemental Table S3; “Materials and Methods”). In addition to the already characterized miPs (Yang et al., 2000; Arata et al., 2006; Noro et al., 2006; Kimura et al., 2008; Magnani and Hake, 2008), we identified homologs of KNATM in monocotyledonous plants.
We inferred maximum parsimony traces of the Myeloid ecotropic viral integration site (Meis)/KNOX miP characters along the phylogenetic tree, which indicate that Meis/KNOX trans-miPs evolved independently in flowering plants and Caenorhabditis elegans, and cis-miPs evolved independently in D. melanogaster and mammals (Fig. 4A). The phylogenetic distribution and nature of TALE miPs suggest that they arose from ancestral TFs independently, and possibly several times, in plant and metazoan lineages. In line with the independent evolution hypothesis, plant and metazoan TALE miPs have opposite effects on their target TFs. For example, the Arabidopsis KNATM miP inhibits its target TF, whereas the D. melanogaster HD-less Hth miP works as a coactivator (Noro et al., 2006; Kimura et al., 2008; Magnani and Hake, 2008). However, we cannot exclude the possibility that miPs might have been lost in several lineages during evolution or that poor genome annotations precluded the discovery of miPs in such lineages.
Figure 4.
Functional and phylogenetic analyses of miPs and target TALE HD TFs in plants and metazoa. A, Phylogenetic distribution of miPs affecting Meis/KNOX TFs (yellow circle), cis-miPs (blue semicircle), and trans-miPs (red semicircle). B, Yeast two-hybrid analysis of the Arabidopsis KNATM miP (bait) and BELL TF (prey) protein interaction assessed with URA3 (–URA) and lacZ (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside [X-gal]) reporter genes. Bootstrap values are based on POX PPIDs (black) and full-length proteins (red). C, Schematic of PNF gene indicating positions of premature stop-codon mutations (red arrows) that allowed PNF lacking the DBD to interact with KNATM in a reverse yeast two-hybrid experiment. ΔPNF, PNF lacking the DBD.
To further investigate evolution of miPs by domain loss, we examined four additional miP/TF couples in plants (ZPRs, SPPR, MINI ZINC FINGERs [MIFs], and RAS). Similar to KNATM, ZPR and SPPR appeared after their target TFs in plants, suggesting that these miPs evolved from the TFs by domain loss (Supplemental Fig. S4). We could not infer the origin of RAS and MIF miPs relative to their target TFs because both miP and TF families are found in Physcomitrella patens but not in Chlamydomonas reinhardtii (Supplemental Fig. S4). More genome sequences, particularly those from Charophytes that are thought to be the green algae that gave rise to land plants (McCourt et al., 2004), are needed to resolve the evolution of MIF and RAS miPs relative to their target TFs. Overall, our data suggest that the regulation of TALE, HD-Zip III, and HD-Zip IV TFs by miPs evolved relatively recently and followed the evolution of the target TFs. Therefore, we provide evidence that at least three classes of miPs associated with transcriptional regulation contributed to biological complexity by domain loss.
Implication of miPs on TALE TF Evolution
Did the emergence of TALE miPs from the TFs affect the function and evolution of the TFs? To address this, we first asked whether the Arabidopsis KNATM miP interacted with all of its putative BELL TF targets. BELL TF family has five pairs of homologs, likely products of the last two rounds of genome-wide duplication, approximately 70 and 30 million years ago (Fig. 4B; Blanc et al., 2003). KNATM selectively interacted with only one BELL TF paralog protein in four of five pairs of BELL paralogs (Fig. 4B). By contrast, large-scale interaction analyses of Arabidopsis KNOX full-length TFs did not show such selectivity for BELL pairs of paralogs (Hackbusch et al., 2005). These data suggest that members of BELL paralogs subfunctionalized away from KNATM regulation independently. To find where selectivity for KNATM lies in BELL TFs, we performed a reverse yeast two-hybrid experiment with POUNDFOOLISH (PNF), a BELL protein that does not interact with KNATM. We randomly mutagenized PNF and selected for mutations that allowed PNF to interact with KNATM. All positive PNF mutants carried a premature stop codon right after the PNF POX PPID (Fig. 4C; Supplemental Fig. S5). The truncated PNF mutant proteins interacted with KNATM as strongly as PNF's paralog PENNYWISE (Fig. 4B). Therefore, TF selectivity for KNATM does not lie in the interaction domain but possibly in protein conformation or in the C-terminal region. Also, KNATM preferentially interacts with the BELL TF that is more highly expressed in each paralogous pair, as detected by microarray analyses (Schmid et al., 2005). BELL subfunctionalization by reduction in expression might have relieved them from purifying selection, allowing for increased protein divergence.
Similar to KNATM, PHASMID SOCKET ABSENT3 (PSA3), a C. elegans TALE miP, interacts with only one of the three PBC TF homologs (Supplemental Fig. S2). One possible interpretation of these results is that KNATM and PSA3 miPs might play a role in regulating protein dosage in the formation of protein complexes. According to the gene dosage balance hypothesis (Papp et al., 2003), the subunit stoichiometry of protein complexes must be maintained for their correct biological function. The subfunctionalization of only one paralog protein in most of KNATM and PSA3 target TF paralog pairs might have arisen to address dosage imbalance in TALE HD TF complexes. Further investigation into the function of diverse miPs will provide more insight into the mechanism(s) that govern their evolution.
DISCUSSION
Here, we provide a systematic framework to study the role of miPs in gene regulation. This framework allowed us to group miPs and putative target TFs in the same functional and evolutionary context, discover novel miPs and biological pathways controlled by miPs, and begin to address possible origins of miPs.
Of the approximately 400 putative miPs in Arabidopsis, most are uncharacterized. However, we identified a few well-known proteins as miPs; these regulate a diverse array of processes but had not been considered previously in the miP context. For example, we found AUXIN/INDOLE3-ACETIC ACID proteins as miPs for AUXIN RESPONSE FACTOR TFs involved in plant hormone auxin signaling, MIF proteins as miPs for zinc finger-HD family of TFs involved in meristem development, and PHYTOCHROME RAPIDLY REGULATED1 (PAR1), PAR2, and LONG HYPOCOTYL IN FAR RED1 as miPs for the basic helix-loop-helix TF family involved in shade avoidance and phytochrome signaling (Ulmasov et al., 1997; Kim et al., 2002; Hu et al., 2008; Galstyan et al., 2011). Therefore, we placed several previously characterized transcriptional regulators under the same umbrella of miPs.
Our designation of miPs as putative regulators of TFs and prediction of their putative TF targets may help elucidate functions of both the miPs and TFs. For example, RAS1 is a protein of unknown molecular function that is involved in salt tolerance during early seedling development (Ren et al., 2010). It was originally identified as a quantitative trait locus (QTL) contributing to variation in seedling greening between Arabidopsis accessions. RAS1 belongs to a small family that has eight members in Arabidopsis, which includes another QTL gene DOG1 (Bentsink et al., 2006). DOG1 controls variation in germination between Arabidopsis accessions. Besides RAS1 and DOG1, the rest of the RAS1/DOG1 family members remain uncharacterized. It is intriguing that both RAS1 and DOG1 were isolated as genes contributing to QTL, which supports the notion that miPs are more rapidly evolving than their TF targets. Because miPs can fine-tune the action of TFs, they may be adaptive in general. It remains to be seen how frequently miPs exist as QTLs in plants.
The conserved protein sequence in the RAS1/DOG1 and TGA family has been designated as the DOG1 domain (Bentsink et al., 2006). This domain is plant specific and is conserved in all sequenced land plants (data not shown). The function of the DOG1 domain is unknown. However, based on the concept of miPs, we predict it to be involved in protein interactions. It is possible that RAS1 and TGA1 may interact physically or, alternatively, interact with a third partner, similar to how BELL TFs interact with the MEINOX domain of both the KNATM miP and KNOTTED TFs (Kimura et al., 2008; Magnani and Hake, 2008). We did not find evidence for physical interactions between RAS1 and TGA1 in yeast or in planta (data not shown), suggesting that RAS1 and TGA1 may share a third partner. We currently do not know if other members of the RAS1/DOG1 family also regulate members of the TGA family. If RAS1 modulates TGA1’s activity via interacting with a third partner, other members of the third partner's family may interact with other members of the RAS1/DOG1 and TGA families.
Genetic evidence shows RAS1 to be a negative regulator of both salt tolerance during early seedling development and defense against bacterial pathogens; mutants carrying a knockdown allele of RAS1, ras1-1, is more tolerant to salt treatment and resistant to bacterial growth (Fig. 3, A and D). Contrarily, TGA1 is a positive regulator of both salt tolerance and pathogen defense; mutants carrying a null allele of TGA1, tga1-1, are more sensitive to both salt treatment and bacterial pathogen growth (Fig. 3, A and D). Interestingly, double mutants carrying both mutations show different responses to salt stress and pathogen attack. Double mutants of tga1-1;ras1-1 are more resistant to salt, similar to ras1-1, whereas the double mutants are more sensitive to pathogen attack, similar to tga1-1. The nature of this apparently opposite behavior of the genetic interactions in the two processes and developmental stages remains to be elucidated. One possibility is that RAS1 and TGA1 may interact with a different partner in response to salt stress during early seedling development and bacterial pathogens in mature leaves. The identification of the possible third partner and the molecular mechanism of the actions of RAS1 and TGA1 may help elucidate the relationships between biotic and abiotic stress tolerance mechanisms in plants.
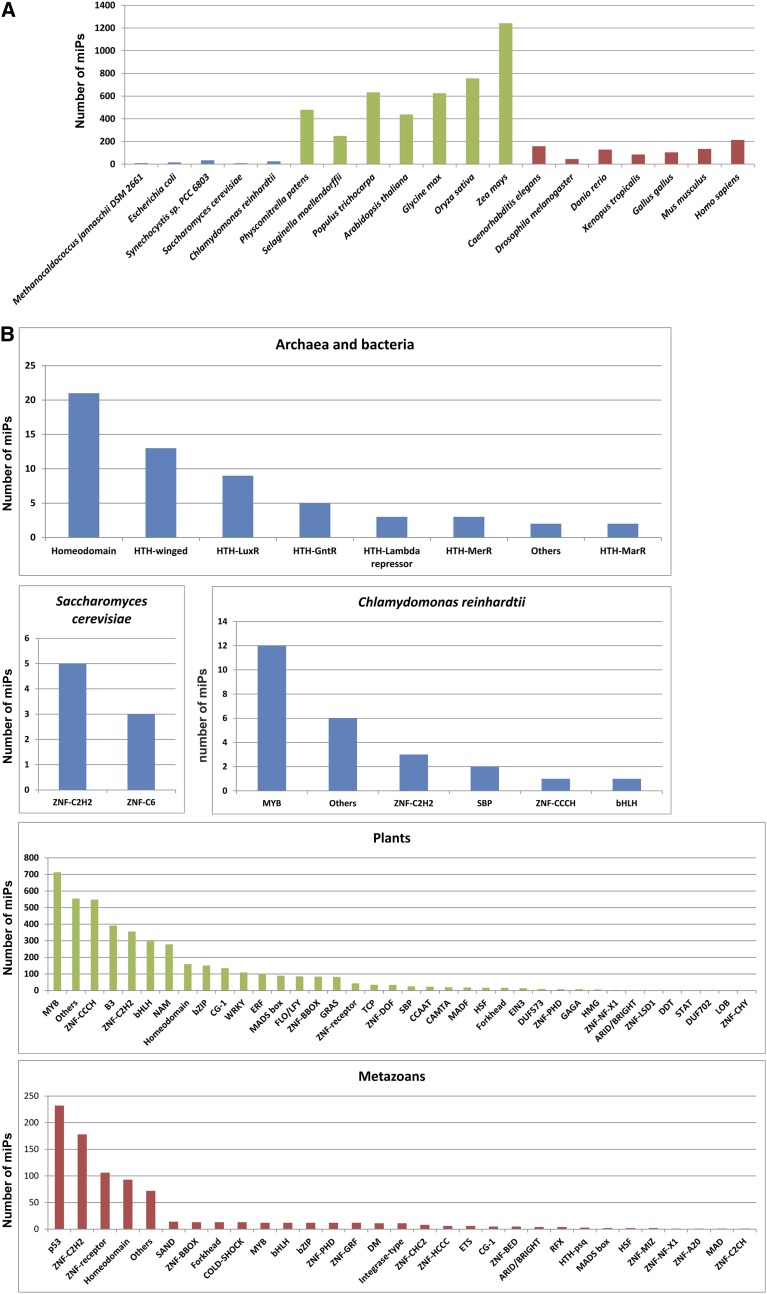
Our study suggests that miP-mediated gene regulation is widespread because it affects several organisms and biological pathways. Therefore, we used the same approach to predict miPs and their target TFs in 19 organisms across three domains of life, including species belonging to archaea, prokaryotes, fungi, plants, and metazoans. We detected miPs in all organisms examined and for almost all TF families (Fig. 5; Supplemental Table S4). A GO enrichment analysis of the predicted target TFs revealed enrichment in developmental (Arabidopsis and D. melanogaster), stress response (Arabidopsis), and metabolic processes (Arabidopsis, Mus musculus, and Homo sapiens; Supplemental Table S2). Our analysis suggests a potentially ubiquitous layer of transcriptional regulation by miPs and lays a foundation for identification and future analysis of novel miPs. Finally, miPs might also offer new tools to modulate TF function in human gene therapy and plant bioengineering (Seo et al., 2012) by exploiting existing miPs or by creating synthetic ones.
Figure 5.
Distribution of putative miPs across taxa and TF families. A, Distribution of putative miPs (expressed in absolute numbers) predicted to affect TFs across microbes (blue bars), plants (green bars), and animals (red bars). B, Distribution of putative miPs across target TF families in archaea and bacteria (Methanocaldococcus jannaschii DSM 2661, Escherichia coli, and Synechocystis sp. PCC 6803), yeast, C. reinhardtii, plants (P. patens, Selaginella moellendorffii, Oryza sativa, Populus trichocarpa, Arabidopsis, Glycine max, and Zea mays), and metazoans (C. elegans, D. melanogaster, Danio rerio, Xenopus tropicalis, Gallus gallus, M. musculus, and H. sapiens).
MATERIALS AND METHODS
Bioinformatics
Supplemental Figure S1 shows the overall flow of the algorithm we used to predict miPs and their putative target TFs, which consists of two major steps: finding proteins that share sequence similarity with TFs and filtering out potential false positives. The handful of characterized miPs are shorter than 200 amino acids in length and are therefore difficult to identify by sequence similarity with their target TFs through a stringent BLAST analysis. To overcome this problem, we created two protein databases for each organism under analysis: one comprising all proteins (allDB), for a more stringent analysis, and the other one including only proteins smaller than 200 amino acids in length (smallDB), for a less stringent search. The algorithm uses TFs as query in two BLASTP searches against the allDB and smallDB with a low (0.01) and high (0.5) e-value cutoffs, respectively. To remove potential false positives from the less stringent BLASTP analysis against smallDB, the algorithm filters the hits that do not match TFs using a reverse BLASTP search, with an e-value of 1E-23, against the allDB. The resulting protein data sets are filtered for two intrinsic miP features; by our definition, miPs do not carry a DBD and are not larger or bear different domains than their target TFs. The algorithm removes proteins carrying a DBD by performing a Position-Specific Iterated BLAST search against the Pfam domain database (Bateman et al., 2004). Furthermore, the algorithm discards proteins longer than their target TFs, with a 10% tolerance, and proteins carrying domains different from their targets by comparing domain architectures using the InterProScan software (Zdobnov and Apweiler, 2001). To further reduce the number of potential false positives, we applied an absolute length filter of 500 amino acids because all known miPs and the average PPID are considerably smaller (Xia et al., 2008).
We chose genomes to analyze based on the completed sequence, phylogenetic position, and genome size. Representatives from all kingdoms were selected. We downloaded archaea, prokaryotic, fungal, and metazoan genome sequences (Methanocaldococcus jannaschii DSM 2661, Escherichia coli strain K-12 [substrain MG1655], Synechocystis sp. PCC 6803, yeast [Saccharomyces cerevisiae], Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Xenopus tropicalis, Gallus gallus, Mus musculus, and Homo sapiens) from the National Center for Biotechnology Information’s Genome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/), whereas algal and plant genomes (Chlamydomonas reinhardtii, Physcomitrella patens, Selaginella moellendorffii, Oryza sativa, Populus trichocarpa, Arabidopsis [Arabidopsis thaliana], and Glycine max), except Zea mays, were downloaded from Phytozome (http://www.phytozome.org). The Z. mays genome sequence was downloaded (the filtered set) from http://www.maizesequence.org/. We downloaded plant TF sequences from Plant TFDB (http://planttfdb.cbi.pku.edu.cn/) and prokaryotic and metazoan TF sequences from the DBD database (http://www.transcriptionfactor.org/). We classified TFs into families as follows: We collected the DBDs from the Interpro database (Hunter et al., 2012) using both manual curation and domain mapping to TF proteins in the 19 species we used in this study. We then classified the DBDs based on conventions used by the DBD database (Kummerfeld and Teichmann, 2006) and BioBase (http://www.biobase-international.com/). In total, we used 579 DBDs from Interpro to classify TFs into 128 families (Supplemental Table S5). Arabidopsis TFs that did not contain DBDs were manually removed.
We performed multiple sequence alignments using Muscle 3.2 and displayed them with Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) using the BLOSUM62 matrix and a 51% threshold for shading (Edgar, 2004). We constructed phylogenetic trees using the neighbor-joining algorithm implemented in the MEGA software suite (Tamura et al., 2007). We used mean character difference, among-site rate variation, and random seed initiation; 10,000 bootstrap replicates were performed followed by identification of the consensus tree.
GO enrichment analyses were conducted using DAVID and a P value of 0.01 as the statistical cutoff (Dennis et al., 2003).
Molecular Biology
Coding sequences were PCR amplified from Arabidopsis and C. elegans cDNA libraries (kindly provided by Dr. Kathy Barton’s laboratory, Carnegie Institution, and Prof. Kang Shen’s laboratory, Stanford University, respectively) and cloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer’s instructions. The SPPR genomic sequence, including the upstream intergenic region, was PCR amplified from Arabidopsis genomic DNA and cloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer’s instructions. The pFWA:GFP marker line was described previously (Kinoshita et al., 2004). For yeast two-hybrid assays, sequences were mobilized from pENTR/D-TOPO into pDEST22 (Invitrogen) and pDEST32 (Invitrogen) according to the manufacturer’s instructions. For overexpression and GUS fusion analyses, sequences were mobilized from pENTR/D-TOPO into pMDC32 and pMDC163, respectively (Curtis and Grossniklaus 2003). β-Glucuronidase activity was assayed as described previously (Jefferson 1989).
Yeast Two-Hybrid Assays
The Invitrogen ProQuest yeast two-hybrid system with Gateway Technology was used according to the manufacturer’s instructions. Positive interactions were assessed with the URA3 (encoding for Orotidine-5′-phosphate decarboxylase) and the lacZ (encoding for β-galactosidase) reporter genes. More than six independent colonies per pair of constructs were tested. Each construct was tested individually, and no self-activation was detected. To assess the interaction strength of protein pairs, the collection of control strains that contains plasmid pairs expressing fusion proteins with a spectrum of interaction strengths was used. +++, ++, and + correspond to interactions similar in strength to those observed with pPC97-Fos/pPC86-Jun, pPC97-CYH2–dDP/pPC86-dE2F, and pPC97-RB/pPC86-E2F1, respectively. For the reverse yeast two-hybrid screen, the PNF coding sequence was mutagenized by PCR with Mutazyme (Stratagene). The PCR products were then cotransformed with pDEST32-PNF and digested with NotI and AscI restriction enzymes into yeast cells carrying pDEST22-ΔKNATMB (Magnani and Hake, 2008).
Plant and Genetic Materials
Seeds of Arabidopsis ecotypes Columbia (Col-0) or Landsberg erecta (Ler) were used for all experiments. The mutant carrying the fwa-1 allele in Ler was previously described (Soppe et al., 2000). Mutants carrying the tga1-1, tga2-2, tga3-1, tga4-1, tga7-1, tga1-1;tga4-1, and tga2-1;tga3-1;tga5-1;tga6-1 alleles in Col-0 were kindly provided by Dr. Xinnian Dong (Duke University). The mutant carrying the ras1-1 allele (SALK_058470C) in Col-0 was obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). Lines homozygous for ras1-1 allele were confirmed by PCR. Lines homozygous for both the ras1-1 and tga1-1 alleles were obtained by crossing the single mutants and confirming genotypes by PCR in the F2 population.
Flowering Time Assays
Arabidopsis Ler plants (wild type, fwa-1, and T3 generation fwa-1;35S:SPPR) were grown in the greenhouse under long-day conditions for the flowering time assays. More than 30 plants for each genotype were analyzed. Days to bolting were determined when the main inflorescence reached 1 cm in length.
Salt Tolerance Assays
Arabidopsis Col-0 seeds (wild type, ras1-1, tga1-1, and ras1-1;tga1-1) were surface sterilized with 15% (v/v) household bleach with approximately 0.2% (v/v) Tween 20, washed five times in sterilized water as described previously (Lee et al., 2010), and sown on Murashige and Skoog medium (MS) plates containing 0.8% agar and 3% (w/v) Suc. Salt plates were supplemented with 150 mm NaCl. After stratification at 4°C for 3 d, the seeds on the agar plates were placed in a growth chamber (22°C, 24 h of light at 40 µE m–2 s–1 fluorescent illumination). Germination rate was determined at 96 h after the plates were placed in the growth chamber. Sixteen days after transfer to the growth chamber, the frequency of green seedlings was counted. Germination assay was conducted in at least nine independent experiments in which 108 to 204 seeds per genotype per condition were assayed for each experiment. Green seedling assay was conducted in nine independent experiments, where 100 to 108 seeds per genotype per condition were assayed for each experiment. Statistical analysis for the germination rate assay was performed using IBM’s SPSS statistics version 20. Briefly, two-way ANOVA (P < 0.05) was performed to determine the effect of genotype and salt treatment on germination rate. To discriminate the means, one-way ANOVA with Tukey’s multiple comparison test (P < 0.05) was performed.
SAR Assays
Arabidopsis Col-0 plants (wild type, ras1-1, tga1-1, and ras1-1;tga1-1) were grown in pots in Pro-Mix soil (Premier Horticulture) in a growth chamber (22°C, 80% relative humidity, 125 µE m–2 s–1 fluorescent illumination) on a 10-h-light/14-h-dark cycle. Fully expanded leaves of 4- to 5-week-old plants were used for the SAR assay. SAR assay was performed as described by Shearer et al. (2012) with modification. Pseudomonas syringae pv tomato DC3000 avirulence Rpt2 [avrRpt2]) and P. syringae pv maculicola ES4326 were grown on nutrient yeast glycerol agar medium at 28°C. Three leaves were hand inoculated by complete infiltration of the leaf tissue with 1 mm MgCl2 or a 1 × 106 colony forming units (cfu) mL–1 suspension of P. syringae pv tomato DC3000 (avrRpt2) in 1 mm MgCl2 using a needleless syringe. Two days later, remaining leaves were infiltrated with a 2 × 105 cfu mL–1 suspension of P. syringae pv maculicola ES4326. Leaf discs per treatment per time point were collected at the time of infiltration and 3 d postinfiltration, ground in 1 mm MgCl2, and spotted on nutrient yeast glycerol agar medium plates in triplicate to determine the bacterial titer. Six biological replicates were used for each experiment, and the experiment was repeated at least three times with similar results. The average bacterial titer ± 95% confidence interval from a representative experiment is reported. The statistical analysis was performed using IBM’s SPSS statistics version 20. Briefly, two-way ANOVA (P < 0.05) was performed to determine the effect of genotype and pretreatment on bacterial growth. To discriminate the means, one-way ANOVA with Tukey’s multiple comparison test (P < 0.05) was performed.
Transgenic Plants
For stable transformation, Agrobacterium tumefaciens strain GV3101 was used to transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). mRNA abundance was analyzed by semiquantitative reverse transcription PCR as previously described (Sambrook and Russell, 2006). SPPR transcripts were amplified using SPPR-F (TAGCAACGCTCGAGTCTCCT) and SPPR-R (TAGCAACGCTCGAGTCTCCT) primers. ACTIN2 (At3g18780) transcript was used as standard and amplified with Actin-forward (TGGTCGTACACCCGGTATTGTGCTGGATT) and Actin-reverse (TGTCTCTTACAATTTCCCGCTCTGCTGTT) primers.
Microscopy
GFP fluorescence in Arabidopsis seeds was detected using a Leica SP5 confocal laser-scanning microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Tables S1 and S4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Flow chart of the algorithm used to predict miPs.
Supplemental Figure S2. Protein-protein interactions between putative miPs and their predicted target HD TFs from Arabidopsis and C. elegans.
Supplemental Figure S3. Scheme of TGA and RAS1 domain architectures.
Supplemental Figure S4. Phylogenetic analyses of miPs in plants.
Supplemental Figure S5. KNATM selectivity for BELL paralog TFs.
Supplemental Table S1. Putative miPs and their predicted target TFs for Arabidopsis.
Supplemental Table S2. GO enrichment analysis.
Supplemental Table S3. Putative miPs predicted to target TALE Meis/KNOX TFs.
Supplemental Table S4. Putative miPs and their predicted target TFs across kingdoms.
Supplemental Table S5. TF classification.
Supplementary Material
Acknowledgments
We thank Flavia Bossi for testing RAS1-TGA1 physical interactions; Meng Xu for gathering protein interaction data from public databases and discussions on statistical analyses; members of the Rhee, Barton, and Evans labs for discussions; José R. Dinneny and Sarah Hake for comments on the manuscript; Kang Shen for providing the C. elegans cDNA library; Xinnian Dong for providing tga1-1, tga2-2, tga3-1, tga4-1, tga7-1, and tga1-1;tga4-1 double mutants and the tga2-1;tga3-1;tga5-1;tga6-1 quadruple mutants; and Kathy Barton for sharing resources.
Glossary
- TF
transcription factor
- PPID
protein-protein interaction domain
- DBD
DNA-binding domain
- miP
microProtein
- HD
homeodomain
- cDNA
complementary DNA
- GO
Gene Ontology
- SAR
systematic acquired resistance
- TALE
three-amino acid loop extension
- QTL
quantitative trait locus
- allDB
database of all proteins
- smallDB
database of proteins smaller than 200 amino acids in length
- Col-0
ecotype Columbia
- Ler
ecotype Landsberg erecta
- MS
Murashige and Skoog
- cfu
colony forming units
- MS
Murashige and Skoog medium
Footnotes
This work was supported by the Carnegie Institution for Science (to S.Y.R., N.d.K., H.-I.N., K.P.), the Life Sciences Research Foundation (Howard Hughes Medical Institute Fellowship to E.M.), the National Science Foundation (grant no. 0929413 to K.P.), and the National Institutes of Health (grant no. 2R01 GM068886–06A1 to M.B.M.).
The online version of this article contains Web-only data.
References
- Alifragis P, Poortinga G, Parkhurst SM, Delidakis C. (1997) A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc Natl Acad Sci USA 94: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata Y, Kouike H, Zhang Y, Herman MA, Okano H, Sawa H. (2006) Wnt signaling and a Hox protein cooperatively regulate psa-3/Meis to determine daughter cell fate after asymmetric cell division in C. elegans. Dev Cell 11: 105–115 [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25: 4173–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S. (2001) Emc, a negative HLH regulator with multiple functions in Drosophila development. Oncogene 20: 8299–8307 [DOI] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, et al. (2013) The BioGRID interaction database: 2013 update. Nucleic Acids Res 41: D816–D823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J, Martinez-Garcia JF. (2011) The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J 66: 258–267 [DOI] [PubMed] [Google Scholar]
- Gatz C. (2013) From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol Plant Microbe Interact 26: 151–159 [DOI] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF. (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 102: 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller K, Rambaldi I, Daniels E, Featherstone M. (2004) Subcellular localization of multiple PREP2 isoforms is regulated by actin, tubulin, and nuclear export. J Biol Chem 279: 49384–49394 [DOI] [PubMed] [Google Scholar]
- Hu W, dePamphilis CW, Ma H. (2008) Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. J Integr Plant Biol 50: 1031–1045 [DOI] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–D312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Lee I. (2006) KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. (2012) The IntAct molecular interaction database in 2012. Nucleic Acids Res 40: D841–D846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X. (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Woo JC, Song PS, Soh MS. (2002) HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J 30: 711–719 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Koenig D, Kang J, Yoong FY, Sinha N. (2008) Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L, Kondrashov AS. (2000) The impact of comparative genomics on our understanding of evolution. Cell 101: 573–576 [DOI] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA. (2006) DBD: a transcription factor prediction database. Nucleic Acids Res 34: D74–D81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. (2010) Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R. (2003) Transcription regulation and animal diversity. Nature 424: 147–151 [DOI] [PubMed] [Google Scholar]
- Magnani E, Hake S. (2008) KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG. (2004) Charophyte algae and land plant origins. Trends Ecol Evol 19: 661–666 [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. (2006) Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev 20: 1636–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Ren Z, Zheng Z, Chinnusamy V, Zhu J, Cui X, Iida K, Zhu JK. (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 107: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF. (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2006) Amplification of cDNA generated by reverse transcription of mRNA. CSH Protoc 2006 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Hong SY, Ryu JY, Jeong EY, Kim SG, Baldwin IT, Park CM. (2012) Targeted inactivation of transcription factors by overexpression of their truncated forms in plants. Plant J 72: 162–172 [DOI] [PubMed] [Google Scholar]
- Shearer HL, Cheng YT, Wang L, Liu J, Boyle P, Després C, Zhang Y, Li X, Fobert PR. (2012) Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol Plant Microbe Interact 25: 1459–1468 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Staudt AC, Wenkel S. (2011) Regulation of protein function by “microProteins.” EMBO Rep 12: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. (1991) The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development 113: 245–255 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. (1992) Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev 6: 2592–2605 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J. (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. (2007) A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Fu Z, Hou L, Han JD. (2008) Impacts of protein-protein interaction domains on organism and network complexity. Genome Res 18: 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hwang CK, D’Souza UM, Lee SH, Junn E, Mouradian MM. (2000) Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J Biol Chem 275: 20734–20741 [DOI] [PubMed] [Google Scholar]
- Zalewski CS, Floyd SK, Furumizu C, Sakakibara K, Stevenson DW, Bowman JL. (2013) Evolution of the class IV HD-zip gene family in streptophytes. Mol Biol Evol 30: 2347–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.