MicroRNA-regulated growth-regulating factors activate expression of specific targets to regulate floral organ development, affecting characteristics such as husk openness and sterile lemma length.
Abstract
Inflorescence and spikelet development determine grain yields in cereals. Although multiple genes are known to be involved in the regulation of floral organogenesis, the underlying molecular network remains unclear in cereals. Here, we report that the rice (Oryza sativa) microRNA396d (OsmiR396d) and its Os Growth Regulating Factor (OsGRF) targets, together with Os Growth Regulating Factor-Interacting Factor1 (OsGIF1), are involved in the regulation of floral organ development through the rice JMJD2 family jmjC gene 706 (OsJMJ706) and crinkly4 receptor-like kinase (OsCR4). Transgenic knockdown lines of OsGRF6, a predicted target of OsmiR396d, and overexpression lines of OsmiR396d showed similar defects in floral organ development, including open husks, long sterile lemmas, and altered floral organ morphology. These defects were almost completely rescued by overexpression of OsGRF6. OsGRF6 and its ortholog OsGRF10 were the most highly expressed OsGRF family members in young inflorescences, and the grf6/grf10 double mutant displayed abnormal florets. OsGRF6/OsGRF10 localized to the nucleus, and electrophoretic mobility shift assays revealed that both OsGRF6 and OsGRF10 bind the GA response element in the promoters of OsJMJ706 and OsCR4, which were reported to participate in the regulation of floral organ development. In addition, OsGRF6 and OsGRF10 could transactivate OsJMJ706 and OsCR4, an activity that was enhanced in the presence of OsGIF1, which can bind both OsGRF6 and OsGRF10. Together, our results suggest that OsmiR396d regulates the expression of OsGRF genes, which function with OsGIF1 in floret development through targeting of JMJ706 and OsCR4. This work thus reveals a microRNA-mediated regulation module for controlling spikelet development in rice.
Spikelets, the basic inflorescence units in rice (Oryza sativa), contain the grains and typically consist of a flower with one lemma, one palea, two lodicules, six stamens, and one pistil (Bommert et al., 2005; Itoh et al., 2005). Many genes have been reported to be involved in spikelet development. The maintenance of meristem organization requires a series of genes, including TONGARI-BOUSHI1, Aberrant Panicle Organization2 (APO2)/Rice FLORICAULA, APO1, Aberrant Spikelet and Panicle1, Oryza sativa Extraordinary Glume1, MOSAIC FLORAL ORGANS1 (MFO1), OsMADS6, and leafy hull sterile1, as well as a few microRNAs (miRNAs), such as OsmiR172 (Aukerman and Sakai, 2003; Chen et al., 2005; Sentoku et al., 2005; Ikeda et al., 2007a, 2007b; Ikeda-Kawakatsu et al., 2009, 2012; Li et al., 2009, 2010a; Ohmori et al., 2009; Cui et al., 2010; Gao et al., 2010; Tanaka et al., 2012; Yoshida et al., 2012). Multiple genes, including G1 (Elongated Empty Glume) and OsMADS34, are required for lemma identity (Yoshida et al., 2009; Gao et al., 2010; Hong et al., 2010), and mutation of key floral regulator genes disrupts the morphology of the palea and lemma, for instance, in the leafy hull sterile1/osmads1 (Jeon et al., 2000; Prasad et al., 2005), mfo1/osmads6 (Ohmori et al., 2009; Li et al., 2010a), floral-organ-number (Li et al., 2007), open beak (Horigome et al., 2009), and depressed palea1/rep1 (Jin et al., 2011) mutants. An Os crinkly4 receptor-like kinase (OsCR4) was recently reported to maintain the interlocking of the palea and lemma by promoting epidermal cell differentiation (Pu et al., 2012). However, how other genes are involved in this process remains poorly understood.
Many miRNAs, along with their regulatory interactions with their targets, have been highly conserved during evolution. For instance, multiple miR396 loci are present in both Arabidopsis (Arabidopsis thaliana; ath-MIR396a–c) and rice (osa-MIR396a–i; Jones-Rhoades and Bartel, 2004). Overexpression of AtmiR396 in Arabidopsis suppresses the expression of its targets, including seven of the nine Growth Regulating Factor (GRF) genes, leading to altered leaf growth by disrupting the coordination of cell division and differentiation (Kim et al., 2003; Horiguchi et al., 2006; Kim and Lee, 2006; Lee et al., 2009; Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). Arabidopsis GRF-Interacting Factor1 (AtGIF1) interacts with AtGRF1, and the atgif1 mutant exhibits narrower leaves and petals (Kim and Kende, 2004; Horiguchi et al., 2011). OsGRFs have been reported as potential regulators of stem growth in rice (van der Knaap et al., 2000; Choi et al., 2004). Down-regulation of OsGRF1 in the rice rhd1 mutant leads to later heading and also, delayed growth and development (Luo et al., 2005). However, less is known about the molecular genetic network mediating miRNA regulation of rice meristem, palea, and lemma identity, which affect the open versus closed state of the spikelets among other characteristics.
In this study, we characterize an OsmiR396d-mediated regulatory network. We show that OsmiR396d and its target OsGRF genes are involved in floral organogenesis. Our data suggest that OsGRF genes encode plant-specific transcription factors that bind the promoters of targets, such as the rice JMJD2 family jmjC gene 706 (OsJMJ706; Sun and Zhou, 2008) and OsCR4 (Pu et al., 2012), to regulate husk opening and floral organ identity. We also show that OsGRFs interact with Os Growth Regulating Factor-Interacting Factor (OsGIF) to influence the activity of their targets.
RESULTS
OsGRF6 Knockdown and OsmiR396d Overexpression both Cause Abnormal Floret Phenotypes
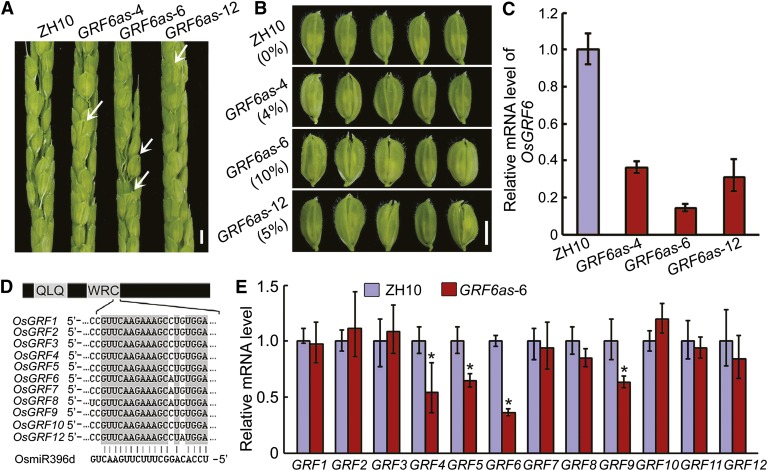
To investigate the biological function of OsGRF6, OsGRF6 antisense transgenic lines (OsGRF6as) were created. The OsGRF6as plants displayed abnormal floret phenotypes, including open husks and long sterile lemmas compared with the wild type (Fig. 1, A and B). Expression levels of OsGRF6 in the three OsGRF6as lines were down-regulated (GRF6as-4, 0.36; GRF6as-10, 0.31; GRF6as-6, 0.14) compared with the wild type (Fig. 1C), and the degree of down-regulation correlated with the percentage (GRF6as-4, 4%; GRF6as-10, 5%; GRF6as-6, 10%) of florets showing defects (Fig. 1, B and C). Proteins in the OsGRF family share sequence similarity in their WRC domain (Fig. 1D), prompting us to test whether OsGRF6 paralogs were also down-regulated. Indeed, OsGRF4, OsGRF5, OsGRF6, and OsGRF9 were all down-regulated in OsGRF6as line 6 (OsGRF6as-6; Fig. 1E).
Figure 1.
Phenotypic and molecular analysis of OsGRF6 antisense transgenic plants. A, Spikelets of OsGRF6 antisense transgenic (OsGRF6as) plants before flowering. White arrowheads indicate florets with open husks or long sterile lemmas. Bar = 3 mm. B, Florets of OsGRF6as plants before flowering. The numbers to the left indicate the ratio of open-husk or long sterile lemma florets. Bar = 3 mm. C, Expression levels of OsGRF6 in the young panicles were detected by qRT-PCR in OsGRF6as plants (n = 3; means ± sds). D, Diagram representing the OsGRF genes and OsmiR396d. The interaction of 11 miR396d-regulated OsGRFs from rice with OsmiR396d is shown, and OsGRF11 was not regulated by OsmiR396d in rice. QLQ and WRC indicate the conserved domains that define the OsGRF family. E, Transcription levels of OsGRF genes in 2-week-old seedlings of ZH10 and OsGRF6as line 6 (OsGRF6as-6) were analyzed by qRT-PCR. *, Significant difference at P ≤ 0.05 compared with ZH10 by Student’s t test (n = 3; means ± sds).
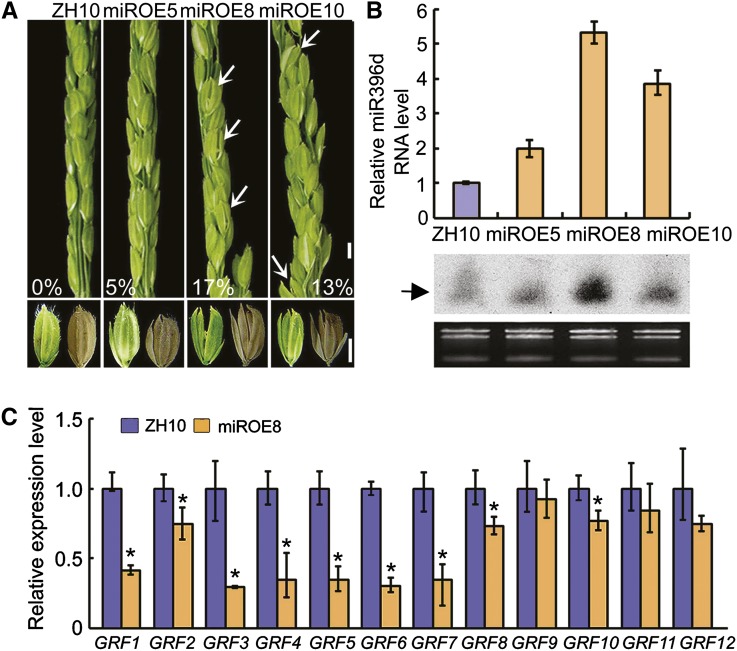
OsGRF6 is one of the predicted target genes of the 21-nucleotide noncoding RNA OsmiR396d (Li et al., 2010b). We, therefore, generated OsMIR396d-overexpressing transgenic (miROE) lines (Supplemental Fig. S1). RNA gel blot and quantitative reverse transcription-PCR (qRT-PCR) assays showed that miR396d transcript levels were increased in several independent miROE lines (Fig. 2B). The spikelets of miROE lines displayed a variety of abnormal phenotypes, including open husks and long sterile lemma florets, compared with the ZhongHua10 (ZH10) wild type (Fig. 2A). In addition, the palea and lemma in the miROE transgenic plants did not lock well together even in the seeds, and the sterile lemmas were longer compared with ZH10 (Fig. 2A). The more enhanced expression of OsmiR396d (miROE5, 2.0; miROE10, 3.9; miROE8, 5.3), the higher ratios of abnormal florets (miROE5, 5%; miROE10, 13%; miROE8, 17%) appeared in the miROE transgenic lines (Fig. 2, A and B), which suggests that the expression levels of OsmiR396d were correlated with the percentage of florets showing defects. There was a higher proportion of abnormal florets (17%) in miROE lines than in OsGRF6as (10%) plants (Figs. 1B and 2A). Correspondingly, the expression levels of OsGRF family members, including OsGRF1, OsGRF2, OsGRF3, OsGRF4, OsGRF5, OsGRF6, OsGRF7, OsGRF8, and OsGRF10, were significantly reduced in miROE compared with OsGRF6as lines (Figs. 1E and 2C). These results suggest that the expression of the OsGRF family is repressed by OsmiR396d to control floret development.
Figure 2.
Phenotypic and molecular analysis of miROE lines. A, Phenotypes of spikelets and seeds in miROE and ZH10 plants. White arrowheads indicate florets with open husks or long sterile lemmas. Numbers indicate the ratio of open-husk or long sterile lemma florets. Bars = 3 mm. B, Expression levels of mature OsmiR396d were detected by qRT-PCR (n = 3; means ± sds) and small RNA blots (arrowhead indicates miR396). C, Transcription levels of OsGRF genes in 2-week-old seedlings of ZH10 and miROE8 were analyzed by qRT-PCR. *, Significant difference at P ≤ 0.05 compared with ZH10 by Student’s t test. (n = 3; means ± sds).
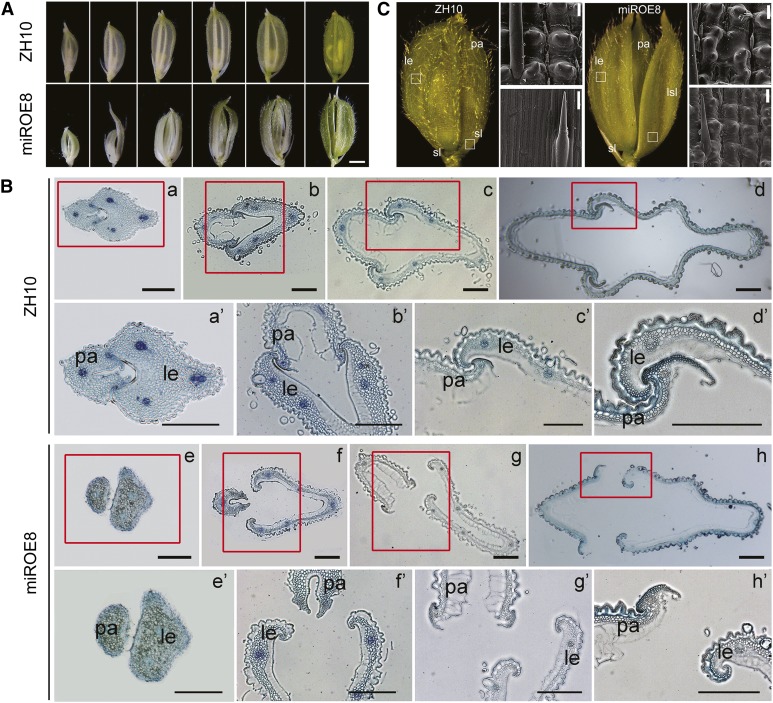
From the spikelet development stage 9 (SP9), the palea and lemma usually grow rapidly by continuous cell division and elongation. During this process, the wild-type lemma interlocked completely with the palea at the top, enclosing the floral organs, whereas an open-husk phenotype appeared in the miROE florets (Fig. 3A). Analysis on the cross sections of florets at different developmental stages showed that the irregular interlocking between palea and lemma was commencing at the very early developmental stages of florets (Fig. 3B). Ordinarily, husk opening is a process in which the lodicules of each floret absorb water, swell, and force apart the lemma and palea. Scanning electron microscopy assays showed no obvious differences in the cells or the structure of lodicules between wild-type and miROE lines (Supplemental Fig. S2, A and B). Histological analysis of lemmas revealed that primary vascular bundles were inclined to one side in some miROE florets, whereas the bundles were in the middle of the lemma in the wild type (Supplemental Fig. S2, C and D). The wild-type florets showed perfect interlocking of the lemma and palea (Supplemental Fig. S2E), whereas self-fused regions of lemma and palea epidermis appeared in the miROE plants (Supplemental Fig. S2F). In florets of the wild type, sterile lemmas contained only sclerenchymatous cells between two epidermal layers, whereas for elongated sterile lemmas in miROE florets, that was not the case (Fig. 3C). Then, scanning electron microscopy analysis indicated that, compared with the smooth, narrow files of cells on the outer epidermal surface of wild-type sterile lemmas (Fig. 3C), epidermal cells of long sterile lemmas in miROE seemed to be transformed into cells with compact small bulges that resembled those bulges observed in wild-type lemma/palea (Fig. 3C). All these findings suggest that overexpression of OsmiR396d, with the resulting down-regulation of OsGRF family members, caused alterations in floret organogenesis and disrupted the interlocking of the palea and lemma.
Figure 3.
Phenotypic analysis of florets in miROE and ZH10 plants. A, Florets of miROE8 and ZH10 plants at different developmental stages. Bar = 1.5 mm. B, Cross sections of interlocking regions of palea and lemma in ZH10 and miROE8 florets at different developmental stages. a′ to d′ or e′ and f′ to h′ indicates the enlargement of tissues in red frames of a to d or e and f to h. le, Lemma; pa, palea. Bars = 200 μm. C, Epidermal surface of le (top) and sterile lemma (sl) or long sterile lemma (lsl; bottom) of florets in ZH10 and miROE8 plants. Bars = 20 μm.
OsGRF6 and OsGRF10 Predominate in Young Inflorescences
Publicly available microarray data show that some members of the OsGRF family, such as OsGRF3, OsGRF6, OsGRF10, and OsGRF11, are predominantly expressed in young inflorescences (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi; Supplemental Fig. S3). By contrast, other putative targets of OsmiR396d, such as LOC_Os08g37520 and LOC_Os06g03980, show weak expression in young inflorescences (Supplemental Fig. S4, A and B). We performed qRT-PCR assays and found that OsGRF4, OsGRF6, and OsGRF10 were the most highly expressed OsGRF genes in young inflorescences (Supplemental Figs. S3 and S4C).
To examine the expression patterns of OsmiR396d and OsGRF6, transgenic rice plants harboring the GUS gene driven by the putative OsmiR396d (1,549 bp) or OsGRF6 (1,569 bp) promoter were generated. For both lines, GUS staining assays showed stronger signals in young florets compared with other tissues, especially in the interlocking region of palea and lemma, the pollen, and the vascular bundles of the lemma (Supplemental Fig. S5, A and B), supporting the idea that OsmiR396d and OsGRF6 might function in floret development.
Overexpression of OsmiR396d-Resistant OsGRF6 Rescues the Phenotypes of OsMIR396d-Overexpressing Lines
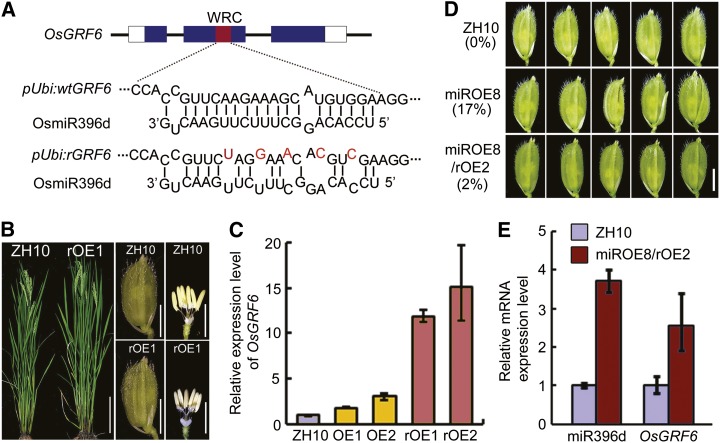
We then created an OsmiR396d-resistant form of OsGRF6 (rOsGRF6) by introducing a synonymous mutation that should prevent its degradation by OsmiR396d (Fig. 4A). Lines overexpressing rOsGRF6 did not show abnormal floret as wild-type ZH10 (Fig. 4B), although the rOsGRF6 mRNA accumulated to higher levels than OsGRF6 mRNA in OsGRF6 transgenic overexpression lines (Fig. 4C). A reduced percentage in open-husk or long sterile lemma florets (down to 2% from 17% in the miROE8 background; Fig. 4D) emerged in the rOsGRF6 overexpression transgenic plants on the miROE genetic background, which was from the crossed lines (Fig. 4, D and E). These findings, combined with the fact that the OsGRF6as lines with reduced levels of OsGRF6 showed similar abnormal florets, suggest that the floret phenotypes in miROE lines might be caused mainly by the decrease in OsGRF activity.
Figure 4.
Phenotypic analysis of OsGRF6-overexpressing transgenic plants. A, Schematic diagram of the OsGRF6 and rOsGRF6 genes as well as OsmiR396d. The interaction between OsGRF6/rOsGRF6 and OsmiR396d is shown. Red letters in rOsGRF6 represent synonymous mutations that prevent targeting for degradation by OsmiR396d. B, rOsGRF6 overexpression plants at heading stage (bar = 20 cm) and the florets before flowering (bar = 3 mm). C, Expression levels of OsGRF6 were detected by qRT-PCR in OsGRF6 OE (OE) and rOsGRF6 OE (rOE) lines (n = 3; means ± sds). D, The abnormal floret phenotypes of miROE plants were almost completely rescued by crossing with rOsGRF6 OE plants. The numbers to the left indicate the ratio of open-husk or long sterile lemma florets. Bar = 3 mm. E, Expression levels of miR396d and OsGRF6 were detected by qRT-PCR in ZH10 and miROE8–crossed rOsGRF6 OE line 2 (miROE8/rOE2) plants (n = 3; means ± sds).
The osgrf6/osgrf10 Double Mutant Has Altered Florets
We tried to find the important genes of the OsGRF family that play roles in floret development by tissue expression analysis, and OsGRF6 as well as OsGRF10 were found to be highly expressing in young inflorescences (Supplemental Figs. S3 and S4C). The osgrf6 (PFG_3A-16508.R) mutant, which has a transfer-DNA insertion in the last exon causing it to lack full-length mRNA for OsGRF6 (Supplemental Fig. S6, A–C), exhibited a semidwarf phenotype compared with the wild type, but no defects were found in the florets (Supplemental Fig. S6D). Floret development also appeared normal in the osgrf10 (PFG_3A-03348.L) mutant, which had an insertion in the last exon of OsGRF10 (Supplemental Fig. S7A), leading to somewhat reduced levels of OsGRF10 mRNA but a more severe decrease in the protein (Supplemental Fig. S7, C and E). The osgrf6/osgrf10 double mutant not only showed a more severe semidwarf phenotype but also had defects in the florets (Fig. 5). About 5% (Fig. 5, D and E) of the florets in the double mutant showed long sterile lemmas (Fig. 5G), missing paleas (Fig. 5H), and abnormal numbers of stamens (Fig. 5, J and K), like those findings in the miROE and OsGRF6as plants. These defects in the florets of the osgrf6/osgrf10 double mutant suggest that OsGRF6 and OsGRF10 function redundantly in floret development.
Figure 5.
Phenotypic analysis of grf6/grf10 double mutant. A, Genomic identification of the grf6/grf10 double mutant. LP, Left primer; RP, right primer; LB, the T-DNA left border primer. DJ (−/−) is the wild type, and +/+ indicates the homozygous mutant. B, Plants of grf6/grf10 double mutant and the DJ wild type at the tillering stage. Bar = 20 cm. C, Inflorescences of grf6/grf10 and the DJ wild type. Bar = 2 cm. D and E, Inflorescences in C are enlarged to show detail. White arrowheads in E indicate open-husk or long sterile lemma florets of the grf6/grf10 double mutant. F and I, Wild-type floret and inner organs of the floret, respectively. G, H, J, and K, Florets and inner organs of florets in the grf6/grf10 double mutant. Bars = 3 mm.
OsGRF6 and OsGRF10 Interact with OsGIF Proteins
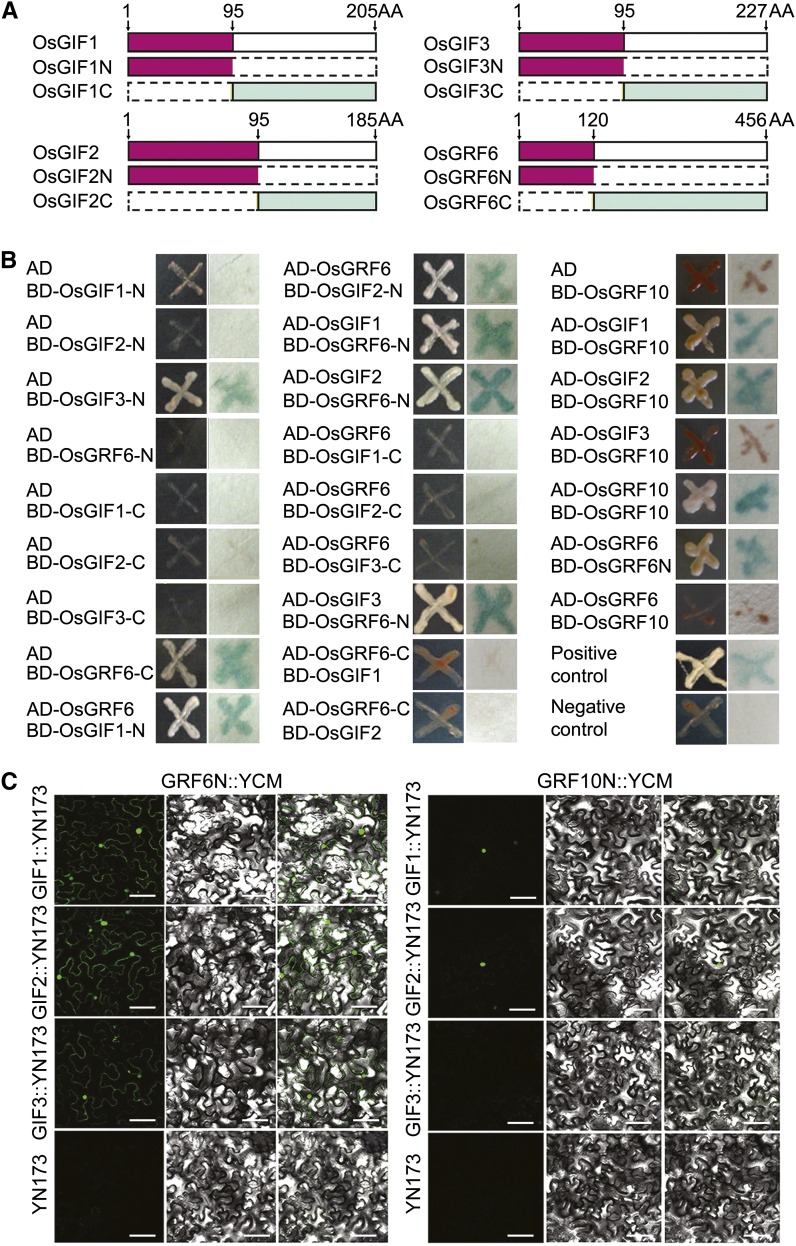
Arabidopsis GIF proteins interact with AtGRF1 (Kim and Kende, 2004), prompting us to test whether this finding is also true of their orthologs in rice. Yeast (Saccharomyces cerevisiae) two-hybrid assays showed that OsGRF6, OsGIF1, OsGIF2, and OsGIF3 interacted at their N-terminal domains, whereas OsGRF10 interacted with OsGIF1 and OsGIF2 but not OsGIF3 (Fig. 6, A and B). These interaction patterns of OsGIF1, OsGIF2, OsGIF3, OsGRF6, and OsGRF10 were confirmed in vivo using bimolecular fluorescence complementation (BiFC) assays, which revealed interactions in the nucleus (Fig. 6C). Additional support for nucleus localization of OsGRF10 was provided by transient transfection assays in rice leaf protoplasts. The fluorescence from an OsGRF10-GFP fusion was found to overlap with 4′,6-diamidino-2-phenylindole staining of the nucleus (Supplemental Fig. S5C).
Figure 6.
Proteins interaction assays. A, Schematic diagrams of OsGRF6, OsGIF1, OsGIF2, and OsGIF3. B, Yeast two-hybrid interaction assays of OsGRF6, OsGRF10, OsGIF1, OsGIF2, and OsGIF3 as well as OsGRF6 and OsGRF10. AH109 cells containing different plasmid combinations were grown on the selective medium SD-Leu Trp His adenine, and a single clone was used for β-galactosidase activity assays. C, BiFC assays to verify the interactions of OsGRF6, OsGRF10, OsGIF1, OsGIF2, and OsGIF3. Tobacco (Nicotiana spp.) leaves cotransformed with GRF6N:YCM and YN173 empty vectors or GRF10N:YCM and YN173 empty vectors were used as negative controls. Yellow fluorescent protein signals were found in GRF6N:YCM and OsGIF1/OsGIF2/OsGIF3:YN173 cotransformed leaves as well as OsGRF10N:YCM and OsGIF1/2:YN173 cotransformed leaves. No signal was found in OsGRF6N:YCM and YN173 empty vectors, OsGRF10N:YCM and YN173 empty vectors, or OsGRF10N:YCM and OsGIF3:YN173 cotransformed leaves. SD, Synthetic Defined; YCM, mutated eYFP C-terminus; YN173, eYFP N-terminal amino acids 1 to 173. Bar = 100 µm.
Genome-Wide Expression Analysis in miROE Plants
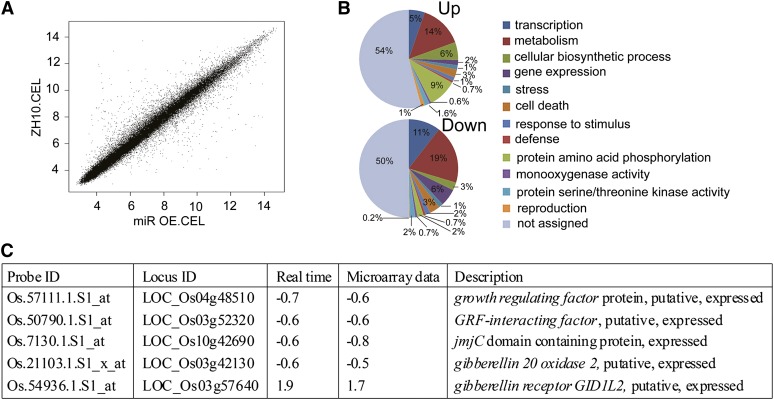
To understand further how OsGRF genes are involved in rice floret development, we used a whole-genome microarray chip approach to analyze differences in expression patterns between miROE8 and wild-type plants. The reliability of the chip assay was confirmed by signal scatter graph (Fig. 7A). In young inflorescences (SP4–SP8) of miROE8 plants, 64 genes were down-regulated and 43 genes were up-regulated compared with the wild type (Supplemental Tables S1 and S2). Gene ontology analysis revealed that genes involved in metabolism or reproduction had significantly altered expression levels in the miROE8 plants compared with the wild type (Fig. 7B). qRT-PCR assays for several genes confirmed the chip data (Fig. 7C; Supplemental Table S3).
Figure 7.
Global analysis of gene expression in young inflorescences of miROE8 compared with ZH10. A, Scatter graph of signals. Only spots with a present signal were used to determine the false-positive rate. B, Predicated functions of the proteins encoded by up-regulated and down-regulated genes in young inflorescences of miROE8 compared with ZH10 on microarray analysis. C, qRT-PCR analysis of relative transcript levels of selected genes from the rice whole-genome DNA microarray experiment (n = 3; means ± sds).
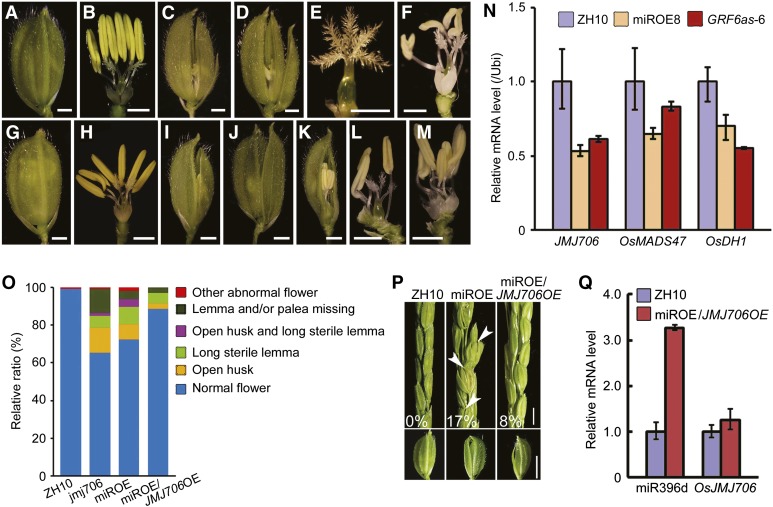
Of note, the jmjC domain-containing gene OsJMJ706, which encodes an H3K9 demethylase (Sun and Zhou, 2008), appeared in the list of down-regulated genes (Fig. 7C). The expression of OsJMJ706 was decreased to −0.6-fold in miROE8 plants. It was consistent with that from the JMJ706 artificial micro-RNA transgenic lines, in which OsJMJ706 expression is the slight reduced (about −0.5-fold) but could cause abnormal floret phenotype (Sun and Zhou, 2008). It might be explained by a complicated regulation network that is possibly involved in the miR396d-targeted OsGRFs. Based on the results, we compared the phenotypes of a jmj706 mutant and miROE8 plants (Fig. 8, A–M). Both miROE8 and jmj706 plants had open husks, long sterile lemmas, and abnormal pistils or stamens in the florets (Fig. 8, A–M). In the jmj706 mutant, there was a high ratio of florets missing lemmas and/or paleas, whereas the ratio was lower in miROE8 plants (Fig. 8O). Expressions of downstream genes of OsJMJ706, such as OsMADS47 and Degenerated Hull1 (OsDH1), are decreased in jmj706 (Sun and Zhou, 2008). OsMADS47 and OsDH1 expression levels were also decreased in miROE8 and OsGRF6as-6 (Fig. 8N). The similarity of these floret phenotypes suggests that OsJMJ706 might be involved in floret developmental regulation mediated by OsmiR396d.
Figure 8.
Phenotypic comparisons of miROE8 and jmj706 mutant plants. A and B, Florets before flowering of ZH10. C to F, Florets before flowering of miROE8 showing open husks (C and D), long sterile lemmas (C and D), abnormal stigmas (E), and abnormal numbers of stamens (F). G and H, Florets before flowering of ZH11. I to M, Florets before flowering of jmj706 showing open husks (I), long sterile lemmas (J), missing lemmas or paleas (K), abnormal numbers of stamens and abnormal stigmas (L), and abnormal lodicules (M). Bars in A to M = 1 mm. N, Transcription levels of JMJ706, OsMADS47, and OsDH1 genes in young inflorescences before flowering of miROE8, GRF6as-6 plants, and ZH10 were analyzed by qRT-PCR (n = 3; means ± sds). O, Statistical analysis of different kinds of florets in ZH10, jmj706, miROE8, and miROE8/JMJ706OE plants. The proportional distributions of the florets are indicated by different colors. P, Spikelet before flowering in ZH10, miROE8, and miROE8/JMJ706OE plants. White arrowheads indicate open-husk or long sterile lemma florets in miROE8 plants. Numbers in the figure indicate the ratios of florets with open husks or long sterile lemmas. Bar = 3 mm. Bottom, The florets taken from ZH10, miROE8, and miROE8/JMJ706OE plants are shown. Bar = 4 mm. Q, Transcription levels of miR396d and OsJMJ706 in young inflorescences of ZH10 and miROE8/OsJMJ706OE plants (n = 3; means ± sds).
Overexpression of OsJMJ706 Partly Rescued the Abnormal Floret Phenotypes in miROE Plants
To test whether the floral defects in miROE plants are caused by reduced expression of OsJMJ706, we performed a genetic complementation test by crossing OsJMJ706 overexpression plants with miROE8 plants. OsJMJ706 expression levels were analyzed in six independent crossed plants (Fig. 8, P and Q; Supplemental Table S4). The percentage of open-husk or long sterile lemma florets in the population decreased from 17% in the florets of miROE plants to 8% in the florets of crossed plants, indicating that overexpression of OsJMJ706 can partly rescue the abnormal floret phenotypes of miROE transgenic plants (Fig. 8P; Supplemental Table S4). These data suggest that OsJMJ706 is genetically functional in floret development mediated by OsmiR396d in rice.
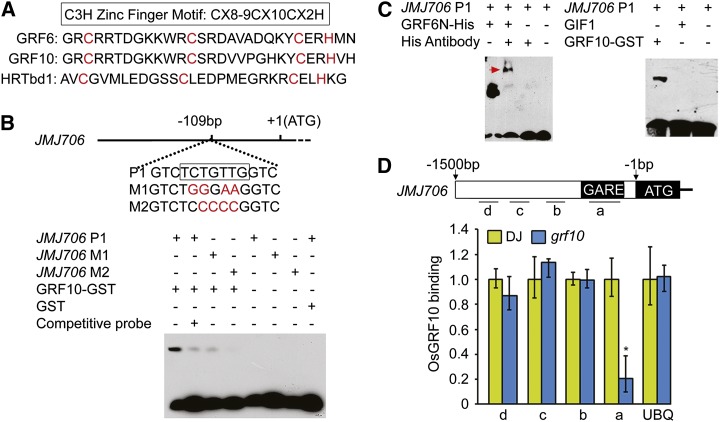
OsJMJ706 Might Be a Direct Target of OsGRFs
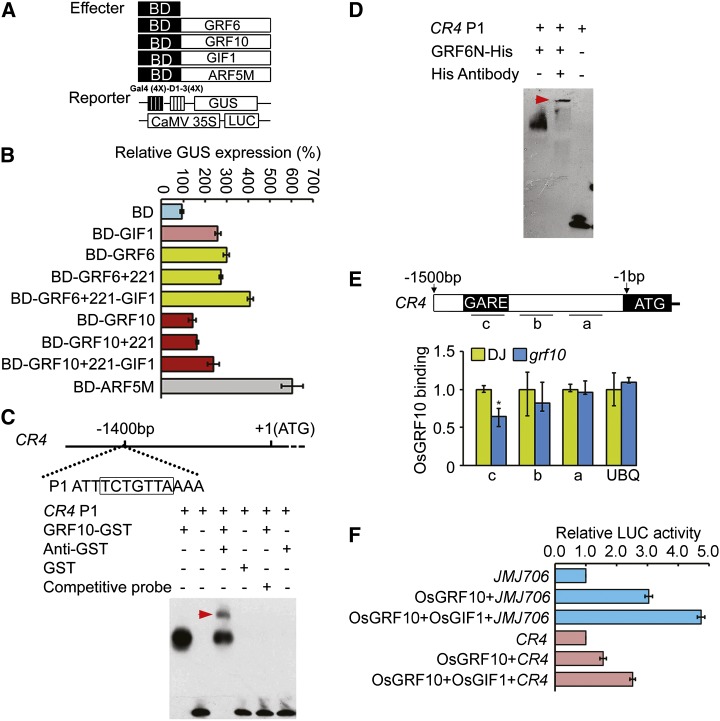
Protein sequence alignment revealed a single C-X8-9-C-X10-C-X2-H–type zinc-binding motif in the WRC domain of OsGRF family proteins (Fig. 9A). The motif corresponded to that in Hordeum Repressor of Transcription (HRT), a barley transcriptional repressor protein that is proposed to bind the GA response element (GARE) motif (Raventós et al., 1998), hinting that the GRFs, as putative transcription factors, may also bind to the GARE motif. Sequence alignments showed that a GARE-like sequence was present at the region −109 to −115 bp from the transcription start site in the promoter of OsJMJ706 (Fig. 9B). This finding, combined with the fact that mutation of OsJMJ706 resulted in similar floret defects like in miROE lines (Fig. 8, A–M), led us to test whether OsJMJ706 was a direct target of GRF family proteins. Electrophoretic mobility shift assays (EMSAs) showed that OsGRF6 and OsGRF10 bound to the P1 sequence in the promoter of OsJMJ706 (Fig. 9, B and C). By contrast, the OsGIF1 protein did not (Fig. 9C). Mutations in the P1 sequence caused reduced binding by OsGRF10. Chromatin immunoprecipitation (ChIP) assays using available antibody for OsGRF10 (Supplemental Fig. S7) showed that sequence region a of OsJMJ706 accumulated more in the cv DongJin (DJ) wild type than in the grf10 mutant (Fig. 9D). These results indicate that OsGRF10 can specifically bind to the GARE in the promoter of OsJMJ706. OsJMJ706, thus, seems to be a direct target of GRF10 in the regulation of floret development.
Figure 9.
OsGRF6 and OsGRF10 proteins can bind to the promoter of OsJMJ706 both in vitro and in vivo. A, Alignment of amino acid sequences of the C3H zinc finger domain of HRTdb1 in barley (Hordeum vulgare) and OsGRF6 and OsGRF10 in rice. B and C, EMSA shows that OsGRF10-GST and OsGRF6N-His fusion proteins can bind to the promoter of OsJMJ706, whereas OsGIF1 proteins cannot. The GARE cis-element 109 bp upstream of the transcription initiation site is shown in the black box in B. P1, M1, and M2 representing the JMJ706 promoter-binding site and the mild and the severe mutations of P1, respectively, were used as biotin-labeled probes. The red arrowhead in C indicates the complex composed of OsGRF6N-His, JMJ706 P1, and His antibody. D, ChIP assay of binding of OsGRF10 to the promoter of OsJMJ706 tested using anti-OsGRF10 antibody. Regions in the promoter of OsJMJ706 analyzed by real-time PCR are shown by short lines marked with letters (a–d; a is the region containing the GARE). The quantification of binding by OsGRF10 is shown in the bar graph, and the promoter of Ubiquitin (UBQ) is the control. *, Significant difference at P ≤ 0.05 compared with DJ by Student’s t test. Data are means ± sds (n = 3).
Transcriptional Activities of OsGRF6 and OsGRF10 Are Enhanced by OsGIF1
GRF proteins were predicated to be transcription factors in Arabidopsis (Kim and Kende, 2004). A transcriptional activation assay was performed using the Gal4 binding domain (BD)-OsGRF6/OsGRF10 fusion protein and a constitutively expressed reporter gene containing four upstream Gal4 DNA-binding sites (Gal4 [4X]-D1-3[4X]-GUS; Fig. 10A). The data showed that both OsGRF6 and OsGRF10 caused activation of the reporter gene expression (Fig. 10B). When OsGIF1-GFP was added to the assays, the activation activities of both OsGRF6 and OsGRF10 were greatly increased compared with either alone (Fig. 10B). The specific binding and activation activity of transcription to JMJ706 was also analyzed (Fig. 10F). Cotransformation of OsGRF10 and OsGIF1 showed that OsGIF1 also can enhance the transactivation activity of OsGRF10 compared with OsGRF10 alone (Fig. 10F). All these data suggest that OsGRF6 or OsGRF10 had transcriptional activation activity and that OsGIF1 can act as the transcriptional coactivator for them.
Figure 10.
OsGRF6 and OsGRF10 proteins have transactivation activity and can bind to the promoter of OsCR4 in vitro and in vivo. A, Schematic representation of the reporter and effector constructs used in the transcription activity assays. B, OsGRF6 and OsGRF10 function as transcription activators, whereas OsGIF1 protein may function as a coactivator. Arabidopsis leaf protoplasts were cotransfected with two reporter genes and one effector gene. Effector genes contain yeast Gal4 BD fused in frame with OsGRF6, OsGRF10, OsGIF1, OsGIF2, or Auxin Response Factor5M (ARF5M; positive control). Data are means of three independent experiments. C and D, EMSA showing that GRF10-GST and GRF6N-His fusion proteins can bind to the promoter of OsCR4. GARE cis-element 1,400 bp upstream of the transcription initiation site is shown in the black box in C. The OsCR4 P1 oligonucleotide was used as the biotin-labeled probe. Red arrowheads indicate the complexes composed of OsGRF10-GST, CR4 P1, and GST antibody or OsGRF6N-His, CR4 P1, and His antibody. E, ChIP assays of the binding of OsGRF10 to the promoter of OsCR4 tested using anti-OsGRF10 antibody. Regions in the promoter of OsCR4 analyzed by real-time PCR are shown by short lines marked with letters (a–c; c is the region containing the GARE). The binding ability of the GARE cis-element was reduced in the grf10 mutant; the promoter of Ubiquitin (UBQ) is the control. *, Significant difference at P ≤ 0.05 compared with DJ by Student’s t test. Data are means ± sds (n = 3). F, Transcriptional activation activity assays in Arabidopsis protoplasts. JMJ706 and CR4 indicate the promoters of OsJMJ706 and OsCR4 genes individually. Data are means of three independent experiments. LUC, Luciferase.
OsCR4 Is Targeted by OsGRFs
OsCR4 maintains the interlocking of the palea and lemma by promoting epidermal cell differentiation in rice (Pu et al., 2012). The open-husk phenotype of OsCR4 RNA interference plants resembles that of the miROE transgenic plants (Fig. 2A). The transcription levels of OsCR4 were down-regulated in the miROE transgenic and osgrf6/osgrf10 double mutant plants (Supplemental Fig. S8 and Fig. 11B), which suggests that expression of OsCR4 may be negatively regulated by miR396d in rice. A GARE was identified at −1,400 bp in the promoter of OsCR4 (Fig. 10C), and EMSA assays showed that both OsGRF10 and OsGRF6 proteins could bind to that region of the OsCR4 promoter (Fig. 10, C and D). ChIP assays using the OsGRF10 antibody showed that OsGRF10 could specifically bind to the GARE of OsCR4 in vivo as well (Fig. 10E). The specific binding and activation activity of OsGRF10 to OsCR4 was also analyzed in the transient system (Fig. 10F). OsGIF1 also can enhance the transactivation activity of OsGRF10 to the promoter of CR4 compared with OsGRF10 alone (Fig. 10F). Therefore, OsCR4 seems to be another target of OsGRF6 and OsGRF10 to regulate floret development.
Figure 11.
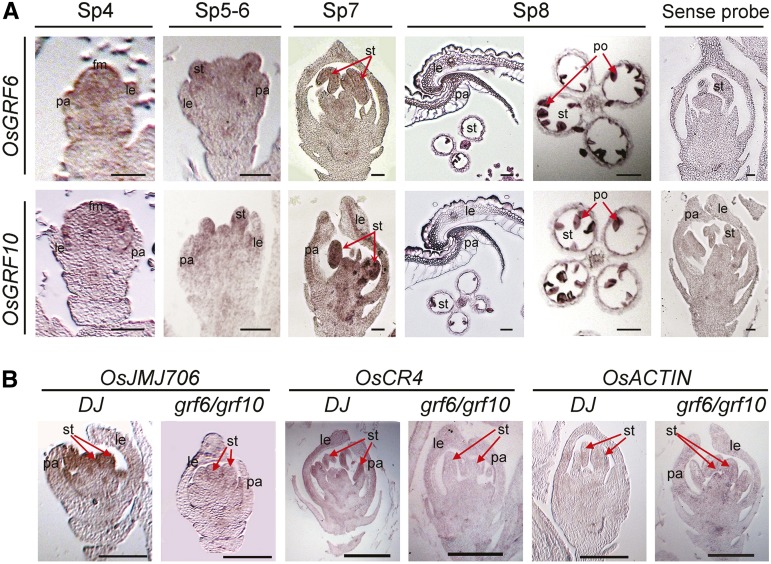
RNA in situ hybridization assays. A, RNA in situ hybridization assays for OsGRF6 and OsGRF10 in florets at different developmental stages in DJ. Red arrows in SP7 and SP8 indicate stamens and pollens individually. fm, Floral meristem; le, lemma; pa, palea; po, pollen; st, stamen. Bars = 50 µm. B, RNA in situ hybridization assays of OsJMJ706 and OsCR4 in florets of DJ and grf6/grf10 double mutants at the overlapping expression regions of OsGRF6 and OsGRF10. Red arrows indicate stamens. OsACTIN was used as the internal control. Bars = 200 µm.
DISCUSSION
The reproductive organs in the Poaceae family (grass) are the basic unit to determine grain yield. Although multiple genes are known to be involved in regulation of the identity of grass-specific organs, such as palea, lemma, lodicules, pistils, and stamens, the molecular mechanism controlling the floral organs has been the subject of a vast and largely inconclusive discussion. It is still of great interest to elucidate new genes involved in the genetic control of this developmental process in rice.
OsmiR396d-Dependent OsGRFs Regulate Floret Development in Rice
miR396 represents one of the most deeply conserved miRNA families in plants, but the miR396d variant, which is distinguished from miR396a, miR396b, and miR396c by differences in three nucleotides, is found only in monocots (Sunkar and Jagadeeswaran, 2008). Transgenic rice plants overexpressing OsmiR396d only occasionally exhibited small leaf phenotypes, like that caused by AtmiR396 in Arabidopsis (Supplemental Fig. S9). By contrast, overexpression of OsmiR396d, with the resulting down-regulation of its targets, frequently led to abnormal florets with open husks or long sterile lemmas (Fig. 2). OsGRF6 and OsGRF10 are two targets of OsmiR396d. Both osgrf6 and osgrf10 single mutants showed semidwarf phenotypes but no floret defects (Supplemental Figs. S6 and S7). The grf6/grf10 double mutant did exhibit defects in floral organ development (Fig. 5), suggesting that OsGRF6 and OsGRF10 function redundantly in floret development.
The floret defects in the miROE plants could be rescued nearly completely by expression of an rOsGRF6 in the miROE background (Fig. 4). In Arabidopsis, AtmiR396 negatively regulates cell proliferation by repressing the expression of AtGRF genes in a dosage-dependent manner (Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). Recently, AtmiR396 was reported to mediate pistil development by suppressing its GRF target genes in Arabidopsis (Liang et al., 2014). Our study reveals that OsGRF genes participate in floret development in a redundant manner in rice, which expands our view on the function of miR396 as well as its GRF target genes.
OsmiR396d Is Essential for Interlocking between Palea and Lemma
From SP9, open-husk phenotypes caused by malformed interlocking between palea and lemma occurred in miROE florets (Fig. 3, A and B; Supplemental Fig. S2F). In stages Sp4 to Sp6, OsGRF6 and OsGRF10 are slightly expressed in the floral meristems (Fig. 11A). At SP7, OsGRF10 was expressed higher in the stamen, palea, and lemma than the expression of OsGRF6, whereas in SP7 and SP8, OsGRF6 and OsGRF10 are expressed highly in the stamens as well as the interlocking regions of palea and lemma (Fig. 11A). OsmiR396d and OsGRF6 shared an expression pattern in the interlocking region of palea and lemma, the pollen, and the vascular bundles of the lemma (Supplemental Fig. S6B). All these data are consistent with the phenotype in the flower. Overexpression of OsmiR396d caused the anomalous numbers of pistils and stamens in the miROE florets (Fig. 8, E and F), which resemble the phenotype in the jmj706 mutant (Fig. 8, L and M; Liu et al., 2009). If we equate palea/lemma with sepals, the open husks in the miROE florets strikingly resemble that of the open sepal phenotype of atgif triple mutant (Lee et al., 2014) and might be a result of the lemma and palea not growing normally enough to reach out together (Supplemental Fig. S2, D and F). Similar open-husk phenotypes also occur in transgenic lines of OsCR4 RNA interference (Pu et al., 2012) and jmj706 mutants (Sun and Zhou, 2008). Consistent with this finding, the expression levels of OsJMJ706 and OsCR4 were decreased in the miROE plants (Fig. 8N; Supplemental Fig. S8).
OsGRF Directly Affects the Transcription Activity of OsJMJ706 and OsCR4 to Regulate Floret Development
The OsGRF family is composed of 12 members in rice (Choi et al., 2004). All OsGRF genes have the conserved QLQ and WRC domains in their N-terminal regions (Fig. 1D; van der Knaap et al., 2000; Kim et al., 2003), and their variable C-terminal regions are rich in Pro, Gln, His, Ala/Gly, and Ser/Thr, characteristics frequently found in transcription factors (Choi et al., 2004). Indeed, the C-terminal region of OsGRF6 showed strong transactivation activity (Figs. 6B and 10B), which was previously reported for OsGRF1 (Choi et al., 2004). OsGRF genes were preferentially expressed in the young inflorescence, with the expression levels of OsGRF6 and OsGRF10 higher than those levels of the other OsGRF family members. OsGRF10 also showed transactivation activity in vivo (Fig. 10B), although its activity was only one-half that of OsGRF6 in the absence of the interacting protein OsGIF1. The promotion of OsGRF6 and OsGRF10 transactivation activity by OsGIF1 supports the idea that OsGIF1 acts as a transcriptional coactivator to regulate the activity of OsGRFs in vivo (Fig. 10, B and F).
OsJMJ706 is required for floral organ development (Sun and Zhou, 2008). In miROE, OsGRF6 antisense transgenic plants, and osgrf6/osgrf10 double mutant plants, expression levels of OsJMJ706 were down-regulated (Figs. 8N and 11B). In addition, the miROE plants and the jmj706 mutant exhibited marked phenotypic overlap in floral defects, including long sterile lemmas, open husks, missing lemmas or paleas, and abnormal numbers of stamens or pistils (Fig. 8, C–F and I–M). Overexpression of OsJMJ706 in miROE plants could partly rescue the abnormal floret phenotypes (Fig. 8P), and OsGRF6 and OsGRF10 have a C-X8-9-C-X10-C-X2-H–type zinc-binding motif (Raventós et al., 1998) that could bind to the GARE sequence in the promoter of OsJMJ706 (Fig. 9, B–D). Combined with the finding that both OsGRF6 and OsGRF10 had stronger transactivation activity in the presence of OsGIF1 (Fig. 10, B and F), this finding suggests that OsGRF transcription factors interact with their coactivator OsGIF1 to promote the transcription of their target genes, such as OsJMJ706, and mediate development of husks and floral organ identity in rice. The AtGIF gene family was just reported to play an essential role in control of male and female reproductive development (Lee et al., 2014), whereas AtmiR396 was reported to mediate pistil development by suppressing its GRF target genes to impair the formation of GRF/GIF cotranscription complex in Arabidopsis (Liang et al., 2014). The functions of AtGRFs as well as AtGIFs in reproductive organ development are similar with our results in rice.
In summary, this work shows that floral development in rice is dependent on OsmiR396d and involves an miR396d/GRF/GIF regulation module. OsGRF genes are negatively regulated by OsmiR396d (Li et al., 2010b). OsGRFs, modulated by their interaction with OsGIF1, directly activate expression of targets, including OsJMJ706 and OsCR4, to regulate floral organ development, affecting characteristics such as husk openness and sterile lemma length. The open-husk trait may be of use for facilitating manipulation of genetic hybridization in breeding. Our elucidation of this miR396d/GRF/GIF regulation module thus provides insights into the mechanism by which floral organ development is controlled and has great potential application for cross breeding in rice molecular breeding programs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All transgenic rice (Oryza sativa) plants were obtained in the rice ssp. japonica ‘Zhonghua10’ background. Both the OsGRF6 and OsGRF10 mutants were obtained from the Rice Functional Genomics Express Database of Korea (http://signal.salk.edu/cgi-bin/RiceGE; Jeong et al., 2002). All rice plants used in our study were grown in either Beijing, China from June to October or Hainan, China from January to May. Young inflorescences from miROE and ZH10 plants before heading were used in the microarray experiments.
Total RNA Isolation and qRT-PCR
Total RNA was extracted from different tissues of rice using the TRIzol RNA extraction kit according to the user manual (Invitrogen). Total RNA (2 µg) was treated with DNaseI (Fermentas) and then reverse transcribed in a total volume of 25 µL with 0.5 µg of oligo(dT)18, 0.75 mm deoxyribonucleotide triphosphate, and 200 units of moloney murine leukemia virus reverse transcriptase (Promega). qRT-PCR analyses were performed on an Mx3000P (Stratagene) Real-Time PCR System using the SYBR Green Master Mix (TOYOBO) according to the manufacturer’s instructions. The expression levels of the samples were normalized to that of Ubiquitin. The gene-specific primers used in the qRT-PCR are described in Supplemental Table S3.
Global Analysis of Gene Expression
Global analysis of gene expression was done using the rice Affymetrix GeneChip, with the wild-type and miROE8 plants independently grown under the same growth conditions. Total RNA was isolated from young inflorescences before flowering (spikelet development stages 4 to 8) with the total RNA extraction kit (TRIzol reagent; Invitrogen). All processes were conducted according to the GeneChip Standard Protocol (Eukaryotic Target Preparation; Affymetrix). Statistical analyses of the microarray data were as previously described (Wang et al., 2008).
Small RNA Analysis
Total RNA was extracted using the TRIzol RNA extraction kit according to the user manual (Invitrogen). Approximately 50 µg of total RNA was separated on 15% (v/v) polyacrylamide denaturing gels and then electrophoretically transferred to Hybond-N+ membranes (GE Healthcare UK Ltd.). Membranes were UV cross linked (Cell Biosciences Alphaimagine HP) and then hybridized with DNA oligonucleotides complementary to the miR396d sequence, which had been end labeled with [γ-32P]ATP by T4 polynucleotide kinase (New England Biolabs). Membranes were prehybridized for at least 3 h and then hybridized overnight at 37°C in ULTRAhyb-Oligo hybridization buffer (Ambion). The membranes were briefly air dried and then exposed to x-ray film for autography at −80°C. Images were acquired by scanning the film.
Alternatively, miR396d levels were determined by stem-loop qRT-PCR as described previously (Chen et al., 2005). The samples were normalized using U6 small nuclear RNA in the stem-loop qRT-PCR. The sequences of the primers used are described in Supplemental Table S3.
Microscopic Observations
Different developmental stages as well as the GUS-staining florets were fixed in solution of 3.7% (v/v) formaldehyde, 50% (v/v) ethanol, and 5% (v/v) acetic acid and embedded in Paraplast Plus (Sigma-Aldrich). Eight-micrometer-thick sections were stained with toluidine blue (no staining for GUS-staining florets) for light microscopic analysis (Zeiss; Guo et al., 2013).
DNA Gel Blot Analysis
Genomic DNAs were isolated from 2-week-old seedlings, and 20 µg of total DNAs was digested with EcoRI or HindIII. The fractionated DNAs were electrophoresed on 0.7% agarose gels and then blotted to nylon membranes. The GUS gene used as the probe was labeled with [α-32P]dCTP. Hybridization was performed as previously described (Xu et al., 2005).
Yeast Two-Hybrid Assays
The complementary DNAs (cDNAs) of OsGRF6, OsGRF10, OsGIF1, OsGIF2, and OsGIF3 were cloned into the pGADT7 and pGBKT7 vectors and then cotransformed into yeast (Saccharomyces cerevisiae) strain AH109. Transformants were screened for growth on medium lacking Leu, Trp, His, and Ade. Recovered individual colonies were used for β-galactosidase filter assays.
BiFC
BiFC assays were performed using a previously described protocol (Waadt et al., 2008). For BiFC assays, the full-length cDNAs of OsGIF1, OsGIF2, and OsGIF3 were cloned into the pSPYNE173 vector, and the 5′ region of the cDNAs of OsGRF6 and OsGRF10 were cloned into the pSPYCE(M) vector. Plasmids were individually electroporated into Agrobacterium spp. (strain GV3101), and the different strains carrying the pSPYNE-OsGIF1/OsGIF2/OsGIF3 and pSPYCE-OsGRF6/OsGRF10 plasmids as well as the helper strain carrying P19 plasmid were coinfiltrated into tobacco (Nicotiana spp.) leaves. Yellow fluorescent protein fluorescence was visualized with a confocal scanning microscope 72 h after infiltration.
Transient Assays for Activation Activity in Vivo
Activation assays were carried out in protoplasts prepared from 4-week-old Arabidopsis (Arabidopsis thaliana) seedlings of the Columbia ecotype grown under short-day conditions (Zhu et al., 2008). The DNA BD from GAL4 was used, and the GAL4 BD-OsGRF6, GAL4 BD-OsGRF10, and GAL4 BD-OsGIF1 fusion proteins could bind to the GAL4 DNA-binding sites of the GUS reporter. The known activator protein ARF5M was used as a positive control. The GUS reporter containing four upstream GAL4 DNA-binding sites (GAL4 [4X]-D1-3[4X]-GUS) as well as the luciferase reporter were cotransformed with GAL4 BD-OsGRF6 into Arabidopsis protoplasts. The GUS activity was quantified as described (Jefferson et al., 1987). A plasmid carrying the luciferase gene under the control of the 35S promoter was used as an internal control to normalize the data for variations in the experiment (Yoo et al., 2005).
The full-length cDNA of either OsGRF10 or OsGIF1 was fused into the plant binary vector221 (pBI221) driven by the 35S promoter to generate pBI221-OsGRF10 or pBI221-OsGIF1, respectively. The plasmid carrying the GUS gene under the control of the 35S promoter was used as a normalization control. Values represent means ± sds of three replicates. In the transient assays, cotransformation of OsGRF10 and OsGIF1 was done for identifying effects of OsGIF1 (Guo et al., 2013).
EMSA
EMSA was performed as described (Ma et al., 2009). The full-length cDNAs of OsGRF10, OsGIF1, and cDNA coding for the N-terminal region containing the DNA-binding domain of OsGRF6 were amplified with gene-specific primers and fused into the EcoRI and SalI sites of the expression vector pGEX-4T-1 or pET-28a+. The constructs were transformed into Escherichia coli BL21 (DE3). Cells were grown at 37°C and induced by the addition of isopropylthio-β-galactoside to a final concentration of 1 mm when the optical density at 600 nm was 0.6. The pGEX-4T-1-OsGRF10 fusion protein was purified with Glutathione Sepharose 4B (GE Healthcare). The GIF1 protein was obtained by digesting pGEX4T-1-GIF1 fusion protein with thrombin. We failed to purify the pET-28a+-GRF6N protein, and the cell lysate of pET-28a+-OsGRF6N was used for EMSA.
Oligonucleotides (Supplemental Table S3) were synthesized (Invitrogen) and labeled using the Biotin 3′ End DNA Labeling Kit (Pierce). We used standard reaction mixtures for EMSA containing 2 µg of purified OsGRF10–glutathione S-transferase (GST) or OsGIF1 protein or the supernatant of OsGRF6N-His cell lysates, 2 µL of biotin-labeled annealed oligonucleotides, 2 µL of 10× binding buffer (100 mm Tris, 500 mm KCl, and 10 mm dithiothreitol [pH 7.5]), 1 µL of 50% (v/v) glycerol, 1 µL of 1% (v/v) Nonidet P-40, 1 µL of 1 m KCl, 1 µL of 100 mm MgCl2, 1 µL of 1 µg/µL poly(dI-dC), and double-distilled water to a final volume of 20 µL. The reactions were incubated at room temperature (25°C) for 20 min, electrophoresed on 10% native polyacrylamide gels, and then transferred to N+ nylon membranes (Millipore) in 0.5× Tris-borate/EDTA buffer at 380 mA at 4°C for 60 min. Biotin-labeled DNA was detected using the LightShift Chemiluminescent EMSA Kit (no. 20148; Pierce). For super shift assays of the OsGRF6N-His or OsGRF10-GST protein, 1 µg of anti-His antiserum or 1 µg of anti-GST antiserum was added to the reaction before the 20-min incubation of the DNA fragments at room temperature.
ChIP Assays
Leaf samples (2 g) of 2-week-old rice seedlings of osgrf10 mutant and the DJ wild type were used for ChIP assays performed as described previously (Sun and Zhou, 2008; Li et al., 2011). The OsGRF10 antibody was used for immunoblot and ChIP analysis. The extracted DNAs were dissolved in 30 μL double-distilled water. Real-time PCR was used to determine the amounts of genomic DNA immunoprecipitated in the ChIP experiments. The primers used in ChIP assays are listed in Supplemental Table S3. The ChIP experiments were replicated three times independently and yielded similar results each time.
Subcellular Localization of OsGRF10
The full-length cDNA sequence of OsGRF10 was amplified with gene-specific primers (Supplemental Table S3). The PCR products were digested with XbaI and KpnI and then ligated into the pBI221 vector, in which the coding sequence of the OsGRF10 gene was fused to the 5′ end of the GFP gene in frame driven by the cauliflower mosaic 35S promoter. The fusion construct as well as the control vector with pBI221 alone were transformed into rice leaf and sheath protoplasts. After incubation of the protoplasts for 16 h in the dark at 30°C, GFP fluorescence was observed using a laser scanning confocal microscope (ZEISS LSM 510 META).
In Situ Hybridization
RNA in situ hybridization was performed as described previously (Xu et al., 2005). Digoxigenin-labeled hybrid probes were transcribed in vitro from cDNA of OsGRF6, OsGRF10, OsJMJ706, OsCR4, and ACTIN with gene-specific primers (Supplemental Table S3). Young florets from DJ and double mutant grf6/grf10 were used for RNA in situ hybridization assays. Slides were photographed under a microscope (Zeiss).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. DNA gel blot assay of miROE-transformed rice plants.
Supplemental Figure S2. Phenotypic analysis of floret inner organs in miROE8 and ZH10.
Supplemental Figure S3. Expression patterns of OsGRF genes in different tissues.
Supplemental Figure S4. Expression patterns of predicted targets of OsmiR396d and OsGRF genes.
Supplemental Figure S5. Expression pattern of OsmiR396d and its target genes.
Supplemental Figure S6. Molecular identification of the osgrf6 mutant.
Supplemental Figure S7. Molecular identification of the osgrf10 mutant.
Supplemental Figure S8. Expression of OsCR4 in young spikelets.
Supplemental Figure S9. Small leaves were occasionally found in miROE plants.
Supplemental Table S1. Microarray analysis of OsmiR396d OE plants (down-regulated genes with │log2 ratio│≥ 2. P value is showed as log2 of fold-change value).
Supplemental Table S2. Microarray analysis of OsmiR396d OE plants (up-regulated genes with │log2 ratio│≥ 2; P value is showed as log2 of fold-change value).
Supplemental Table S3. Primers used in this work.
Supplemental Table S4. Statistical analysis of different kinds of florets as well as expression level of JMJ706 in miROE and miROE/JMJ706OE crossed plants.
Supplementary Material
Acknowledgments
We thank Daoxiu Zhou (Institut de Biotechnologie des Plantes, Université Paris Sud 11, Orsay, France) and Yu Zhao (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China) for gifts of the jmj706 mutant and the JMJ706-overexpressing plants; Rongxi Jiang, Yuda Niu, and Xiaoxia Wang (Institute of Botany, Chinese Academy of Sciences) for assistance in rice gene transformation; and Wei Luo and Bo Wang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the rice hybridization and help in rice field management.
Glossary
- BD
binding domain
- BiFC
bimolecular fluorescence complementation
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- DJ
cv DongJin
- EMSA
electrophoretic mobility shift assays
- GST
glutathione S-transferase
- miRNA
microRNA
- qRT
quantitative reverse transcription
- miROE
OsMIR396d-overexpressing transgenic
Footnotes
This work was supported by the National Natural Sciences Foundation of China (grant nos. 31270331 and 31121065 to the innovation team).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aukerman MJ, Sakai H. (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46: 69–78 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Kim JH, Kende H. (2004) Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol 45: 897–904 [DOI] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z. (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D. (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SY, Xu YY, Liu HH, Mao ZW, Zhang C, Ma Y, Zhang QR, Meng Z, Chong K. (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong LL, Qian Q, Zhu KM, Tang D, Huang ZJ, Gao L, Li M, Gu MH, Cheng ZK. (2010) ELE restrains empty glumes from developing into lemmas. J Genet Genomics 37: 101–115 [DOI] [PubMed] [Google Scholar]
- Horigome A, Nagasawa N, Ikeda K, Ito M, Itoh JI, Nagato Y. (2009) Rice open beak is a negative regulator of class 1 knox genes and a positive regulator of class B floral homeotic gene. Plant J 58: 724–736 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. (2006) Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J Plant Res 119: 37–42 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H. (2011) ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 52: 112–124 [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh JI, Nagato Y. (2012) ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J 69: 168–180 [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Yasuno N, Oikawa T, Iida S, Nagato Y, Maekawa M, Kyozuka J. (2009) Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol 150: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y. (2007a) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Itoh M, Kyozuka J, Nagato Y. (2007b) Analyses of ABERRANT PANICLE ORGANIZATION1 (APO1) gene regulating the spikelet number in rice. Plant Cell Physiol 48: S52 [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, et al. (2000) leafy hull sterile1 Is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An SY, Kang HG, Moon S, Han JJ, Park S, Lee HS, An KS, An GH. (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Luo Q, Tong HN, Wang AJ, Cheng ZJ, Tang JF, Li DY, Zhao XF, Li XB, Wan JM, et al. (2011) An AT-hook gene is required for palea formation and floral organ number control in rice. Dev Biol 359: 277–288 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi DS, Kende H. (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH. (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH. (2009) The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol 151: 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Wynn AN, Franks RG, Hwang YS, Lim J, Kim JH. (2014) The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev Biol 386: 12–24 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Jia R, Yin C, Zong J, Kong H, Zhang D. (2010a) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li H, Xue D, Gao Z, Yan M, Xu W, Xing Z, Huang D, Qian Q, Xue Y. (2009) A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J 57: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang J, Qian Q, Xu Y, Zhang C, Xiao J, Du C, Luo W, Zou G, Chen M, et al. (2011) Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu P, Zhang H, Peng H, Zhang Q, Wang X, Wu X. (2007) Characterization and identification of a novel mutant fon(t) on floral organ number and floral organ identity in rice. J Genet Genomics 34: 730–737 [DOI] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang WX, Sunkar R. (2010b) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu DQ. (2014) Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol 164: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DM, Song Y, Chen ZX, Yu DQ. (2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Luo AD, Liu L, Tang ZS, Bai XQ, Cao SY, Chu CC. (2005) Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. J Integr Plant Biol 47: 745–752 [Google Scholar]
- Ma QB, Dai XY, Xu YY, Guo J, Liu YJ, Chen N, Xiao J, Zhang DJ, Xu ZH, et al. (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U. (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Pu CX, Ma Y, Wang J, Zhang YC, Jiao XW, Hu YH, Wang LL, Zhu ZG, Sun DY, Sun Y. (2012) Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J 70: 940–953 [DOI] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J. (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273: 23313–23320 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Kato H, Kitano H, Imai R. (2005) OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Genet Genomics 273: 1–9 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhou DX. (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105: 13679–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY. (2012) The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Kim JH, Kende H. (2000) A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol 122: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X. (2011) miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 62: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu Y, Zhang C, Ma Q, Joo SH, Kim SK, Xu Z, Chong K. (2008) OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YY, Wang XM, Li J, Li JH, Wu JS, Walker JC, Xu ZH, Chong K. (2005) Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol Biol 57: 773–784 [DOI] [PubMed] [Google Scholar]
- Yoo JH, Park CY, Kim JC, Heo WD, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, Kang YH, et al. (2005) Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY. (2012) Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J 70: 327–339 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Suzaki T, Tanaka W, Hirano HY. (2009) The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci USA 106: 20103–20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ, et al. (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA 105: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.