A mutated master regulator of zygotic embryogenesis is essential for creating somatic embryos and enhancing asexual propagation in Kalanchoë.
Abstract
Kalanchoë daigremontiana reproduces asexually by generating numerous plantlets on its leaf margins. The formation of plantlets requires the somatic initiation of organogenic and embryogenic developmental programs in the leaves. However, unlike normal embryogenesis in seeds, leaf somatic embryogenesis bypasses seed dormancy to form viable plantlets. In Arabidopsis (Arabidopsis thaliana), seed dormancy and embryogenesis are initiated by the transcription factor LEAFY COTYLEDON1 (LEC1). The K. daigremontiana ortholog of LEC1 is expressed during leaf somatic embryo development. However, KdLEC1 encodes for a LEC1-type protein that has a unique B domain, with 11 unique amino acids and a premature stop codon. Moreover, the truncated KdLEC1 protein is not functional in Arabidopsis. Here, we show that K. daigremontiana transgenic plants expressing a functional, chimeric KdLEC1 gene under the control of Arabidopsis LEC1 promoter caused several developmental defects to leaf somatic embryos, including seed dormancy characteristics. The dormant plantlets also behaved as typical dormant seeds. Transgenic plantlets accumulated oil bodies and responded to the abscisic acid biosynthesis inhibitor fluridone, which broke somatic-embryo dormancy and promoted their normal development. Our results indicate that having a mutated form of LEC1 gene in K. daigremontiana is essential to bypass dormancy in the leaf embryos and generate viable plantlets, suggesting that the loss of a functional LEC1 promotes viviparous leaf somatic embryos and thus enhances vegetative propagation in K. daigremontiana. Mutations resulting in truncated LEC1 proteins may have been of a selective advantage in creating somatic propagules, because such mutations occurred independently in several Kalanchoë species, which form plantlets constitutively.
Asexual reproduction is the simplest form of reproduction, occurring in many plants and animals. Various members of the Kalanchoë genus reproduce asexually through the ectopic formation of plantlets directly from differentiated tissues in the leaf (Garcês et al., 2007). These ectopic plantlets can be formed constitutively in some species or induced in response to various environmental cues and stresses (Garcês and Sinha, 2009). Previously, we have shown that leaf plantlet formation among the constitutive Kalanchoë plantlet-forming species such as Kalanchoë daigremontiana occurs by coopting both organogenesis and embryogenesis programs into the leaves (Garcês et al., 2007). K. daigremontiana somatic embryos develop symmetrically along the leaf margins in serrations, following a developmental program that resembles zygotic embryogenesis. Mature plantlets detach from the mother leaf and grow into new plants. We previously showed that the embryogenic LEAFY CONTYLEDON1 (LEC1) ortholog KdLEC1 is expressed in both somatic and zygotic embryos of K. daigremontiana (Garcês et al., 2007). LEC1 is known as an embryonic key regulator that is required for normal embryo development during early morphogenesis and to initiate and/or maintain the maturation phase and inhibit precocious embryo germination late in embryogenesis (Meinke, 1992; Meinke et al., 1994; West et al., 1994; Lotan et al., 1998; Vicient et al., 2000; Kwong et al., 2003; Braybrook and Harada, 2008). Furthermore, the loss-of-function mutation of LEC1 results in embryos that do not undergo developmental arrest and are nonviable because they are desiccation intolerant (Meinke, 1992; Meinke et al., 1994; West et al., 1994; Lotan et al., 1998; Vicient et al., 2000). lec1 embryos lack the protein and lipid bodies characteristic of wild-type embryos and show abnormal anthocyanin accumulation (Vicient et al., 2000). LEC1 is a member of CCAAT box-binding factors (CBFs) known as HEME-ACTIVATED PROTEIN3 (HAP3) (Lotan et al., 1998). The B domain of HAP3 subunits, including LEC1, contains conserved amino acid residues that account for their interaction with other CBF subunits and for DNA-binding activity of the CBF complex (Li et al., 1992; Xing et al., 1993; Sinha et al., 1996). In Arabidopsis (Arabidopsis thaliana), the B domain of LEC1 is necessary and sufficient for its activity in embryogenesis (Lee et al., 2003). However, KdLEC1 has a nonfunctional truncated B domain, which differs from the other LEC1-type proteins (Garcês et al., 2007). Among the Kalanchoë species that reproduce asexually, those with a mutated LEC1 protein produce plantlets constitutively and do not produce viable seeds. By contrast, Kalanchoë species that produce plantlets only upon stress induction have an intact LEC1 gene and are able to produce viable seed (Garcês et al., 2007). We have shown that a chimeric KdLEC1 protein, which comprised the KdLEC1 A and C domain and 11 LEC1-LIKE (L1L) residues at the 3′ end of the KdLEC1 B domain, was functional and able to complement the lec1 mutation by conferring desiccation tolerance to lec1-1 mutant seeds (Garcês et al., 2007). Therefore, we have speculated that the loss of LEC1 function in the clade of Kalanchoë species that forms plantlets constitutively appears to have been of a selective advantage in creating somatic propagules, because such mutations occurred in parallel at least twice within this clade (Garcês et al., 2007). Because the survival of leaf somatic embryos in these species is not affected by the loss of LEC1 function, due to the bypass of seed desiccation, we asked whether transforming K. daigremontiana plants with this functional chimeric KdLEC1 protein could help us understand the origin of the mutated KdLEC1 protein. Here, we show that transforming K. daigremontiana with a functional KdLEC1 gene inhibited and/or arrested leaf embryo development. Moreover, this functional KdLEC1 gene caused leaf embryos to accumulate oil bodies, which are normally found in seed embryos before germination. This suggests that the functional KdLEC1 imposes seed dormancy characteristics to these transgenic leaf embryos and adversely affects vegetative propagation in these plants.
LEC genes interact with plant hormone such as abscisic acid (ABA) and GA, establishing and responding to the high ratio of ABA to GA that is characteristic of the maturation phase of zygotic embryogenesis (Braybrook and Harada, 2008). In Arabidopsis zygotic embryogenesis, ABA levels increase coincidently with initiation of the maturation phase, remaining high throughout the maturation phase. The ABA levels decline late in maturation and reach a minimum in developing seedlings (Hays et al., 2001; Braybrook and Harada, 2008). By contrast, GA levels are low throughout the maturation phase but increase during germination (Hays et al., 2001; Ogawa et al., 2003). Furthermore, consistent with their roles in zygotic embryogenesis, the ABA-to-GA ratio is also known to affect somatic embryogenesis (SE). ABA treatments promote SE, while GA has an antagonistic role in SE in carrot (Daucus carota) cells (Tokuji and Kuriyama, 2003; Kikuchi et al., 2006). In addition, in many instances, seed dormancy can be released by applying fluridone, an inhibitor of carotenoid synthesis that causes a decrease in endogenous ABA levels (Yoshioka et al., 1998; Grappin et al., 2000). Application of fluridone to the embryos prior to ABA accumulation is proven to reduce ABA levels and prevent onset of embryo dormancy (Fong et al., 1983; Le Page-Degivry et al., 1990; Yoshioka et al., 1998). We have used fluridone to prevent ABA accumulation in dormant seed-like transgenic leaf embryos and investigated whether reducing ABA could break the dormancy in these defective leaf embryos. Here, we show that fluridone rescues the leaf embryo developmental defects caused by functional KdLEC1 in K. daigremontiana. Taken together, our results indicate that having a mutated form of LEC1 gene in K. daigremontiana (KdLEC1) is essential to bypass the leaf embryo dormancy and thus to generate viable plantlets.
RESULTS
Functional KdLEC1 Inhibits Leaf Plantlet Development
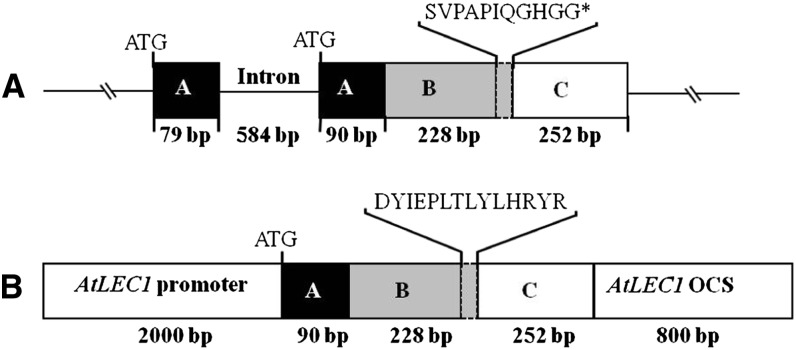
In our previous study, the native truncated KdLEC1 gene (Fig. 1A) was unable to rescue lec1 mutants (Garcês et al., 2007), while a functional KdLEC1 gene (Fig. 1B) was sufficient to rescue the Arabidopsis lec1 mutant phenotypes and confer desiccation tolerance to lec1-1 mutant seeds. To determine the role of KdLEC1 in asexual reproduction (SE and plantlet formation), we transformed K. daigremontiana with this functional KdLEC1 gene under the control of Arabidopsis LEC1 promoter and terminator (Fig. 1B). A total number of 23 transgenic plants showed abnormal plantlet phenotypes, and the presence of the neomycin phosphotransferase II (NPTII) transgene was confirmed in nine of those plants by Southern-blot analysis (data not shown). Among these transgenic plants analyzed, eight lines displayed severe phenotypes that affected the asexual process of reproduction.
Figure 1.
Schematic illustration of K. daigremontiana native and chimeric functional KdLEC1 protein structures. A, KdLEC1 native genomic clone (1,233 bp). The A, B, and C domains are represented by black, gray, and white boxes, respectively. The dashed, small gray box located at the 3′ end of the B domain represents the KdLEC1 unique amino acids. KdLEC1 gene has a 20-nucleotide deletion in the C-terminal region of the B domain that caused the addition of 11 amino acids (SVPAPIQGHGG) and a premature stop codon (*) in the B domain, resulting in a truncated form of the LEC1 protein. B, Chimeric KdLEC1 functional protein. The second exon of KdLEC1 A domain plus B and C domains were inserted into the LEC1 expression cassette. In this construct, only the KdLEC1 B domain 3′ end unique sequence (SVPAPIQGHGG) was substituted by L1L 3′ end sequence (DYIEPLTLYLHRYR).
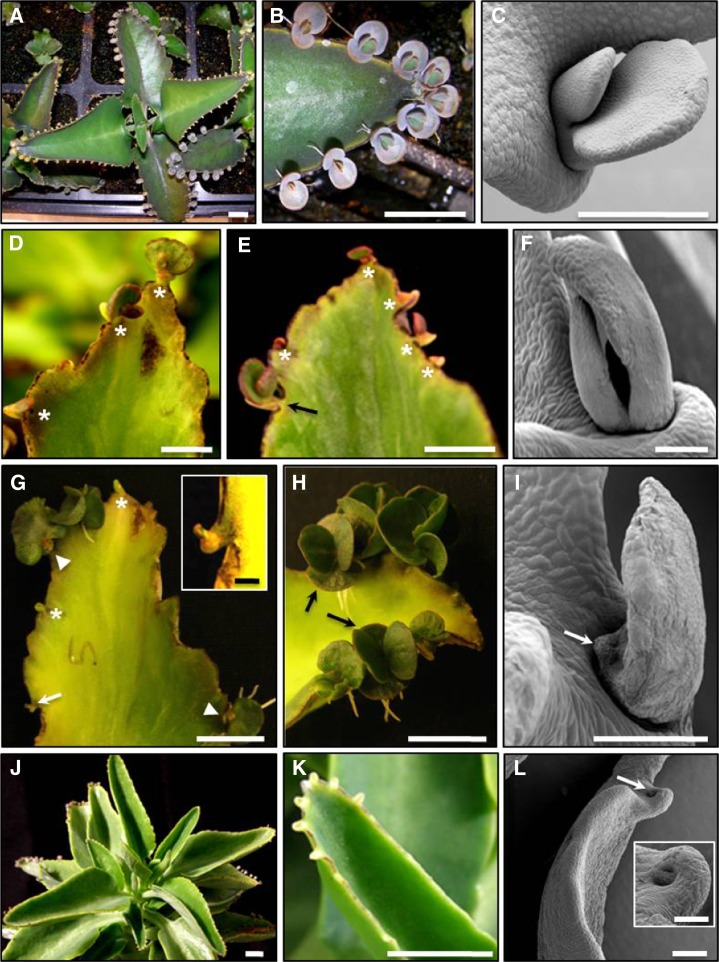
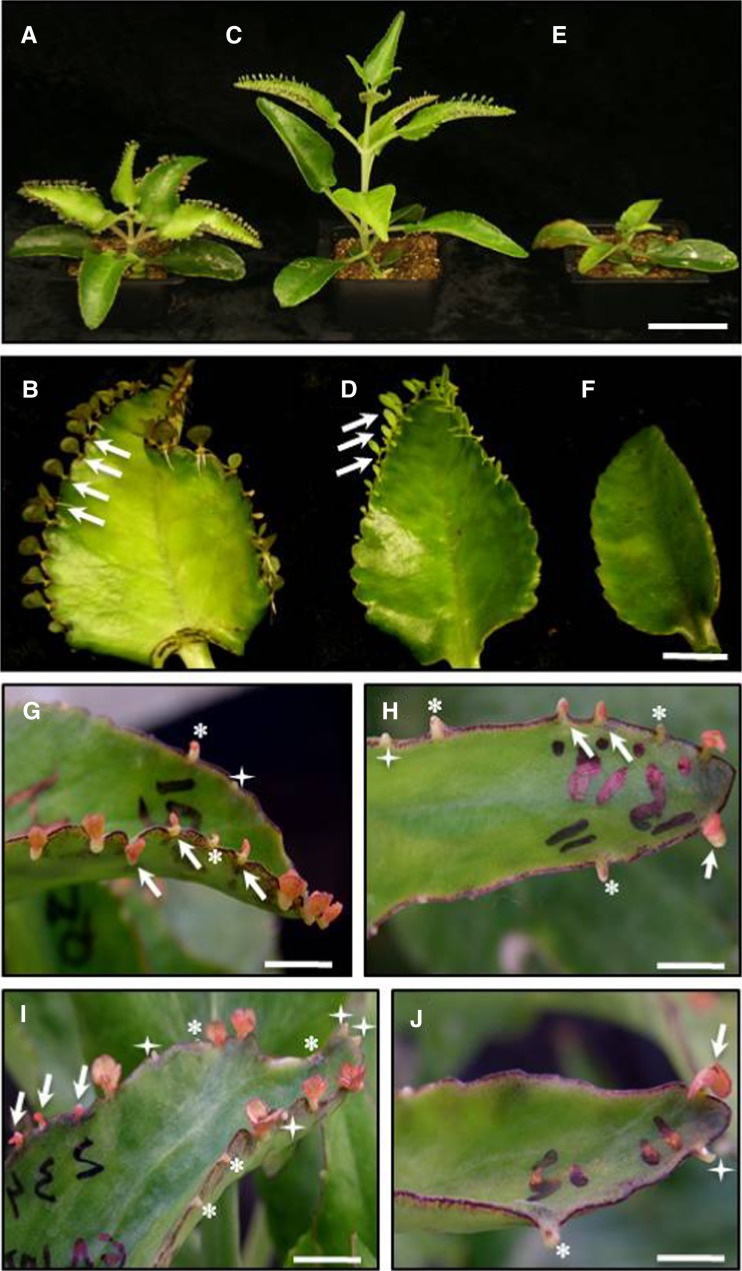
Restoration of KdLEC1 function resulted in several abnormalities during plantlet and leaf development (Fig. 2, A–I). The majority of the transgenic plants produced plantlets that arrested after forming only the first two cotyledon-like leaves (Fig. 2, D–G). In some cases, plantlets had abnormal anthocyanin accumulation (Fig. 2E) or produced only a single leaf (Fig. 2, G–I). In the strongest cases, plantlet formation was completely aborted (Fig. 2G, inset) or totally absent (empty pedestals; Fig. 2, G and K). In addition, the leaves of transgenic plants produced fewer serrations and pedestals, resulting in smoother leaf margins (Fig. 2, J–L). In lines with the most dramatic phenotypes, leaf pedestals were restricted to the tip of leaves (Fig. 2K). These plants also showed plantlet abortion at the earliest stages of development (Fig. 2L). Overall results show that expressing the functional KdLEC1 inhibits leaf plantlet development, disrupting the vegetative propagation process in this species.
Figure 2.
Phenotypic alterations of functional KdLEC1 transgenic plants. A, Nontransformed plant. B, Detail of a nontransformed leaf from A showing wild-type leaf plantlet development. C, SEM image of a wild-type plantlet developing on the leaf pedestal. D, Leaf of Kd2 transgenic plant showing reduced plantlet number (stars). E, Leaf of Kd4 transgenic plant with plantlets showing abnormal morphology or arrested development as well as anthocyanin accumulation (stars). Note that in rare cases, wild-type-looking plantlets were also formed (arrow). F, SEM image of an abnormal Kd4 transgenic plantlet. G, Kd5 transgenic leaf showing arrested (arrow and inset) or nonexistent plantlet development (empty pedestals, stars). In other plantlets, only one leaf was developed (arrowheads). H, Leaf of a Kd6 transgenic plant showing that few plantlets developed; however, of these, some were similar to wild-type plantlets (arrows). I, SEM image of a Kd5 transgenic plantlet showing aborted cotyledon-like leaf (arrow). J, Kd9 transgenic plant showing that pedestals were reduced in number and restricted to the tips of the leaves. K, Detail of a Kd9 transgenic leaf showing smooth leaf margins with empty pedestals at the tip of the leaf. L, SEM image of a Kd9 transgenic leaf showing empty pedestal (arrow and inset). Bars = 500 µm (SEM images) and 1 cm (others).
Functional KdLEC1 Reduced Leaf Serrations and Pedestal Formation
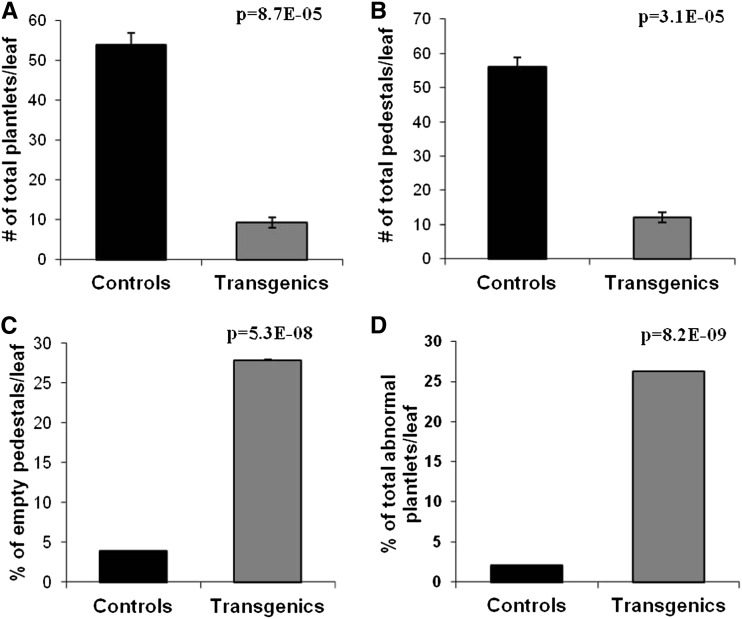
Given that wild-type plantlets always develop on pedestals in leaf serrations, we asked whether inhibition of plantlet formation in KdLEC1 transgenic plants was due to the loss of pedestal formation or to the early abortion of plantlet development itself after the formation of pedestal. Statistical analysis performed on a total of 23 transgenic plants and four nontransformed plants showed that the total number of plantlets per leaf was drastically reduced in the transgenic plants (P = 0.000087; Fig. 3A) and that this was correlated with a drastic reduction in the number of serrations and pedestals per leaf (P = 0.000031; Fig. 3B) compared with nontransformed leaves. However, detailed analysis revealed that the reduction of plantlet number was not only related to the reduction of pedestal number, but also due to the increase of empty pedestals or pedestals that formed plantlets that aborted early in development (P = 0.000000053; Fig. 3C). The expression of a KdLEC1 with a functional B domain in these transgenic plants also caused an increase in the number of abnormal-looking plantlets compared with control plants (P = 0.0000000082; Fig. 3D). In conclusion, functional KdLEC1 expression inhibited serration formation and consequently pedestal formation, which, in turn, reduced drastically the number of plantlets per leaf.
Figure 3.
Statistical analysis of the phenotypic alterations observed on leaves of functional KdLEC1 transgenic versus nontransformed plants. The number of plantlets (A) and pedestals (B) per leaf was drastically reduced in the transgenic plants. C, Transgenic plants showed an increased number of pedestals that do not form plantlets (empty pedestals). D, The number of plantlets with abnormal phenotypes was drastically increased in the transgenic plants. Data are reported as mean values ± sd (n = 23 for transgenic plants and n = 4 for nontransformed plants). Means of the two groups of plants were compared using the Student’s t test and were considered significantly different when P ≤ 0.001.
Functional KdLEC1 Conferred Seed Dormancy Characteristics to Leaf Embryos
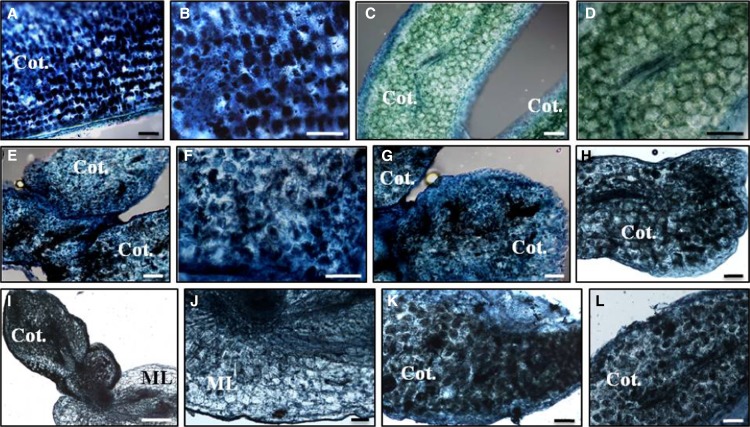
The precocious arrest of somatic embryos in KdLEC1 transgenics might result from the inappropriate initiation of the seed dormancy program in leaves. One characteristic of seed dormancy is the accumulation of oil bodies during seed maturation. We looked for oil body accumulation in wild-type and transgenic plantlets. Oil bodies were not detected in wild-type plantlets but were found in several transgenic plantlets (Fig. 4, E–L), similar to the positive controls (Fig. 4, A and B; pumpkin [Cucurbita maxima] seeds). This suggests that expressing a functional KdLEC1 gene in these plants was sufficient to induce seed dormancy characteristics in transgenic plantlets and disrupt vegetative propagation in this species.
Figure 4.
Detection of oil bodies stained with Sudan IV Black B in wild-type and transgenic functional KdLEC1 somatic embryos. A to B, Section of zygotic pumpkin cotyledon (positive control). B, Detail of cells in A showing oil body deposition (black spots). C to D, Section of a K. daigremontiana wild-type somatic embryo. D, Close view of C showing the absence of oil bodies in wild-type cotyledon cells. E to L, Sections of Kd2 (E and F), Kd4 (G), Kd5 (H), Kd6 (I and J), Kd7 (K), and Kd9 (L) transgenic somatic embryos. F, Close view of a cotyledon region in E. Note oil body deposition in these cells was similar to B. I, Section of Kd6 transgenic somatic embryo still attached to the mother leaf (ML). J, Close view of the ML in I showing the absence of oil bodies in the ML tissues. Bars = 10 μm.
Functional KdLEC1 Gene Is Expressed in Transgenic Leaf Margins
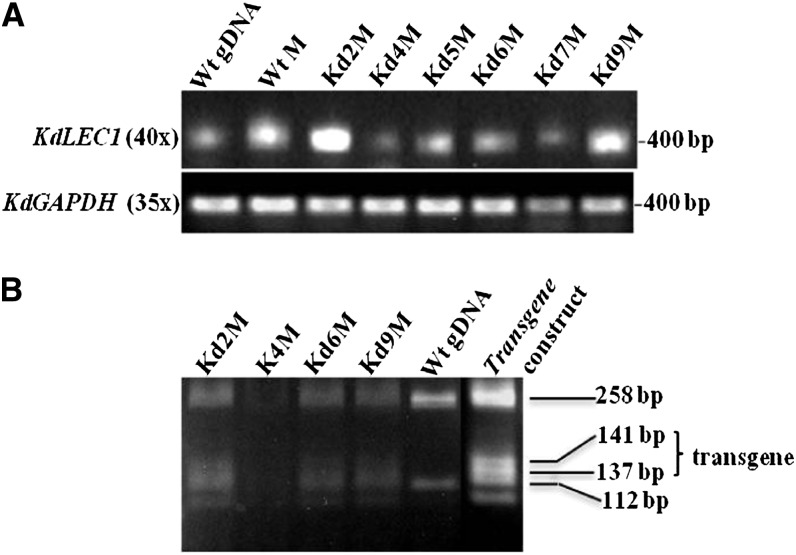
To determine the expression levels of truncated KdLEC1 (native) and functional KdLEC1 (transgene) in transgenic lines, we performed semiquantitative reverse transcription (RT)-PCR. RT-PCR results showed that the KdLEC1 expression was up-regulated in the leaf margins of the transgenic plants. However, these high levels of KdLEC1 expression represent both native and functional KdLEC1 gene expression. To differentiate the native from the transgene KdLEC1 transcripts, RT-PCR products were digested with Hpy991, which restriction sites are polymorphic between the native and functional KdLEC1 3′ end of the B domain. Hpy991 digestion profiling revealed that functional KdLEC1 transgene-specific bands (141 and 137 bp) and both transgene and native KdLEC1-specific bands (258 and 112 bp) were present in the KdLEC1 transcripts from the transgenic plants (Fig. 5B). These results indicate that the functional KdLEC1 transgene as well as endogenous KdLEC1 transcripts were expressed in all the transgenic plant leaf margins (Kd2, Kd4, Kd6, and Kd9). Together, these results suggest that the functional KdLEC1 gene under the regulation of Arabidopsis LEC1 promoter and terminator is being constitutively expressed in the K. daigremontiana leaf margins, causing the phenotypes observed in these transgenic lines.
Figure 5.
Semiquantitative RT-PCR analysis of endogenous (native gene) and functional KdLEC1 (transgene). A, RNA expression levels for native KdLEC1 in nontransformed plant tissues (wild type [Wt]) and in functional KdLEC1 transgenic plants (Kd). B, Digestion profiles of transgenic transcripts and nontransformed native KdLEC1 genomic DNA (gDNA). All KdLEC1 transgene transcripts shared two unique bands (141 and 137 bp), as did the transgene construct (positive control), showing that the functional KdLEC1 was expressed in all the transgenic plants analyzed. On the other hand, both transgenic and nontransformed plants show two common bands (258 and 112 bp), which correspond to endogenous KdLEC1 expression profile. RNA expression levels were normalized to KdGAPDH mRNA in A. M, Margins from third-stage leaf development (1.5-cm-long leaves).
GA3 Applications to Functional KdLEC1 Transgenic Plants Did Not Rescue Abnormal or Arrested Plantlet Development
Exogenous application of GAs (usually GA3 or GA4+7) is known to release embryo dormancy in seeds (Nuñez-Elisea and Davenport, 1998; Grappin et al., 2000; Kirmizi et al., 2010). We asked whether GA3 or uniconazole, a growth retardant that inhibits exclusively the biosynthetic pathway of GAs (Izumi et al., 1984), would affect leaf plantlet development in wild-type and arrested KdLEC1 transgenic plantlets. The application of uniconazole to K. daigremontiana wild-type plants (Fig. 6A) induced dwarfism (Fig. 6E) and total inhibition of leaf plantlet development (Fig. 6F). By contrast, the application of GA3 to K. daigremontiana wild-type plants resulted in the elongation of both plant (Fig. 6C) and leaf plantlets (Fig. 6D, arrows). These results confirmed that GA3 and uniconazole have antagonistic effects in wild-type K. daigremontiana plants and plantlets.
Figure 6.
Effect of GA3 and uniconazole (GA3 inhibitor) on wild-type and functional KdLEC1 transgenic plants. A and B, Nontreated wild-type plant (A) and leaf (B), showing normal plant and plantlet (arrows) development. C and D, The wild type treated with 50 µm GA3, showing elongated plant (C) and plantlets (D). E to F, Wild-type plant treated with 30 µm uniconazole showing plant dwarfism (E) and no plantlet formation (F). G, Functional KdLEC1 Kd4 transgenic leaf without treatment. Most leaves formed arrested (stars) or morphologically abnormal plantlets (arrows) and empty pedestals (four-point star). H to J, Functional KdLEC1 transgenic leaves treated with 50 (H) and 100 μm (I and J) GA3. GA3 applications were not able to rescue abnormal (arrows) or arrested (stars) plantlet development. Bars = 5 cm (A, C, and E) and 1 cm (B, D, and F–J).
LEC genes have been implicated in repressing GA activity in seed embryos (Curaba et al., 2004; Braybrook and Harada, 2008). We determined whether the arrest of somatic embryos in functional KdLEC1 transgenics was also mediated through GA3. If this is the case, the exogenous application of GA3 to these plantlets should restore their normal development. However, GA3 applications were not able to rescue the development of neither abnormal nor arrested plantlets. Although two GA3 concentrations (50 and 100 µm) were applied to transgenic Kd4 plantlets, none of them had an effect on plantlet development (Fig. 6, I and J). It is known that a fine balance of ABA to GA ratios seems to be required to promote the maturation phase, germination, and seedling growth (Gutierrez et al., 2007; Yamaguchi et al., 2007; Braybrook and Harada, 2008). These results suggest that factors other than the overall amount of GA3 levels, such as ABA levels, in transgenic leaf plantlets might regulate their abnormal or arrested development.
ABA Application Had No Effect on Releasing Leaf Plantlet Dormancy, while Reducing ABA Levels by Fluridone Restored Their Normal Development
ABA is thought to play a role in promoting embryo maturation and inhibiting germination (Le Page-Degivry and Garello, 1992; Gutierrez et al., 2007; Santos-Mendoza et al., 2008). To determine whether ABA promotes dormancy in leaf embryos, we treated leaves of functional KdLEC1 transgenic plants with the ABA biosynthesis inhibitor fluridone (Fig. 7). Fluridone applications had no immediate effects on the preexisting developmentally abnormal and dormant seed-like transgenic Kd2 leaf embryos (Fig. 7, C and D) compared with the controls (Fig. 7, A and B). Neither 30 nor 100 µm fluridone could rescue the dormancy phenotypes. However, 1 month after stopping the fluridone treatments, a dramatic effect on breaking leaf embryo dormancy was observed in the newly formed leaves, particularly with the 100 µm fluridone concentration (Fig. 7, E and F). A drastic improvement in plantlet development was seen in the newly formed leaves (Fig. 7E, L1) when compared with older leaves (Fig. 7E, L2) of the same plant. This is consistent with the fluridone effect on seed embryo dormancy breaking prior to ABA accumulation (Fong et al., 1983; Le Page-Degivry et al., 1990; Yoshioka et al., 1998). While fluridone is known to have multiple pleiotropic effects on the plant, our results show that fluridone can restore normal plantlet development, likely due to systemic ABA biosynthesis inhibition prior to its accumulation. Interestingly, ABA applications failed to aggravate the transgenic dormant seed-like phenotype and did not affect normal wild-type plantlet development (data not shown). One explanation for this result is that the window of developmental time during which ABA can impose dormancy is narrow and was missed during our experiments (Fong et al., 1983; White et al., 2000).
Figure 7.
Effect of fluridone treatments on functional KdLEC1 transgenic leaves. A and B, Nontreated transgenic plant (A) and leaf (B) with abnormal or arrested plantlets. C to F, Functional KdLEC1 Kd2 transgenic leaves treated with 30 (C and D) and 100 μm (E and F) fluridone. Neither 30 nor 100 µm fluridone treatments could immediately rescue the dormancy phenotypes of plantlets. E, Fluridone treatment was able to break plantlet dormancy on newly formed leaves (L1, arrows), while no effect was observed on fluridone-treated preexisting leaves (L2, stars). F, Detail of another leaf similar to L1 leaf in E showing well-developed plantlets due to fluridone applications (arrows). Note the bleaching effect in these leaves caused by fluridone treatments. Bars = 1 cm (A–F).
DISCUSSION
The survival of leaf somatic embryos in K. daigremontiana species is not affected by the loss of LEC1 function due to the bypass of seed desiccation. Therefore, we asked whether transforming K. daigremontiana plants with a functional chimeric KdLEC1 protein (Fig. 1B) could help explain the origin of the mutated KdLEC1. Here, we show that among the functional KdLEC1 transgenic plants, most lines displayed severe phenotypes that affected normal vegetative propagation (Figs. 2 and 3). Although few leaf embryos continued to grow similarly to the wild type, repairing KdLEC1 protein caused a drastic reduction in the total number of embryos formed per leaf in most transgenic plants (Fig. 3A). Because somatic embryos always form on pedestals in K. daigremontiana leaf serrations, a correlation between plantlet reduction and absence of pedestal formation was seen in transgenic plants. In lines with the most dramatic phenotypes, leaf serrations and pedestals were still formed, and yet somatic embryos failed to develop (Fig. 3C). Most of the leaf embryos formed in these transgenic plants were abnormal or arrested in development (dormant seed-like phenotype), and in some extreme cases, leaf embryos were aborted early in development (Fig. 3D). These results are consistent with other studies that have shown that ectopic-expressing LEC genes promote somatic embryo development and that, particularly for 35S::LEC1, seedlings arrest as embryo-like seedlings and fail to develop further (Lotan et al., 1998; Stone et al., 2001, 2008; Santos-Mendoza et al., 2005). Here, we show that contrary to nontransformed leaf embryos, functional KdLEC1 expression under the control of LEC1 promoter and terminator in K. daigremontiana resulted in dormant seed-like leaf embryos. Transgenic leaf embryos accumulated storage compounds, such as oil bodies similar to those of pumpkin seeds, at early developmental stages (Fig. 4). Interestingly, functional LEC1 gene could not rescue seed viability (data not shown). This may be due to the fact that the region of the Arabidopsis B domain used in the transgene was not sufficient to restore the function of KdLEC1 in zygotic embryogenesis and that other embryogenic genes as well as KdLEC1 are also involved in the loss of seed viability in this species. Over the course of evolution, mutations may have accumulated in other genes that affect embryo viability and may not be under selective pressures to purge these mutations.
Semiquantitative RT-PCR results showed that KdLEC1 expression levels were high in all the functional KdLEC1 transgenic lines analyzed, especially in Kd2 and Kd9, during leaf plantlet development when compared with nontransformed wild-type tissues (Fig. 5A). However, these high levels of KdLEC1 expression represent the expression levels of both transgene and endogenous KdLEC1 gene. Moreover, Hpy991 digestion results of these RT-PCR products showed that both transgenic and endogenous KdLEC1 transcripts were expressed in functional KdLEC1 transgenic leaf margins. Together, these results show that the functional KdLEC1 transgene is expressed in all the transgenic plant leaf margins analyzed and indicate that the phenotypes observed on plantlet development of the transgenic plants are due to the expression of the functional KdLEC1 gene under the regulation of LEC1 promoter and terminator. These results are consistent with other analysis showing that ectopic expression of LEC1, FUSCA3 (FUS3), and LEC2 genes induces the accumulation of lipid and protein reserves characteristic of developing seeds in cells of vegetative and reproductive tissues (Lotan et al., 1998; Stone et al., 2001, 2008; Santos-Mendoza et al., 2005). Moreover, seed-specific overexpression of maize (Zea mays) LEC1 (ZmLEC1) resulted in an average 35% increase in seed oil but reduced germination and vegetative growth in the field (Shen et al., 2010)
In Arabidopsis, the seed maturation program is controlled by a network of transcription factors that includes the B3 domain transcription factors (ABA-INSENSITIVE3 [ABI3], FUS3, and LEC2; Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001) and two LEC1-type HAP3 family CCAAT-binding factors, LEC1 and L1L (Lotan et al., 1998; Kwong et al., 2003). Given that these transcription factors have strong positive roles in promoting seed-specific programs, mechanisms have evolved to ensure that their expression is suppressed during vegetative growth. These results provide strong support for the conclusion that repairing KdLEC1 in K. daigremontiana plants is sufficient to induce seed dormancy characteristics to the transgenic plantlets and consequently affect plantlet germination and the vegetative propagation process in this species. The first suppressor of LEC genes to be characterized was PICKLE (PKL), which encodes a putative chromatin-remodeling factor belonging to the CHROMODOMAIN/HELICASE/DNA-BINDING3 (CHD3) family and regulates transition from embryonic to vegetative development (Ogas et al., 1999). In agreement with LEC overexpression phenotypes, pkl mutants accumulate storage macromolecules and give rise to embryogenic calli in culture. These embryonic characteristics are enhanced by GA synthesis inhibitors and repressed by exogenous GA treatments (Ogas et al., 1997), providing additional support for an already-described antagonistic interaction between the LEC transcription factors and GA (Curaba et al., 2004; Braybrook and Harada, 2008). We showed that GA3 and uniconazole, a growth retardant that inhibits exclusively the biosynthetic pathway of GAs (Izumi et al., 1984), have antagonistic effects in wild-type K. daigremontiana plants. It is known that GA levels are low throughout the maturation phase in both zygotic and somatic embryos but increase during embryo germination (Hays et al., 2001; Ogawa et al., 2003). This effect was evident in wild-type K. daigremontiana plants when applications of uniconazole resulted in the total suppression of leaf embryo development in all leaves. Our results showed that GA3 applications had no effect on transgenic leaf embryo developmental abnormalities. Although GA has been described to stimulate germination of dormant seeds in some species, there are many instances where GA alone is ineffective (Nonogaki, 2006), suggesting that GA is necessary but not sufficient for breaking the dormancy (Finkelstein et al., 2008).
The ectopic expression of LEC1 causes the activation of LEC2, FUS3, and ABI3 genes in Arabidopsis (Kagaya et al., 2005). Thus, the LEC genes may act in concert to regulate maturation programs, likely helping to establish and respond to the high ratio of ABA to GA that characterizes the maturation phase of embryogenesis (Le Page-Degivry and Garello, 1992; Gutierrez et al., 2007; Braybrook and Harada, 2008; Santos-Mendoza et al., 2008). ABA is thought to have several roles in promoting embryo maturation and inhibiting embryo germination. In fact, in addition to PKL, the maize VIVIPAROUS1 and its Arabidopsis ortholog, ABI3, designated as VIVIPAROUS1/ABI3-LIKE (VAL) factors (Jia et al., 2013), are also implicated in maintaining repression of the embryogenic genes in the vegetative tissues. Our results show that repairing KdLEC1 protein was sufficient to transform leaf embryos into a dormant seed-like developmental state, followed by the accumulation of storage compounds characteristic of seed maturation phase. The application of fluridone directly to preexisting abnormal and dormant leaf embryos did not improve their development, indicating that fluridone is ineffective at rescuing embryo dormancy after high ABA levels are established. However, a striking effect was seen when new leaves were formed in these same plants (Fig. 7, E and F). The inhibition of de novo ABA biosynthesis by fluridone in the newly formed leaves of these plants caused a complete break of leaf embryo dormancy and induced their “germination.” Our results are in agreement with two other reports, which showed that the applications of fluridone to developing embryos, before the increase of endogenous ABA levels, prevented both ABA accumulation and development of embryo dormancy (Fong et al., 1983; White et al., 2000). On the other hand, none of the exogenous ABA applications could aggravate the transgenic dormant seed-like phenotype, nor did they affect normal wild-type leaf embryo development (data not shown). This indicates that ABA alone is not sufficient to induce dormancy to K. daigremontiana leaf embryos. In fact, the ABA-deficient maize mutant kernels are viviparous and precociously germinate in spite of a large contribution of ABA from the maternal plant (White et al., 2000). These results have led to the supposition that a threshold level of ABA at the appropriate time is required to block viviparous development (Fong et al., 1983; White et al., 2000).
All together, our results strongly suggest that repairing KdLEC1 in K. daigremontiana transgenic plants imposed dormancy to leaf embryos, affecting vegetative propagation in these plants, likely through increased ABA levels in leaves. Given that the functional KdLEC1 gene drastically altered K. daigremontiana asexual propagation, it is likely that the switch from sexual to asexual reproduction in the evolution of this species was triggered by truncation of the KdLEC1 protein. Mutations resulting in truncated LEC1 proteins could be of a selective advantage in creating somatic propagules. This is supported by the fact that such mutations occurred independently (at least twice) in species that propagate solely sexually within the clade of Kalanchoë genus (Garcês et al., 2007). The occurrence of the mutated KdLEC1 also appears to facilitate coevolution in which the loss of sexual reproduction coincides with the gain of plantlet formation in these species. The mutated KdLEC1 gene may have lowered the efficiency of zygotic embryogenesis by making these embryos desiccation intolerant, while it allowed plantlets to bypass the seed-like embryo dormancy program and assure their survival. It is also likely that once sexual reproduction was abrogated, the absence of selective pressure to maintain the various components of the process caused multiple mutations that make the entire SE program more stable. We previously showed that in addition to KdLEC1, another embryogenic marker, KdFUS3, is also expressed during leaf embryo development (Garcês et al., 2007). Future studies on the role of other embryogenic genes, such as KdFUS3, as well as developmental cues regulating these genes in the leaves will provide further insights in the evolution of such an efficient and adaptive asexual propagation trait.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Kalanchoë daigremontiana wild-type and transgenic plants were grown in the greenhouse at 29°C in a photoperiod of 16 h/8 h. The first whole leaves counting from the top of the plants were used for plant transformation. One-year-old transgenic plants were used for statistical analysis. Three-week-old plantlets from transgenic and nontransformed plants were grown in the conditions described above and were used for hormone treatments.
Vector Construction and Plant Transformation
To investigate the effects of repairing K. daigremontiana KdLEC1 protein in sexual and asexual reproduction, we transformed these plants with the same construct described in Garcês et al. (2007). This construct contains the K. daigremontiana native A, starting with the second ATG, and the whole C domain of KdLEC1 protein. In addition, the 12 unique amino acids of KdLEC1 truncated B domain (SVPAPIQGHGGZ) were substituted for the corresponding specific residues of Arabidopsis (Arabidopsis thaliana) L1L B domain (DYIEPLTLYLHRYR) to reconstitute a complete B domain (Fig. 1B). This construct is driven by the Arabidopsis LEC1 promoter and terminator (Gleave, 1992; Lee et al., 2003) and was cloned in the Gateway pMDC100 cassette C1, which carries the NPTII gene conferring kanamycin resistance to positive transformants. The Agrobacterium tumefaciens LBA4404 strain carrying this construct was used to transform K. daigremontiana leaf explants according to the protocol described in Garcês et al. (2007) and Garcês and Sinha (2009). The NPTII gene was used to select transformed plants as well as to probe the Southern blots of transgenic plants to confirm independent lines.
Expression Analysis
Total RNA was isolated from different tissues by using RNeasy Mini Kit (Qiagen). Leaf margins at the third stage of development (1.5-cm-long leaves; Garcês et al., 2007) were collected for the wild type and for six independent transgenic plants and frozen in liquid nitrogen and stored at –80°C for total RNA extractions. The KdLEC1 RNA expression levels were assessed using semiquantitative RT-PCR. First strand complementary DNA was generated from 5 µg DNase-treated RNA for each sample in a 40-µL reaction volume by using SuperScript II reverse transcriptase (Invitrogen Life Technologies) according to Untergasser protocol specifications (Untergasser, 2008).
Given that the sequence difference between native and functional KdLEC1 genes are only 14 nucleotides at the 3′ end of the B domain, the KdLEC1 RNA expression levels of the native and functional were determined using the same set of primers: KdLEC1 (KdLEC1AF1/KdLEC1CR2, 5′-CAGCAACGGCGCCGAGTGTTC-3′/5′-ATGATGATGCCCCATATGAAACGTC-3′). K. daigremontiana GLYCERALDEHYDE3-PHOSPHATE DEHYDROGENASE (KdGAPDH) was used as the constitutively expressed control gene (KdGAPDH1/KdGAPDH2, 5′-GGAGCAGAGATAACAACCTTC-3′/5′-TCCATTCATCAACACAGACTAC-3′). Typically, for RT-PCR analysis, each 20-µL PCR mixture contained 2 µL of complementary DNA (as template), 1.25 units of Taq DNA Polymerase, PCR buffer, and 50 mm MgCl2 (Promega), 2.5 mm each deoxyribonucleotide triphosphate (Invitrogen Life Technologies), and 10 mm each primer. DNA fragments were amplified by using the following conditions: 3 min at 94°C, 35 to 40 cycles of 30 s at 94°C, 30 s at 60°C, 40 s at 72°C, and 5 min at 72°C. To differentiate between native and functional KdLEC1 RNA expression levels, both nontransformed and transgene RT-PCR products were digested with Hpy991 restriction enzyme.
Statistical Analysis
The total number of plantlets and pedestals were collected for both transformed and nontransformed plants. Data were collected from 23 transgenic plants versus four nontransformed plants. Means of the two groups of plants were compared using the Student’s t test and were considered significantly different when P ≤ 0.001. Statistical analyses were performed using the R statistical programming environment.
Scanning Electron Microcopy (SEM) Analysis
Samples for SEM were fixed and observed as described by Garcês et al. (2007) and Garcês and Sinha (2009). Electronic images were obtained directly from the SEM with a Hitachi S-3500 N scanning electron microscope (Hitachi Science Systems) at 5 to 20 kV accelerating voltage and processed with Adobe Photoshop version 6.0 (Adobe Systems).
Microscopy and Oil Bodies Detection
Early developmental stages of transformed and nontransformed plantlets were collected and sectioned using the vibratome (Vibratome Series 1000 Sectioning System, Technical Products International). Fresh sections were placed on slides and were incubated in 70% (v/v) ethanol for a few seconds at RT, followed by staining with 0.07% (w/v) Sudan IV and Sudan Black B solution in 70% (v/v) ethanol for 10 min for oil bodies detection. Sections were washed in 50% (v/v) ethanol for 1 min at RT and mounted in 100% (v/v) glycerine (Ruzin, 1999). Pumpkin (Cucurbita maxima) seed sections were used as a positive control for the oil bodies detection. Samples were viewed with a Nikon Eclipse E600 microscope equipped with a RT Color Spot (Diagnostic Instruments) digital camera. Electronic images were acquired by using SPOT image program version 3.5 (Diagnostic Instruments) and processed with Adobe Photoshop version 6.0 (Adobe Systems).
Hormone Treatments
To test the effect of several plant hormones with well-known roles in embryo maturation, germination, and seedling growth, we have performed the following assays in K. daigremontiana plantlet development. (1) First, we have investigated the effect of both GA3 and of uniconazole, an inhibitor of GAs in wild-type plantlets (untransformed plants). To do this, we sprayed 1-month-old wild-type plants with a 50 µm GA3 solution made from a 10 mm GA3 stock solution (Sigma). These plants were also treated with 30 µm uniconazole [(E)-l-(4-chlorophenyl)-4,4-dimethyl-2-(l,2,4-triazole-l-yl)-l-penten-3-ol; Sumagic, Valent Corporation] solution, and as a control assay, plants were sprayed with water (1% [v/v] Tween 20 in water). Both GA3 and uniconazole solutions were made in water, and treatments were performed for a total period of 2 months. (2) Different leaves from a repaired KdLEC1-transformed (Kd4) plant were treated with 50 and 100 µm GA3 solutions. To study their effect on the developmental abnormal and arrested transgenic plantlets, we applied five drops per plantlet using a plastic Pasteur pipette every 2 d for 1 month. Control assays were also applied to plantlets of nontransformed plants, with similar early development stages to transgenic plantlets. Applications were performed to a total of four leaves per plant, and each treatment was repeated in five plants. Leaves were labeled with a marker to facilitate the experiment and the applications. (3) The same experimental design described above was also performed for ABA (PhytoTechnology Laboratories) and for fluridone (l-methyl-3-phenyl-5-[3-(trifluoromethyl)phenyl]-4(1H)-pyridinone; PhytoTechnology Laboratories), an ABA inhibitor. Untransformed and functional KdLEC1-transformed plants were treated for 1 month with 30 and 100 µm ABA as well as with 30 and 100 µm fluridone. Phenotypes were recorded at the end of the treatment and again 1 month later. Both solutions were made from 1 mg mL–1 stock solutions, and ABA solution was made in water and fluridone in dimethyl sulfoxide.
All plants used on these assays were 1 year old, and 1% (v/v) of Tween 20 (Sigma) was added to all solutions to increase their permeability. All stock solutions were protected from light and saved at 4°C for up to 6 months.
Glossary
- ABA
abscisic acid
- CBF
CCAAT box-binding factor
- SE
somatic embryogenesis
- RT
reverse transcription
- SEM
scanning electron microscopy
Footnotes
This work was supported by the National Science Foundation (grant nos. GEPR 0820854 and IBN 0344743 to N.R.S.).
Articles can be viewed online without a subscription.
References
- Braybrook SA, Harada JJ. (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G. (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136: 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Fong F, Smith JD, Koehler DE. (1983) Early events in maize seed development: 1-methyl-3-phenyl-5-(3-(trifluormethyl)phenyl)-4-(1H)-pyridinone induction of vivipary. Plant Physiol 73: 899–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcês H, Sinha N (2009) The ‘mother of thousands’ (Kalanchoë daigremontiana): a plant model for asexual reproduction and CAM studies. In Emerging Model Organism. A Laboratory Manual, Vol 2. Cold Spring Harbor Laboratory Press, New York [DOI] [PubMed] [Google Scholar]
- Garcês HM, Champagne CE, Townsley BT, Park S, Malhó R, Pedroso MC, Harada JJ, Sinha NR. (2007) Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc Natl Acad Sci USA 104: 15578–15583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M. (2000) Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210: 279–285 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Hays DB, Mandel RM, Pharis RP. (2001) Hormones in zygotic and microspore embryos of Brassica napus. Plant Growth Regul 35: 47–58 [Google Scholar]
- Izumi K, Yamaguchi I, Wada A, Oshio H, Takahashi N. (1984) Effects of a new plant growth retardant (E)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1,2,4 triazol-1-yl)-1-penten-3-ol (S-3307) on the growth and gibberellin content of rice plants. Plant Cell Physiol 25: 611–617 [Google Scholar]
- Jia H, McCarty DR, Suzuki M. (2013) Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol 163: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Sanuki N, Higashi K, Koshiba T, Kamada H. (2006) Abscisic acid and stress treatment are essential for the acquisition of embryogenic competence by carrot somatic cells. Planta 223: 637–645 [DOI] [PubMed] [Google Scholar]
- Kirmizi S, Guleryuz G, Arslan H, Sakar FS. (2010) Effects of moist chilling, gibberellic acid, and scarification on seed dormancy in the rare endemic Pedicularis olympica (Scrophulariaceae). Turk J Bot 34: 225–232 [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page-Degivry MT, Barthe P, Garello G. (1990) Involvement of endogenous abscisic acid in onset and release of Helianthus annuus embryo dormancy. Plant Physiol 92: 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page-Degivry MT, Garello G. (1992) In situ abscisic acid synthesis: a requirement for induction of embryo dormancy in Helianthus annuus. Plant Physiol 98: 1386–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ. (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Mantovani R, Hooft van Huijsduijnen R, Andre I, Benoist C, Mathis D. (1992) Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res 20: 1087–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto MA, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Miséra S. (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Meinke DW. (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H. (2006) Seed germination: the biochemical and molecular mechanisms. Breed Sci 56: 93–105 [Google Scholar]
- Nuñez-Elisea R, Davenport TL. (1998) Gibberellin and temperature effects on dormancy and shoot morphogenesis of mango (Mangifera indica L.). Sci Hortic (Amsterdam) 77: 11–21 [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C. (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin SE (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L. (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC. (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Kim IS, Sohn KY, de Crombrugghe B, Maity SN. (1996) Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol Cell Biol 16: 328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci USA 105: 3151–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuji Y, Kuriyama K. (2003) Involvement of gibberellin and cytokinin in the formation of embryogenic cell clumps in carrot (Daucus carota). J Plant Physiol 160: 133–141 [DOI] [PubMed] [Google Scholar]
- Untergasser A (2008) cDNA synthesis using superscript II. Untergasser's Lab. http://www.untergasser.de/lab/protocols/cdna_synthesis_superscript_ii_v1_0.htm (July 3, 2013)
- Vicient CM, Bies-Etheve N, Delseny M. (2000) Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J Exp Bot 51: 995–1003 [DOI] [PubMed] [Google Scholar]
- West MAL, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CN, Proebsting WM, Hedden P, Rivin CJ. (2000) Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol 122: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Fikes JD, Guarente L. (1993) Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J 12: 4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Nambara E (2007) Seed Development, Dormancy, and Germination, Vol 27. Blackwell Publishing, Oxford [Google Scholar]
- Yoshioka T, Endo T, Satoh S. (1998) Restoration of seed germination at supra optimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol 39: 307–312 [Google Scholar]