Transgenes specifically targeting fatty acid synthesis, triacylglycerol synthesis, and triacylglycerol breakdown lead to an additive effect on seed oil content in Arabidopsis.
Abstract
Increasing the yield of oilseed crops is an important objective for biotechnologists. A number of individual genes involved in triacylglycerol metabolism have previously been reported to enhance the oil content of seeds when their expression is altered. However, it has yet to be established whether specific combinations of these genes can be used to achieve an additive effect and whether this leads to enhanced yield. Using Arabidopsis (Arabidopsis thaliana) as an experimental system, we show that seed-specific overexpression of WRINKLED1 (a transcriptional regulator of glycolysis and fatty acid synthesis) and DIACYLGLYCEROL ACYLTRANSFERASE1 (a triacylglycerol biosynthetic enzyme) combined with suppression of the triacylglycerol lipase SUGAR-DEPENDENT1 results in a higher percentage seed oil content and greater seed mass than manipulation of each gene individually. Analysis of total seed yield per plant suggests that, despite a reduction in seed number, the total yield of oil is also increased.
Vegetable oils (triacylglycerols [TAGs]) are an important part of our bioeconomy. They make a significant contribution to human and livestock nutrition but also provide feedstock for the production of various industrial chemicals, including biodiesel (Durrett et al., 2008; Dyer et al., 2008). The volume of vegetable oil that is produced globally has increased very substantially in recent decades and now is in excess of 150 million metric tons per year (http://www.fas.usda.gov/data/oilseeds-world-markets-and-trade). There is an urgent need to improve the yield of oil crops to meet societies’ ever-growing demand for this renewable feedstock (Lu et al., 2011). The genetic architecture of yield in oilseed crops is complex (Würschum et al., 2012). Incremental improvements continued to be made through conventional breeding, combined with better crop management (Weselake et al., 2009). However, one approach that biotechnologists are exploring is the potential of transgenic technologies to boost the metabolic flux of carbon to oil within the developing seed, thereby partitioning more of the plant’s carbon to this product (Weselake et al., 2009).
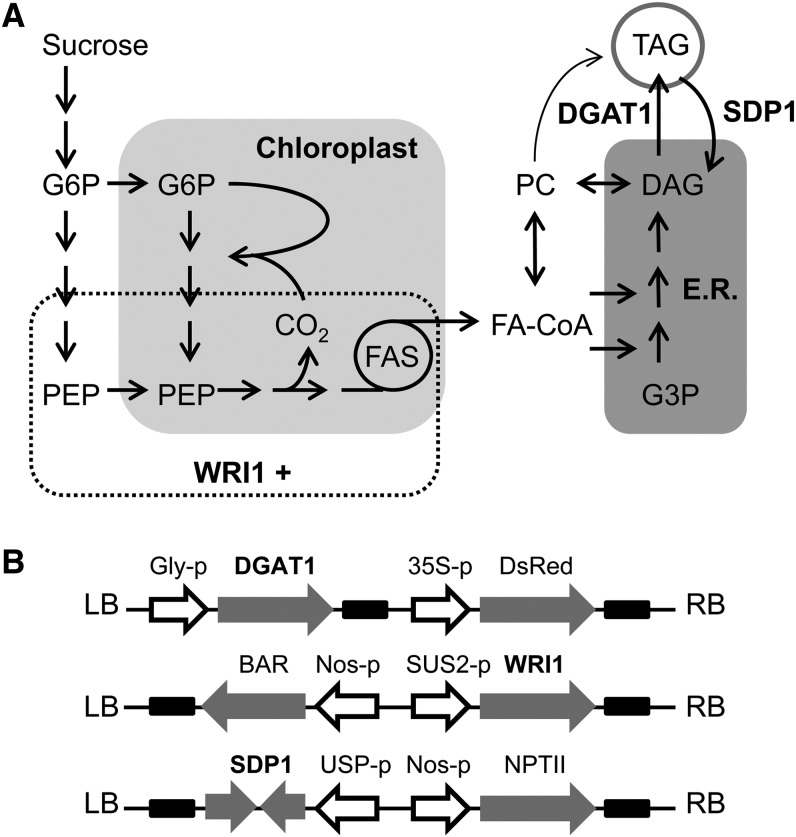
The pathway for the conversion of Suc to oil in developing seeds, such as those of the model plant Arabidopsis (Arabidopsis thaliana), is complex and involves multiple subcellular compartments (Schwender et al., 2004; Durrett et al., 2008; Fig. 1A). Briefly, Suc from maternal tissues is imported into the cell and converted to glucose-6-phosphate (G6P) via either a Suc synthase- or invertase-dependent pathway (Barratt et al., 2009). The G6P is then metabolized by glycolysis to pyruvate, in either the cytosol or chloroplast, with transport between the compartments primarily at the level of G6P and phosphoenolpyruvate (Schwender et al., 2004). Pyruvate within the chloroplast is converted to acetyl-CoA by the pyruvate dehydrogenase complex and used to make long-chain fatty acids utilizing a multisubunit acetyl-CoA carboxylase and type II fatty acid synthase (Durrett et al., 2008). In the presence of light, a proportion of the CO2 lost during the conversion of pyruvate to acetyl-CoA is also refixed via a Rubisco bypass (Schwender et al., 2004). Fatty acids are exported from the chloroplast and activated to acyl-CoAs, which serve as the substrates for the esterification of glycerol-3-phosphate (G3P) at the endoplasmic reticulum to form glycerolipids (Durrett et al., 2008). Most of the diacylglycerol (DAG) produced by this pathway is transiently converted to phosphatidylcholine (PC) before it is used to make TAG (Bates et al., 2012). Fatty acids are also rapidly shuttled in and out of PC from the acyl-CoA pool via an acyl-editing cycle that allows them to be modified and redistributed (Bates et al., 2012). Finally, DAG acyltransfereases esterify DAG to make TAG using either acyl-CoA (e.g. DIACYLGLYCEROL ACYL TRANSFERASE1 [DGAT1]) or PC (PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE1) as an acyl donor (Zhang et al., 2009).
Figure 1.
Simplified diagrams illustrating the pathway for oil metabolism in developing Arabidopsis seeds (A) and three T-DNA constructs designed to manipulate different stages in the pathway by seed-specific overexpression of WRI1, DGAT1, and RNAi suppression of SDP1 (B). E.R., Endoplasmic reticulum; FA-CoA, fatty acyl-CoA; FAS, fatty acid synthase; LB, left border; PEP, phosphoenolpyruvate; RB, right border.
A number of studies have established that seed oil content can be increased by changing the expression levels of individual enzymes involved in oil metabolism (Roesler et al., 1997; Zou et al., 1997; Jako et al., 2001; Vigeolas et al., 2007; Kelly et al., 2013a; Kim et al., 2014). However, the impact of this approach is theoretically limited. The majority of control that is exerted over the rate of carbon flux through a metabolic pathway rarely lies predominantly with one step and is usually distributed between many (Fell, 1997). Top-down metabolic control analysis, performed on developing seeds, suggests that this is the case for oil synthesis (Weselake et al., 2008; Tang et al., 2012). Constraint-based metabolic modeling also predicts that seed oil content is sensitive to multiple reactions (Schwender and Hay, 2012). An alternative approach to targeting individual enzymes is to manipulate the expression of transcriptional master regulators that govern the expression of multiple enzymes in the metabolic pathway (Broun, 2004; Grotewold, 2008). This too has proven to be an effective means to enhance seed oil content (Cernac and Benning, 2004; Shen et al., 2010). However, this approach is less precise and can lead to unwanted side effects when the chosen regulator impacts physiologically important processes other than the chosen metabolic pathway (Century et al., 2008; Shen et al., 2010).
Given these constraints, one rational approach to metabolic pathway engineering is to stack genes (i.e. simultaneously alter the expression of multiple genes) and test empirically whether combinations provide a better effect. Surprisingly, there are currently no academic reports of gene stacking to enhance seed oil content, although several recent studies have successfully applied this approach to manipulate oil metabolism in leaves (Sanjaya et al., 2011; Fan et al., 2013; Kelly et al., 2013b; Vanhercke et al., 2013, 2014; Winichayakul et al., 2013). In general these studies suggest that a push-pull-protect strategy may be effective (Fig. 1A). For example, expression of the WRINKLED1 (WRI1) transcription factor can be used to drive glycolysis and fatty acid synthesis (Cernac and Benning, 2004), expression of DIACYLGLYCEROLACYL TRANSFERASE1 (DGAT1) can be used to draw fatty acids into TAG (Jako et al., 2001), and turnover can also be minimized by disrupting lipolysis (Fan et al., 2013; Kelly et al., 2013b; Vanhercke et al., 2013; Winichayakul et al., 2013). Here, we investigate the effects of stacking these gene modifications on seed oil content and yield in Arabidopsis.
RESULTS AND DISCUSSION
Metabolic control over oil production in developing seeds resides in both the production of fatty acids and their utilization for TAG assembly (Weselake et al., 2008; Tang et al., 2012). Previous studies have shown that overexpression of the WRI1 transcription factor activates the expression of multiple genes associated with lower glycolysis and fatty acid synthesis (Baud et al., 2007; To et al., 2012) and can enhance oil content in seeds of Arabidopsis (Cernac and Benning, 2004). However, WRI1 has little influence over the expression of genes involved in TAG assembly and packaging (Baud et al., 2007; To et al., 2012). The key enzyme responsible for TAG assembly is encoded by DGAT1 (Katavic et al., 1995; Zhang et al., 2009), and overexpression of this gene can also enhance seed oil content (Jako et al., 2001).
To test whether combined expression of WRI1 and DGAT1 can have an additive effect on seed oil content, and also whether this might be further augmented by blocking TAG turnover by RNA interference (RNAi) suppression of the lipase SUGAR-DEPENDENT1 (SDP1; Kelly et al., 2013a; Kim et al., 2014), transfer DNA (T-DNA) constructs were designed for the manipulation of each gene under the control of a seed-specific promoter (Fig. 1B). For WRI1 overexpression, the Arabidopsis SUCROSE SYNTHASE2 (SUS2) promoter was chosen. This promoter drives strong expression with a developmental pattern that overlaps that of WRI1 (Baud et al., 2004; Winter et al., 2007; Angeles-Núñez and Tiessen, 2012) and has previously been used to drive the expression of a WRI1 homolog from maize (Zea mays; OVULE DEVELOPMENT PROTEIN2) without adverse effects on seed germination (Meyer et al., 2010). The glycinin promoter from soybean (Glycine max) was used for DGAT1 overexpression because it drives strong expression throughout embryo maturation (Fatihi et al., 2013), and an SDP1 RNAi construct was placed under the control of the UNKNOWN SEED PROTEIN (USP) promoter from Vicia faba, which is also active throughout embryo maturation (Kelly et al., 2013a).
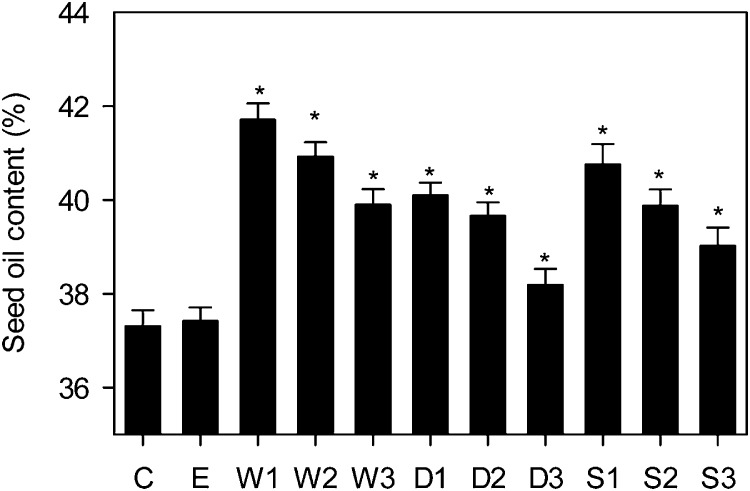
Each construct was transformed into Arabidopsis Columbia-0 (Col-0) plants using Agrobacterium tumefaciens, and approximately 40 transformants were screened for elevated seed percentage oil content in the T2 generation using low-resolution time domain NMR spectroscopy (Hobbs et al., 2004). For each construct, transformants exhibited a range in percentage seed oil content (Supplemental Fig. S1). Transgenic lines with the highest apparent percentage oil content were then taken to the T3 generation, and three homozygous single-insert lines were identified for each construct by segregation analysis. Twelve individual plants of the wild type, an empty vector control, and the WRI1 (W1–W3), DGAT1 (D1–D3), and RNAi SDP1 (S1–S3) lines were grown under controlled conditions in a random block design experiment, and the percentage oil content was measured (Fig. 2). The selected W, D, and S lines all exhibited statistically significant (P < 0.05) increases in percentage seed oil content versus either the wild type or the empty vector control. WRI1 overexpression tended to result in the highest percentage seed oil content. However, the use of different promoters means that it is not possible to draw any strong conclusion concerning the comparative strength of effect of each transgene.
Figure 2.
Seed oil contents of independent transgenic lines expressing WRI1, DGAT1, or RNAi SDP1 under the control of seed-specific promoters. Values are means ± se of measurements on seeds from individual plants (n = 12) of each genotype grown in controlled conditions. Asterisks denote statistically significant differences from the wild type (lsd test), where significant differences between genotypes were detected (F test). C is the wild type, E is an empty vector control, and W, D, and S are independent homozygous T3 lines expressing the WRI1, DGAT1, and RNAi SDP1 transgenes.
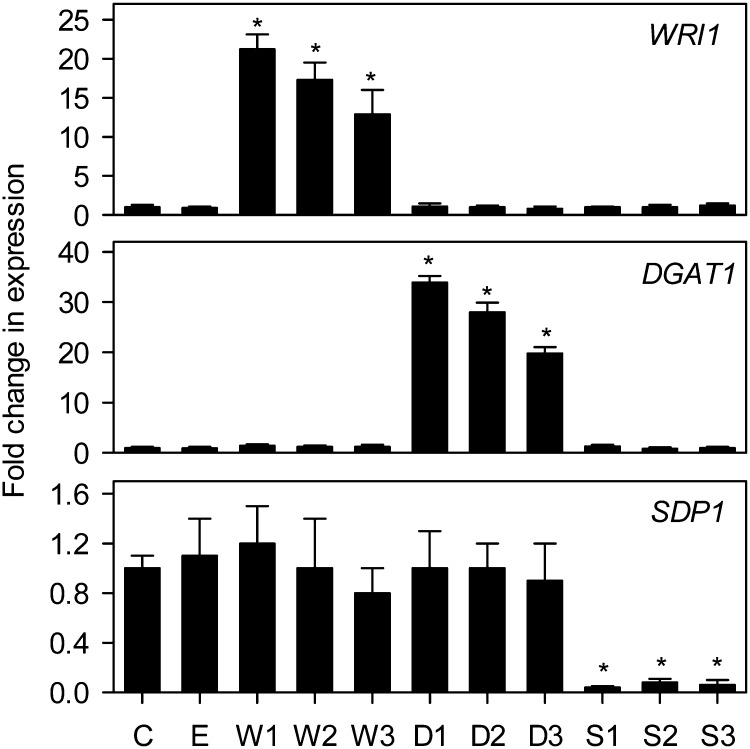
To confirm that expression of the target genes (WRI1, DGAT1, and SDP1) was altered in the transgenic lines, quantitative reverse transcription-PCR analysis was performed on developing siliques containing seeds in developmental stages 6 to 9 (Winter et al., 2007; Fig. 3). WRI1 and DGAT1 transcripts were increased between approximately 10- and 35-fold in W and D lines, and SDP1 transcripts were reduced by more than 10-fold in S lines. In each case, the individual line with the largest change in transcript abundance also had the highest percentage seed oil content (Figs. 2 and 3). No obvious reciprocal effects were observed on the expression of any one of the three target genes by the expression of another gene (Fig. 3). This finding is consistent with other studies that suggest that WRI1 is not a significant regulator of DGAT1 expression (Baud et al., 2007; To et al., 2012).
Figure 3.
Analysis of WRI1, DGAT1, and SDP1 expression in developing seeds of transgenic lines. The values are means ± se of measurements taken using quantitative reverse transcription-PCR on developing siliques from four plants of each genotype and are expressed as a proportion of the wild type. 18S expression was used as a control for normalization. Asterisks denote statistically significant differences from the wild type (lsd test), where significant differences between genotypes were detected (F test). C is the wild type, E is an empty vector control, and W, D, and S are independent homozygous T3 lines expressing the WRI1, DGAT1, and RNAi SDP1 transgenes.
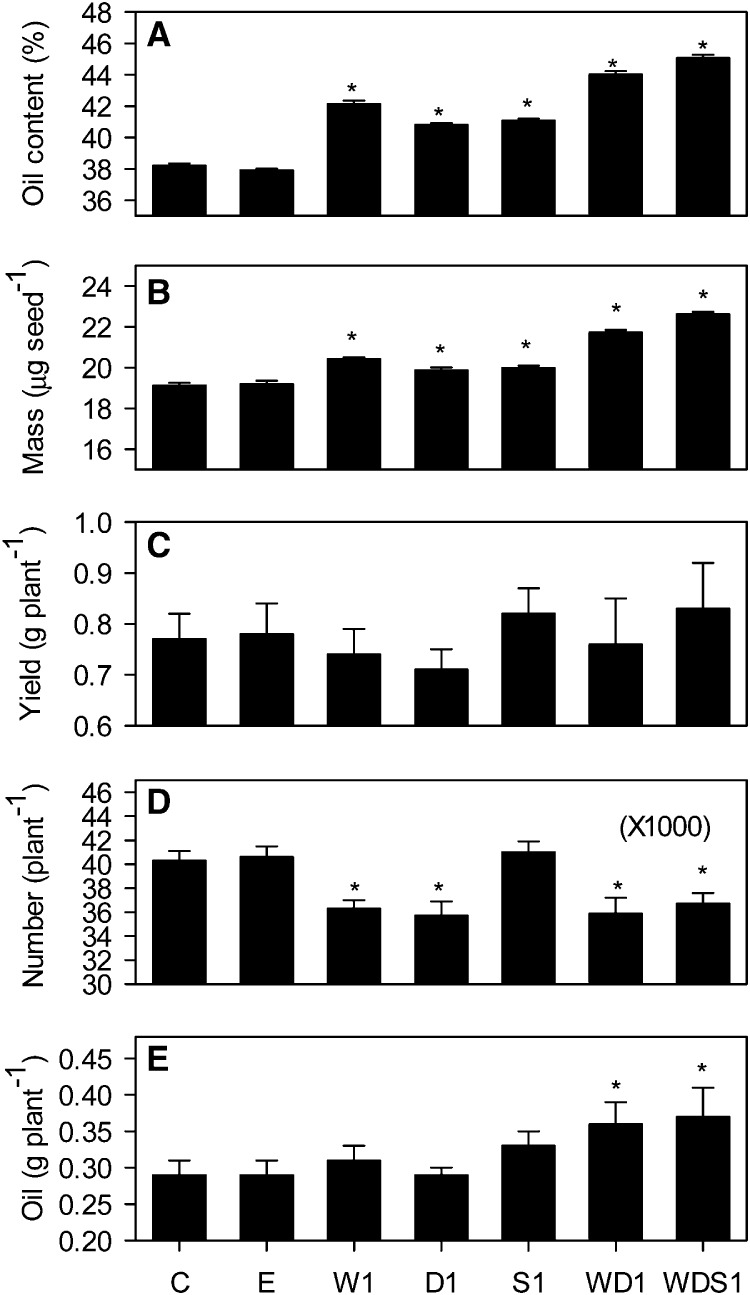
The constructs used for WRI1, DGAT1 overexpression, and SDP1 RNAi each carried different selectable markers (BAR [bialaphos resistance], DsRed [red fluorescent protein] and NPTII [neomycin phosphotransferase II], respectively), allowing the selection of double and triple homozygous lines following genetic crossing between W1, D1, and S1 transgenic lines and segregation analysis. Twelve individual plants of each of these genotypes were grown under controlled conditions in a random block design experiment, and the percentage oil content of seed from each plant was measured (Fig. 4A). When the WD1 transgenic line expressing both WRI1 and DGAT1 was examined, it not only exhibited significantly higher percentage seed oil content (P < 0.05) than the wild type or the empty vector control but also higher percentage oil content than W1, D1, or S1 (Fig. 4A). The percentage seed oil content in WD1 was approximately 44%, which is an approximately 1.2-fold increase over the wild type. Furthermore, when the WDS1 transgenic line expressing WRI1, DGAT1, and RNAi SDP1 was compared, the percentage seed oil content was significantly (P < 0.05) greater than in WD1 (Fig. 4A). When the experiment was performed on the next generation, the results were repeated (Supplemental Fig. S2). These data suggest that engineering multiple steps in the TAG metabolic pathway can lead to an additive effect on percentage seed oil content, which is heritable. Analysis of seed fatty acids using gas chromatography showed that there were only very minor compositional changes in W1, D1, S1, WD1, and WDS1 lines (Table I), while total fatty acid content varied as a percentage of seed weight in the same manner as was found for oil using time domain NMR (Table I; Fig. 4A). Small but statistically significant (P < 0.05) decreases in seed percentage protein and Suc content were also detected in most lines that exhibited increased percentage fatty acid/oil content (Table I). This may reflect competition between the pathways for substrates and cofactors (Schwender and Hay, 2012). The lack of a strong effect on seed fatty acid composition is consistent with what has been reported for single-gene manipulations of WRI1, DGAT1, and SDP1 (Jako et al., 2001; Cernac and Benning, 2004; Kelly et al., 2013a).
Figure 4.
Individual and combined effects of altering WRI1, DGAT1, and SDP1 expression on seed percentage oil content (A), average seed mass (B), seed yield (C), seed number (D), and oil yield (E). Values are means ± se of measurements on seeds from individual plants (n = 12) of each genotype grown in controlled conditions. Asterisks denote statistically significant differences from the wild type (lsd test), where significant differences between genotypes were detected (F test). C is the wild type, E is an empty vector control, and W, D, and S are independent homozygous lines expressing the WRI1, DGAT1, and RNAi SDP1 transgenes individually and in combination.
Table I. Storage reserve composition of seeds with altered WRI1, DGAT1, and SDP1 expression.
Values are means of measurements on seeds from individual plants (n = 4–10) of each genotype grown in controlled conditions. se are not shown but are less than 5% of the mean for all values. For individual fatty acids, they are a percentage of total, and for total fatty acids, protein, and Suc, they are a percentage of seed weight. C, Wild type; E, empty vector control; and W, D, and S are independent homozygous lines expressing the WRI1, DGAT1, and RNAi SDP1 transgenes individually and in combination. Asterisks denote statistically significant differences from the wild type (lsd test), where significant differences between genotypes were detected (F test).
| Compound | C | E | W1 | D1 | S1 | WD1 | WDS1 |
|---|---|---|---|---|---|---|---|
| 16:0 | 8.3 | 8.2 | 8.4 | 8.0 | 8.4 | 8.1 | 7.9 |
| 18:0 | 3.3 | 3.4 | 3.3 | 3.1 | 3.3 | 2.9 | 2.9 |
| 18:1 | 13.9 | 14.1 | 13.8 | 15.6 | 14.0 | 15.6 | 15.8 |
| 18:2 | 28.5 | 28.7 | 28.5 | 28.1 | 28.4 | 28.4 | 28.6 |
| 18:3 | 19.9 | 19.7 | 19.3 | 19.6 | 20.0 | 19.5 | 19.4 |
| 20:1 | 20.4 | 20.2 | 20.1 | 20.7 | 20.6 | 20.3 | 20.4 |
| Total fatty acids | 37.6 | 36.9 | 41.8* | 39.7* | 40.1* | 42.9* | 44.3* |
| Protein | 21.1 | 18.8 | 19.3 | 18.1* | 18.3* | 16.4* | 17.7* |
| Suc | 3.4 | 3.2 | 3.1* | 3.1* | 2.8* | 2.9* | 3.0* |
In addition to seed storage reserve content, additional traits were examined in W1, D1, S1, WD1, and WDS1 lines (Fig. 4). Analysis of 1,000 seed weight suggested that the expression of WRI1, DGAT1, or RNAi SDP1, using seed-specific promoters, also leads to a significant increase (P < 0.05) in average seed mass (Fig. 4B). By capturing all the seeds produced by each plant, it was also possible to determine total seed yield in grams per plant (Fig. 4C) and therefore to derive both total seed number per plant (Fig. 4D) and total oil yield in grams per plant (Fig. 4E). It is noteworthy that the variation observed in total seed yield between individual plants of the same genotype was larger than the variation in 1,000 seed weight or percentage oil content (Fig. 4). However, the combined data suggest that seed number per plant is significantly (P < 0.05) decreased in W1, D1, WD1, and WDS1 lines when compared with the wild type or the empty vector control (Fig. 4D). Despite this, the total oil yield per plant is significantly (P < 0.05) increased in the WD1 and WDS1 lines (Fig. 4E). These data suggest that an increase in seed oil content in Arabidopsis Col-0 can lead to an increase in plant oil yield under the experimental conditions used in this study.
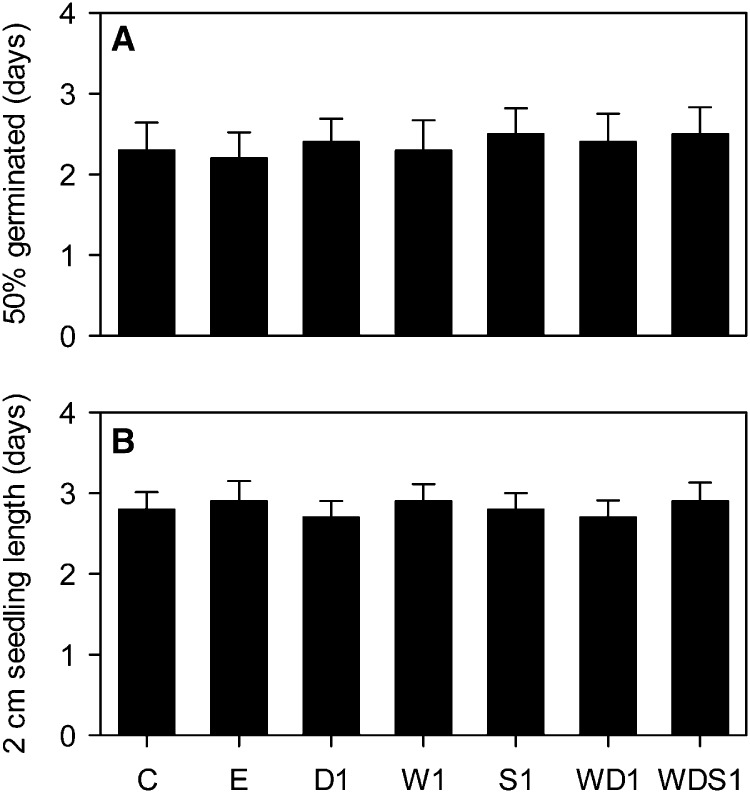
Seed vigor (i.e. rapid uniform germination and strong seedling growth) is an important agronomic trait (Finch-Savage et al., 2010). Manipulation of WRI1, DGAT1, and SDP1 expression each has the potential to negatively impact seed vigor (Lu and Hills, 2002; Eastmond, 2006; Sanjaya et al., 2011). To determine whether the seed-specific expression strategy employed in this study adversely affects germination rate and initial seedling (root and shoot) growth rate (Finch-Savage et al., 2010), they were measured using W1, D1, S1, WD1, and WDS1 lines (Fig. 5). The average time for 50% of seeds of each genotype to germinate was not significantly different from those of the wild type and the empty vector control (P > 0.05) when seeds were imbibed with water and placed on vertical agar plates with one-half-strength Murashige and Skoog salts at 20°C with constant low light (Fig. 5A). The initial rates of seedling growth (measured at the time between germination and 2 cm in length) in these conditions were also not significantly different (P > 0.05) from those in the wild type and the empty vector control (Fig. 5B). Coexpression of WRI1, DGAT1, and RNAi SDP1, therefore, does not appear to be detrimental to seed vigor under laboratory conditions. Furthermore, no abnormal growth phenotypes were observed throughout the life cycle of the plants.
Figure 5.
Individual and combined effects of altering WRI1, DGAT1, and SDP1 expression on germination rate (A) and early seedling growth rate (B). Values are means ± se of measurements on seeds from individual plants (n = 6) of each genotype grown in controlled conditions. The times required for 50% of the seeds to germinate (A) and for seedlings then to reach 2 cm in length (B) were scored. No significant differences between genotypes were detected (F test). C is the wild type, E is an empty vector control, and W, D, and S are independent homozygous lines expressing the WRI1, DGAT1, and RNAi SDP1 transgenes individually and in combination.
CONCLUSION
This study shows that certain transgene manipulations, with a positive effect on oil accumulation in developing seeds, can be combined to give an additive effect. Overexpression of WRI1 or DGAT1 has been reported to increase seed oil content (Jako et al., 2001; Cernac and Benning, 2004). However, WRI1 predominantly targets the expression of genes associated with glycolysis and fatty acid synthesis (Baud et al., 2007; To et al., 2012), while DGAT1 catalyzes the final step in the assembly of TAG from DAG and acyl-CoA (Katavic et al., 1995). Top-down metabolic control analysis studies suggest that a significant proportion of control over TAG synthesis lies within both portions of the metabolic pathway (Weselake et al., 2008; Tang et al., 2012). The additive effect of overexpression of these two genes supports this conclusion. In addition to TAG biosynthetic capacity, TAG turnover can occur during late seed development and can reduce the final oil content of seeds (Kelly et al., 2013a; Kim et al., 2014). Therefore, it is consistent that combining WRI1 and DGAT1 overexpression with the suppression of SDP1 can result in a further increase in percentage seed oil content.
As a strategy for metabolic pathway engineering, stacking these transgenes might be an effective means to enhance seed oil content in crops. Arabidopsis is only a model species and has not been subjected to artificial selection for agronomic traits such as seed oil content or yield. However, WRI1 and DGAT1 overexpression have been reported individually to boost seed oil content in maize (Zheng et al., 2008; Shen et al., 2010) and oilseed rape (Brassica napus; Weselake et al., 2008; Wu et al., 2014). Furthermore, suppression of SDP1 can also enhance seed oil content in oilseed rape (Kelly et al., 2013a) and Jatropha curcas (Kim et al., 2014). There is also potential for stacking additional genes to obtain a further increase in seed oil content. For example, G3P dehydrogenase and lysophosphatidic acid acyltransferase activities also contribute to glycerolipid synthesis, and overexpression of each has been reported to enhance seed oil content (Zou et al., 1997; Vigeolas et al., 2007). In the case of G3P dehydrogenase, WRI1 might already enhance activity, since members of the WRI gene family are required for the full expression of GLYCEROL-3-PHOSPHATE DEHYDROGENASEc1 in Arabidopsis seeds (To et al., 2012).
Although, under our experimental conditions, we provide evidence that an increase in seed oil content can translate into greater oil yield, the relationship is very likely to be less than proportionate. No significant increase in seed yield (P > 0.05) could be detected in any of the engineered lines, but a significant (P < 0.05) reduction in seed number was apparent in most lines. A tradeoff between seed size/oil content and seed number is consistent with the hypothesis that seed oil production is limited by both source and sink metabolism (Fatihi et al., 2013). In crops, the balance of source-sink limitation is known to change with growth conditions, such as field planting density (Habekotté, 1997). Therefore, the extent to which engineering an increase in sink strength can enhance seed or oil yield would also be expected to depend on environment. Ultimately, greater seed oil content (either per seed or as a percentage of seed weight) only has economic value if it is not offset by a reduction in total yield per hectare. Further work will be required to determine the efficacy of this form of multigene metabolic engineering in crops under field conditions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type (Col-0) Arabidopsis (Arabidopsis thaliana) seeds were obtained from the European Arabidopsis Stock Centre (University of Nottingham). The seeds were surface sterilized, plated on agar plates containing one-half-strength Murashige and Skoog salts (Sigma-Aldrich), and imbibed in the dark for 4 d at 4°C. The plates were then placed in a growth chamber set to 16 h of light (23°C)/8 h of dark (18°C), photosynthetic photon flux density of 250 μmol m−2 s−1, and 70% relative humidity. After 5 d, seedlings were transplanted to moist Levingtons F2 compost in 7-cm2 pots. Individual plants were arranged into a 12-block, one-way randomized design with one plant of each genotype per block and randomized within each block. When plants began to flower, a 60-cm stick was inserted in each pot, and the pot was placed inside an upright rectangular transparent perforated plastic bag (60 × 6 cm). The bag was sealed around the lower stem prior to seed shedding to ensure that all the seeds from each plant were retained.
Creation of DNA Constructs and Arabidopsis Transformation
WRI1 (At3g54320), DGAT1 (At2g19450), and approximately 1.7 kb of the SUS2 promoter (At5g49190) were amplified by PCR from Arabidopsis complementary DNA or genomic DNA using the following primer pairs: WRI1 (5′-GCCTCGAGATGAAGAAGCGCTTAACCACTTCC-3′ and 5′-GCCTCGAGTTATTCAGAACCAACGAACAAGCCC-3′), DGAT1 (5′-CGTCTAGAATGGCGATTTTGGATTCTGCTGGCG-3′ and 5′-CGCTCGAGTCATGACATCGATCCTTTTCGGTTCATCAGG-3′), and SUS2 promoter (5′-AAGGATCCATGTCGACTACAAAGCGCCAAGGAGATA-3′ and 5′-CGCTCGAGCAAGCTACAGTGAATTTAAA-3′). The products were cloned into pCR 2.1-TOPO using the TOPO TA Cloning Kit from Invitrogen. DGAT1 was excised using XbaI and XhoI and inserted into pBinGlyRed3 using T4 DNA Ligase (Invitrogen). The SUS2 promoter was excised using BamHI and XhoI and inserted into pBinGlyBar1. The pBinGlyBar1 vector was then digested with XhoI, and WRI1 was inserted 3′ of the SUS2 promoter. A hairpin RNAi construct containing a 300-bp region of SDP1 (460–760 bp 3′ of ATG) fused to the Vicia faba USP promoter (Kelly et al., 2013a) was created by gene synthesis and inserted into pGWB13 (Karimi et al., 2002) using Gateway cloning (Invitrogen). The three constructs were transformed into Agrobacterium tumefaciens strain GV3101 by heat shock and into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transformants containing T-DNA insertions were selected by DsRed expression (detected with a fluorescence microscope) or herbicide or antibiotic resistance.
Transcript Analysis
DNase-treated total RNA was isolated from stages 6 to 9 developing Arabidopsis siliques (Winter et al., 2007) using the RNeasy kit from Qiagen with modifications described by Eastmond (2006). The synthesis of single-stranded complementary DNA was carried out using SuperScript II RNase H− reverse transcriptase from Invitrogen. Quantitative real-time PCR was performed as described by Rajangam et al. (2013). The primer pairs used for real-time PCR were QWRI1 (5′-AAACGAGCCAAAAGGGCTAAG-3′ and 5′-GGGCTTGTCGGGTTATGAGA-3′), QDGAT1 (5′-TGGATTCTGCTGGCGTTACTAC-3′ and 5′-AGCCTATCAAGATCGACGAACTCT-3′), QSDP1 (5′-AAATGGCTTACCGGAGGAAGTT-3′ and 5′-TGAGCCCATTCCTCATAAGTCA-3′), and Q18S (5′-TCCTAGTAAGCGCGAGTCATC-3′ and 5′-CGAACACTTCACCGGATCAT-3′).
Seed Storage Reserve Analysis
Seed oil and moisture contents were measured by low-resolution time domain NMR spectroscopy using a Minispec MQ20 (Bruker) fitted with a robotic sample-handling system (Rohasys). The oil and moisture calibration was constructed according to the manufacturer’s instructions using nine approximately 0.5-g oilseed rape (Brassica napus) seed samples ranging between approximately 5% and 10% moisture content and between approximately 30% and 55% oil content (r2 > 0.99). Seed percentage oil content values were adjusted to 9% water content. Seed fatty acid content was determined by gas chromatographic analysis of fatty acid methyl esters following direct transmethylation according to the method of Browse et al. (1986). Pentadecanoin was added to the samples as an internal control to allow quantification. Seed storage protein and Suc contents were determined using the methods of Baud et al. (2002).
Seed Germination and Seedling Growth Assays
The seeds were plated on agar plates containing one-half-strength Murashige and Skoog salts (Sigma-Aldrich). The plates were then placed vertically in a growth chamber set to 20°C with constant low light (photosynthetic photon flux density of 10 μmol m−2 s−1). The average times to 50% germination and from radicle emergence to 2-cm seedling length were measured.
Statistical Analyses
The number of replicates (n) and the se are shown for most measurements. One-way ANOVA was used to assess the differences between genotypes for measurements of seed percentage oil content and seed weight. Following significant (P < 0.05) F test results, means of interest were compared using the appropriate lsd value at the P = 0.05 level of significance on the corresponding degrees of freedom. The GenStat (VSN International) statistical system was used for these analyses.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY254038 (WRI1), AJ238008 (DGAT1), and AY136470 (SDP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Seed oil content of T2 transgenic lines.
Supplemental Figure S2. Heritability of the effect on seed percentage oil content.
Supplementary Material
Acknowledgments
We thank Eve Shaw (University of Warwick, Coventry, UK) and Fiona Bryant (Rothamsted Research, Harpenden, UK) for assistance with plant growth and harvesting and Edgar Cahoon (University of Nebraska, Lincoln) for providing the pBinGlyRed3 and pBinGlyBar1 vectors used in this study.
Glossary
- G6P
glucose-6-phosphate
- G3P
glycerol-3-phosphate
- DAG
diacylglycerol
- PC
phosphatidylcholine
- RNAi
RNA interference
- T-DNA
transfer DNA
- Col-0
Columbia-0
- TAG
triacylglycerol
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (Institute Strategic Program grant and grant no. BB/E022197/1).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Angeles-Núñez JG, Tiessen A. (2012) Regulation of AtSUS2 and AtSUS3 by glucose and the transcription factor LEC2 in different tissues and at different stages of Arabidopsis seed development. Plant Mol Biol 78: 377–392 [DOI] [PubMed] [Google Scholar]
- Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA 106: 13124–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J, Lu C. (2012) Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol 160: 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Vaultier MN, Rochat C. (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55: 397–409 [DOI] [PubMed] [Google Scholar]
- Broun P. (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7: 202–209 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. (2008) Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol 147: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Benning C, Ohlrogge J. (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J 54: 593–607 [DOI] [PubMed] [Google Scholar]
- Dyer JM, Stymne S, Green AG, Carlsson AS. (2008) High-value oils from plants. Plant J 54: 640–655 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ. (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yan C, Zhang X, Xu C. (2013) Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 25: 3506–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A, Zbierzak AM, Dörmann P. (2013) Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds. Plant Physiol 163: 973–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA (1997) Understanding the Control of Metabolism. Portland Press, London [Google Scholar]
- Finch-Savage WE, Clay HA, Lynn JR, Morris K. (2010) Towards a genetic understanding of seed vigour in small-seeded crops using natural variation in Brassica oleracea. Plant Sci 179: 582–589 [Google Scholar]
- Grotewold E. (2008) Transcription factors for predictive plant metabolic engineering: are we there yet? Curr Opin Biotechnol 19: 138–144 [DOI] [PubMed] [Google Scholar]
- Habekotté B. (1997) Options for increasing seed yield of winter oilseed rape (Brassica napus L.): a simulation study. Field Crops Res 54: 109–126 [Google Scholar]
- Hobbs DH, Flintham JE, Hills MJ. (2004) Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol 136: 3341–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC. (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L. (1995) Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Shaw E, Powers SJ, Kurup S, Eastmond PJ. (2013a) Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol J 11: 355–361 [DOI] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ. (2013b) The SUGAR-DEPENDENT1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Yang SW, Mao HZ, Veena SP, Yin JL, Chua NH. (2014) Gene silencing of Sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol Biofuels 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Hills MJ. (2002) Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiol 129: 1352–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Napier JA, Clemente TE, Cahoon EB. (2011) New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol 22: 252–259 [DOI] [PubMed] [Google Scholar]
- Meyer K, Damude HG, Everard JD, Ripp KG, Stecca KL, inventors. January 4, 2010. Use of a seed specific promoter to drive odp1 expression in cruciferous oilseed plants to increase oil content while maintaining normal germination. International Patent Application No. PCT/US2010/029609 [Google Scholar]
- Rajangam AS, Gidda SK, Craddock C, Mullen RT, Dyer JM, Eastmond PJ. (2013) Molecular characterization of the fatty alcohol oxidation pathway for wax-ester mobilization in germinated jojoba seeds. Plant Physiol 161: 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J. (1997) Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol 113: 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya, Durrett TP, Weise SE, Benning C. (2011) Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol J 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J, Hay JO. (2012) Predictive modeling of biomass component tradeoffs in Brassica napus developing oilseeds based on in silico manipulation of storage metabolism. Plant Physiol 160: 1218–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC. (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Guschina IA, O’Hara P, Slabas AR, Quant PA, Fawcett T, Harwood JL. (2012) Metabolic control analysis of developing oilseed rape (Brassica napus cv Westar) embryos shows that lipid assembly exerts significant control over oil accumulation. New Phytol 196: 414–426 [DOI] [PubMed] [Google Scholar]
- To A, Joubès J, Barthole G, Lécureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S. (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T, El Tahchy A, Liu Q, Zhou XR, Shrestha P, Divi UK, Ral JP, Mansour MP, Nichols PD, James CN, et al. (2014) Metabolic engineering of biomass for high energy density: oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol J 12: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T, El Tahchy A, Shrestha P, Zhou XR, Singh SP, Petrie JR. (2013) Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett 587: 364–369 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Waldeck P, Zank T, Geigenberger P. (2007) Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J 5: 431–441 [DOI] [PubMed] [Google Scholar]
- Weselake RJ, Shah S, Tang M, Quant PA, Snyder CL, Furukawa-Stoffer TL, Zhu W, Taylor DC, Zou J, Kumar A, et al. (2008) Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. J Exp Bot 59: 3543–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PB, Harwood JL. (2009) Increasing the flow of carbon into seed oil. Biotechnol Adv 27: 866–878 [DOI] [PubMed] [Google Scholar]
- Winichayakul S, Scott RW, Roldan M, Hatier JH, Livingston S, Cookson R, Curran AC, Roberts NJ. (2013) In vivo packaging of triacylglycerols enhances Arabidopsis leaf biomass and energy density. Plant Physiol 162: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Liu ZH, Hu ZH, Huang RZ. (March 3, 2014) BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J Integr Plant Biol http://dx.doi.org/10.1111/jipb.12158 [DOI] [PubMed] [Google Scholar]
- Würschum T, Liu W, Maurer HP, Abel S, Reif JC. (2012) Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor Appl Genet 124: 153–161 [DOI] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, et al. (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40: 367–372 [DOI] [PubMed] [Google Scholar]
- Zou J, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC. (1997) Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 9: 909–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.