An Arabidopsis Heat Shock Factor affects tolerance to salt as well as other abiotic stresses, forms homodimers dependent on the redox regulation, interacts with MAP kinases, and alters the expression of a large set of stress-induced genes.
Abstract
Heat shock factors (HSFs) are principal regulators of plant responses to several abiotic stresses. Here, we show that estradiol-dependent induction of HSFA4A confers enhanced tolerance to salt and oxidative agents, whereas inactivation of HSFA4A results in hypersensitivity to salt stress in Arabidopsis (Arabidopsis thaliana). Estradiol induction of HSFA4A in transgenic plants decreases, while the knockout hsfa4a mutation elevates hydrogen peroxide accumulation and lipid peroxidation. Overexpression of HSFA4A alters the transcription of a large set of genes regulated by oxidative stress. In yeast (Saccharomyces cerevisiae) two-hybrid and bimolecular fluorescence complementation assays, HSFA4A shows homomeric interaction, which is reduced by alanine replacement of three conserved cysteine residues. HSFA4A interacts with mitogen-activated protein kinases MPK3 and MPK6 in yeast and plant cells. MPK3 and MPK6 phosphorylate HSFA4A in vitro on three distinct sites, serine-309 being the major phosphorylation site. Activation of the MPK3 and MPK6 mitogen-activated protein kinase pathway led to the transcriptional activation of the HEAT SHOCK PROTEIN17.6A gene. In agreement that mutation of serine-309 to alanine strongly diminished phosphorylation of HSFA4A, it also strongly reduced the transcriptional activation of HEAT SHOCK PROTEIN17.6A. These data suggest that HSFA4A is a substrate of the MPK3/MPK6 signaling and that it regulates stress responses in Arabidopsis.
Heat shock factors (HSFs) are transcriptional regulators that mediate the activation of large set of genes induced by high temperature or other stress conditions. HSFs are important regulators of cellular stress responses in plants and animals (Kotak et al., 2007; Akerfelt et al., 2010; Scharf et al., 2012). HSFs recognize the heat stress elements (HSEs), conserved motifs in promoters of heat-induced targets, such as heat shock protein (HSP) genes (Pelham, 1982; Wing et al., 1989; Akerfelt et al., 2010; Scharf et al., 2012). The yeast (Saccharomyces cerevisiae) and Drosophila melanogaster genomes encode a single HSF, whereas mammals have four HSF genes (Akerfelt et al., 2010). Plants possess diverse families of HSFs that are encoded by 21 genes in Arabidopsis (Arabidopsis thaliana) and 52 loci in soybean (Glycine max; Nover et al., 2001; Scharf et al., 2012). Remarkable differences in their transcriptional regulation indicate that plant HSFs underwent considerable functional diversification (von Koskull-Döring et al., 2007).
HSFs share a conserved modular structure. The N-terminal DNA-binding domain is responsible for the HSE recognition in the promoters of HSF target genes. The hydrophobic heptad repeat region for oligomerization (HRA/B) located close to the DNA-binding domain mediates trimerization, which is prerequisite for their transcription factor activity (Nover et al., 2001; Kotak et al., 2004; Anckar and Sistonen, 2011; Scharf et al., 2012). Accumulating evidence suggests that activation of plant and mammalian HSFs is regulated by a similar mechanism, but the regulation of plant HSFs is much less understood compared with their mammalian counterparts (Akerfelt et al., 2010; Scharf et al., 2012). In the absence of stress, human HSF1 is inactive and forms a cytoplasmic complex with the heat shock protein HSP90. Accumulation of denatured proteins during heat stress consumes HSP90 and releases HSF1 from the complex, which then undergoes phosphorylation, sumoylation, and trimerization prior to its nuclear import. Subsequent binding of HSF1 to the HSE facilitates its interaction with the RNA polymerase II complex and initiation of transcription of target genes (Akerfelt et al., 2010; Anckar and Sistonen, 2011).
Certain HSFs are thought to function as molecular peroxide sensors that respond to alterations in levels of reactive oxygen species (ROS) during stress by conformational change and multimer formation, leading to subsequent transcriptional activation of their target genes (Miller and Mittler, 2006). ROS accumulate in response to biotic and abiotic stress and represent important signaling molecules in the coordination of stress acclimation pathways (Foyer and Noctor, 2005; Miller et al., 2010; Suzuki et al., 2012). Generation of ROS signals, in particular hydrogen peroxide (H2O2), during heat and oxidative stress mediates the activation of various HSFs and induction of their downstream target genes (Miller and Mittler, 2006; Volkov et al., 2006; Kotak et al., 2007). Specific HSFs are involved in signaling cross talk with key components of ROS signaling, including the mitogen-activated protein (MAP) kinases MPK3 and MPK6 and members of the zinc-finger transcription factor families, and thus also contribute to transcriptional control of a large battery of oxidative stress-responsive genes (Davletova et al., 2005b; Miller and Mittler, 2006; Pucciariello et al., 2012; Evrard et al., 2013).
Due to considerable redundancy of Arabidopsis HSF genes, only few hsf mutants display identifiable phenotypic changes (Scharf et al., 2012). Most functional data rely on overexpression or combination of multiple mutations of HSFs (Liu et al., 2011). Based on these studies, the HSFA1 subfamily is defined as a master regulator of heat stress responses (Mishra et al., 2002; Liu et al., 2011), whereas HSFA2, which is also induced by high light intensity and osmotic and oxidative stress, is proposed to act in multiple stress-signaling pathways (Nishizawa et al., 2006; Banti et al., 2010). HSFA1 and HSFA2 form heterodimers resulting in synergistic transcriptional activation of target genes (Chan-Schaminet et al., 2009), and overexpression of HSFA2 was shown to confer tolerance to several stress stimuli (Banti et al., 2010). HSFA3 is activated by dehydration-responsive element-binding protein 2A, which is likely involved in the coordination of drought and heat stress signaling and activation of both water and heat stress-responsive genes (Sakuma et al., 2006; Yoshida et al., 2008). The function of the HSFA4 group is not well known. HSFA4A of wheat (Triticum aestivum) and rice (Oryza sativa) were reported to increase cadmium tolerance (Shim et al., 2009). HSFA4 from tomato (Solanum lycopersicum) was shown to activate heat stress-induced genes and interact with HSFA5-generating inactive heterooligomers. HSFA4 and HSFA5 belong to a separate group within the class A HSFs, sharing similarities in the DNA-binding and oligomerization domain and C-terminal conserved sequence motifs (Baniwal et al., 2004; Kotak et al., 2004; Baniwal et al., 2007). In the rice spotted leaf7 mutant, one base substitution in the DNA-binding domain of HSFA4D resulted in the appearance of necrotic lesions on leaves, suggesting stress hypersensitivity (Yamanouchi et al., 2002). Arabidopsis HSFA4A was implicated in regulation of responses to high light and oxidative stress by regulating the transcription of ASCORBATE PEROXIDASE1 (APX1) and ZAT12 genes (Davletova et al., 2005a). These observations supported the hypothesis that the HSFA4 group functions as antiapoptotic factors through regulation of plant ROS homeostasis (von Koskull-Döring et al., 2007). While most HSFs act as activators of gene expression, HSFB and HSFA5 function as repressors, which inhibit other HSFs, including HSFA4, in interaction with general transcription factors (Czarnecka-Verner et al., 2004; Baniwal et al., 2007). HSFs are also known to respond to upstream Ca2+ and ROS signaling and modulate transcription of heat-, abscisic acid-, and salicylate-responsive genes (Scharf et al., 2012).
Here, we report on the functional characterization of HSFA4A, which was identified as a factor conferring salt tolerance when expressed in an estradiol-inducible fashion in Arabidopsis cells. Whereas inactivation of HSFA4A by an insertion mutation enhances salt sensitivity and lipid peroxidation, estradiol induction of HSFA4A confers salt tolerance, reduces lipid peroxidation and H2O2 accumulation, and decreases the sensitivity to oxidative agents. Nuclear accumulation of HSFA4A is enhanced by salt treatment in parallel with ROS accumulation and reduced by Ala replacement of three Cys residues. HSFA4A interacts with and is phosphorylated in vitro by the MAP kinases MPK3 and MPK6 on three residues, Ser-309 being the major, which, when mutated, diminishes the transactivation of the heat shock protein gene HSP17.6A. Based on these data, we propose that HSFA4A may regulate plant responses to salt and oxidative stress as a substrate of MPK3/MPK6.
RESULTS
Estradiol Induction of HSFA4A Confers Salt Tolerance in Cultured Cells
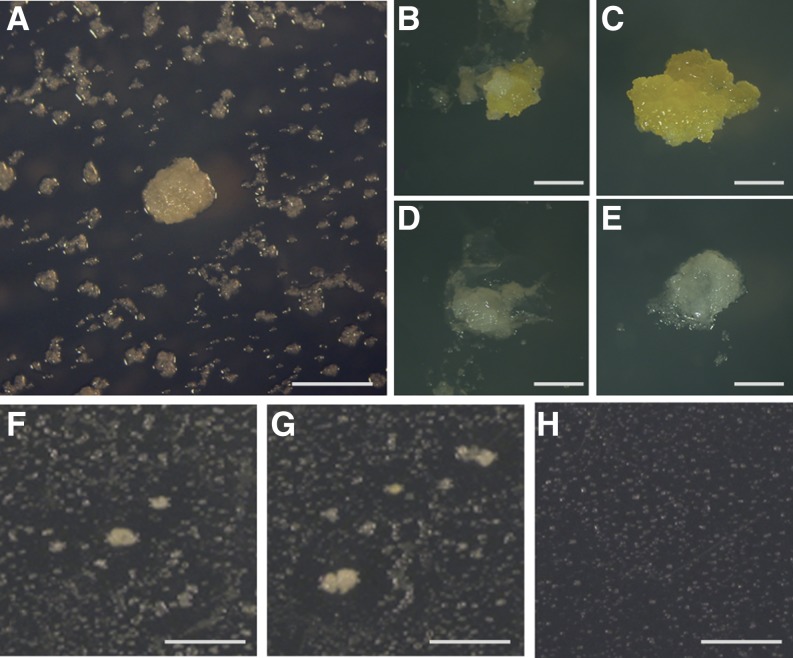
The previously described Conditional Overexpression System (COS; Papdi et al., 2008) was used to perform large-scale Agrobacterium spp.-mediated transformation of an Arabidopsis cell culture (Supplemental Fig. S1). From three separate transformation experiments, we obtained a total of 1.2 million hygromycin-resistant cell colonies, from which 290 microcalli showed various levels of viability and growth in the presence of 175 mm NaCl and 5 µm estradiol (Fig. 1A; Supplemental Fig. S1; Supplemental Table S1). When selected calli were further cultured on salt-containing medium, four of the transformed calli showed salt tolerance in the presence of estradiol, while they were sensitive in the absence of the inducer (Fig. 1, B–E, depicts an example of estradiol-dependent salt tolerance in a transformed colony and in wild-type cell culture). The genes were isolated from the COS expression vector by PCR amplification using DNA samples prepared from these salt-tolerant transformants. Sequencing of the amplified complementary DNAs (cDNAs; Supplemental Table S2) indicated that two transformants (lines no. 1 and 7) carried two transfer DNA (T-DNA) inserts with full-length or truncated cDNAs encoding NITRILASE2 (NIT2), HSFA4A, CYTOCHROME B5 ISOFORM D, and MINICHROMOSOME MAINTENANCE2, whereas cDNAs of single COS vector inserts in the other two lines derived from the XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE11 and GLUTATHIONE S-TRANSFERASE9 genes. As HSFA4A corresponds to an uncharacterized member of the Arabidopsis HSF family (von Koskull-Döring et al., 2007), which shows high sequence identity with other HSFs only in its DNA-binding domain (Supplemental Fig. S2), we addressed the question whether the observed salt tolerance phenotype was conferred by estradiol-inducible overexpression of the HSFA4A cDNA. In fact, we found that, in contrast to NIT2, Agrobacterium spp.-mediated retransformation of HSFA4A cDNA by the estradiol-inducible pER8GW vector (Papdi et al., 2008) into Arabidopsis cells revealed that chemically induced expression of HSFA4A is sufficient to confer salt tolerance and sustain growth of calli in the presence of 175 mm NaCl (Fig. 1, F–H).
Figure 1.
Identification of salt-tolerant COS-transformed Arabidopsis calli. A, Screening of COS-transformed cell culture on MS medium containing 175 mm NaCl and 5 µm estradiol. B and C, Growth of transformed line number 1 on high-salt medium without (B) and with estradiol (C). D and E, Wild-type Arabidopsis calli on high-salt medium in the absence (D) or presence (E) of estradiol. F and G, pER8GW-HSFA4A-transformed Arabidopsis colonies growing on high-salt medium containing 5 µm estradiol. Transformation frequency was approximately 5% in this experiment. H, Transformed cells are unable to grow on high-salt medium in the absence of estradiol. Bar = 5 mm. [See online article for color version of this figure.]
Effects of HSFA4A on Growth and Abiotic Stress Tolerance in Transgenic Plants
HSFA4A is a yet uncharacterized member of the Arabidopsis HSF family but has closely related members in a broad range of plant species showing high sequence identity with other HSFs in its DNA-binding domain (von Koskull-Döring et al., 2007; Scharf et al., 2012; Supplemental Fig. S2). To demonstrate that HSFA4A confers stress tolerance also in plants, the estradiol-inducible pER8GW-HSFA4A construct was introduced into Arabidopsis (ecotype Columbia [Col-0]) plants.
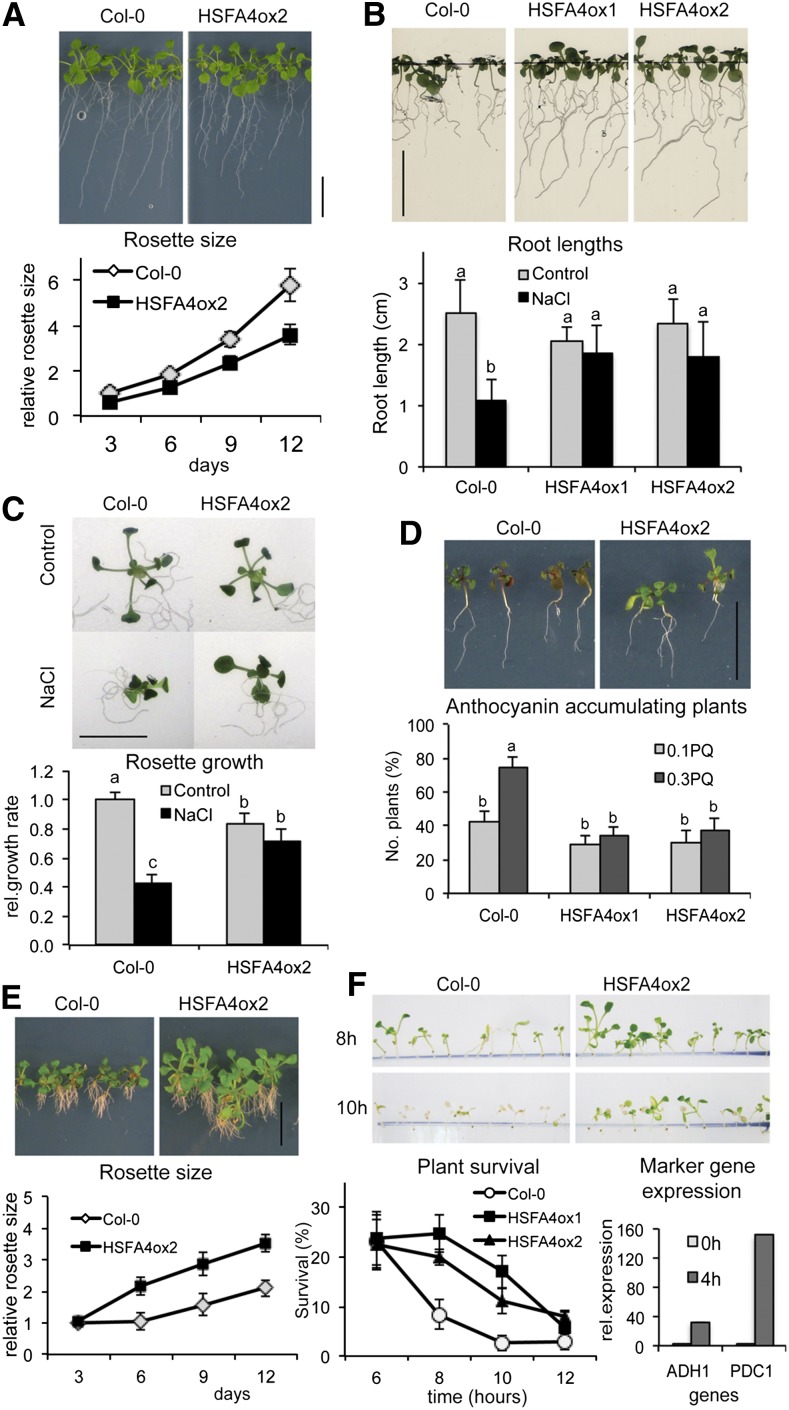
Overexpression of HSFA4A in transgenic lines resulted in 20% to 30% reduction of plant growth compared with wild-type plants on one-half-strength Murashige and Skoog (MS) media supplemented with 5 µm estradiol (Fig. 2A). However, when grown in the presence of salt and estradiol, root growth was inhibited by 60% in the wild type, but only a reduction of 10% to 20% was detected in HSFA4A overexpressing (HSFA4ox) plants (Fig. 2B). Similarly, the average rosette growth of the wild type was reduced by more than 50%, while growth of the HSFA4ox2 line was only slightly inhibited by salt (Fig. 2C).
Figure 2.
HSFA4A overexpression confers stress tolerance to Arabidopsis. A, Growth of Col-0 wild-type and HSFA4A-overexpressing (HSFA4ox2) plants on one-half-strength MS media with estradiol (control media). Graph shows growth of relative rosette sizes. B, Root growth of Col-0, HSFA4ox1, and HSFA4ox2 plants on high-salt medium (100 mm NaCl, 12 d) supplemented with estradiol. Graph shows average root lengths after 12 d of growth. C, Rosette growth of Col-0 and HSFA4ox2 plants on control and high-salt medium (100 mm NaCl) for 12 d. Graph shows relative growth rates calculated from linear trend of average rosette sizes measured in each time point, where 1 equals growth of wild-type plants in control media. All media contained estradiol. D, Paraquat-triggered anthocyanin accumulation in Col-0 and HSFA4ox2 plantlets (0.3 µm paraquat, 12 d). Graph shows percentage of anthocyanin-accumulating plants treated with 0.1 or 0.3 µm paraquat for 12 d in the presence of estradiol. E, Effect of H2O2 on seedling development of wild-type and HSFA4ox2 plants (12 d on 5 mm H2O2 with estradiol). Relative rosette size graph indicates growth of wild-type and HSFA4ox2 plants calculated from the increase of rosette size in a given time point, relative to the average rosette size of each line at the beginning of the treatment (day 0). F, Tolerance to anoxia. Seedlings were treated with estradiol and subjected to anoxia, and survival was scored 7 d after recovery. Average survival rates of plantlets after 6, 8, 10, and 12 h of anoxia are shown on left graph. Expression of anoxia-induced marker genes ADH1 (AT1G77120) and PDC1 (AT4G33070) in anoxia-treated wild-type seedlings (right graph) in control (0 h) and anoxic (4 h) conditions. All treatments included 5 µm estradiol. For stress treatments, 5-d-old in vitro-germinated seedlings were transferred to different culture media. Bars = 1 cm. Error bars indicate ses, and different letters show significant differences at P < 0.05 (Duncan’s test).
Because ROS generation and signaling is known to play an important role in mounting salt tolerance (Miller et al., 2010; Huang et al., 2012), we also examined the effects of H2O2, paraquat, and anoxia on the growth of plants overexpressing HSFA4A. HSFA4ox2 seedlings grew better and showed less anthocyanin accumulation than the wild type on media containing estradiol and either 0.1 or 0.3 µm paraquat (N,N-dimethyl-4,4-bipyridinium dichloride), a herbicide that generates oxygen radicals and promotes anthocyanin accumulation (Fig. 2D; Kytridis and Manetas, 2006). When grown in the presence of 5 mm H2O2 and estradiol, the HSFA4ox2 line developed 30% to 40% larger rosettes and displayed less root damage than the wild type (Fig. 2E). In addition, overexpression of HSFA4A conferred better survival rate under anoxia. Following 8 and 10 h of anoxia, only 8% and 3% of the wild type survived compared with 20% to 25% and 12% to 17% of HSFA4ox plants, respectively (Fig. 2F). Activation of anoxia-responsive marker genes ALCOHOL DEHYDROGENASE1 (ADH1) and PYRUVATE DECARBOXYLASE1 (PDC1) in wild-type plants confirmed low oxygen conditions in this experiment (Fig. 2F).
HSFA4A Overexpression Reduces Oxidative Damage
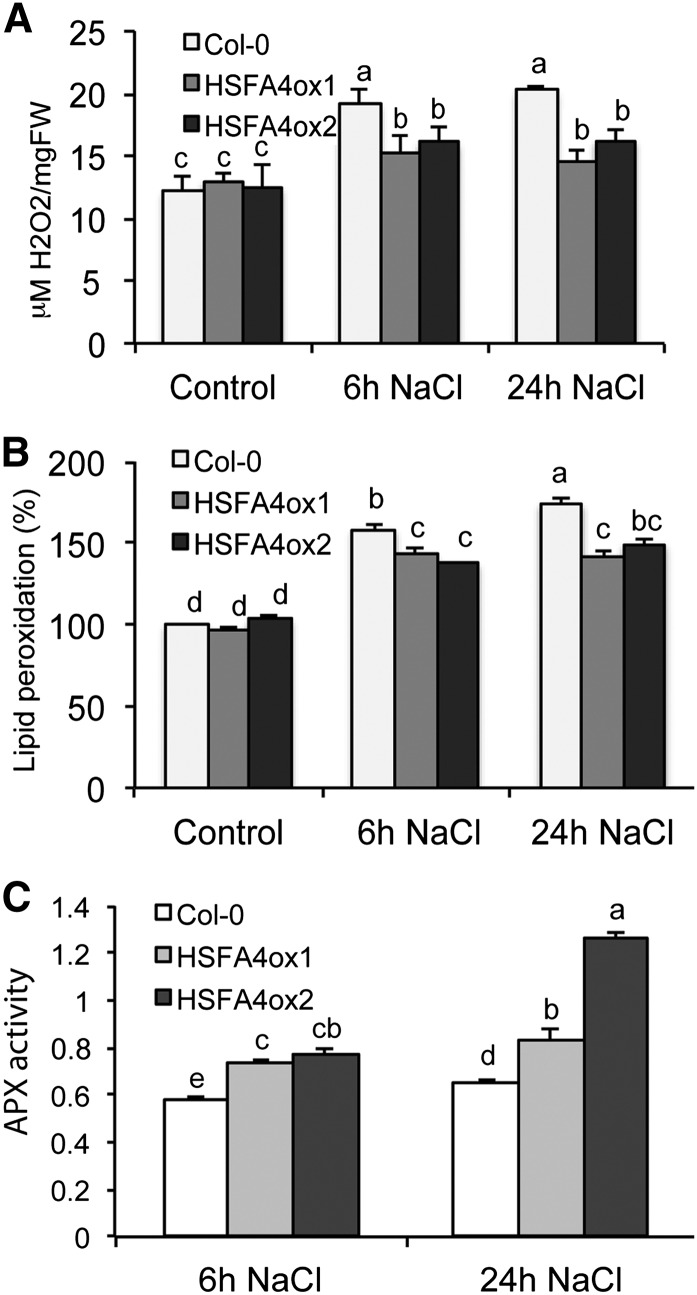
Enhanced tolerance of HSFA4ox plants to H2O2 and paraquat suggested a potential increase of antioxidant capacity. We found that estradiol induction of HSFA4A lowered the levels of salt-induced H2O2 accumulation and lipid peroxidation. Whereas under normal growth condition H2O2 levels were comparable in wild-type and HSFA4ox plants, treatment with 150 mm NaCl for 6 and 24 h increased the amount of H2O2 by 70% in the wild type but only by 20% and 30% in HSFA4ox plants (Fig. 3A). Furthermore, the end products of lipid peroxidation generated by peroxide-triggered oxidative damage and measured by malondialdehyde were elevated in the wild type by 60% to 70% following salt treatment compared with a 30% to 40% increase in HSFA4ox plants (Fig. 3B). In correlation with lower accumulation of H2O2, HSFA4ox plants displayed higher APX activities at 24 h of salt treatment, which suggested enhanced removal of H2O2 through oxidation of ascorbate to dehydroascorbate (Fig. 3C).
Figure 3.
HSFA4A overexpression reduces oxidative damage during salt stress. A, H2O2 accumulation in Col-0 wild-type, HSFA4ox1, and HSFA4ox2 plants treated with 150 mm NaCl for 6 and 24 h. B, Lipid peroxidation in salt-stressed plants calculated by malondialdehyde accumulation. One hundred percent corresponds to lipid damage in wild-type nonstressed plants. C, APX activities in salt-treated plants. In vitro-grown plantlets were used for the experiments at age of 14 d. Error bars indicate ses, and different letters show significant differences at P < 0.05 (Duncan’s test). Five micromolar estradiol was included in all treatments. FW, Fresh weight.
Inactivation of HSFA4A Confers Hypersensitivity to Salt
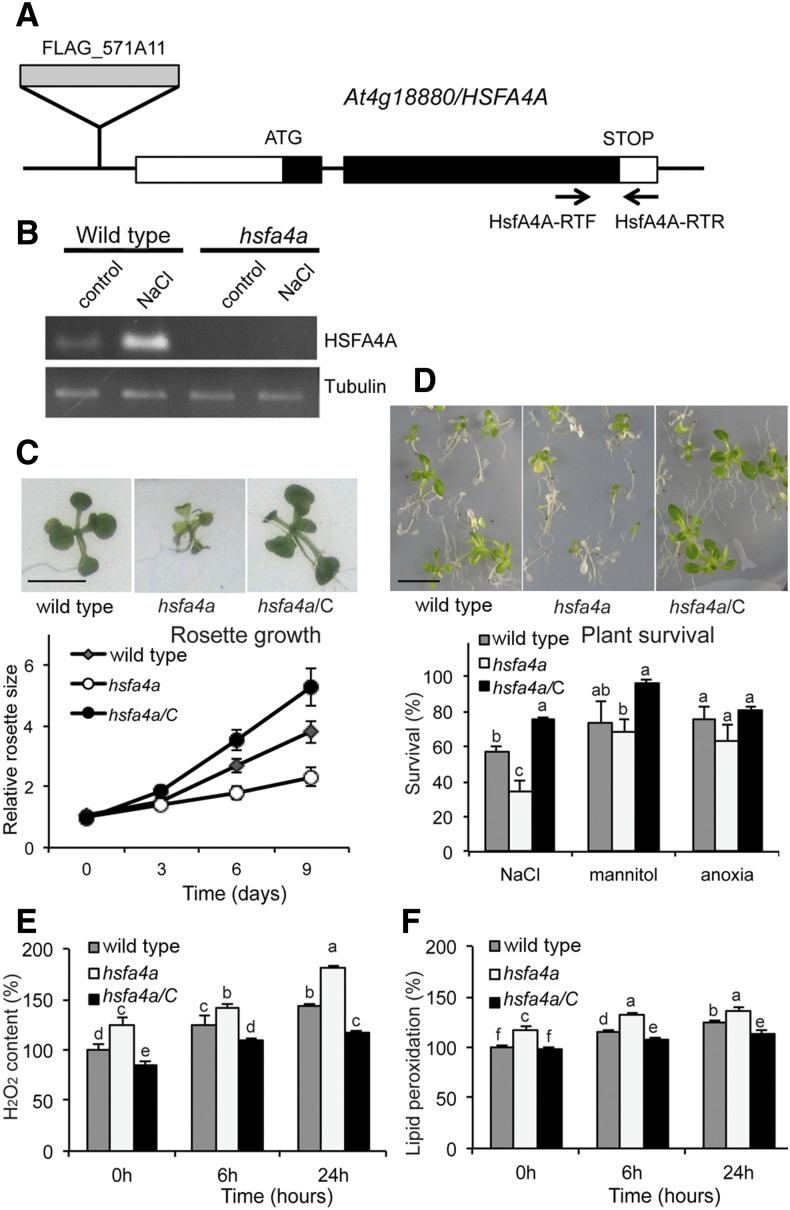
Characterization of the FLAG_571A11 T-DNA insertion mutant, which carries a T-DNA insert in the promoter at position -671 of the ATG of the HSFA4A (At4g18880) gene (Fig. 4A) provided further evidence for the involvement of HSFA4A in salt tolerance. Transcription of HSFA4A was induced by salt treatment in wild-type plants, while in the hsfa4a mutant, the T-DNA insertion abolished both basic and salt-inducible expression of HSFA4A (Fig. 4B). In standard growth conditions, growth and fertility of the wild type and hsfa4a mutant were indistinguishable. In the presence of 100 mm NaCl, the hsfa4a mutant showed 40% reduction of rosette growth compared with the wild type. The salt tolerance of hsfa4a could be genetically complemented by constitutive expression of HSFA4A (Fig. 4C). Whereas the hsfa4a mutant showed reduced growth and viability in the presence of salt, its responses to osmotic stress and anoxia did not reveal significant differences compared with the wild type (Fig. 4D). We detected enhanced H2O2 accumulation and higher lipid peroxidation rate in response to salt treatment in the hsfa4a mutant compared with the wild type, and this molecular phenotype could also be genetically complemented by constitutive expression of HSFA4A (Fig. 4, E and F).
Figure 4.
The hsfa4a T-DNA insertion mutant is hypersensitive to salinity. A, Position of FLAG_57A11 T-DNA insertion in the At4g18880 gene and primers used for reverse transcription (RT)-PCR. B, RT-PCR analysis of HSFA4A transcription in wild-type and hsfa4a mutant seedlings treated with 100 mm NaCl for 6 h. C, Rosettes of the wild type, hsfa4a mutant, and complemented mutant (hsfa4a/C). Five-day-old seedlings were transferred to and grown on medium supplemented by 100 mm NaCl for 9 d. Graph shows relative rosette sizes on 100 mm NaCl, where 1 corresponds to size of wild-type seedlings on day 0. D, Survival of wild-type, hsfa4a mutant, and hsfa4a/C plants on high-salt medium (200 mm NaCl, 5 d). Viability of plants was scored 7 d after stress recovery. Graph shows survival rates after high-salt, osmotic (600 mm mannitol, 5 d), and anoxia treatments (8 h; see Fig. 2E). E and F, Accumulation of H2O2 (E) and lipid peroxidation rates (F) in plants subjected to salt stress (150 mm NaCl, 6 and 24 h); for these assays, whole seedlings were used. Bars = 1 cm. [See online article for color version of this figure.]
Transcriptional Regulation of HSFA4A
Similarly to salt, H2O2 and paraquat treatments stimulated HSFA4A transcription within 1 h, leading to a peak of transcript levels at 6 h and decline of expression at 24 h. By contrast, heat stress increased HSFA4A mRNA levels at 1 and 6 h but was highest at 24 h (Supplemental Fig. S3A). H2O2 also elevated HSFA4A mRNA within 1 h and remained high for 96 h but never reached as high level as estradiol-induced mRNA of the overexpression HSFA4A construct (Supplemental Fig. S3B). According to transcript profiling data deposited at the eFP Browser database (http://bar.utoronto.ca/efp_arabidopsis; Winter et al., 2007), HSFA4A expression is moderately increased by heat but more dramatically stimulated by salt, osmotic and cold stress, UV-B irradiation, Pseudomonas spp. infection, and effectors N-terminal 22-amino acid fragment of flagellin and Harpin HrpZ (Supplemental Figs. S4 and S5). HSFA4A changes upon stress were noticeably distinct in roots compared with shoots for a number of stresses, as was previously noted (Kilian et al., 2007).
To visualize spatial pattern of HSFA4A expression during development, the activity of a pHSFA4A-GUS gene construct was analyzed in various organs. The HSFA4A promoter proved to be active in a broad range of organs and cell types, including leaves, cotyledons, hypocotyls, and roots, expressed in stipules and trichomes, and abundant in growing tissues, such as in young leaves, the meristematic zone of root tips, and the elongation zone of lateral roots (Supplemental Fig. S6).
Identification of HSFA4A-Regulated Genes
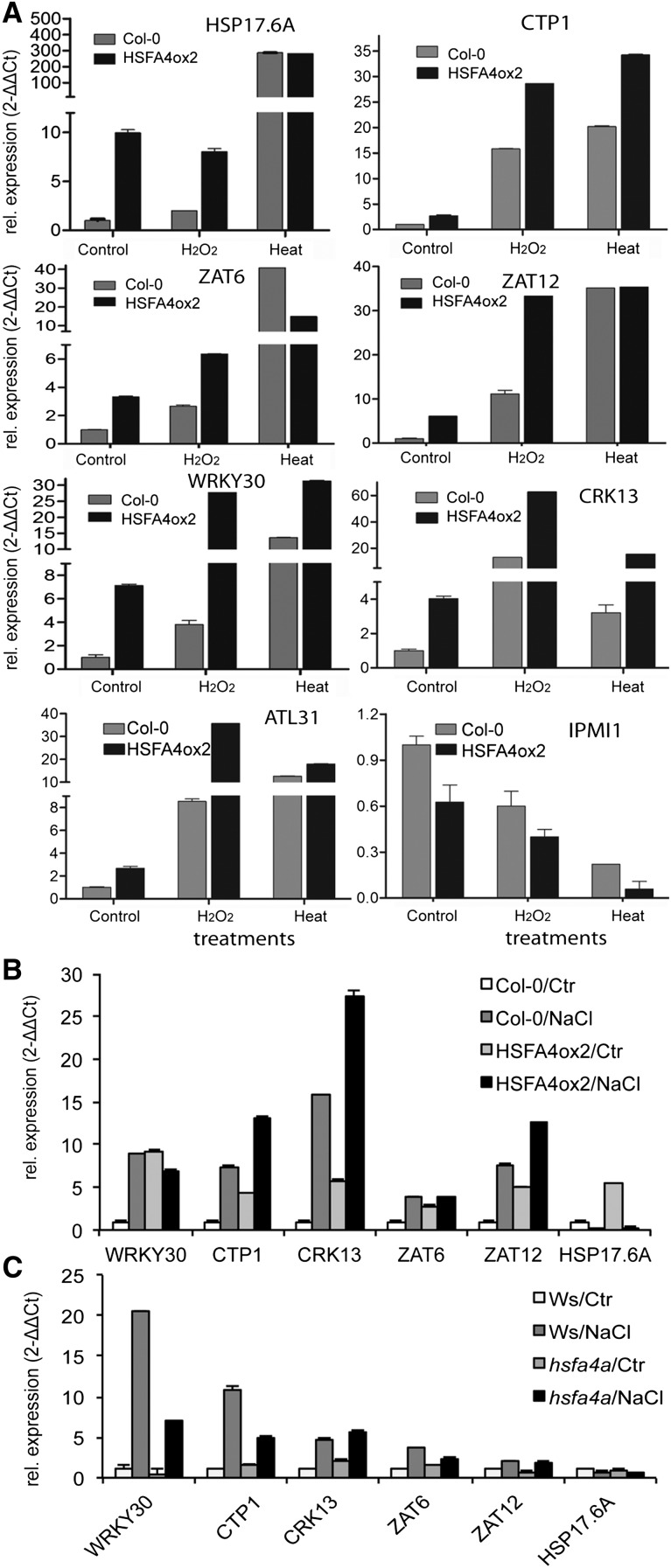
To screen for genes showing differential transcription due to HSFA4A overexpression and to relate this to ROS responses, we performed an RNA sequencing (RNA-Seq) transcript profiling experiment (Gene Expression Omnibus accession GSE40735). We used wild-type and HSFA4Aox2 plants that were treated with 5 µm estradiol in the presence or absence of 1 mm H2O2 for 6 h. We did three pairwise comparisons for differentially expressed genes: (1) wild type ± H2O2, (2) wild type versus HSFA4Aox2, and (3) wild type versus HSFA4Aox2 in the presence of H2O2 (Supplemental Fig. S7; Supplemental Data S1). Gene Ontology analysis of the up- and down-regulated gene categories obtained in transcript profiling experiment are summarized in Supplemental Figure S8, A to C. We selected genes that were common in these three comparisons (Supplemental Table S3) and annotated to stress responses for quantitative reverse transcription (qRT)-PCR analysis and ended up with a set of eight HSFA4A-regulated genes: small heat shock protein HSP17.6A (AT5G12030), copper transporter CTP1 (AT5G52760), C2H2 zinc finger proteins ZAT6 (AT5G04340) and ZAT12 (AT5G59820), transcription factor WRKY30 (AT5G24110), cysteine-rich receptor kinase-like protein CRK13 (AT4G23210), and ubiquitin ligase ARABIDOPSIS TOXICOS EN LEVADURA31 (ATL31; AT5G27420) were all up-regulated, while the isopropylmalate isomerase IPMI1 (AT3G58990) gene was reduced. Expression of HSF4Aox2 up-regulated genes could be further elevated by H2O2, while heat stress did not changed the induction of most of these genes. The HSF4Aox2 down-regulated gene IPMI1 was repressed by H2O2 and heat treatments (Fig. 5A). Transcription of most HSFA4A-induced genes, such as WRKY30, CTP1, and ZAT12, was also enhanced by salt stress (Fig. 5B). The hsfa4a mutation reduced salt-induced expression of WRKY30, CTP1, and ZAT6 but did not alter transcription of the CRK13, ZAT12, and HSF17.6A genes compared with the wild type (Fig. 5C). In conclusion, HSFA4A modulated the transcription of a subset of heat-, H2O2-, and salt-responsive genes.
Figure 5.
HSFA4A regulates the expression of a set of stress-related genes. A, Transcript analysis of eight HSFA4A target genes in HSFA4ox2 and Col-0 wild-type plants using qRT-PCR: HSP17.6A (AT5G12030), CTP1 (AT5G52760), ZAT6 (AT5G04340), ZAT12 (AT5G59820), WRKY30 (AT5G24110), CRK13 (AT4G23210), ATL31 (AT5G27420), and IPMI1 (AT3G58990). Plants were treated with 1 mm H2O2 or heat stress (37°C) for 6 h. All plants were treated with 5 μm estradiol. B and C, HSFA4A-regulated genes can be induced by salt. B, Expression of selected HSFA4A-regulated genes in HSFA4ox2 and Col-0 plants in control conditions (Ctr) or treated with 100 mm NaCl for 6 h (all plants were treated with 5 μm estradiol). C, Expression of the selected genes in hsfa4a mutant and wild-type plants (ecotype Wassilewskija [Ws]) in control conditions (Ctr) or treated with 100 mm NaCl for 6 h. Plants were 2 weeks old when subjected to treatments; whole seedlings where used. Transcript abundance was calculated with the 2−ΔΔCt method according to Fohgrub and Kempken (2012).
Next, we examined whether the promoters of HSFA4A-responsive genes carry consensus heat shock response elements. Among the 57 genes activated by HSFA4A and by H2O2 in both Col-0 and HSFA4Aox background, 33 promoters carried one or more conserved HSEs, whereas putative HSFB-binding sites were identified in 12 promoters (Supplemental Table S3). By contrast, 11 promoters with HSE elements and four with HSFB-binding sites were uncovered in the 42 genes that showed analogous down-regulation by HSFA4A and H2O2 (Supplemental Table S4). HSE elements and putative binding sites for HSFB, Trihelix, and MYC_MYB transcription factors were more frequent in the promoters of HSFA4A and H2O2 up-regulated genes, while promoter sequences of down-regulated genes showed an enrichment of predicted GROWTH-REGULATING FACTOR, C2C2(Zn)GATA, MADS, and homeodomain/leucine-zipper transcription factor sites (Supplemental Table S5). According to coexpression analysis (hierarchical clustering) with the Genevestigator database, 80% of these genes are enhanced in various abiotic and biotic stress conditions in the ROS-overproducing catalase2 (cat2) and fluorescent in blue light (flu) mutants but down-regulated during germination, while 75% of genes down-regulated by HSFA4A and H2O2 are repressed by several abiotic stress stimuli, including salicylic acid, but induced during germination (data not shown).
Previous studies demonstrated that different plant HSFs can cross regulate the expression of other HSF genes (Scharf et al., 2012). In our RNA-Seq data, from 16 HSF genes, 14 showed similar regulation, whereas transcription of HSFA8 was increased and HSFA6B reduced by estradiol induction of HSFA4A (Supplemental Fig. S9).
HSFA4A Shows Nuclear Redox State-Dependent Homomeric Interaction
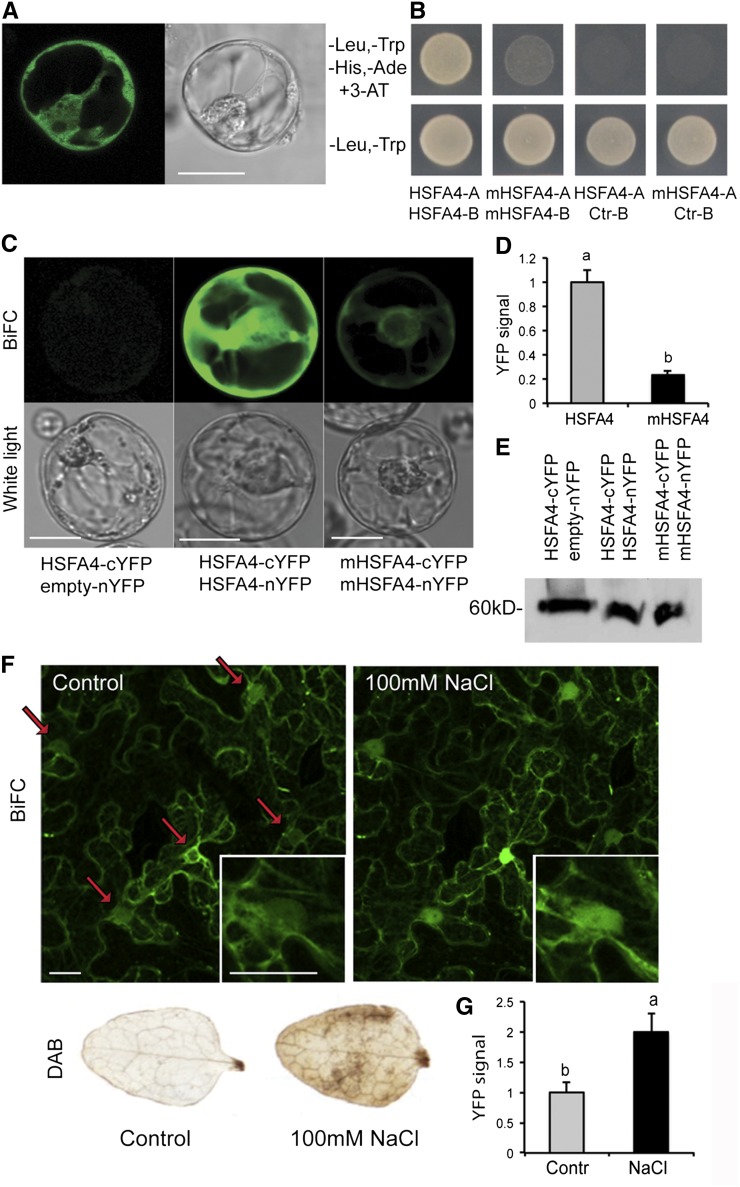
Similarly to other plants HSFs (Scharf et al., 2012), HSFA4A in C-terminal fusion with the yellow fluorescent reporter protein (YFP) showed cytoplasmic and nuclear localization when expressed in Arabidopsis protoplasts under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Fig. 6A). During their activation preceding nuclear import, animal HSFs undergo trimerization, which is thought to be stimulated through oxidation of their Cys residues by H2O2, accumulating during heat stress (Ahn and Thiele, 2003). To examine whether HSFA4A shares the conserved feature of homomeric interaction, HSFA4A was fused to both the activation domain (AD) or DNA-binding domain (BD) of the transcription factor GAL4 and expressed in yeast by the pGAD and pGBT9 vectors, respectively. Only cells carrying both vectors grew on selective 3-amino-1,2,4-triazole medium, indicating homomeric interaction of HSFA4A-AD and HSFA4A-BD proteins (Fig. 6B). Arabidopsis HSFA4A carries six Cys residues, three of which (C229, C267, and C295) are conserved in Brassicaceae spp. (Supplemental Fig. S2). The homomeric interaction in the yeast two-hybrid (Y2H) assay was abolished when these three conserved Cys residues were replaced with Ala by site-directed mutagenesis of HSFA4A cDNA (Fig. 6B; AD fusion, mHSFA4-A and BD fusion, mHSFA4-B).
Figure 6.
Localization and dimerization of HSFA4A in plant cells. A, Intracellular localization of HSFA4A-YFP fusion protein. B, Homodimeric interaction of HSFA4A in Y2H system. Yeast growth on selective medium (-Leu, -Trp, -His, -Ade, and 3-amino-1,2,4-triazole [3-AT]) indicates interaction of the assayed proteins. HSFA4-A and HSFA4-B indicate HSFA4A fused to activation and DNA-binding domains of GAL4, respectively. mHSFA4-A and mHSFA4-B indicate triple Cys mutant HSFA4A (C229A, C267A, and C295A). Ctr-B indicates empty vector with DNA-binding domain. C, BIFC in Arabidopsis protoplasts: transient expression of HSFA4A-nYFP and empty cYFP vectors (left), HSFA4A-nYFP and HSFA4A-cYFP (middle), and triple Cys mutant mHSFA4A-nYFP and mHSFA4A-cYFP (right). D, Quantitative evaluation of YFP signals in protoplasts transfected by wild-type (HSFA4) and triple Cys mutant HSFA4A-YFP gene constructs (mHSFA4). E, Western detection of HSFA4-cYFP in protoplasts of the transient BiFC experiment. The GFP-specific antibody (Roche) detects the C-terminal fragment of YFP. F, BIFC in tobacco cells. Transient expression of HSFA4A-nYFP and HSFA4A-cYFP in tobacco mesophyll cells. Leaves were treated with 100 mm NaCl for 20 min (right). Arrows indicate positions of nuclei, and insert shows enlarged image of a cell nucleus. Right and left sections display the same microscopic field before and after salt treatment. 3,3′-diaminobenzidine (DAB), H2O2 accumulation in control and salt-treated tobacco leaves visualized by DAB staining, which was used to monitor the ROS production in salt-treated tobacco leaves. G, Quantitative evaluation of YFP signal in nuclei of 20 transfected tobacco cells. Microphotographs in all figures show representative images. Bars = 20 μm. Error bars indicate ses, and different letters show significant differences at P < 0.05 (Duncan’s test).
To detect homomeric interaction of HSFA4A in plant cells, we created HSFA4A-nYFP and HSFA4A-cYFP fusions with the N- and C-terminal domains of split YFP, respectively, and expressed either in Arabidopsis protoplasts or in Agrobacterium spp.-infiltrated tobacco (Nicotiana benthamiana) leaves. Due to reconstitution of split YFP by interaction of HSFA4A-nYFP and HSFA4A-cYFP proteins, bimolecular fluorescence complementation (BiFC) was observed in both assays (Fig. 6, C and F). In parallel experiments, using similar fusions with mHSFA4A carrying the Cys-to-Ala replacements, the BiFC signal intensity was 75% lower compared with wild-type HSFA4A (Fig. 6, C and D), indicating that the Cys residues are functionally important to aid in vivo homomeric interaction of HSFA4A. Control immunoblotting with anti-GFP antibody, which also detects YFP, revealed that reduced BiFC signal intensity detected with the mHSFA4A constructs was not due to different levels of expression or stability of wild-type and mutant versions of HSF4A (Fig. 6E). Treatment of tobacco leaves with 100 mm NaCl led to ROS accumulation and increased the BiFC signal intensity 2-fold by parallel enhancement of nuclear localization of interacting HSFA4A-nYFP and HSFA4A-cYFP proteins (Fig. 6, F and G).
HSFA4A Interacts with and Is Phosphorylated by MPK3 and MPK6
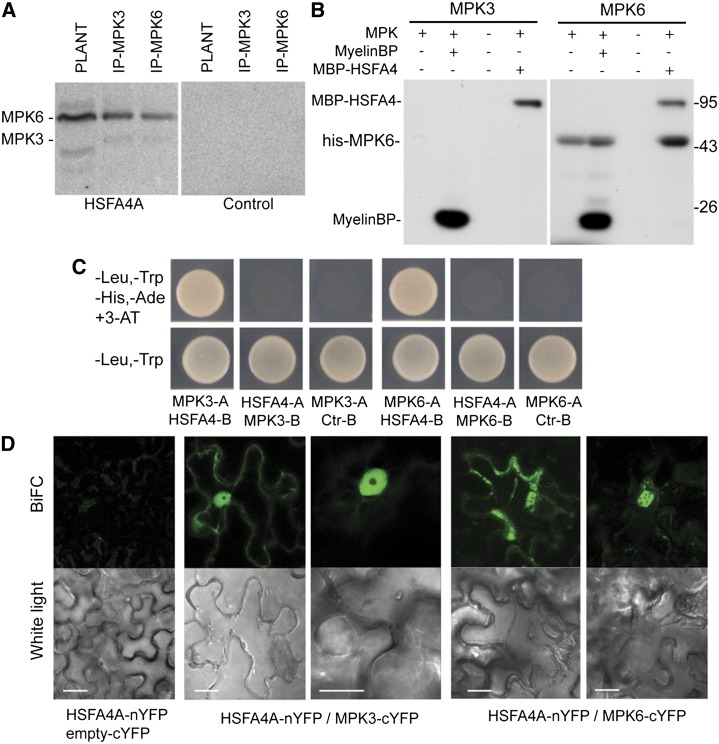
In yeast and mammals, phosphorylation by either MAP kinases or the glycogen synthase kinase3 is required for activation or repression of HSFs (Chu et al., 1996; Kim et al., 1997; Hashikawa and Sakurai, 2004). In Arabidopsis, the MAP kinases MPK3 and MPK6 were implicated in ROS signaling (Liu et al., 2010). Therefore, we examined whether HSFA4A can be a substrate for the ROS-stimulated MPK3 and MPK6 through their immunoprecipitation from wild-type plant protein extracts followed by in-gel kinase assays with purified HSFA4A. Due to cross reactivity of commercial antibodies, both MPK3 and MPK6 were immunoprecipitated by the anti-MPK3 and anti-MPK6 IgGs but could nevertheless be identified by their different mobilities. The in-gel assays indicated that HSFA4A was used as a phosphorylation substrate by both MPK3 and MPK6, though the two kinases showed slightly different activities (Fig. 7A). To corroborate these results, His6-tagged forms of MPK3 and MPK6 were purified and used in kinase assays with HSFA4A, which was purified by the help of an N-terminal maltose-binding protein (MBP) tag. HSFA4A was phosphorylated both by MPK3 and by MPK6 to a similar level as the commonly used MAP kinase substrate myelin basic protein (MyelinBP Fig. 7B).
Figure 7.
Phosphorylation and interaction of HSFA4A with MAP kinases. A, Phosphorylation of HSFA4A substrate with immunoprecipitated MPK3 and MPK6 kinases analyzed by in-gel kinase assay. Anti-MPK3 and anti-MPK6 antibodies (Sigma) partially recognize both kinases. PLANT indicates extracts obtained from wild-type Arabidopsis (Landsberg erecta). Control indicates gel without HSFA4A substrate. B, In vitro phosphorylation of purified HSFA4A by MPK3 and MPK6. MyelinBP was used as artificial substrate in positive control reaction. His-tagged MPK3/MPK6 was used in phosphorylation reactions. HSFA4A was tagged with maltose-binding protein (MBP-HSFA4). C, Interaction of HSFA4A with MPK3 and MPK6 in Y2H system. A indicates proteins fused to the activation domain, B indicates proteins fused to the DNA-binding domain, and Ctr indicates empty vector with the DNA-binding domain. D, Interaction of HSFA4A-nYFP, MPK6-cYFP, and MPK3-cYFP in A. tumefaciens-transformed tobacco leaf cells. BIFC indicates interacting proteins. Bars = 20 μm.
MAP kinases are known to interact with their substrates (Dóczi et al., 2012). To detect HSFA4A-MPK3 or -MPK6 interactions, we performed Y2H assays. MPK3 and MPK6 showed Y2H interactions when HSFA4A was part of the Gal4 DNA-binding domain and the kinases were fused to the Gal4 activation domain (Fig. 7C). Formation of HSFA4A-MPK3/MPK6 complexes was subsequently confirmed in planta. HSFA4A-nYFP, carrying the N-terminal domain of split YFP, showed strong BiFC interaction in the nuclei and occasionally in the cytoplasm of tobacco leaf epidermal cells MPK3-cYFP and MPK6-cYFP harboring the C terminus of split YFP (Fig. 7D).
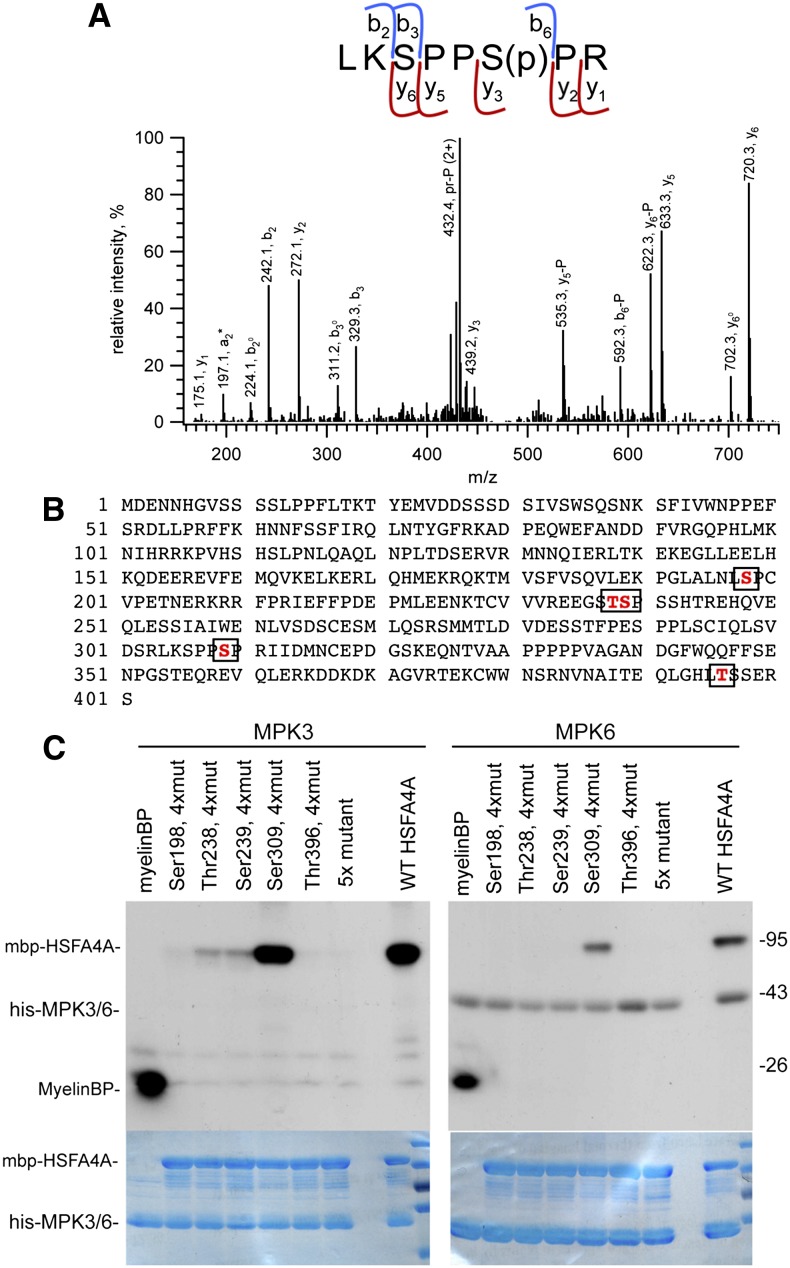
To identify the phosphorylation sites, purified MBP-HSFA4A was phosphorylated in vitro by MPK3 and digested with trypsin, and the peptide mixture was analyzed by mass spectrometry. Liquid chromatography-tandem mass spectrometry analysis of the tryptic digests confirmed the presence of the HSFA4A protein with high sequence coverage (>90%). Mass spectrometry analysis of the TiO2-enriched samples revealed the presence of multiple phosphopeptides enabling the assignment of four phosphorylation sites (Fig. 8B). Ser-198 and Thr-238 or Ser-239 were found to be phosphorylated exclusively in the MPK3-treated samples (incomplete fragmentation of the corresponding phosphopeptide prevented exact assignment for the latter site). Two further sites, Ser-309 (Fig. 8A) and Thr-396, were found to be phosphorylated both in the control and the kinase-treated samples. However, the level of phosphorylation is clearly elevated by MPK3. Comparison of the mass spectrometry peak areas of the corresponding precursor peptides in the control and the MPK3-treated samples indicated about 5-fold increase for Ser-309 phosphorylation, represented by LKSPPS(Phospho)PR, mass-to-charge ratio (m/z) 481.247 (2+), [304–311] of HSFA4A, and approximately 10-fold increase for Thr-396 phosphorylation, represented by NVNAITEQLGHLT(Phospho)SSERS, m/z 1018.476 (2+), [384–401] of HSFA4A, in the kinase-treated samples (Supplemental Data S2).
Figure 8.
Identification of HSFA4A phosphorylation sites. A, Identification of HSFA4A phosphorylation site by mass spectrometry. The collision-induced dissociation (CID) spectrum of m/z 481.247 (2+) representing the phosphorylated [304–311] sequence of HSFA4A (Uniprot ID O49403). Fragment ions b3 (unmodified) and y5 and y6 (phosphorylated) unambiguously prove that the site of modification is Ser-6, i.e. Ser-309 of HSFA4A. P denotes the 98-D neutral loss of the phosphate from the corresponding fragment ions. Observed peptide backbone cleavages are indicated in the sequence. B, Amino acid sequence of HSFA4A with MPK3 phosphorylation sites boxed. C, Phosphorylation of wild-type and mutant HSFA4A by MPK3 and MPK6. 5x mutant indicates all identified Ser and Thr residues changed to Ala, and 4xmut indicates all Ser and Thr changed to Ala, except the indicated one. MyelinBP was used in positive control reactions. Top section shows phosphorylation reaction, and bottom section shows Coomassie-stained gels indicating equal loading. [See online article for color version of this figure.]
Ala exchange of all five candidate phosphorylation sites completely abolished HSFA4A phosphorylation by MPK3 and MPK6 (Fig. 8C). HSFA4A mutated at four predicted phosphosites, except Ser-309 was phosphorylated almost as well as wild-type HSFA4A by MPK3 and MPK6. By contrast, when unaltered Ser-198, Ser-238, or Thr-239 residues were combined with Ala replacements of other four predicted phosphosites, phosphorylation of HSFA4A by MPK3 was strongly reduced but remained still detectable (Fig. 8C). Consequently, this combinatorial analysis of phosphosite Ala replacements indicated that, in in vitro conditions, Ser-309 is the most preferred site in HSFA4A for phosphorylation by both MPK3 and MPK6.
Phosphorylation of HSFA4A Enhances Transient Transactivation of HSP17.6A Transcription
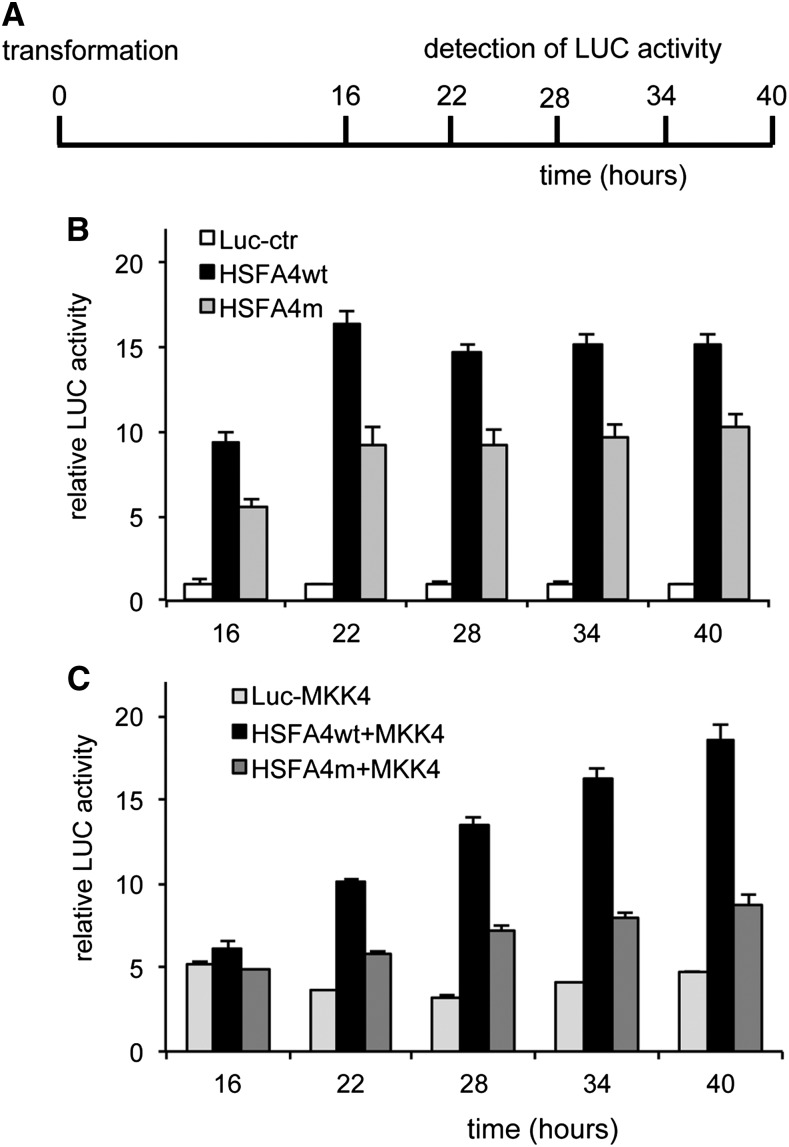
To assay the in vivo function of the MPK3 and MPK6 phosphosites, we designed a cell-based promoter activation assay by HSFA4A on the HSP17.6A promoter-reporter gene construct. HSP17.6A promoter sequences 2 kb upstream of translation start were fused to a firefly luciferase (LUC+) reporter gene and cotransformed into Arabidopsis protoplasts along with CaMV 35S promoter-cDNA constructs expressing either wild-type or Ser-309Ala mutant HSFA4A (HSFA4m). By monitoring luciferase activity at different time points up to 40 h after transformation (Fig. 9A), we observed that transactivation by wild-type HSFA4A increased the activity of pHSP17.6A-LUC reporter construct 15-fold compared with basic LUC levels. In comparison, HSFA4m carrying the Ser-309Ala replacement showed 30% to 40% lower transactivation of the reporter gene (Fig. 9B). The MAP kinase kinase4 (MKK4) is known to activate MPK3 and MPK6 (Asai et al., 2002; Kim et al., 2011). Next, we transformed cells with constitutive active MKK4EE to activate downstream MAP kinases MPK3 and MPK6 (Bartels et al., 2009) together with either the wild type or Ser-309Ala mutant of HSFA4A, which led to a steady increase of pHSP17.6A-LUC transactivation reaching about 18-fold increase upon 40 h with the wild type but was around 50% less with the Ser-309Ala mutant form of HSFA4A (Fig. 9C). These results suggested that MPK3 and MPK6 activation increases the HSFA4A transactivation function in vivo, while mutating the Ser-309 to nonphosphorylatable Ala abrogates it.
Figure 9.
Activation of pHSP17.6A-LUC reporter construct in transient expression system. A, Scheme of the experiment. Arabidopsis protoplasts were transformed with the LUC reporter construct and the wild-type HSFA4A (HSFA4wt) or Ser-309 mutant HSFA4A (HSFA4m) under the control of the CaMV 35S promoter at 0 h. Luciferine was added at 16 h, and LUC activity was detected 16, 22, 28, 34, and 40 h after transformation. (B) Trans activation of HSP17A-LUC with wild type and mutant HSFA4A constructs. C, Transactivation of HSP17.6A by HSFA4A constructs and constitutively active MKK4EE. In both graphs, normalized LUC activities are shown, where activities of control (Luc-ctr) is 1.
DISCUSSION
HSFA4A Confers Salt Tolerance in the COS Selection System
The HSF transcription factor family has 21 members in Arabidopsis, with considerable functional redundancy (Scharf et al., 2012). Due to overlapping regulatory functions of HSFs, the classical genetic approach based on characterization of individual gene mutations has a relatively low resolution power (Liu et al., 2011). Overexpression of individual HSFs, on the other hand, could alter tissue, cell type, or developmental stage-specific functions, combinatorial interactions, and transcriptional regulation of HSFs, leading to biased conclusions on their function. Therefore, in this work, we used a combination of knockout mutation with chemically induced overexpression, site-specific mutagenesis, in vivo and in vitro protein interactions and kinase assays, mass spectrometric and mutational analysis of phosphorylation sites, transcript profiling, and various physiological tests to gain insights into regulatory functions of Arabidopsis HSFA4A, one of the hitherto noncharacterized members of the functionally redundant HSF family.
HSFA4A was identified in a large-scale screening for cDNAs that confer tolerance to salinity when expressed in an estradiol-inducible manner in Arabidopsis cells. From 1.2 million microcalli transformed with an estradiol-inducible cDNA expression library, we recovered four transformants capable of proliferating in the presence of 175 mm NaCl. One of these expressed the full-length cDNA-encoded HSFA4A, from which recurrent expression of HSFA4A in calli and seedlings recapitulated the enhanced salt tolerance phenotype. A possible role of HSFA4A in modulation of salt tolerance was corroborated by the observation that while its overexpression enhanced salt tolerance, the hsfa4a knockout mutation conferred sensitivity to salt stress. Subsequently, we showed that transcription of HSFA4A is inducible by salt, H2O2, paraquat, and heat treatments, which are all known to induce ROS accumulation. In addition, mining the depository of public transcript profiling data indicated that HSFA4A transcription is also stimulated by drought, cold, UV-B light, hypoxia, and ozone (Davletova et al., 2005a; Gadjev et al., 2006), as well as several pathogens and their effectors (Libault et al., 2007), and the flu mutation characterized by singlet oxygen production in the light (op den Camp et al., 2003; Gadjev et al., 2006). This suggested that HSFA4A is a target of a ROS-mediated signal transduction pathway. By contrast, expression of HSFA4A is inhibited by mutations that disrupt salicylic acid biosynthesis and salicylic acid-related pathogen defense (e.g. salicylic acid induction deficient2 phytoalexin deficient4 nonexpresser of PR1 genes1 and enhanced disease susceptibility1 Chaouch et al., 2010; Straus et al., 2010).
Study of the effects of estradiol-inducible HSFA4A overexpression revealed that induction of HSFA4A confers enhanced tolerance not only to NaCl, but also to osmotic stress, paraquat, H2O2, and anoxia, which generate oxidative stress. It is thus not surprising that potential HSFA4A orthologs from wheat and rice were reported to confer cadmium tolerance when expressed in yeast and transgenic rice, and the putative rice hsfa4a mutant was found to be hypersensitive to cadmium (Shim et al., 2009). In comparison, overexpression of HSFA2 enhances salt and osmotic stress tolerance (Ogawa et al., 2007), while HSFA3 confers drought tolerance (Yoshida et al., 2008), suggesting that particular members of the HSF family are implicated in responses to various environmental stress conditions. Analogously to overproduction of C-REPEAT/DRE BINDING FACTOR transcription factors, which confer tolerance to cold stress (Achard et al., 2008), estradiol induction of HSFA4A thus conferred some growth inhibitory effect in the absence of stress while decreased growth inhibition under salt stress.
Despite activation of HSFA4A by ROS-generating stress conditions, the hsfa4a mutant does not show enhanced sensitivity to osmotic stress and anoxia, indicating that other HSFs likely compensate for the lack of HSFA4A to mount sufficient tolerance to these stresses. The salt-stressed hsfa4a mutant displays enhanced H2O2 accumulation and lipid peroxidation compared with the wild type, suggesting that oxidative stress tolerance is lowered by inactivation of HSFA4A. On the other hand, enhanced oxidative stress tolerance of HSFA4Aox plants can partly be due to increased APX1 activity leading to lower H2O2 content and reduced lipid peroxidation. These data suggest that HSFA4A is involved in the modulation of ROS metabolism and responses to oxidative stress. HSF-dependent regulation of Apx genes has been previously documented (Panchuk et al., 2002; Li et al., 2005; Banti et al., 2010), underlining the importance of HSFs as mediators of oxidative stress responses.
HSFA4A-Regulated Genes
We identified genes that are controlled by HSFA4A and peroxide signals and are annotated to respond to diverse range of stresses, such as heat, salt, heavy metals, and/or pathogens. qRT-PCR analysis confirmed that HSFA4A is involved in transcriptional activation of HSP17.6A, ZAT6, ZAT12, CTP1, WRKY30, and CRK13, which play pivotal roles in the regulation heat, salt, oxidative, osmotic, heavy metal, pathogenic, and starvation stress responses (Sun et al., 2001; Davletova et al., 2005b; Devaiah et al., 2007; Libault et al., 2007; Peng et al., 2012). Most of these genes are up-regulated by salinity, indicating that they are involved in defenses to salt stress. Similarly to H2O2, ZAT12 stimulates the induction of the APX1 gene in response to light stress, providing a feedback regulatory circuit for H2O2 removal (Davletova et al., 2005b). WRKY30 is induced by superoxide anions generated in damaged chloroplasts (Scarpeci et al., 2008), whereas ZAT6 is activated by osmotic and salt stress (Liu et al., 2013). These results suggest that HSFA4A can act in common pathways with these transcription factors. The expression of several APX and HSP genes is known to be controlled by HSF complexes in response to H2O2-derived signals during heat stress (Volkov et al., 2006). Similarly, HSFA4A seems to modulate transcription of a set of target genes involved in mounting defense to abiotic and biotic stress stimuli. Data obtained from RNA-Seq pointed to a substantial overlap between the HSFA4A-regulated gene set and those deregulated in mutants enhancing ROS production (e.g. cat2 and flu; op den Camp et al., 2003; Gadjev et al., 2006). Promoters of many HSFA4A-regulated genes contain one or more HSE motifs, suggesting that HSFA4A and/or other HSFs might directly control their transcription
Homomeric Interaction of HSFA4A
Activation of HSFs involves the formation of homotrimers, which is required for their nuclear import and high-affinity binding to conserved HSEs in the promoters of target genes (Anckar and Sistonen, 2011). Our yeast and BiFC protein interaction studies illustrate that HSFA4A forms homodimers (or trimers) in both yeast and plant cells, which is stimulated by salt treatment. Similarly, Arabidopsis HSFA1A and HSFA1B were reported to form homo- and heterodimers, which is thought to be essential for transcriptional activation of their target genes (Li et al., 2010). Repression of the activity of HSFA4 through interaction of HSFA5 was reported in tomato and Arabidopsis (Baniwal et al., 2007). Such interaction can be responsible for the regulation of HSFA4 in other species as well. As transcript levels of HSFA5 were not affected significantly by enhanced HSFA4A expression, balance between the two factors was probably changed, leading to HSFA4A excess in the overexpressing plants. Importance of proper control of HSFA4A activity can be illustrated by the fact that HSFA4Aox plants displayed growth deficit in nonstress conditions when compared with the wild type. Oligomerization of mammalian HSFs in response to oxidative stress is thought to be mediated by H2O2-dependent oxidation of Cys residues (Ahn and Thiele, 2003). Although oligomerization domains were shown to be essential for interactions between HSFA1a and HSFA1b in Arabidopsis (Li et al., 2010) and HSFA1 and HSFA2 in tomato (Chan-Schaminet et al., 2009), the importance of conserved Cys residues in homomeric interactions of plant HSFs has not been clearly demonstrated. Our data support a likely role of Cys residues in dimerization of Arabidopsis HSFA4A, because exchanging conserved Cys residues to Ala greatly reduces the formation of HSFA4A dimers without affecting protein stability. However, the fact that these residues are conserved in HSFA4A homologs in Arabidopsis-related species but not in HSFA4A proteins of orange (Citrus sinensis) grapevine (Vitis vinifera), and poplar (Populus spp.) suggests that formation of redox-sensitive disulfide bonds of Cys residues is probably not a sole requirement for HSFA4A dimerization. Therefore, the prediction that plant HSFs function as molecular sensors of ROS signals (Miller and Mittler, 2006) remains to be revisited by further studies.
Phosphorylation of HSFA4A by Interacting MPK3 and MPK6 Kinases
Our data demonstrate that HSFA4A interacts with MPK3 and MPK6 in both yeast and plant cells and that these kinases phosphorylate HSFA4A as substrate in vitro. Phosphorylation of heat shock factors by MAP kinases was reported in several organisms, including mammalian cells (Chu et al., 1996; Kim et al., 1997; Chen et al., 2001), yeast (Hashikawa and Sakurai, 2004), and plants (Link et al., 2002; Evrard et al., 2013). While phosphorylation repressed HSF1-dependent transcriptional activation in human cells (Chu et al., 1996), multimerisation of yeast HSF in connection with hyperphosphorylation enhanced induction of target genes (Hashikawa and Sakurai, 2004). Heat-activated MAP kinases were shown to transduce heat stress signals by phosphorylating HSFs in tomato or promoting HSP gene expression in tobacco (Link et al., 2002; Suri and Dhindsa, 2008). Recently, phosphorylation and activation of Arabidopsis HSFA2 by MPK6 was reported (Evrard et al., 2013). MPK3- and MPK6-dependent phosphorylation of HSFA4A and their physical interaction therefore correlate with earlier observations indicating that some HSFs can be substrates for MAP kinases. We identified Ser-309 as the preferential MPK3 and MPK6 phosphorylation site in HSFA4A in vitro, which is located between two activator domains. This residue is conserved in closely related homologs of Eutrema salsugineum and Brassica napus but not in more distantly related plants, such as orange, grapevine, or poplar. Whether HSFA4A is phosphorylated on Ser-309 site after MPK3 and MPK6 activation in vivo remains to be established, but these experiments are challenging (Dephoure et al., 2013). To overcome this limitation, we decided to perform in vivo functional studies by site-directed mutagenesis. We established that Ser-309 phosphorylation of HSFA4A can have a regulatory function in plant cells because exchange of this Ser for Ala reduces the transactivation of pHSP17.6A-LUC reporter by HSFA4A in a cell-based transient transformation assay in cell culture-derived protoplasts. Furthermore, the constitutively active form of MKK4 stimulates transactivation of the HSP17.6A promoter-driven LUC expression, both alone and in combination with HSFA4A. These results suggest that HSFA4A could act in concert with the MKK4 MPK3/MPK6 phosphorylation cascade controlling the transcription of HSP17.6A. However, further studies are needed to characterize the function of MAP kinase-mediated HSFA4A phosphorylation in transcription regulation of other target genes.
It is therefore intriguing that MPK3 and MPK6 were identified as central regulators of plant innate immunity, hypoxia, and salt and osmotic stress responses, which control cross talk between different stresses, hormonal signals, and second messengers, such a ROS (Droillard et al., 2002; Teige et al., 2004; Chang et al., 2012; Rasmussen et al., 2012; Smekalova et al., 2013). MAP kinases were reported to phosphorylate other transcription factors such as WRKY33, which is involved in regulation of ethylene biosynthesis (Li et al., 2012), or MYB44, which regulates abscisic acid sensitivity (Nguyen et al., 2012b). MPK3 and MPK6 can form in vivo complexes with ZAT10/salt tolerance zinc finger, regulating plant defense responses (Mittler et al., 2006; Nguyen et al., 2012a), while under salt and osmotic stress, MPK6 interacts with and phosphorylates ZAT6 (Liu et al., 2013). Thus, MPK3/MPK6 appear to regulate a wide range of biotic and abiotic defense responses mediated by ROS signals and coordinate the activity of various transcription factors such as WRKY, MYB, ZAT, and HSF that, in turn, control the transcription of a large set of target genes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col-0) plants were grown in sterile conditions using one-half-strength MS culture medium under controlled conditions with an 8-h-light/16-h-dark light cycle at 22°C and 100 μE m–2 s–1 light intensity. For HSFA4A transcription activation studies, in vitro-grown wild-type plants were incubated in liquid one-half-strength MS medium as control and one of the following additives: 1 mm H2O2, 1 μm paraquat, 150 mm NaCl, or 300 mm mannitol. Heat treatments were performed at 37°C in a growth chamber (70% relative humidity, 100 μE m–2 s–1). The knockout Arabidopsis line (FLAG_571A11) containing a T-DNA insertion in the promoter region of the HSFA4A gene was obtained from the Institut National de la Recherche Agronomique Versailles T-DNA collection (ecotype Wassilewskija; http://urgv.evry.inra.fr/FLAGdb). Homozygous hsfa4a mutant line was generated by PCR-based genotyping using gene-specific and T-DNA-specific primers (Supplemental Table S6). Tolerance to stress conditions, such as salinity and osmotic and oxidative stress, was tested in vitro conditions as described (Verslues et al., 2006; Papdi et al., 2010). For details, see Supplemental Materials and Methods S1.
Transformation and Screening of Arabidopsis Cell Cultures
Root-derived Arabidopsis cell suspension culture (Mathur et al., 1995) was used for large-scale transformation and screening. Cell suspension culture was transformed with the COS cDNA library as described (Papdi et al., 2008; Rigó et al., 2012). Briefly, 25 mL of cell suspension was infected with Agrobacterium tumefaciens culture of the COS cDNA library and cocultivated for 2 d under continuous shaking. Subsequently, cells were collected by centrifugation, washed with and subcultured in liquid culture medium, and supplemented by 400 mg L–1 Claforan and 15 mg L–1 hygromycin. Transformed cell cultures were subcultured at weekly intervals with 10× volume of fresh culture medium. The hygromycin-resistant cell culture was plated onto 100 petri dishes (150-mm diameter) containing agar-solidified callus culture medium (Mathur et al., 1995) supplemented with 400 mg L–1 Claforan, 5 μm estradiol, and 175 mm NaCl. Transformation efficiency was estimated by plating an aliquot of transformed cell suspension on medium supplemented with 400 mg L–1 Claforan and 15 mg L–1 hygromycin. After 3 weeks of culture, growing microcalli were split and moved to fresh agar plates containing 175 mm NaCl with or without 5 μm estradiol. Calli growing on estradiol-containing plates were used for rescuing cDNAs carried by the T-DNA inserts.
Analysis of Physiological Parameters
H2O2 content and lipid peroxidation were determined in 2-week-old plants as described (Zsigmond et al., 2012). APX activity was assayed according to Nakano and Asada (1981). Histochemical detection of GUS enzyme activity in pHSFA4A-GUS-expressing plants was performed in five independent transgenic lines as reported (Jefferson et al., 1987). All experiments were repeated at least twice. Anthocyanin accumulation in paraquat-treated seedlings was scored according to Kortstee et al. (2011; see Supplemental Materials and Methods S1).
Molecular Methods
HSFA4A was isolated by PCR amplification of cDNA insert from genomic DNA of the selected A1 callus. Nucleotide sequence of the PCR fragment was determined, and the fragment was cloned in pDONR201 vector and subsequently inserted into the plant expression vector pER8GW as described (Papdi et al., 2008; Rigó et al., 2012). Full-length cDNA of MPK3 and MPK6 was cloned from an Arabidopsis cDNA library as described in Supplemental Materials and Methods S1. Gene cloning, in vitro-targeted mutagenesis, and vector construction is described in Supplemental Materials and Methods S1.
Total RNA was isolated from plant tissues using the Tri Reagent method (Chomczynski and Sacchi, 1987). cDNA templates were prepared from DNase-treated RNA. Transcript analysis was made with qRT-PCR or semiquantitative RT-PCR as reported (Papdi et al., 2008; for details, see Supplemental Materials and Methods S1). Sequences of PCR primers are listed in Supplemental Table S6. Each experiment was repeated at least twice.
For transcript profiling and gene expression analysis, 2-week-old Col-0 and HSFA4Aox2 plants were treated with 5 µm estradiol in the absence or presence of 1 mm H2O2 for 6 h in liquid one-half-strength MS medium. Total RNA was isolated with RNeasy Kit (Qiagen), RNA quality and quantity was measured on Bioanalyzer (Agilent Technologies) and Qubit (Life Technologies), and total RNA samples were processed using the SOLiD Total RNA-Seq Kit (Life Technologies), according to the manufacturer’s instructions. RNA samples were processed using the SOLiD Total RNA-Seq Kit (Life Technologies), and the templates were sequenced on a SOLiD 5500xl instrument using the 50-base sequencing chemistry. Basic bioinformatic analysis of the RNA-Seq data was performed with Genomics Workbench 4.7.2 (CLC Bio). RNA-Seq reads were mapped onto the Arabidopsis reference genome (The Arabidopsis Information Resource 10), and the number of sequencing reads generated from each sample was converted into reads per kilobase of exon model per million mapped reads (Mortazavi et al., 2008; see Supplemental Materials and Methods S1).
Gene Ontology analysis was performed with the Gene Ontology annotation search tool of The Arabidopsis Information Resource database (http://www.arabidopsis.org/tools/bulk/go/index.jsp). Coexpression analysis and clustering was made with Genevestigator (http://www.genevestigator.com/gv; Zimmermann et al., 2004). Identification of predicted promoter elements was done with AthaMap (http://www.athamap.de; Steffens et al., 2005) and Promomer (http://bar.utoronto.ca; Toufighi et al., 2005) tools.
Y2H Assay
For Y2H analyses, yeast (Saccharomyces cerevisiae) PJ69-4a was cotransformed with pGAD424 and pGBT9 vectors carrying the cloned inserts as described (James et al., 1996). Transformants were grown on appropriate dropout media to select for transformed cells as well as to monitor the activation of the Adenine requiring2 and/or Histidine3 reporter genes in the presence of 1 mm 3-aminotriazol.
Protein Localization and Detection
In vivo microscopic observations were made with an Olympus confocal laser-scanning microscope. For transient expression, Arabidopsis protoplasts were isolated from cell suspension, and polyethylene glycol-mediated transformation was performed as described (Mathur et al., 1995). Typically, 105 protoplasts were transformed with 20 μg plasmid DNA of 35S-HSFA4A-nYFP and 35S-HSFA4A-cYFP constructs or their mutant versions and cultured for 24 h prior to fluorescence observation or western-blot analysis. The average value of YFP intensity per area of mHSFA4A-n/cYFP-expressing protoplasts was normalized to HSFA4A-n/cYFP-expressing cells, measuring at least 20 cells for each BiFC combination. Every experiment was repeated three times. For western-blot analysis, protoplasts transformed with 35S-HSFA4A-cYFP or 35S-mHSFA4A-cYFP were harvested and boiled with SDS sample buffer. Protein samples were separated by 10% (w/v) SDS-PAGE and western blotting was performed as described (Zsigmond et al., 2008) using anti-GFP mouse monoclonal antibody (Roche) for the detection of cYFP-tagged proteins. BiFC assays in tobacco (Nicotiana benthamiana) were performed as described (Lumbreras et al., 2010). Tobacco leaves were transfected with equal volumes of combined A. tumefaciens solutions carrying 35S-HSFA4A-n/cYFP, 35S-MPK6-n/cYFP, and 35S-MPK3-n/cYFP or 35S-n/cYFP control constructs. YFP-derived fluorescence was monitored 24 h after infiltration. Stress treatments were performed by application of saline solution (100 mm NaCl in one-half-strength MS medium) on immobilized infiltrated leaves. Fluorescence was monitored in 20-min intervals in the same microscopic field. Reconstituted YFP signal intensities were measured with the ImageJ software.
In Vitro and In-Gel Kinase Assays
The kinases (His6-MPK3 and His6-MPK6) and the substrate proteins (His6-HSFA4A, MBP-HSFA4A, and MBP-tagged mutant versions of HSFA4A) were expressed in Escherichia coli strain BL21(DE3) Rosetta (Novagen) and purified by affinity chromatography on nickel-nitrilotriacetic acid agarose affinity (Qiagen) or Amylose resin (New England Biolabs) following the manufacturer’s instructions. In vitro phosphorylation assays with purified His6-MPK3 or His6-MPK6 and MBP-HSFA4A, MBP-tagged mutant versions of HSFA4A, Myelin Basic Protein (MyelinBP; Sigma), or the MBP tag were carried out with 1 μg His6-MPK3/MPK6 in 20 μL kinase buffer (25 mm Tris-HCl, pH 7.5, 20 mm MgCl2, 1 mm dithiothreitol, and 5 µCi [γ-32P]ATP) containing 2 μg MBP-HSFA4A constructs, 2 μg MyelinBP, or 2 μg MBP as substrate at 24°C for 30 min. Reactions were boiled with SDS sample buffer, and size was separated in 12% (w/v) SDS-PAGE. Gels were stained with PageBlue Protein Staining Solution (Thermo Scientific), dried, and subjected to autoradiography (Nguyen et al., 2012a). In-gel kinase assay and MPK3/MPK6 protein immunoprecipitation were performed as previously described (Lumbreras et al., 2010). Fifty micrograms of crude plant proteins and MPK3 and MPK6 precipitates were extracted from wild-type plants (ecotype Landsberg erecta) and loaded onto a 10% (w/v) SDS-PAGE gel containing 0.5 mg mL–1 His6-HSFA4A. The control polyacrylamide gel was assayed without substrate.
Identification of HSFA4A Phosphorylation Sites
MBP-HSFA4A was phosphorylated by His6-MPK3 in a kinase reaction performed in the presence of 200 µm cold ATP. As control, a parallel reaction without kinase was included. Samples were resolved by SDS-PAGE as described, and MBP-HSFA4A protein bands were excised after gel staining with PageBlue Protein Staining Solution. One-dimensional SDS-PAGE gel samples containing the recombinant HSFA4A protein were treated with the general in-gel digestion protocol (reduction and alkylation of Cys side chains followed by tryptic digestion for 4 h; for detailed protocol, see http://msf.ucsf.edu/ingel.html). Approximately 20% of the resulting peptide mixture was analyzed directly, and the remaining 80% was subjected to TiO2 enrichment (Larsen et al., 2005) prior to mass spectrometry analysis on an Orbitrap Elite mass spectrometer (Thermo Scientific). Phosphopeptides were identified by database search, and all identifications were inspected manually. For detailed experimental setup, see Supplemental Data S1 and S2.
Transient Expression Assays
Protoplasts were transformed with 10 μg plasmid DNA of pHSP17.6A-LUC construct and 20 μg plasmid DNA of 35S-HSFA4A-cYFP (HSFwt) or 35S-Ser-309HSFA4A-cYFP (HSFm). When indicated, 10 μg of pK2GW7-mycMKK4EE were included. All transformations were performed in triplicates. After 16-h culture, protoplasts were transferred to a dark 96-well microtiter plate, sharing each single transformation into two wells containing 3 mm d-luciferin solution. Luciferase activity was determined with an automated luminometer (TopCount NXT; Perkin-Elmer) for 24 h as described (Palágyi et al., 2010).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AEE84101, ESQ55497, ADX69244, XP_002267171, EEE97421, and XP_006467594.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic illustration of COS screening and cloning strategy.
Supplemental Figure S2. Multiple protein sequence alignment of the Arabidopsis HSFA4A and related transcription factors from five plant species.
Supplemental Figure S3. Regulation of HSFA4A transcription.
Supplemental Figure S4. Transcriptional activation of the HSFA4A gene by abiotic stress conditions.
Supplemental Figure S5. Transcriptional activation of HSFA4A by biotic stress.
Supplemental Figure S6. Spatial expression of pHSFA4A-GUS in transgenic plants.
Supplemental Figure S7. HSFA4A regulates the expression of a large set of target genes.
Supplemental Figure S8. Gene Ontology categorization of gene sets that were up- or down-regulated by H2O2 or HSFA4A or both.
Supplemental Figure S9. Expression profiles of Arabidopsis heat shock factors in wild-type and HSFA4A-overexpressing plants.
Supplemental Table S1. Summary of results of cell suspension transformation with the COS library.
Supplemental Table S2. Arabidopsis cDNAs identified in COS-transformed cell cultures.
Supplemental Table S3. Genes up-regulated by HSFA4A and H2O2.
Supplemental Table S4. Genes down-regulated by HSFA4A and H2O2.
Supplemental Table S5. Predicted transcription factor-binding sites in promoter regions of HSFA4A- and H2O2-induced or -repressed genes.
Supplemental Table S6. List of oligonucleotides used in this study.
Supplemental Materials and Methods S1. Gene cloning, RNA-seq analysis, mass spectrometry, analysis of stress tolerance, and gene expression.
Supplemental Data S1. Compiled RNA-Seq transcript profiling data.
Supplemental Data S2. Mass spectrometry data showing phosphorylation of HSFA4A peptides.
Supplementary Material
Acknowledgments
We thank Annamári Király and Mihály Dobó for assistance in growing the plants, Ferhan Ayaydin and Ágnes Cséplő for assistance in microscopy, László Kozma-Bognár for assistance in measuring luciferase activity, Elena Nájar for help in kinase assays, Montserrat Pagès for critical discussions, and anonymous reviewers for reading, suggestions, and corrections to improve the manuscript.
Glossary
- BiFC
bimolecular fluorescence complementation
- HSE
heat stress element
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- MAP
mitogen-activated protein
- COS
Conditional Overexpression System
- cDNA
complementary DNA
- T-DNA
transfer DNA
- Col-0
ecotype Columbia
- MS
Murashige and Skoog
- RNA-Seq
RNA sequencing
- RT
reverse transcription
- CaMV
Cauliflower mosaic virus
- Y2H
yeast two-hybrid
- m/z
mass-to-charge ratio
Footnotes
This work was supported by Országos Tudományos Kutatási Alap (grant no. K–81765), Hungary-Serbia Cross-border Co-operation Programme (project no. HUSRB/1002/214/036), EU–FP6 Marie Curie Action (Fellowship no. FP6–020232–2), EU–FP7 Marie Curie (Fellowship no. FP7–330713), and European Cooperation in Science and Technology action FA0605.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SG, Thiele DJ. (2003) Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev 17: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. (2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80: 1089–1115 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, et al. (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29: 471–487 [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Chan KY, Scharf KD, Nover L. (2007) Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem 282: 3605–3613 [DOI] [PubMed] [Google Scholar]
- Banti V, Mafessoni F, Loreti E, Alpi A, Perata P. (2010) The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 152: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf KD. (2009) Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J Biol Chem 284: 20848–20857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R, Jang CJ, Branco-Price C, Nghiem P, Bailey-Serres J. (2012) Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol Biol 78: 109–122 [DOI] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, Mhamdi A, Vandorpe M, Langlois-Meurinne M, Van Breusegem F, Saindrenan P, Noctor G. (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Tsai SH, Shen SC, Lin JK, Lee WR. (2001) Alternative activation of extracellular signal-regulated protein kinases in curcumin and arsenite-induced HSP70 gene expression in human colorectal carcinoma cells. Eur J Cell Biol 80: 213–221 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. (1996) Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem 271: 30847–30857 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Pan S, Salem T, Gurley WB. (2004) Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol 56: 57–75 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. (2005a) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005b) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Gould KL, Gygi SP, Kellogg DR. (2013) Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell 24: 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóczi R, Okrész L, Romero AE, Paccanaro A, Bögre L. (2012) Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci 17: 518–525 [DOI] [PubMed] [Google Scholar]
- Droillard M, Boudsocq M, Barbier-Brygoo H, Laurière C. (2002) Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett 527: 43–50 [DOI] [PubMed] [Google Scholar]
- Evrard A, Kumar M, Lecourieux D, Lucks J, von Koskull-Döring P, Hirt H. (2013) Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fohgrub U, Kempken F. (2012) Molecular analysis of fungal gene expression upon interkingdom competition with insects. Methods Mol Biol 944: 279–286 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa N, Sakurai H. (2004) Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol Cell Biol 24: 3648–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF. (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39: 969–987 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim J, Nueda A, Meng YH, Dynan WS, Mivechi NF. (1997) Analysis of the phosphorylation of human heat shock transcription factor-1 by MAP kinase family members. J Cell Biochem 67: 43–54 [DOI] [PubMed] [Google Scholar]
- Kim SH, Woo DH, Kim JM, Lee SY, Chung WS, Moon YH. (2011) Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem Biophys Res Commun 412: 150–154 [DOI] [PubMed] [Google Scholar]
- Kortstee AJ, Khan SA, Helderman C, Trindade LM, Wu Y, Visser RG, Brendolise C, Allan A, Schouten HJ, Jacobsen E. (2011) Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res 20: 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P. (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Kytridis VP, Manetas Y. (2006) Mesophyll versus epidermal anthocyanins as potential in vivo antioxidants: evidence linking the putative antioxidant role to the proximity of oxy-radical source. J Exp Bot 57: 2203–2210 [DOI] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jørgensen TJ. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 4: 873–886 [DOI] [PubMed] [Google Scholar]
- Li C, Chen Q, Gao X, Qi B, Chen N, Xu S, Chen J, Wang X. (2005) AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci China C Life Sci 48: 540–550 [DOI] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Doll J, Weckermann K, Oecking C, Berendzen KW, Schöffl F. (2010) Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins. Eur J Cell Biol 89: 126–132 [DOI] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Link V, Sinha AK, Vashista P, Hofmann MG, Proels RK, Ehness R, Roitsch T. (2002) A heat-activated MAP kinase in tomato: a possible regulator of the heat stress response. FEBS Lett 531: 179–183 [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY. (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu XM, Kim KE, Kim KC, Nguyen XC, Han HJ, Jung MS, Kim HS, Kim SH, Park HC, Yun DJ, et al. (2010) Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species. Phytochemistry 71: 614–618 [DOI] [PubMed] [Google Scholar]
- Liu XM, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K, Kim MC, Yun DJ, Chung WS. (2013) Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Commun 430: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Vilela B, Irar S, Solé M, Capellades M, Valls M, Coca M, Pagès M. (2010) MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J 63: 1017–1030 [DOI] [PubMed] [Google Scholar]
- Mathur J, Koncz C, Szabados L. (1995) A simple method for isolation, liquid culture, transformation and regeneration of Arabidopsis thaliana protoplasts. Plant Cell Rep 14: 221–226 [DOI] [PubMed] [Google Scholar]
- Miller G, Mittler R. (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 98: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nakano Y, Asada K. (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22: 867–880 [Google Scholar]
- Nguyen XC, Hoang MH, Kim HS, Lee K, Liu XM, Kim SH, Bahk S, Park HC, Chung WS. (2012a) Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem Biophys Res Commun 423: 703–708 [DOI] [PubMed] [Google Scholar]
- Nguyen XC, Kim SH, Lee K, Kim KE, Liu XM, Han HJ, Hoang MH, Lee SW, Hong JC, Moon YH, et al. (2012b) Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep 31: 737–745 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T. (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palágyi A, Terecskei K, Adám E, Kevei E, Kircher S, Mérai Z, Schäfer E, Nagy F, Kozma-Bognár L. (2010) Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol 153: 1834–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F. (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L. (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Leung J, Joseph MP, Salamó IP, Szabados L. (2010) Genetic screens to identify plant stress genes. Methods Mol Biol 639: 121–139 [DOI] [PubMed] [Google Scholar]
- Pelham HR. (1982) A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30: 517–528 [DOI] [PubMed] [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236: 1485–1498 [DOI] [PubMed] [Google Scholar]
- Pucciariello C, Banti V, Perata P. (2012) ROS signaling as common element in low oxygen and heat stresses. Plant Physiol Biochem 59: 3–10 [DOI] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J. (2012) MAP kinase cascades in Arabidopsis innate immunity. Front Plant Sci 3: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigó G, Papdi C, Szabados L. (2012) Transformation using controlled cDNA overexpression system. Methods Mol Biol 913: 277–290 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM. (2008) Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol Biol 66: 361–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y. (2009) Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21: 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smekalova V, Doskocilova A, Komis G, Samaj J. (2013) Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol Adv 32: 2–11 [DOI] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R. (2005) AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucleic Acids Res 33: W397–W402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren van Themaat E, Bartsch M, Parker JE. (2010) Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 62: 628–640 [DOI] [PubMed] [Google Scholar]
- Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N. (2001) At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 27: 407–415 [DOI] [PubMed] [Google Scholar]
- Suri SS, Dhindsa RS. (2008) A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ 31: 218–226 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Mullineaux PM, Schöffl F. (2006) Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol Biol 61: 733–746 [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wing D, Koncz C, Schell J. (1989) Conserved function in Nicotiana tabacum of a single Drosophila hsp70 promoter heat shock element when fused to a minimal T-DNA promoter. Mol Gen Genet 219: 9–16 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. (2002) A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci USA 99: 7530–7535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Sakuma Y, Todaka D, Maruyama K, Qin F, Mizoi J, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2008) Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368: 515–521 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsigmond L, Rigó G, Szarka A, Székely G, Otvös K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. (2008) Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol 146: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsigmond L, Szepesi A, Tari I, Rigó G, Király A, Szabados L. (2012) Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant Sci 182: 87–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.