Rice His phosphotransfer proteins function as positive regulators of the cytokinin signaling pathway and play different roles in salt and drought tolerance in rice.
Abstract
Cytokinin plays an important role in plant development and stress tolerance. Studies of Arabidopsis (Arabidopsis thaliana) have demonstrated that cytokinin acts through a two-component system that includes a histidine (His) kinase, a His phosphotransfer protein (HP), and a response regulator. Phylogenetic analyses have revealed the conservation of His kinases but lineage-specific expansion of HPs and response regulators in rice (Oryza sativa). However, whether the functions of rice HPs have diverged remains unknown. In this study, two rice authentic HPs (OsAHP1 and OsAHP2) were knocked down simultaneously via RNA interference (RNAi), and the transgenic OsAHP-RNAi plants exhibited phenotypes expected for a deficiency in cytokinin signaling, including dwarfism with reduced internode lengths, enhanced lateral root growth, early leaf senescence, and reduced tiller numbers and fertility under natural conditions. The OsAHP-RNAi seedlings were also hyposensitive to exogenous cytokinin. Furthermore, OsAHP-RNAi seedlings were hypersensitive to salt treatment but resistant to osmotic stress relative to wild-type plants. These results indicate that OsAHPs function as positive regulators of the cytokinin signaling pathway and play different roles in salt and drought tolerance in rice.
Cytokinin is one of the most important phytohormones, well known for regulating many aspects of plant growth and development, including cell division, shoot initiation and differentiation, apical dominance, chloroplast biogenesis, leaf senescence, vascular differentiation, photomorphogenesis, gravitropism, fertility, and seed development (Mok and Mok, 2001; Mizuno, 2004; Ferreira and Kieber, 2005; Sakakibara, 2005; Aloni et al., 2006; Müller and Sheen, 2007). Studies of Arabidopsis (Arabidopsis thaliana) have shown that cytokinin signal transduction involves a multistep two-component system composed of a sensor His kinase (HK), a His phosphotransfer protein (HP), and a response regulator (RR; Hwang and Sheen, 2001). The binding of cytokinin to the cyclase/HK-associated sensing extracellular domain of a membrane-localized receptor HK leads to autophosphorylation of the kinase at a conserved His residue; the phosphoryl group is subsequently transferred to an Asp in the C-terminal receiver domain of the HK. An HP then accepts the phosphoryl group from the HK via the conserved His residue and translocates from the cytoplasm to the nucleus, where it likely delivers the phosphoryl group to an RR at an Asp residue (Hwang and Sheen, 2001; Hutchison and Kieber, 2002; Kieber, 2002; Kakimoto, 2003; To and Kieber, 2008). There are two types of RRs: type A RRs are cytokinin primary responsive genes and negative regulators of cytokinin signaling, while type B RRs are transcription factors and positive regulators of cytokinin signaling (To et al., 2004). Most of our knowledge of the cytokinin signaling pathway is derived from studies of Arabidopsis loss-of-function mutants. The cytokinin receptor triple mutant ahk2,3,4 (Nishimura et al., 2004; Riefler et al., 2006), the His-containing HP quintuple mutant ahp1,2,3,4,5 (Hutchison et al., 2006), and the type B RR triple mutant arr1 arr10 arr12 (Argyros et al., 2008) all show reduced sensitivity to cytokinin and display pleiotropic phenotypes.
An increasing body of evidence, mostly from studies of Arabidopsis, suggests an important role for cytokinin in the regulation of environmental stress responses (Ha et al., 2012). Prolonged drought or extensive salt stress was reported to be associated with the down-regulation of active cytokinin levels (Nishiyama et al., 2011). The constitutive down-regulation of cytokinin levels by the overexpression of cytokinin oxidase (CKK) or by the inactivation of the cytokinin biosynthetic gene isopentenyl transferase results in drought- and salt stress-tolerant phenotypes (Werner et al., 2010; Nishiyama et al., 2011). AHK1 was recognized as a positive regulator of osmotic stress signaling because AHK1 overexpressors exhibited enhanced stress tolerance whereas the ahk1 mutant was sensitive to osmotic stress (Tran et al., 2007; Wohlbach et al., 2008). By contrast, loss-of-function mutations in the cytokinin receptors AHK2 and AHK3 (ahk2 ahk3) or type B ARRs (arr1 arr12) improved tolerance to cold, salt, and drought (Tran et al., 2007; Mason et al., 2010). Individual mutations in arr5, arr6, or arr7 resulted in enhanced cold tolerance, suggesting that type A ARR5, ARR6, and ARR7 negatively regulate cold stress responses (Jeon et al., 2010). Cytokinin has been reported to increase shoot sodium accumulation and thus reduce salt tolerance through the type B regulators ARR1 and ARR12 (Mason et al., 2010). Recently, Arabidopsis AHP2, AHP3, and AHP5 have been demonstrated to be redundantly involved in inducing type A ARR expression in response to cold (Jeon and Kim, 2013) and to function as redundant negative regulators of drought stress responses (Nishiyama et al., 2013). These studies demonstrated that in Arabidopsis, cytokinin signaling negatively affects stress responses. On the other hand, a stress- and maturation-inducible isopentenyl transferase gene increased drought tolerance in tobacco (Nicotiana tabacum) plants, likely due to the cytokinin-mediated inhibition of stress-induced senescence and reactive oxygen species production (Rivero et al., 2007).
In rice (Oryza sativa), there are six cytokinin-response His protein kinases (OsHK1–OsHK6), five HPs (two authentic His-containing phosphotransfer [AHP] proteins [OsAHP1 and OsAHP2] and three pseudo His-containing phosphotransfer [PHP] proteins [OsPHP1–OsPHP3]), and a number of RRs (OsRRs; Du et al., 2007; Schaller et al., 2007). A phylogenetic analysis revealed distinct clusters of OsAHPs, OsPHPs, and OsRRs in rice and convergent clusters of HKs in rice and Arabidopsis (Tsai et al., 2012). The functions of these genes in cytokinin signaling, plant development, and abiotic stress responses remain to be elucidated. In this study, RNA interference (RNAi) was performed to silence the expression of OsAHP1 and OsAHP2 simultaneously. The transgenic OsAHP-RNAi plants exhibited multiple developmental phenotypes and hyposensitivity to exogenous cytokinin. Furthermore, the OsAHP-RNAi seedlings were hypersensitive to salt stress but tolerant to osmotic stress relative to wild-type plants. Our results demonstrate that OsAHP1 and OsAHP2 not only function as positive regulators of the cytokinin signaling pathway but also play specific roles in stress tolerance in rice.
RESULTS
OsAHPs Play an Important Role in Plant Growth and Development
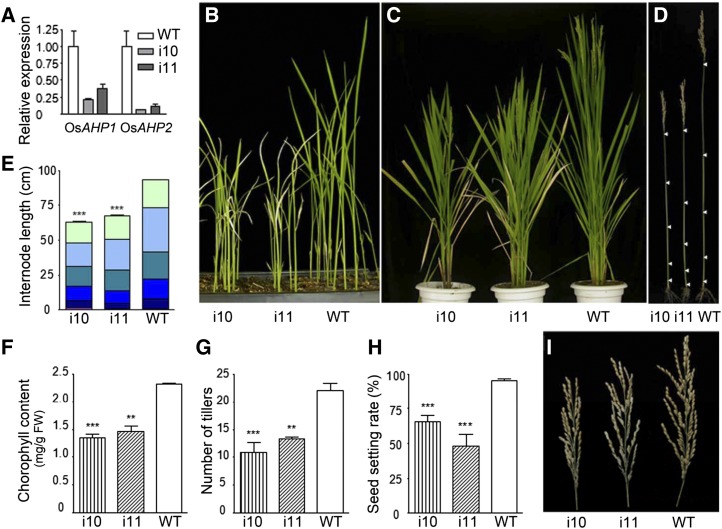
To understand the function of OsAHPs, we first generated an RNAi construct using pTCK303 (Wang et al., 2004), which harbors a 416-bp fragment of the OsAHP2 coding sequence in the reverse and forward directions under the control of the Ubiquitin promoter (Supplemental Fig. S1A). The sequence shares 80% identity with that of OsAHP1 but only 47% to 48% identity with that of OsPHP1 to OsPHP3 (Supplemental Fig. S1B). We then obtained several independent transgenic OsAHP-RNAi lines and selected two of them (i10 and i11) showing representative phenotypes and severe reduction in the RNA levels of both OsAHP1 and OsAHP2 (Fig. 1A). The OsAHP-RNAi plants showed pleiotropic phenotypes during growth and development. They were shorter than the wild type at the seedling and mature stages (Fig. 1, B and C) and exhibited a reduction in the length of each internode (Fig. 1, D and E). In addition, early leaf senescence was observed in the OsAHP-RNAi lines from the seedling stage to the heading stage (Fig. 1, B and C), as indicated by a reduced chlorophyll content compared with wild-type plants (Fig. 1F). The tiller number in the OsAHP-RNAi plants was also reduced to nearly one-half that in wild-type plants (Fig. 1G), and the seed setting rate was also reduced (Fig. 1, H and I). These phenotypes indicate an important role for OsAHPs in plant growth and development.
Figure 1.
Phenotypes of OsAHP-RNAi transgenic plants. Two RNAi lines, i10 and i11, were used as representatives to compare with the wild type (WT). A, RNA levels of OsAHP1 and OsAHP2 in OsAHP-RNAi transgenic lines as determined by qRT-PCR. Os18S was used as an internal reference. B and C, OsAHP-RNAi transgenic plants showed semidwarf and early leaf senescence phenotypes at the seedling stage (B) and the heading stage (C). D, Primary culms at the mature stage. Arrowheads indicate the nodes. E, Measurement of internode lengths for the plants shown in D (n ≥ 9). F, Chlorophyll contents of the rice leaves at tillering stage (n = 3). FW, Fresh weight. G and H, Statistics for the number of tillers (G) and seed setting rate (H) in the OsAHP-RNAi and wild-type lines at the mature stage (n ≥ 4). I, Panicles of OsAHP-RNAi and wild-type lines. Error bars represent sd. Asterisks indicate that the value is significantly different from the value for the wild type as determined by Student’s t test (***P < 0.001, ** P < 0.01).
The mRNA expression levels of three OsPHPs (OsPHP1–OsPHP3) were also detected separately in shoots and roots. Our results indicate reduced expression of all three genes in OsAHP-RNAi shoots and slightly reduced expression in OsAHP-RNAi roots as compared with the wild type (Supplemental Fig. S1C).
OsAHPs Are Required for Cytokinin Responses
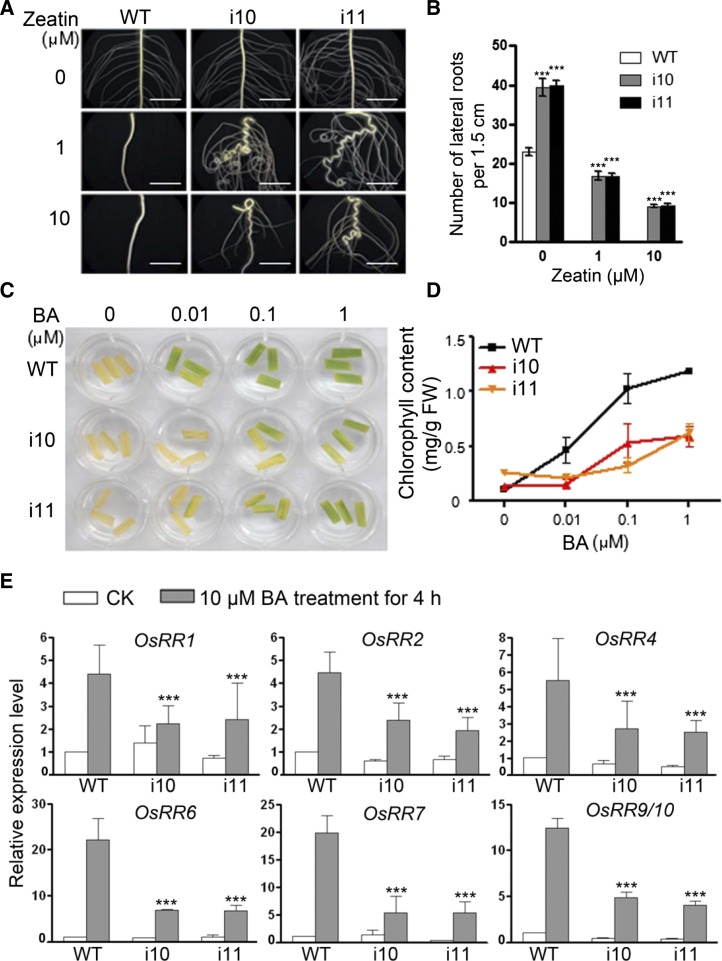
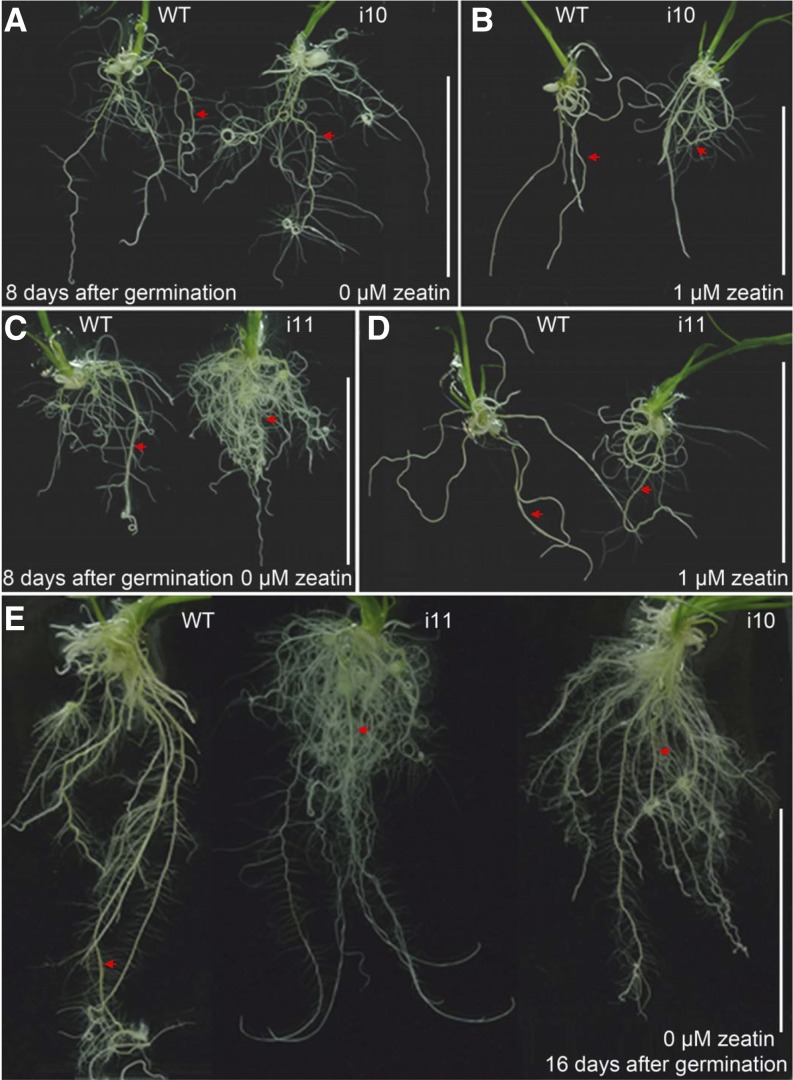
To determine whether OsAHPs are required for cytokinin responses, we examined the sensitivity of OsAHP-RNAi plants to cytokinin treatment. Cytokinin inhibits lateral root formation (Rani Debi et al., 2005). Thus, we examined the effects of exogenous trans-zeatin on the emergence of lateral roots in the OsAHP-RNAi seedlings. In the absence of exogenous zeatin, more lateral roots were formed in the OsAHP-RNAi seminal roots (Fig. 2, A, top row, and B) and adventitious roots (Fig. 3, A, C, and E) compared with wild-type seedlings. In the presence of 1 and 10 μm zeatin, no lateral roots emerged from the wild-type seminal roots (Fig. 2, A, middle and bottom rows, and B) and adventitious roots (Fig. 3, B and D), whereas many lateral roots were generated in the OsAHP-RNAi plant roots, indicating that the OsAHP-RNAi seedlings were less sensitive to the inhibitory effect of endogenous and exogenous cytokinin on lateral root formation. On the other hand, the seminal roots of the OsAHP-RNAi seedlings elongated normally on regular medium, reaching a length similar to that in the wild type (Fig. 3, A, C, and E, arrows). On zeatin-containing medium, however, the seminal roots of the OsAHP-RNAi lines became severely curled (Fig. 2A), indicating greater impairment in gravitropism in response to cytokinin compared with the wild type. These results demonstrate that OsAHPs play important roles in root development.
Figure 2.
OsAHP-RNAi seedlings show reduced sensitivity to exogenous cytokinin (CK). A and B, The inhibitory effects of exogenous trans-zeatin on lateral root generation. Roots of 10-d-old seedlings were grown on 1/2 MS medium with or without the indicated concentrations of zeatin (A); the lateral root number produced on the seminal root (1.5 cm in length) in each seedling was counted (B). Error bars represent sd (n = 12). Bars = 5 mm. C and D, The delayed effects of exogenous BA on dark-induced leaf senescence. Leaves were treated with or without BA in the dark for 45 h (C) and then analyzed for chlorophyll retention (D). Two parallel samples for each treatment were measured, and three repeats were performed. The data are given as means ± sd. FW, Fresh weight. E, Expression of type A RR genes in response to BA treatment. Twelve-day-old seedlings were used for qRT-PCR assays; the mRNA level from wild-type (WT) plants without BA treatment was set at 1. Os18S was used as an internal control. The data are given as means ± sd of three biological replications. Asterisks indicate that the value is significantly different from the value for the wild type as determined by Student’s t test (***P < 0.001).
Figure 3.
Comparison of root phenotypes between wild-type (WT) and OsAHP-RNAi seedlings. Seedlings were grown on 1/2 MS medium with or without 1 μm trans-zeatin for the indicated number of days. Arrows indicate the seminal roots. Bars = 5 cm. [See online article for color version of this figure.]
Cytokinin also delays dark-induced leaf senescence (Hutchison et al., 2006). When kept in the dark for 45 h, wild-type leaf segments lost their chlorophyll and turned pale, whereas treatment with 6-benzyladenine (BA) prevented this dark-induced loss of chlorophyll (Fig. 2, C and D). However, dark-induced leaf senescence in OsAHP-RNAi leaves was less inhibited by the application of cytokinin, indicating that OsAHPs are required for the cytokinin-mediated inhibition of senescence.
Cytokinin signaling activates the expression of type A RR genes (Jain et al., 2006). Cytokinin treatment of 12-d-old rice seedlings for 4 h induced the expression of all six type A OsRR genes, but the fold change was obviously reduced in the OsAHP-RNAi plants compared with wild-type plants (Fig. 2E). These results demonstrate both an essential role for OsAHPs in primary cytokinin signal transduction and that they are positive regulators of the cytokinin signaling pathway in rice.
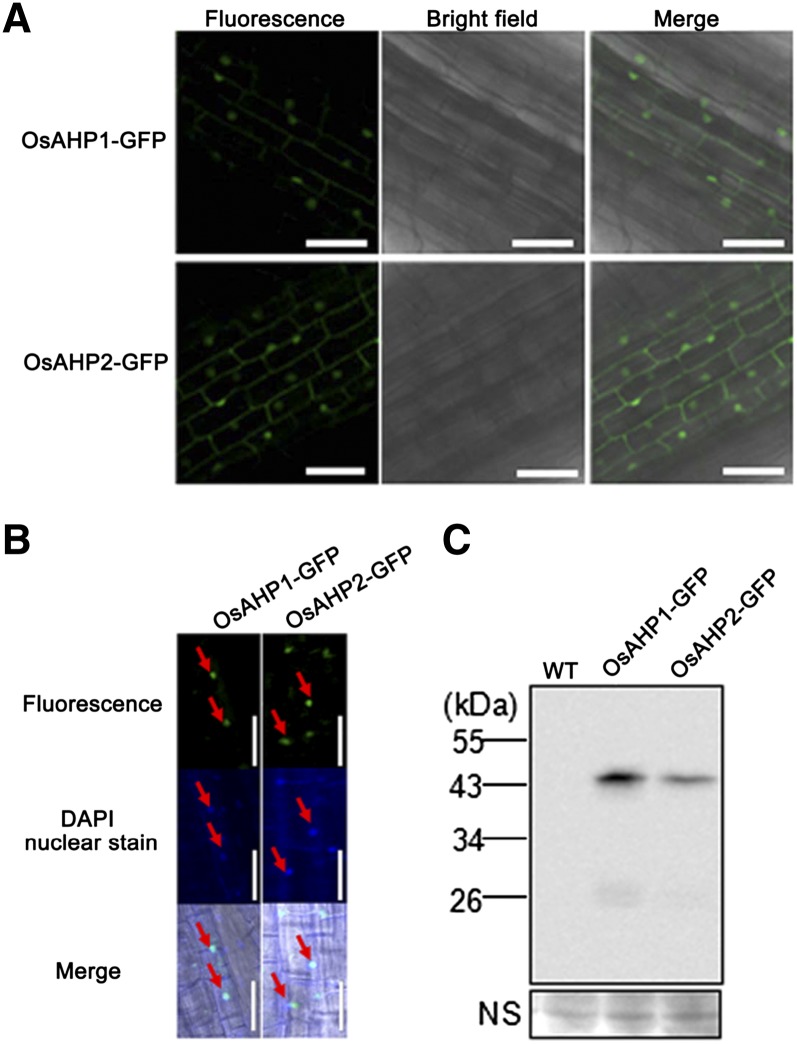
In Arabidopsis, HPs (AHPs) are present in both the nucleus and the cytosol, which allows them to transfer a phosphoryl group from the plasma membrane-localized AHKs to the mainly nucleus-localized ARRs (Punwani et al., 2010). Thus, the subcellular localization of OsAHP1 and OsAHP2 was further investigated in root cells of 2×35S::OsAHP1-GFP and 2×35S::OsAHP2-GFP transgenic rice, respectively. Fluorescence was observed in the cytoplasm and nucleus (Fig. 4A). An overlay with 4′,6-diamidino-2-phenylindole (DAPI) staining further confirmed the nuclear localization of the GFP fusion proteins (Fig. 4B, arrows). Western blotting using an extract of transgenic rice roots expressing OsAHP1-GFP or OsAHP2-GFP and anti-GFP antibodies revealed one band of 43 kD but not one of 27 kD (corresponding to the Mr of GFP alone; Fig. 4C), indicating that the fluorescence signals represent the OsAHP1/2-GFP fusion protein but not the cleavage of GFP. Such subcellular localization is similar to that observed in rice protoplasts (Tsai et al., 2012) and is consistent with a role for OsAHPs in rice cytokinin signal transduction.
Figure 4.
Nuclear and cytosolic localization of OsAHP1 and OsAHP2. A, Confocal images of root cells from transgenic rice harboring 2×35S::OsAHP1-GFP or 2×35S:OsAHP2-GFP. Bars = 50 μm. B, DAPI staining of A. Arrows indicate the overlay of the fluorescent signal with the nuclear DAPI signal. Bars = 50 μm. C, Western-blot assay of GFP fusion proteins in 2×35S::OsAHP1-GFP or 2×35S::OsAHP2-GFP transgenic rice roots using anti-GFP antibodies. NS, Nonspecific band used as a loading reference; WT, wild type.
OsAHP-RNAi Seedlings Are Tolerant to Osmotic Stress But Hypersensitive to Salt Stress
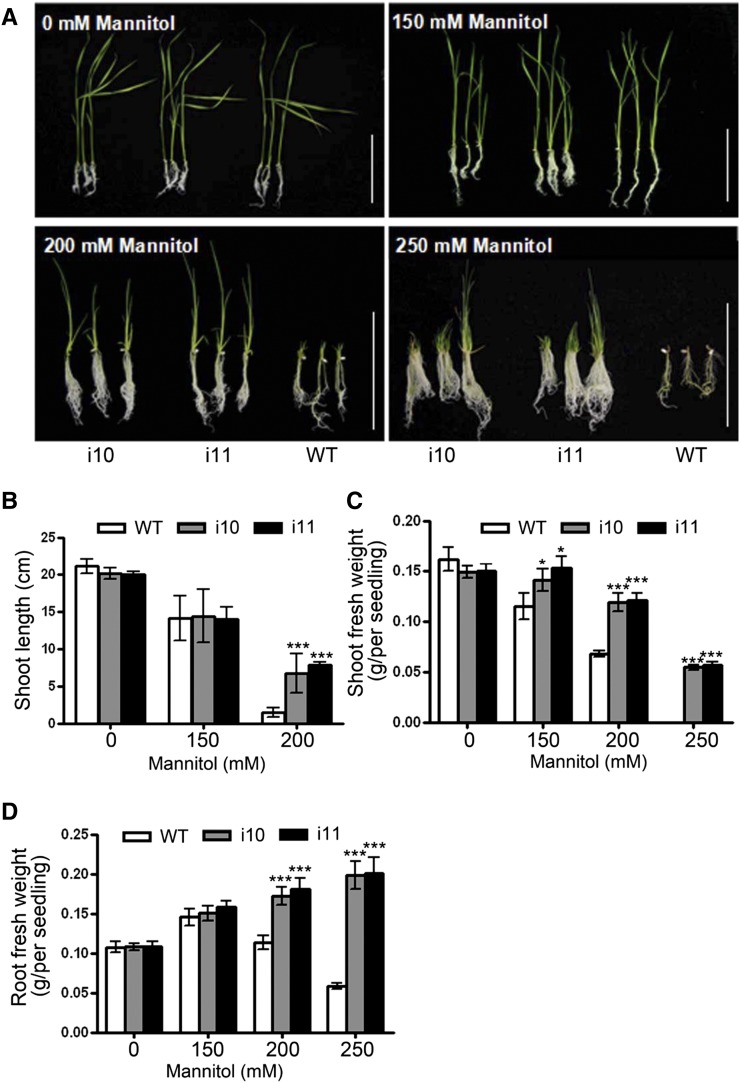
In Arabidopsis, it has been reported that ahp2,3,5 mutants showed increased tolerance to drought and salt stress (Nishiyama et al., 2013). To investigate whether OsAHP1/2 silencing alters the osmotic response, OsAHP-RNAi seedlings were grown in one-half-strength Murashige and Skoog (1/2 MS) medium containing different concentrations of mannitol. As shown in Figure 5, as the mannitol concentration increased, shoot elongation in wild-type seedlings was inhibited while that in the OsAHP-RNAi lines was much less inhibited (Fig. 5, A and B). When the mannitol concentration was increased to 250 mm, wild-type seedling growth was totally inhibited at a very early stage while the OsAHP-RNAi lines were still able to grow well and had a stronger root system (Fig. 5, A, C, and D). These results indicate that OsAHPs play similar roles to Arabidopsis AHPs in negatively regulating the osmotic stress response.
Figure 5.
OsAHP-RNAi seedlings are resistant to osmotic stress. A, Sixteen-day-old OsAHP-RNAi and wild-type (WT) seedlings grown on 1/2 MS medium containing 0, 150, or 200 mm mannitol and 30-d-old seedlings grown on 250 mm mannitol-containing medium. Bars = 10 cm. B to D, Measurement of the shoot lengths (B) and shoot (C) and root (D) fresh weights of 16-d-old seedlings on medium with different mannitol concentrations. The experiment was repeated three times. The data are given as means ± sd (n = 12). Asterisks indicate a significant difference from the wild type by Student’s t test (***P < 0.001, *P < 0.05). [See online article for color version of this figure.]
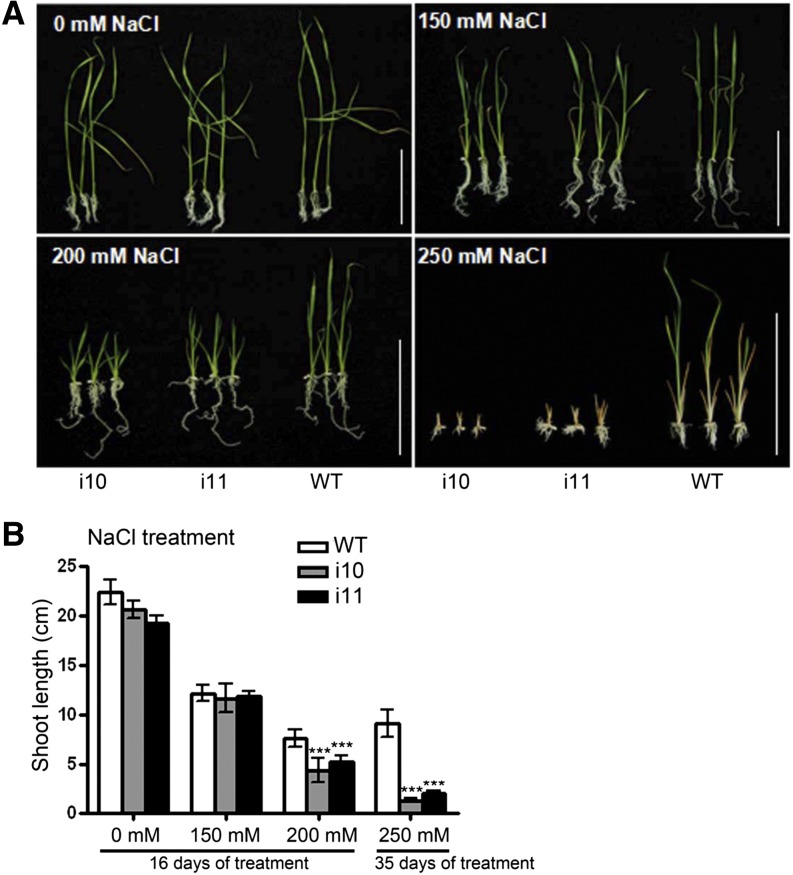
We next investigated the growth of OsAHP-RNAi seedlings under salt stress. As the NaCl concentration increased, shoot elongation was inhibited more severely in OsAHP-RNAi seedlings than in wild-type plants. When the NaCl concentration was increased to 250 mm, OsAHP-RNAi seedling growth stopped completely at a very early stage while the wild-type seedlings were still able to grow to some extent (Fig. 6). These results demonstrate that OsAHPs play a positive role in salt stress tolerance in rice, in contrast to their homologs in Arabidopsis (Nishiyama et al., 2013).
Figure 6.
OsAHP-RNAi seedlings show increased sensitivity to NaCl. A, Sixteen-day-old OsAHP-RNAi and wild-type (WT) seedlings grown on 1/2 MS medium containing 0, 150, or 200 mm NaCl and 35-d-old seedlings grown on 250 mm NaCl-containing medium. Bars = 10 cm. B, Shoot lengths of the seedlings shown in A. The experiment was repeated three times. Error bars represent sd (n = 12). Asterisks indicate a significant difference from the wild type by Student’s t test (***P < 0.001). [See online article for color version of this figure.]
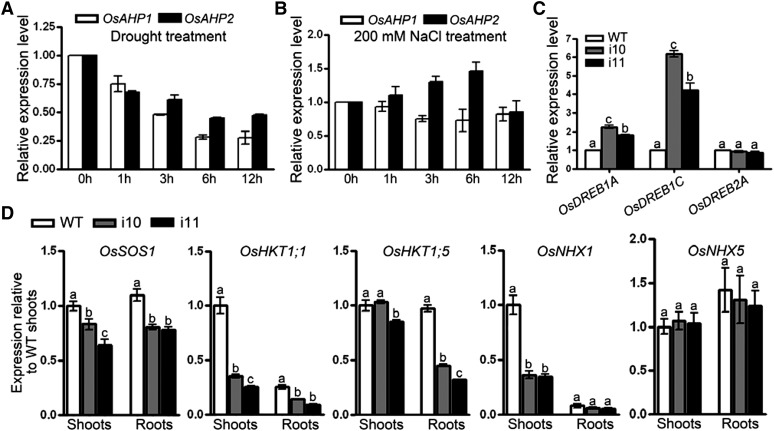
To explain the performance of the OsAHP-RNAi seedlings under conditions of osmotic and salt stress, we first examined the expression of OsAHPs in response to drought and salt treatment by quantitative real-time (qRT)-PCR. Our results indicate that drought inhibited the expression of OsAHP1 and OsAHP2 while salt slightly induced OsAHP2 (Fig. 7, A and B). Further assessment of drought response regulatory genes revealed that the mRNA level of dehydration responsive element-binding protein1C (OsDREB1C) was dramatically increased while that of OsDREB1A was slightly increased in OsAHP-RNAi plants compared with wild-type plants (Fig. 7C). In contrast, several Na+-transporter genes, including rice salt overly sensitive1 (OsSOS1), High-Affinity K+ Transporters (OsHKT1;1 and OsHKT1;5), and Na+/H+ exchangers (OsNHX1), but not OsNHX5, were down-regulated in the shoots or roots of OsAHP-RNAi plants as compared with the wild type (Fig. 7D). These data suggest that cytokinin signaling differentially regulates genes involved in ion stress and osmotic stress in rice.
Figure 7.
The responses of OsAHPs to drought or salt treatment and the expression of stress-related genes in OsAHP-RNAi lines. A and B, qRT-PCR was performed to analyze the OsAHP1 and OsAHP2 mRNA levels in response to the time course of treatment of 12-d-old seedlings with drought (A) and 200 mm NaCl (B). C and D, qRT-PCR assay of OsDREB1A, OsDREB1C, and OsDREB2A expression in 12-d-old OsAHP-RNAi seedlings (C) and of OsSOS1, OsHKT1;1, OsHKT1;5, OsNHX1, and OsNHX5 mRNA levels in 14-d-old OsAHP-RNAi seedlings (D). All values are relative to wild-type (WT) shoots. The data are means of three replicates and were compared by a one-way ANOVA and Duncan’s multiple range test. Different letters (a–c) indicate significant differences (P < 0.05) between lines.
DISCUSSION
The cytokinin signal transduction pathway has been extensively studied in Arabidopsis (Hutchison and Kieber, 2002). The presence of homologous components suggests that the cytokinin signaling pathway is conserved, while experimental evidence for the functions of OsAHPs has not been reported. Furthermore, cytokinin has been shown to play important roles in stress responses in dicots; however, the roles of cytokinin and each signaling component in these stress responses remain poorly characterized in rice. Using RNAi to silence two OsAHP genes, we found that OsAHPs play essential roles not only in cytokinin signaling and development in rice plants but also in the response to abiotic stresses. Our results further reveal distinct roles for OsAHPs, in contrast to Arabidopsis AHPs, in salt and osmotic stress tolerance.
OsAHP1 and OsAHP2 Are Positive Regulators of the Cytokinin Pathway and Function Redundantly in the Regulation of Various Developmental Processes
A sequence alignment and motif analysis revealed two putative functional AHP genes, OsAHP1 and OsAHP2, and three PHP genes, OsPHP1 to OsPHP3, in rice (Tsai et al., 2012). OsAHP1 and OsAHP2, but not OsPHP1 to OsPHP3, contain a His in the highly conserved motif XHQXKGSSXS, which is required for phosphoryl group transfer from the cytokinin receptor HK to the RR (Du et al., 2007). A single osahp1 mutant caused by a Tos17 insertion was indistinguishable from the wild type under normal growth conditions (Ito and Kurata, 2006), suggesting function redundancy between OsAHP1 and OsAHP2. Indeed, OsAHP1 and OsAHP2 share 71% protein sequence identity and have the same subcellular localization (Fig. 4, A and B); moreover, the expression patterns of the two genes overlapped in almost every tissue, as revealed by qRT-PCR (Supplemental Fig. S2A). The phenotypes observed in OsAHP-RNAi rice plants should be due to the combined knockdown of both OsAHP1 and OsAHP2. The strong expression of OsAHP2 in root, especially in root stele, and in lamina joint, leaf blade, basal node, and spikelet (Supplemental Fig. S2, B–H) was almost completely knocked out, while OsAHP1 expression was reduced to a low level (Fig. 1A; Supplemental Fig. S1C). This expression pattern could only maintain weak cytokinin signaling but is not enough to complement OsAHP2 function, resulting in multiple developmental defects (Fig. 1) as well as a reduction (but not a complete deficiency) in cytokinin responses (Fig. 2). These phenotypes are consistent with those of some Arabidopsis cytokinin pathway mutants. For example, ahk2 ahk3 cre1, ahp1,2,3,4,5, and arr1 arr10 arr12 all showed reductions in shoot development and chlorophyll content; moreover, the ahk2 ahk3 cre1 triple mutant was severely sterile (Nishimura et al., 2004; Mason et al., 2005; Hutchison et al., 2006; Riefler et al., 2006; Argyros et al., 2008). Transgenic rice plants overexpressing OsRR6, a type A RR that negatively regulates cytokinin signaling, also displayed dwarfism with poorly developed panicles (Hirose et al., 2007). However, the Arabidopsis mutant ahp1,2-2,3,4,5, in which all five AHPs were knocked out, was seedling lethal (Deng et al., 2010). This effect is extremely severe compared with the developmental phenotypes observed for the OsAHP-RNAi rice plants in this study.
The reduced expression of three OsPHPs in the OsAHP-RNAi plants (Supplemental Fig. S1C) may be the result of transcriptional down-regulation due to knocking down cytokinin signaling but is not the result of direct RNAi, since the target sequence and RNAi fragment share no more than 20 successive nucleotides (Supplemental Fig. S1B). In Arabidopsis, AHP6 is a PHP that functions as an inhibitor of cytokinin signaling (Mähönen et al., 2006). If OsPHPs act as inhibitors of cytokinin signaling (like AHP6), then a reduction in their mRNA expression should lead to enhanced cytokinin signaling and a net balance between positive and negative regulation in OsAHP-RNAi plants. Nevertheless, the exact functions of OsPHP1 to OsPHP3 remain unknown, and the contribution of OsPHP1 to OsPHP3 to the phenotypes of the OsAHP-RNAi plants was not direct. Therefore, the phenotypes we observed are likely the direct result of reduced OsAHP1 and OsPHP2 expression.
The Distinct Roles of OsAHPs in Rice Root Development
The OsAHP-RNAi plants displayed enhanced growth of lateral roots, which were less sensitive to cytokinin inhibition (Figs. 2, A and B, and 3, A–D). This observation is consistent with the findings in Arabidopsis that cytokinin and cytokinin signaling negatively regulate lateral root development. In Arabidopsis, the overexpression of the cytokinin oxidase AtCKX, a cytokinin-degrading enzyme, to reduce the endogenous cytokinin level (Werner et al., 2003, 2010) or the knockout of two of the three cytokinin receptors, ahk2 and ahk3 (Riefler et al., 2006), and two of five HPs, ahp2,3, and ahp3,5, respectively (Hutchison et al., 2006), to turn down cytokinin signaling, all enhanced lateral root growth. Furthermore, reduced sensitivity to the inhibitory effects of cytokinin on lateral root formation was observed in the ahp1,2,3 triple mutant (Hutchison et al., 2006).
However, unlike the Arabidopsis ahp2,3,5-2 triple and ahp1,2,3,4,5 quintuple mutants, which produced a very short and narrow primary root with a defective vascular system but generated normal adventitious roots, and the ahp1,2,3 triple mutant, which had longer roots and less sensitivity to BA inhibition (Hutchison et al., 2006), the OsAHP-RNAi plants produced long seminal roots similar to adventitious roots, and the elongation of these roots was not significantly affected, as compared with wild-type root, by 1 μm zeatin, which is enough to inhibit lateral root formation (Fig. 3). This result indicates that the lateral root initiation is more sensitive to cytokinin inhibition than root elongation in rice. Root development, architecture, and gravitropism are regulated by the interplay of three hormonal signals: cytokinin, auxin, and ethylene (Aloni et al., 2006). Cytokinin affects root elongation through ethylene signaling but affects root meristem size and activity through modulating asymmetric auxin distribution (Ruzicka et al., 2009). Lateral root initiation depends on the local auxin gradient to trigger founder cell specificity (Dubrovsky et al., 2008). Tropic growth is also preceded by the auxin gradient (Esmon et al., 2006). However, cytokinin level and signaling influence cell-to-cell auxin transport by changing the transcription of PIN auxin efflux carrier genes (Ruzicka et al., 2009). AHP6, a cytokinin signaling repressor in Arabidopsis, is required for patterning the lateral root primordium by influencing the localization of Pin-formed1 (PIN1; Moreira et al., 2013). In this study, lateral root inhibition, and the disruption in gravitropism observed in OsAHP-RNAi roots exposed to cytokinin, may have resulted from an alteration in the distribution of the auxin gradient (Kushwah et al., 2011). OsPIN1b, OsPIN1c, and OsPIN9 were found expressed in rice root (Wang et al., 2009) in a similar way to OsAHP2.
In a complementation test, OsAHP2 overexpression partially rescued the growth of the Arabidopsis ahp1,2,3,4,5 quintuple mutant (Supplemental Fig. S3A), in which the expressed OsAHP2-GFP fusion protein was localized to the nucleus, cytosol, and cell surface (Supplemental Fig. S3B), similar to the localization in rice plants (Fig. 4A). Our results suggest a conserved role for OsAHP2 in cytokinin signal transduction, but the molecular function of OsAHP2 may not be exactly equal to that of AHP1 to AHP5, given the limited protein sequence identity between them (35%–45%; Tsai et al., 2012), which may lead to the different roles of OsAHPs from Arabidopsis AHPs in primary root development. In fact, even Arabidopsis AHP1, AHP2, AHP3, AHP4, and AHP5 have different roles in root elongation, lateral root formation, and primary root development in response to BA treatment (Hutchison et al., 2006).
Together, our results provide direct evidence for the important role of OsAHPs in cytokinin signal transduction in rice root development.
OsAHP1 and OsAHP2 Have Opposing Effects on Plant Sensitivity to Osmotic and Salt Stresses in Rice
Many studies of dicots have documented the important role of cytokinin in the responses of plants to abiotic stresses (Albacete et al., 2008; Ghanem et al., 2008; Argueso et al., 2009). Recently, Arabidopsis AHP2, AHP3, and AHP5 were demonstrated to be negative regulators of the drought stress response (Jeon and Kim, 2013; Nishiyama et al., 2013). Here, OsAHP1 and OsAHP2 were found to also act as negative regulators of the osmotic stress response. The OsAHP-RNAi rice plants displayed strong osmotic tolerance with a strong root system under 250 mm mannitol treatment (Fig. 5, A and D). Dehydration suppressed the RNA expression of OsAHPs (Fig. 7A), and the knockdown of OsAHPs led to the up-regulation of some, but not all, OsDREBs (Fig. 7C). Other drought resistance genes, including LATE EMBRYOGENESIS ABUNDANT, were unaffected (data not shown). Since the root system determines the water accessibility of plants, and usually dehydration-tolerant rice plants have a deeper and more highly branched root system (Lynch, 1995), we presume that the increased osmotic tolerance of the OsAHP-RNAi plants under conditions of osmotic stress was conferred mainly by the enhanced lateral roots (Fig. 3) and induced crown roots, which can greatly improve water uptake (Fig. 5A).
However, the OsAHP-RNAi seedlings showed hypersensitivity to salt treatment, with greater inhibition of shoot growth compared with wild-type rice (Fig. 6), in contrast to the enhanced salt tolerance of Arabidopsis ahp mutants (Nishiyama et al., 2013). In Arabidopsis, cytokinin appears to have both positive (Elkeltawi and Croteau, 1987; Gadallah, 1999; Iqbal and Ashraf, 2005; Iqbal et al., 2006) and negative (Tran et al., 2007) effects on salt tolerance, suggesting that the role of cytokinin is complex and that its influence depends on a variety of factors. The salt overly sensitive (SOS) signaling system increases plant salt tolerance by activating Na+-extrusion antiporters (Shi et al., 2002; Martínez-Atienza et al., 2007); the high-affinity K+ transporter1;1 (OsHKT1;1), which mediates inward Na+ transport (Jabnoune et al., 2009), protects young leaves from ion toxicity by the accumulation of Na+ and Cl− in old leaves (Wang et al., 2012); and Na+/H+ antiporter genes (OsNHXs) play roles in the compartmentalization of Na+ into vacuoles (Fukuda et al., 2011). These are all functional genes required to prevent sodium toxicity. In Arabidopsis, cytokinin acts through the transcription factors ARR1 and ARR12 to regulate the transport of Na+ ions from the roots to the shoot by modulating the expression of AtHKT1;1 (Mason et al., 2010). A global transcriptional analysis of Arabidopsis ahp2,3,5 leaves under drought stress revealed the up-regulation of several key salt stress-responsive genes, including HKT1, SOS3, and SOS4 (Nishiyama et al., 2013), suggesting a central role for cytokinin in regulating Na+ ion homeostasis. In rice, however, OsAHP2 was slightly induced by salt stress (Fig. 7B). OsAHPs are required for the expression of OsSOS1, OsHKT1;1, and OsNHX1 (Fig. 7D) and function as positive regulators of salt tolerance.
The opposing response to osmotic and ion stresses by the same transgenic plants was also found in Arabidopsis seedlings overexpressing MYB52, a transcription factor involved in abscisic acid and stress responses; the seedlings were drought tolerant but salt sensitive (Park et al., 2011), probably due to the regulation of different target genes. The enhanced osmotic resistance and reduced salt resistance exhibited by the OsAHP-RNAi plants should reflect the differences in root system architecture required for the adaptation to drought stress and ion stress. The enhancement of lateral roots by reducing cytokinin signaling may improve water uptake under conditions of drought, but it increases the uptake of ions under high-salt conditions. Even worse, some Na+/H+ antiporter genes are down-regulated, leading to disturbed ion homeostasis, growth inhibition, and severe shoot bleaching. Similar results were reported for transgenic tobacco and Arabidopsis plants overexpressing the root-specific cytokinin-degrading enzyme CKX3. Due to the reduced cytokinin level, the plants developed a stronger root system and showed increased drought tolerance, but they accumulated more heavy metals in the shoot from contaminated soils (Werner et al., 2010).
The distinct effects of OsAHP-RNAi on osmotic and salt stress responses reveal a complex interplay between cytokinin signaling and the specific stress response pathways that have evolved in rice. Our studies demonstrate the evolutionary divergence of the role of OsAHPs in stress responses and provide important information for engineering stress-resistant crops.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ‘Nipponbare’) seedlings were grown in a greenhouse under the following conditions: 28°C/25°C, 16 h of light/8 h of dark, and 50% humidity. The rice plants were transferred to a field under natural conditions for long-term cultivation. Arabidopsis (Arabidopsis thaliana ecotype Columbia) was grown under long-day conditions (16 h of light/8 h of dark) at 22°C.
Vector Construction and Plant Transformation
To construct the RNAi vector, the 416-bp coding sequence of OsAHP2 was amplified from complementary DNA (cDNA) using the primer set AHPRNAiF and AHPRNAiR (Supplemental Table S1) and inserted into pTCK303 with SpeI and SacI sites (forward direction) and BamHI and KpnI sites (reverse direction; Wang et al., 2004). To construct ProOsAHP2::GUS, approximately 2.0 kb of the OsAHP2 promoter (−1,978 to −1 bp from the ATG) was amplified from genomic DNA and inserted into pCAMBIA1391Z. To construct 2×35S::OsAHP1-GFP and 2×35S::OsAHP2-GFP, the complete open reading frames of OsAHP1 and OsAHP2 without stop codons were amplified from cDNA and inserted into pMDC83. The constructs were introduced into Agrobacterium tumefaciens strain EHA105, and transgenic rice plants were generated through A. tumefaciens-mediated transformation (Yang et al., 2004). 2×35S::OsAHP2-GFP was also introduced into A. tumefaciens strain GV3101, and Arabidopsis transformation was carried out by the floral dip method (Clough and Bent, 1998). All primer sequences are listed in Supplemental Table S1.
Chlorophyll Content Measurement
The second leaf from the top of wild-type and OsAHP-RNAi transgenic plants at the tillering stage was collected and cut into 5-mm-long pieces, then the chlorophyll was extracted with 80% (v/v) acetone for 24 h under darkness and the content was determined by spectrophotometric measurement of the absorbance at 645 and 663 nm as described previously (Mackinney, 1941).
Seedling Cytokinin-Response Assays
For the root phenotype assay, rice seeds were sterilized with 1% (v/v) NaClO, and uniform seedlings at 2 d after germination were grown in 1/2 MS medium containing 0.3% (w/v) phytagel supplemented with or without the indicated concentrations of trans-zeatin (Sigma). The plate was kept at a 30° angle in a light chamber set at 26°C for the indicated number of days to allow vertical root growth. The lateral root number produced on the seminal root 1.5 cm from the root base was counted. For the chlorophyll retention assay, the third leaf of 14-d-old seedlings was incubated in a hydroponic culture solution containing the indicated concentration of BA (Sigma) in the dark for 45 h, then collected for chlorophyll extraction and measurement. Each treatment was repeated three times. To examine cytokinin-induced gene expression, 12-d-old seedlings were treated with or without 10 μm BA in hydroponic culture solution for 4 h, and then the roots were harvested for OsRR mRNA analysis.
qRT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Five hundred nanograms of total RNA was used as the template for cDNA synthesis. Real-time PCR was subsequently performed to quantify the cDNA using TaKaRa SYBR Premix Ex Taq in an ABI PRISM 7500 real-time PCR instrument (Applied Biosystems). Os18S or OsACTIN1 was used as an internal control to normalize all data. The gene-specific primers used for qRT-PCR are listed in Supplemental Table S2.
GUS Staining (Histochemical GUS Assay)
Plant material was prefixed in 90% acetone for 1 h at –20°C, washed twice with staining buffer without 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and infiltrated with staining solution (100 mm sodium phosphate buffer, pH 7.0, 10 mm sodium EDTA, 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, 0.1% Triton X-100, 0.1 mg mL−1 chloramphenicol, and 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid) under a vacuum for 20 min and incubated at 37°C for 1 to 24 h. Chlorophyll was extracted by passing the sample through increasing concentrations of ethanol. The samples were cleared as described previously (Malamy and Benfey, 1997).
Subcellular Localization
Roots of transgenic rice or Arabidopsis harboring 2×35S::OsAHP1-GFP or 2×35S::OsAHP2-GFP were used for observation. GFP fluorescence was observed using a confocal laser scanning microscope (LSM 510; Carl Zeiss). The nucleus was stained with DAPI and then visualized at an excitation wavelength of 358 nm.
Western-Blot Analysis
Total protein was extracted with 2× SDS protein sample buffer, separated by 12% SDS-PAGE, and transferred to nitrocellulose membranes for blotting with anti-GFP antibodies. Anti-rabbit IgG (H+L)-HRP Conjugate (Bio-Rad) was used as the secondary antibody. The bound antibodies were visualized using the ECL Western Blotting Analysis System (GE Healthcare).
Abiotic Stress Treatments
For the growth assay, plants were grown on 1/2 MS medium containing 0.3% phytagel with various concentrations of NaCl or mannitol for 16 d, or for 30 and 35 d at the highest concentration (as indicated), and used for growth measurements. For stress-induced OsAHP1 and OsAHP2 expression, 12-d-old rice seedlings were immersed in a hydroponic culture solution containing 200 mm NaCl or exposed to dry conditions on Parafilm for the indicated times. The roots of the plants were then harvested for RNA extraction.
Sequence data from this article can be found in the GenBank/RiceGE database (http://signal.salk.edu/cgi-bin/RiceGE) under the following accession numbers: OsAHP1 (Os08g44350), OsAHP2 (Os09g39400), OsPHP1 (Os01g54050), OsPHP2 (Os05g09410), OsPHP3 (Os05g44570), OsRR1 (Os04g36070), OsRR2 (Os02g35180), OsRR4 (Os01g72330), OsRR6 (Os04g57720), OsRR7 (Os07g26720), OsRR9 (Os11g04720), OsRR10 (Os12g04500), OsDREB1A (Os09g0522200), OsDREB1C (Os06g0127100), OsDREB2A (Os01g0165000), OsSOS1 (Os12g44360), OsHKT1;1 (Os06g0701700), OsHKT1;5 (Os01g20160), OsNHX1 (Os07g47100), OsNHX5 (Os09g0286400), Os18S (AF069218), OsACTIN1 (Os03g0718100), and AtUBQ5 (At3g62250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The coding sequence alignment among OsAHPs and OsPHPs, and the relative expression of OsAHPs and OsPHP in OsAHP-RNAi and wild-type rice seedlings.
Supplemental Figure S2. Expression patterns of OsAHP1 and OsAHP2 in rice.
Supplemental Figure S3. OsAHP2 overexpression partially rescued the Arabidopsis ahp1,2,3,4,5 mutant phenotype.
Supplemental Table S1. Primers used for gene cloning.
Supplemental Table S2. Primers used for qRT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Zhi-Yong Wang (Carnegie Institution for Science, Stanford University) for critical reading and editing of the manuscript; Dr. Jianru Zuo (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the Arabidopsis ahp1,2,3,4,5 mutant; and Dr. Kang Chong (Institute of Botany, Chinese Academy of Sciences) for providing the pTCK303 vector. We also thank the anonymous reviewers for their helpful comments, which improved this article greatly.
Glossary
- RNAi
RNA interference
- BA
6-benzyladenine
- DAPI
4′,6-diamidino-2-phenylindole
- 1/2 MS
one-half-strength Murashige and Skoog
- qRT
quantitative real-time
- cDNA
complementary DNA
Footnotes
This work was supported by the National Key Program on the Development of Basic Research in China (grant no. 2012CB114201), the Outstanding Researcher Program of Hebei Province (grant no. sprco46), and the National Nature Science Foundation of China (grant nos. 30671075 and 30800701).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Albacete A, Ghanem ME, Martínez-Andújar C, Acosta M, Sánchez-Bravo J, Martínez V, Lutts S, Dodd IC, Pérez-Alfocea F. (2008) Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J Exp Bot 59: 4119–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R, Aloni E, Langhans M, Ullrich CI. (2006) Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot (Lond) 97: 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Kieber JJ. (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ 32: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng Y, Dong H, Mu J, Ren B, Zheng B, Ji Z, Yang WC, Liang Y, Zuo J. (2010) Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. (2007) The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics 89: 697–707 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkeltawi N, Croteau R. (1987) Salinity depression of growth and essential oil formation in spearmint and marjoram and its reversal by foliar applied cytokinin. Phytochemistry 26: 1333–1334 [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103: 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira FJ, Kieber JJ. (2005) Cytokinin signaling. Curr Opin Plant Biol 8: 518–525 [DOI] [PubMed] [Google Scholar]
- Fukuda A, Nakamura A, Hara N, Toki S, Tanaka Y. (2011) Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233: 175–188 [DOI] [PubMed] [Google Scholar]
- Gadallah M. (1999) Effects of kinetin on growth, grain yield and some mineral elements in wheat plants growing under excess salinity and oxygen deficiency. Plant Growth Regul 27: 63–74 [Google Scholar]
- Ghanem ME, Albacete A, Martínez-Andújar C, Acosta M, Romero-Aranda R, Dodd IC, Lutts S, Pérez-Alfocea F. (2008) Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J Exp Bot 59: 3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2012) Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci 17: 172–179 [DOI] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ. (2002) Cytokinin signaling in Arabidopsis. Plant Cell (Suppl) 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Iqbal M, Ashraf M. (2005) Presowing seed treatment with cytokinins and its effect on growth, photosynthetic rate, ionic levels and yield of two wheat cultivars differing in salt tolerance. J Integr Plant Biol 47: 1315–1325 [Google Scholar]
- Iqbal M, Ashraf M, Jamil A. (2006) Seed enhancement with cytokinins: changes in growth and grain yield in salt stressed wheat plants. Plant Growth Regul 50: 29–39 [Google Scholar]
- Ito Y, Kurata N. (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382: 57–65 [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, et al (2009) Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol 150: 1955–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. (2006) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Kim J. (2013) Arabidopsis Response Regulator1 and Arabidopsis Histidine Phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol 161: 408–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, et al (2010) A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem 285: 23371–23386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Kieber JJ. (2002) Cytokinins. The Arabidopsis Book 1: e0063, /10.1199/tab.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwah S, Jones AM, Laxmi A. (2011) Cytokinin interplay with ethylene, auxin, and glucose signaling controls Arabidopsis seedling root directional growth. Plant Physiol 156: 1851–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinney G. (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140: 315–322 [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ. (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE. (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 64: 753–763 [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. (2004) Plant response regulators implicated in signal transduction and circadian rhythm. Curr Opin Plant Biol 7: 499–505 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC. (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Moreira S, Bishopp A, Carvalho H, Campilho A. (2013) AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PLoS ONE 8: e56370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2007) Arabidopsis cytokinin signaling pathway. Sci STKE 2007: cm5. [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Leyva-Gonzalez MA, Ha CV, Fujita Y, Tanaka M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K, Herrera-Estrella L, et al (2013) Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc Natl Acad Sci USA 110: 4840–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Kang JY, Kim SY. (2011) Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol Cells 31: 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. (2010) The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J 62: 473–482 [DOI] [PubMed] [Google Scholar]
- Rani Debi B, Taketa S, Ichii M. (2005) Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J Plant Physiol 162: 507–515 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E. (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2005) Cytokinin biosynthesis and regulation. Vitam Horm 72: 271–287 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu SH, Wu P, et al (2007) Nomenclature for two-component signaling elements of rice. Plant Physiol 143: 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, Schaller GE, Kieber JJ. (2012) Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol 158: 1666–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang M, Guo R, Shi D, Liu B, Lin X, Yang C. (2012) Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol 12: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P. (2009) Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2: 823–831 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K. (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. (2008) Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20: 1101–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, Lu T. (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167: 281–288 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.