Characteristics of vitamin B12-mediated gene regulation in algae provide insight into the evolution of vitamin B12 auxotrophy.
Abstract
Photosynthetic microalgae play a vital role in primary productivity and biogeochemical cycling in both marine and freshwater systems across the globe. However, the growth of these cosmopolitan organisms depends on the bioavailability of nutrients such as vitamins. Approximately one-half of all microalgal species requires vitamin B12 as a growth supplement. The major determinant of algal B12 requirements is defined by the isoform of methionine synthase possessed by an alga, such that the presence of the B12-independent methionine synthase (METE) enables growth without this vitamin. Moreover, the widespread but phylogenetically unrelated distribution of B12 auxotrophy across the algal lineages suggests that the METE gene has been lost multiple times in evolution. Given that METE expression is repressed by the presence of B12, prolonged repression by a reliable source of the vitamin could lead to the accumulation of mutations and eventually gene loss. Here, we probe METE gene regulation by B12 and methionine/folate cycle metabolites in both marine and freshwater microalgal species. In addition, we identify a B12-responsive element of Chlamydomonas reinhardtii METE using a reporter gene approach. We show that complete repression of the reporter occurs via a region spanning −574 to −90 bp upstream of the METE start codon. A proteomics study reveals that two other genes (S-Adenosylhomocysteine hydrolase and Serine hydroxymethyltransferase2) involved in the methionine-folate cycle are also repressed by B12 in C. reinhardtii. The strong repressible nature and high sensitivity of the B12-responsive element has promising biotechnological applications as a cost-effective regulatory gene expression tool.
Vitamins are essential nutritional components of the human and animal diet and can lead to an array of deficiency diseases if lacking (Martin et al., 2011). However, it is not just animals that require an external source of vitamins for growth, and in fact, a widespread and complex distribution of vitamin requirements exists across the tree of life (Helliwell et al., 2013). These compounds are vital as cofactors for enzymes of central metabolism, but evidence suggests that there may be a selective advantage to outsource the production of these compounds when an external supply is available through the diet or biotic interactions (Croft et al., 2005; Giovannoni, 2012; Helliwell et al., 2013). Microalgae are diverse photosynthetic eukaryotes in which vitamin auxotrophy is prevalent, with 22%, approximately 5%, and more than 50% of surveyed species requiring thiamine (vitamin B1), biotin (B7), and cobalamin (B12), respectively (Croft et al., 2005, 2006). Improved methodology for the quantification of thiamine and cobalamin in environmental samples revealed a heterogenous distribution of these vitamins in the oceans, with generally very low levels free in solution in the photic zone (Sañudo-Wilhelmy et al., 2012). It has also been shown, through fertilization experiments, that vitamins are limiting for algal growth in several marine environments (Gobler et al., 2007). Therefore, it is intriguing that many oligotrophic species such as Ostreococcus tauri are nonetheless vitamin auxotrophs (Worden et al., 2009; Helliwell et al., 2011) and so must be able to obtain a ready source of these organic micronutrients from their surroundings. In the case of cobalamin, the biosynthesis of this complex tetrapyrrole is restricted to prokaryotes, and specific biotic interactions between algae and bacteria have been shown to take place for B12 acquisition (Croft et al., 2005; Kazamia et al., 2012).
Many genes encoding enzymes of vitamin metabolism are down-regulated by an external supply of these organic micronutrients, giving rise to the suggestion that such feedback inhibition loops can drive gene loss, leading to vitamin auxotrophy (Helliwell et al., 2013). In the case of cobalamin, the loss of the B12-independent methionine synthase gene (METE) throughout the algal lineages means that species must then depend on the activity of a second isoform of this enzyme (METE), which requires B12 as a cofactor (Croft et al., 2005; Helliwell et al., 2011). Auxotrophy for vitamin B12 shows no phylogenetic relationship, and even strains of the same species have different B12 requirements (Tang et al., 2010). In algae that have both METE and METH, such as Chlamydomonas reinhardtii and Phaeodactylum tricornutum, it has been shown that METE transcript levels are repressed by the presence of B12 (Croft et al., 2005; Helliwell et al., 2011). Prolonged repression of the gene by a reliable source of vitamin B12, therefore, could lead to the accumulation of mutations and, eventually, gene loss. Support for this model comes from the discovery of METE pseudogenes in the B12-dependent green algae Volvox carteri and Gonium pectorale (Helliwell et al., 2011).
Gene regulation by B12 has been widely studied in bacteria. In Escherichia coli and Salmonella typhimurium, a transacting protein, MetR, positively regulates metE (Cai et al., 1989; Maxon et al., 1989). In contrast, in many other bacteria, B12-mediated regulation of gene expression occurs via ligand-responsive genetic control elements, known as riboswitches. These sequences are found in the 5′ untranslated region (UTR) of regulated mRNAs and bind the ligand directly. Changes in the secondary structure of the mRNA result in altered gene expression, for example by masking the ribosome-binding site (Winkler et al., 2002). B12-responsive riboswitches are widespread within the prokaryote lineages and have been identified in genes involved in B12 transport and biosynthesis (Nahvi et al., 2004) as well as the metE gene of Mycobacterium tuberculosis (Warner et al., 2007). In mammalian cell lines, where METH is translationally up-regulated by B12 supplementation, another mechanism has been reported. There is a 70-bp region in the 5′ UTR just upstream of the start codon, which acts as an internal ribosome entry site, or IRES (Oltean and Banerjee, 2003), the secondary structure of which is influenced by B12. However, electrophoretic mobility shift assays revealed that translational activation in the presence of B12 also requires an auxiliary protein, indicating that it is unlikely to be via a riboswitch (Oltean and Banerjee, 2005).
Recently, a combined proteomic and transcriptomic study identified several B12-responsive genes in addition to METE in the diatoms P. tricornutum (B12 independent) and Thalassiosira pseudonana (B12 dependent; Bertrand et al., 2012). These genes include CBA1, which has been suggested to encode a novel cobalamin transporter. However, the means by which B12-mediated gene regulation occurs in algae remains unknown. Certainly, no B12-specific riboswitches have been identified in eukaryotes to date, although thiamine pyrophosphate riboswitches have been characterized in thiamine biosynthesis genes, including in C. reinhardtii (Croft et al., 2007). Recently, the thiamine pyrophosphate riboswitch in the THI4 gene was incorporated into constructs upstream of the nucleus-encoded chloroplast protein2 (NAC2) gene encoding a regulator of chloroplast translation. Introduction of the plasmid into C. reinhardtii nac2 mutants resulted in transformants in which complementation by the NAC2 transgene was repressed by thiamine (Ramundo et al., 2013). This construct also included a 1.2-kb region upstream of the METE gene, and the authors observed some reduction in NAC2 complementation by exogenous vitamin B12. However, to understand how B12 regulates METE gene expression, a more in-depth study is required. Here, we investigate the effect of B12 on gene expression in different species of algae and define the responsive element of the C. reinhardtii METE gene, providing valuable information on the likely ecological and environmental context necessary for the gene to be lost. As a result, one can begin to address the question of why B12 auxotrophy is so prominent within the many algal lineages.
RESULTS
Defining the Regulation of METE by B12
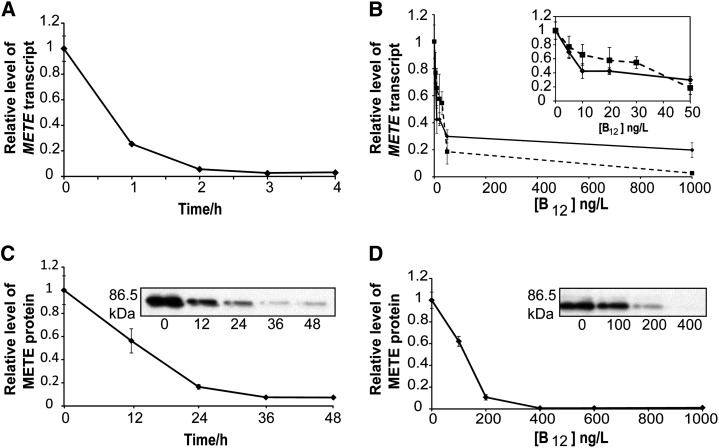
To understand the regulation of METE in C. reinhardtii, the time scale and concentration dependence of B12 repression was investigated using reverse transcription (RT)-quantitative real-time PCR (qPCR). The addition of vitamin B12 resulted in a significant reduction of METE transcript level within 2 h, with an estimated half-life of about 45 min (Fig. 1A). By varying the concentration of B12, as little as 50 ng L−1 could reduce the transcript level to about 30% of that seen in the absence of B12 (Fig. 1B, solid line), although it should be pointed out that even with 1,000 ng L−1 (a concentration that supports the growth of algal B12 auxotrophs; Bertrand et al., 2012; Kazamia et al., 2012), some transcript can still be detected. The same trend was observed for the marine diatom P. tricornutum (Fig. 1B, dashed line). To establish if the transcript levels correlated with the METE protein, western-blot analysis was carried out using polyclonal antibodies against C. reinhardtii METE (Schneider et al., 2008). A band of the expected size (approximately 86.5 kD) was detected in protein extracts prepared from C. reinhardtii grown without B12 (Fig. 1C). After the addition of vitamin B12 (1,000 ng L−1), the METE protein took over 24 h to drop to an undetectable level, with a half-life of approximately 18 h (Fig. 1C), in other words much slower than the transcript (Fig. 1A). Furthermore, over 200 ng L−1 vitamin B12 was required for the METE protein to reach an undetectable level (Fig. 1D). Similar lack of correlation between the responses of transcript and protein levels has been observed for environmental samples (Bertrand et al., 2013).
Figure 1.
The role of vitamin B12 on METE expression. A, RT-qPCR analysis of METE expression in C. reinhardtii over time after the addition of vitamin B12 (1,000 ng L−1) normalized to expression at time 0 (i.e. prior to B12 addition). Transcript expression was quantified and normalized using RACK1 as a housekeeping gene (Mus et al., 2007; Dubini et al., 2009). Values are means and error bars denote the range (n = 2). B, METE transcript abundance after the addition of different concentrations of vitamin B12 for C. reinhardtii (as described above; solid line) and the marine diatom P. tricornutum (as normalized to Histone H4, 30S Ribosomal protein S1, and TATA box-binding protein as housekeeping genes [Siaut et al., 2007]; dashed line). Values are means and error bars denote the range (n = 2). C, METE protein abundance after the addition of vitamin B12 (1,000 ng L−1; as normalized to the quantity of loaded protein calculated from a Coomassie Brilliant Blue-stained gel). Values are means and se (n = 3). D, METE protein abundance after the addition of different concentrations of vitamin B12. Values are means and se (n = 3).
Characterization of the Vitamin B12-Responsive Element of METE in C. reinhardtii
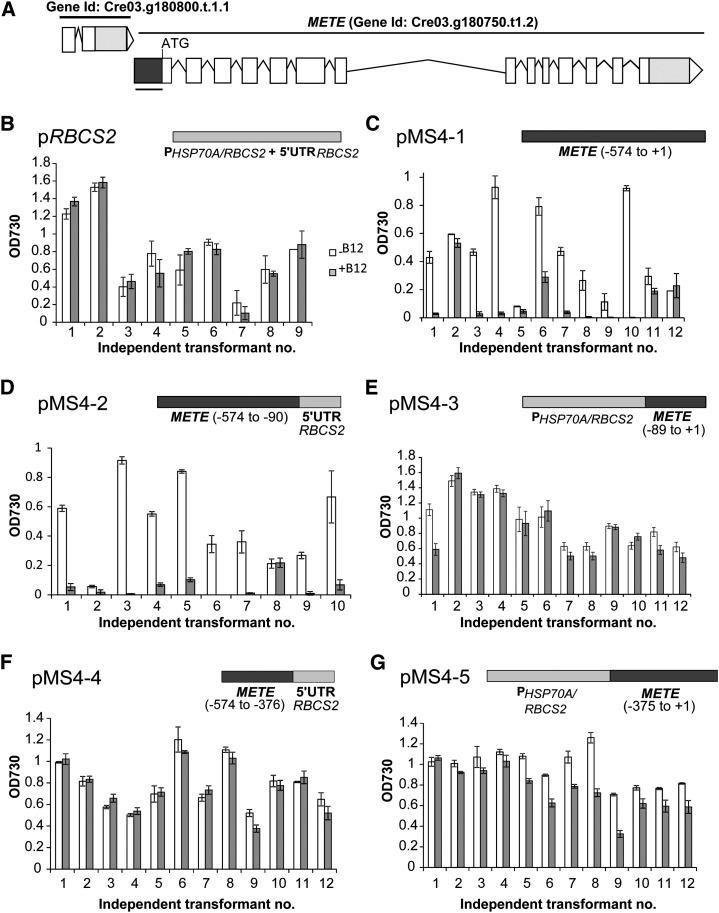
To identify the B12 regulatory region of C. reinhardtii METE, we used a reporter gene approach. Regions upstream of the METE translational start but downstream of the preceding gene (Fig. 2A) were joined to the dual reporter gene sh.BLE-GFP encoding the Streptoalloteichus hindustanus zeocin-binding protein (Sh.Ble) fused to GFP (Supplemental Fig. S1). Plasmid pMS4-1 contains the METE upstream region from −574 to the start codon, whereas pMS4-2 contains the same region but with the −89 to +1 region replaced with the 5′ UTR of the C. reinhardtii Ribulose Bisphosphate Carboxylase/Oxygenase Small Subunit2 (RBCS2) (a control gene that is not repressed by B12). The −89 position was chosen on the basis of EST data from the genome database showing that the longest transcripts extend to this position. This suggests that −89 marks the start of the METE 5′ UTR, with the promoter element upstream. Plasmid pMS4-3 has the converse arrangement, with the HSP70A/RBCS2 promoter element fused to the 89-bp METE 5′ UTR. Finally, pRBCS2 contains both the HSP70A/RBCS2 promoter and the RBCS2 5′ UTR, with no METE sequence, serving as a negative control in terms of B12-responsiveness. The plasmids were introduced into C. reinhardtii cell wall-less mutant15 (cw15), and transformants were selected on the basis of zeocin resistance. Approximately the same number of transformants were obtained for each construct, confirming that the METE −574 to −90 upstream sequence was able to drive expression of the reporter gene.
Figure 2.
Probing the vitamin B12 regulatory element of METE using a reporter gene approach. A, Genome browser view of the METE gene model, downstream of the preceding gene (gene identifier Cre03.g180800.t1.1). B to G, Effects of vitamin B12 (1,000 ng L−1) on growth (optical density at 730 nm [OD730]) at 96 h in the presence of zeocin (10 µg mL−1) for independent transformant lines containing reporter constructs: pRBCS2 (B), pMS4-1 (C), pMS4-2 (D), pMS4-3 (E), pMS4-4 (F), and pMS4-5 (G). White columns, Without B12 treatment; gray columns, with B12. Error bars denote se (n = 3).
We then tested the responses of several independent transformants for each construct in the absence and presence of B12 (1,000 ng L−1). As expected, growth in zeocin of the lines transformed with the control plasmid pRBCS2 was unaffected by B12 (Fig. 2B). In contrast, the majority of transformants containing pMS4-1 and pMS4-2 were unable to grow in medium containing both zeocin and B12 (Fig. 2, C and D). This indicates that the upstream region of METE from −574 to −90 is sufficient for repression by B12. For those transformants that were not responsive, a likely explanation comes from the fact that nuclear transformation in C. reinhardtii occurs via random insertion (through nonhomologous end joining; Tam and Lefebvre, 1993), which can result in rearrangements/partial deletion of the transforming DNA. It is possible, therefore, that the METE sequence has been lost in some of the recovered transformants, and sh.BLE expression is due to integration downstream of actively expressed endogenous genes. For 15 out of 16 transformants containing pMS4-3 (which has the METE 5′ UTR only), there was no response to B12 (Fig. 2E), and although in one case there was some growth reduction in zeocin, it was only partial. To assess whether the presence of the −89 to +1 region enhances sensitivity to vitamin B12, we performed a B12 dosage experiment using representative transformants for pMS4-1 and pMS4-2. In both cases, the response was identical, with a complete suppression of growth induced with 100 ng L−1 B12 (Supplemental Fig. S2). Two further constructs were also analyzed in this study: pMS4-4 (−574 to −376) and pMS4-5 (−375 to +1). Lines transformed with these plasmids showed no or little responsiveness to vitamin B12 (Fig. 2, G and F, respectively), indicating that the B12-responsive element has most likely been disrupted in these constructs.
METE Is Not Repressed in Species That Lack METH
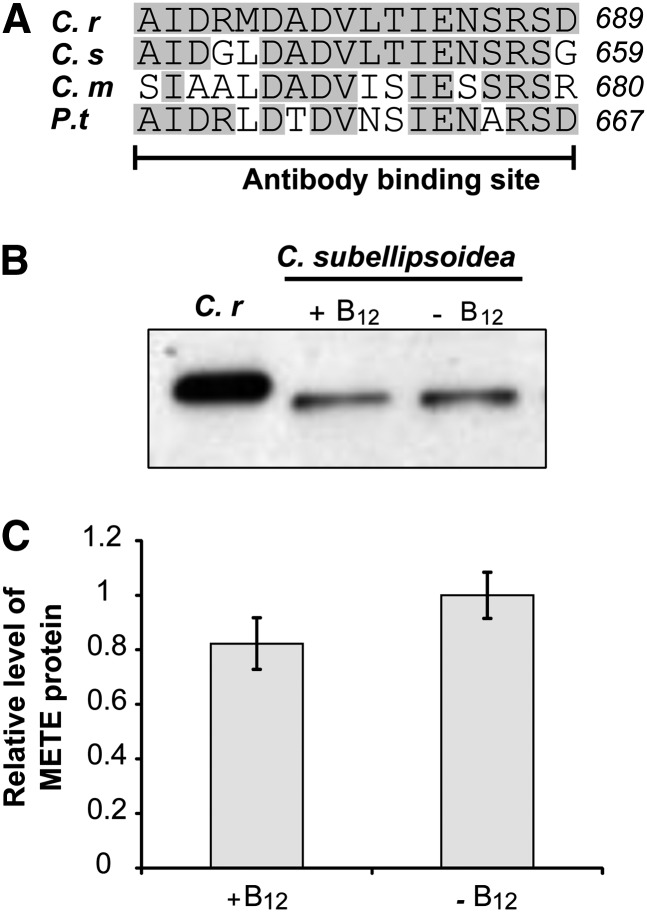
Repression of METE by vitamin B12 has been shown previously in C. reinhardtii and the marine diatom P. tricornutum (Helliwell et al., 2011; Bertrand et al., 2012), both of which also encode METH. However, there are examples of species that have METE only, such as the freshwater green alga Coccomyxa subellipsoidea and the thermophilic red alga Cyanidioschyzon merolae (Helliwell et al., 2011). In these species, we hypothesized that METE would not respond to B12 in the same way as in C. reinhardtii, because repression by vitamin B12 would leave the cells devoid of Met synthase activity altogether. Comparison of the METE amino acid sequence of C. subellipsoidea, C. merolae, and P. tricornutum with the C. reinhardtii METE antibody-binding site reveals sequence conservation between species (Fig. 3A). Therefore, we tested the C. reinhardtii METE antibody (Schneider et al., 2008) on protein samples extracted from these species. The predicted molecular mass of METE from C. subellipsoidea is 82 kD, and a band slightly smaller than that of C. reinhardtii METE (predicted size of 86.5 kD) was detected for samples extracted from C. subellipsoidea (Fig. 3B). Given that METE was present in both the sample extracted from cells grown with 10 µg L−1 B12 and those grown without (Fig. 3B) and at a similar level (Fig. 3C), we conclude that METE is not repressed by B12 in C. subellipsoidea. No bands were detectable on western blots of protein extracts from C. merolae or P. tricornutum, indicating that these proteins are not recognized by the antibody, probably on account of decreased conservation in the region of the antibody-binding site (Fig. 3A). Using an RT-PCR approach, however, we did detect a METE transcript with complementary DNA (cDNA) prepared from C. merolae, and this was present whether the cells were grown with B12 even at 10 µg L−1 (Supplemental Figs. S3 and S4). Thus, in two different algal groups, chlorophytes (C. subellipsoidea) and rhodophytes (C. merolae), METE repression by B12 does not occur when METH is absent.
Figure 3.
METE is expressed in the presence of vitamin B12 in C. subellipsoidea, which lacks METH. A, Comparison of the METE amino acid sequences of C. subellipsoidea (C.s), C. merolae (C.m), and P. tricornutum (P.t) with the C. reinhardtii (C.r) METE antibody-binding site (Schneider et al., 2008). Shared residues are shaded gray. B, METE abundance in C. subellipsoidea in the presence (10 µg L−1) and absence of vitamin B12 using western blotting with the C. reinhardtii METE antibody (Schneider et al., 2008). C, Quantification of METE levels in C. subellipsoidea in the presence (10 µg L−1) and absence of B12. Values are means and se (n = 3).
Exploring the Roles of Other Factors Involved in METE Regulation in C. reinhardtii
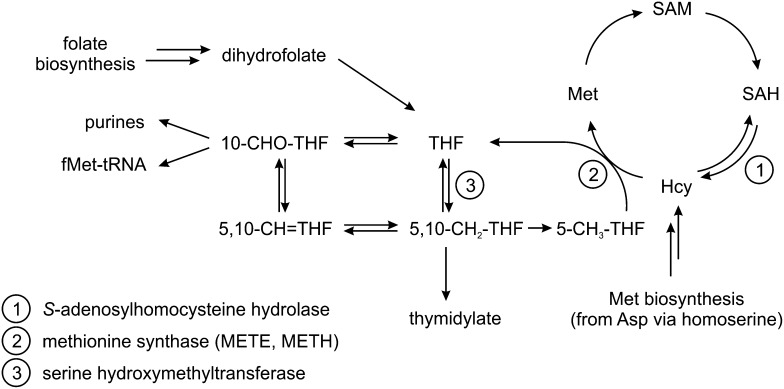
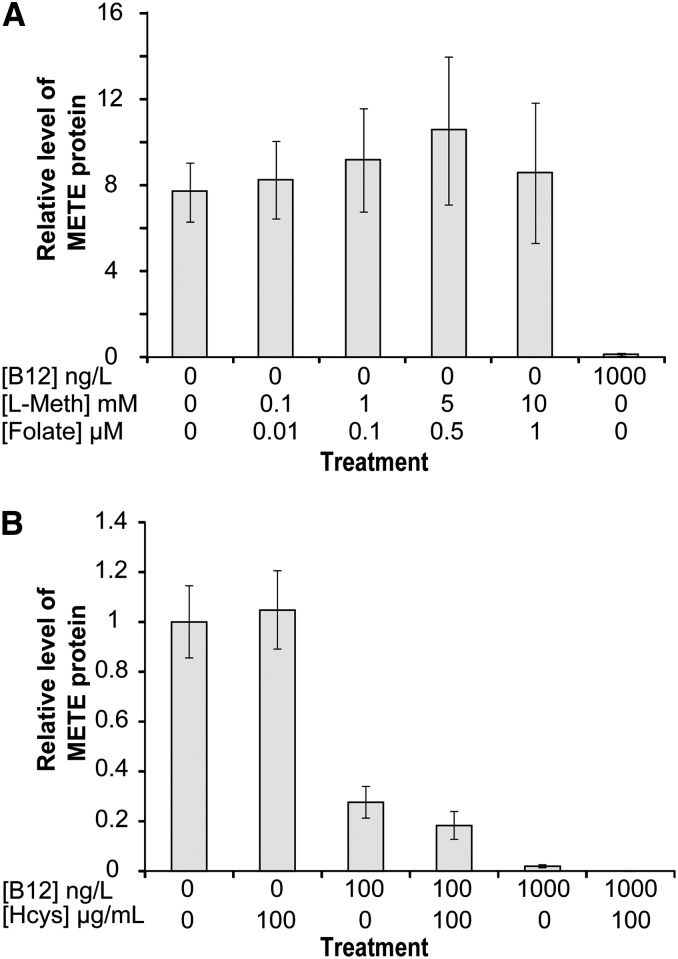
Next, we investigated if other factors regulate C. reinhardtii METE. Met synthase catalyzes the synthesis of Met from homocysteine and methyl-tetrahydrofolate through the transfer of a methyl group from methyl-tetrahydrofolate (which in turn generates tetrahydrofolate; Fig. 4). This is an essential reaction in C1 metabolism, and in animals, which have only METH, lack of B12 as cofactor causes changes in the balance of intracellular folate derivatives. This ultimately leads to a phenomenon known as folate trapping (Scott and Weir, 1994). In E. coli, which has both METE and METH, Met has a negative regulatory effect on METE expression (Milner et al., 1969; Weissbach and Brot, 1991), whereas homocysteine has a positive regulatory role in E. coli, S. typhimurium, and Ralstonia solanacearum (Urbanowski and Stauffer, 1989a, 1989b; Plener et al., 2012). Croft et al. (2005) showed that, in the absence of B12, Met and folate can support the growth of the B12-dependent green alga Lobomonas rostrata when supplemented at concentrations of 10 mm and 1 µm, respectively. To determine if these metabolites have an effect on METE protein abundance in C. reinhardtii, western-blot analysis was carried out on samples extracted from cultures supplemented with different concentrations of Met and folate. No changes were observed with either Met or folate (Fig. 5A). To assess the effect of homocysteine, we included this metabolite at 100 µg mL−1 (0.74 mm) to C. reinhardtii cultures grown under three different B12 treatments: no B12, 100 ng L−1 B12, and 1,000 ng L−1 B12. Again, no effect on METE protein content was observed by western-blot analysis (Fig. 5B). Therefore, based on these data, we conclude that, unlike in E. coli, homocysteine, Met, and folate have no role in the regulation of METE in C. reinhardtii.
Figure 4.
Simplified scheme of C1 metabolism. 10-CHO-THF, 10-Formyl-tetrahydrofolate; 5,10-CH+-THF, 5,10-methenyl-tetrahydrofolate; 5,10-CH2-THF, 5,10-methylene-tetrahydrofolate; 5-CH3-THF, 5-methyl-tetrahydrofolate; Hcy, homocysteine; SAH, S-adenosylhomocysteine.
Figure 5.
Effects of C1 metabolites on METE protein abundance. A, METE protein in response to different concentrations of l-Met (l-Meth) and folate (48 h). B, METE protein in response to dl-homocysteine (Hcys) in different B12 treatments (48 h). Values are means and se (n = 3).
Proteomics Analysis of the Effect of Vitamin B12 on Algal Metabolism
Having defined more clearly the effect of B12 on METE expression and investigated the role of other factors, we took a proteomics approach to examine the effect of B12 on C. reinhardtii metabolism more generally. C. reinhardtii was grown in either the absence or presence of B12, at 1,000 ng L−1 to ensure complete repression of METE. Proteins were extracted from cultures grown to exponential phase (n = 4) and analyzed by two-dimensional difference gel electrophoresis (2D-DIGE; Unlü et al., 1997; Lilley and Friedman, 2004). The eight individual samples were each labeled with Cy5, and then mixed with a Cy3-labeled standard made of equal amounts of each sample. The Cy3/Cy5 mix was separated by two-dimensional gel electrophoresis, followed by silver staining. A total of 1,190 spots were found to be present in at least 12 of the 16 spot maps, and these were analyzed by comparison of the Cy3 and Cy5 intensities. Of these, just 11 spots (0.9%) demonstrated differential abundance of at least 2-fold. Strikingly, all are from proteins that are down-regulated in the presence of vitamin B12 (P ≤ 0.01; Supplemental Fig. S5; Supplemental Tables S1 and S2).
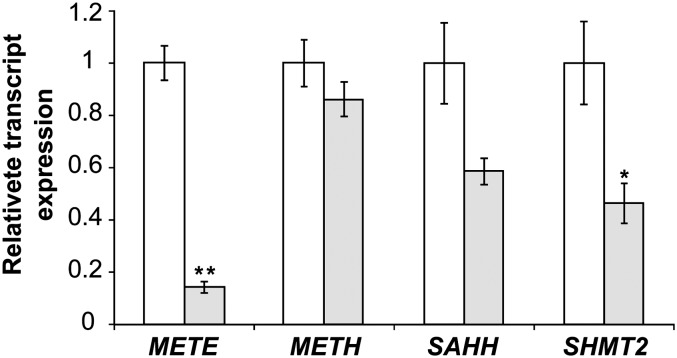
From these 11 hits, there were three groups of three adjacent spots that have the same molecular masses (Supplemental Fig. S5), implying that they are different isoforms of the same protein. One spot from each of these groups was excised from the gels and identified by tandem mass spectrometry (MS/MS; details of spot identification are given in Supplemental Table S1). The two larger groups of three spots were identified as METE. When corrected for the different abundance of each putative isoform, this protein is down-regulated 5.9-fold overall. A possible modification of METE, which might affect mobility leading to multiple spots, is phosphorylation, since this has been predicted previously for C. reinhardtii (Kurvari et al., 1995; Schneider et al., 2008), and METE from tobacco (Nicotiana tabacum) appeared as seven isoforms on a two-dimensional gel (Moscatelli et al., 2005). The third group of adjacent spots, down-regulated an average of 2.5-fold, was serine hydroxymethyltransferase2 (SHMT2), but MS/MS data did not give any information on the nature of these isoforms. Of the two individual spots, one, down-regulated 2.43-fold, was S-adenosylhomocysteine hydrolase (SAHH), while the last (marked with an asterisk in Supplemental Fig. S5) could not be identified. Interestingly, all three identified proteins are neighbors in the metabolic network of C1 compounds (Fig. 4): SAHH is part of the activated methyl cycle and catalyzes the hydrolysis of S-adenosylhomocysteine to yield homocysteine, the substrate of Met synthase, while SHMT catalyzes the reversible transfer of a C1 group from Ser to tetrahydrofolate. RT-qPCR showed that expression ratios for METE, SAHH, and SHMT2 are similar at the transcript level compared with the protein level (Fig. 6; Supplemental Table S3), thus validating the 2D-DIGE approach. Similarly, B12 was confirmed to not affect transcript abundance of METH, as evidenced previously (Croft et al., 2005).
Figure 6.
Investigating other B12-responsive genes in C. reinhardtii. Transcript abundance is shown when C. reinhardtii is grown in the presence or absence of vitamin B12 (1,000 ng L−1). Transcripts were quantified using RT-qPCR with RACK1 as the reference gene (Mus et al., 2007; Dubini et al., 2009). For each gene, the result was normalized by expression in the absence of B12. *P ≤ 0.05, **P ≤ 0.001 compared with expression in the absence of B12 (two-tailed Student’s t test). Values are means and se (n = 3).
Having identified two further genes that respond to vitamin B12 in C. reinhardtii, we used the bioinformatics tool MEME (Multiple EM for Motif Elicitation; Bailey and Elkan, 1994) to search for common sequence motifs shared between the 1-kb upstream regions of the SAHH and SHMT2 genes and the METE (−574 to −90) regulatory element. The METH upstream region was used as a negative control sequence (Bailey and Elkan, 1994). This approach identified one common sequence motif, CTGCGCTG (E-value of 1.6E + 005), present in the SAHH, SHMT2, and METE sequences. However, since the reporter construct pMS4-4 (−574 to −376) contains this motif, and regulation by B12 in this construct is impaired, this motif alone is not sufficient for B12-mediated transcriptional regulation. Understanding exactly how these genes are regulated in the presence of B12, and whether the mode by which this occurs is conserved, remains an exciting prospect.
DISCUSSION
Our study of vitamin B12-mediated regulation of gene expression in algae has provided important insights into the mechanism of this process and its biological consequences. We have defined in detail the response of the C. reinhardtii METE gene to vitamin B12 and shown that it is not sensitive to other C1 metabolites (folate, Met, or dl-homocysteine), and we also found two other targets of B12 repression, SAHH and SHMT, which are both involved in the C1 cycle. In addition, we have successfully isolated a B12-responsive element located upstream of the transcription start of METE, which is sufficient to control the expression of a reporter construct in transformed lines of C. reinhardtii, with the same responsiveness to exogenous B12 as the endogenous METE gene. Since the METE 5′ UTR does not confer this responsiveness, it is unlikely that METE is regulated by a B12 riboswitch as it is in bacteria (Nahvi et al., 2004; Warner et al., 2007) or by an IRES as in mammals (Oltean and Banerjee, 2003, 2005).
The tight repression of METE exhibited in C. reinhardtii is also observed in the unrelated diatom P. tricornutum, which also possesses METH (Croft et al., 2005; Helliwell et al., 2011; Bertrand et al., 2012). This implies that there is a strong cellular advantage to having the gene switched off when possible (i.e. in the presence of B12). In E. coli, MetH has a more than 100-fold higher turnover number than MetE (González et al., 1992), and in the absence of the vitamin, MetE is the second most abundant protein (Karlin et al., 2001), presumably to allow efficient C1 metabolism. Intriguingly, a recent study of thermal tolerance in C. reinhardtii found that nonlethal heat shock treatment resulted in the repression of METE gene transcription (Xie et al., 2013), and cells could only survive if supplied with B12, presumably to allow METH to function. Reduction in METH levels using artificial microRNA (amiRNA) suppressed thermal tolerance. This suggests that having both isoforms of Met synthase provides a degree of environmental flexibility. On the other hand, if there is a reliable and abundant external source of B12, and these conditions prevail over a long period of time, the gene may be lost entirely, which is the derived state of many algal species spanning multiple diverse phyla (Croft et al., 2005; Helliwell et al., 2011). However, natural levels of B12 are reportedly less than 10 ng L−1 (10 pm) in freshwater (Kurata, 1986) and perhaps up to 70 ng L−1 in seawater (Carlucci, 1970; Gobler et al., 2007), with more recent measurements indicating a lower range in the photic zone of the Southern Ocean and North Atlantic Ocean (Panzeca et al., 2009; Sañudo-Wilhelmy et al., 2012). The extracellular concentration of B12 necessary to repress the METE transcript (Fig. 1B) far exceeds the amount of free cobalamin estimated to be present in characterized freshwater and certain marine ecosystems. How, then, has B12 auxotrophy become so common in the algal lineages? There are several examples where algae obtain vitamin B12 via a symbiotic relationship with bacteria (Croft et al., 2005; Wagner-Döbler et al., 2010; Kazamia et al., 2012). A plausible explanation, therefore, is that provision of B12 by associated bacteria could lead to METE loss, which, in turn, would bind the organisms further together. Indeed, this mechanism may have played a role in shaping the evolution of vitamin auxotrophy more generally (Helliwell et al., 2013).
In this regard, it is also relevant that neither Met, nor homocysteine influences METE expression in C. reinhardtii, which is in contrast to several bacterial species studied (Urbanowski and Stauffer, 1989a, 1989b; Weissbach and Brot, 1991; Plener et al., 2012). Since, in the absence of B12, the growth of the B12-dependent alga L. rostrata, a close relative of C. reinhardtii, can be supplemented by Met and folate (at concentrations of 10 mm and 1 µm, respectively), it seems plausible that these compounds do enter the C. reinhardtii cell. Therefore, our data suggest that exogenous levels of vitamin B12 are likely to be the primary factor governing the loss of METE. In contrast to C. reinhardtii and P. tricornutum, in species that lack METH (i.e. C. subellipsoidea and C. merolae; Fig. 3), vitamin B12 does not affect the expression of METE. This suggests that there must have been selection for an alteration in how METE is regulated, either prior to or shortly after the loss of METH in these species. It is noteworthy that C. merolae, a thermophilic red alga, has METE but not METH, since in C. reinhardtii, the latter enzyme has a higher thermotolerance compared with METE (Xie et al., 2013). In the hot sulfurous springs in which C. merolae lives, temperatures can exceed 45°C, with pH being as low as 1.5 (Matsuzaki et al., 2004). It is quite likely, therefore, that B12 degrades under such conditions and that C. merolae does not encounter vitamin B12 in its environment at all, which could explain its loss of METH in this species. Presumably, selection on METE to enhance thermotolerance must also have occurred. As an aside to this, the fact that METE is not repressed by B12 in all algae must also be considered when utilizing METE as a biomarker for evaluating the B12 status of microbial communities in the open field as has been proposed (Bertrand et al., 2013).
Besides probing METE regulation, we also identified further genes that are responsive to vitamin B12 in C. reinhardtii, using a 2D-DIGE proteomics approach. The most pronounced regulation was found for METE, which was 7-fold more abundant in the absence of B12 (Supplemental Table S3). Two other proteins were identified as down-regulated by B12, SAHH and SHMT2, which catalyze adjacent reactions in C1 metabolism (Fig. 4). The SHMT protein was also observed to increase in the marine diatom P. tricornutum in response to B12 depletion (Bertrand et al., 2012), but to our knowledge this is the first report of SAHH being responsive to vitamin B12. Raised levels of these two enzymes are likely to increase the flux of metabolites toward Met synthase (Fig. 4; Supplemental Fig. S5). Therefore, the probable purpose of this regulation pattern is to ensure sufficient Met and S-adenosylmethionine levels in the cytosol when the less efficient METE protein has to operate. The strikingly few changes in global gene expression we detected illustrate how C. reinhardtii subtly adjusts its metabolism to switch from a nonutilizing to a B12-utilizing organism.
Our findings present a significant contribution to the understanding of B12-specific gene regulation and metabolism in algae. Additionally, this work has unexpected applications for algal biotechnology. Inducible gene expression systems are highly desirable for the controlled expression of genes of interest. However, existing platforms are generally regulated by cues that are toxic to the cell and/or physiologically intrusive, such as the upstream elements of the Nitrate Reductase, Cytochrome C6, and Carbonic Anhydrase1 genes (Schroda, 2006), which are sensitive to nitrate limitation, toxic metals (including copper, cobalt, and nickel), and CO2, respectively. Given the minimal effect of vitamin B12 on global protein expression in C. reinhardtii, the novel B12 regulatory system we have identified is likely to have many fewer indirect metabolic effects. Moreover, the micronutrient quantities (as low as 200 ng L−1) sufficient for the complete repression of transgene expression enables cost-effective scale up. For example, just 0.2 g of vitamin B12 would be sufficient to regulate gene expression in 106 L of culture. The combined assets of high sensitivity and metabolic specificity make this an apt gene expression tool for industrial purposes.
MATERIALS AND METHODS
Strains and Growth Conditions
Escherichia coli DH5α was grown in lysogeny broth medium and used in all cloning steps. Details of source and growth conditions for Chlamydomonas reinhardtii strains 12 and cw15 (cw15, mt−), Coccomyxa subellipsoidea C-169, Phaeodactylum tricornutum CCAP 1052/1B, and Cyanidioschyzon merolae M10 are provided in Supplemental Table S4.
RNA Extraction and RT-qPCR
Total RNA was extracted as described by Helliwell et al. (2011) and subsequently treated with the Ambion Turbo DNase-Free Kit to remove genomic DNA. RNA was reverse transcribed into cDNA with SuperScript II (Invitrogen). Details of the primers and specific amplification conditions used for RT-qPCR are given in Supplemental Table S5. For each gene, primers were designed for different exons so that genomic DNA contamination could be assessed. For P. tricornutum, RT-qPCR was carried using Qiagen SYBR Green MasterMix using 4 ng of cDNA and 0.1 µm of each primer per reaction. Unique single products were verified by melt-curve analysis. No amplification was detected in no reverse transcriptase control samples, confirming the absence of genomic DNA. Cycle threshold values of two technical replicates were calculated using Rotor-Gene Q Software version 2.02, and primer efficiency was calculated with LinRegPCR (Ramakers et al., 2003). The expression of each gene was calculated by raising the efficiency of each primer pair to the negative cycle threshold value and then normalizing this by dividing by the mean expression of reference genes (Histone H4, 30S Ribosomal protein S1, and TATA box-binding protein). Representative RT-qPCR samples were additionally checked by agarose gel electrophoresis. Similar protocols were followed for C. reinhardtii, except that the SYBR Green JumpStart Taq ReadyMix kit (Sigma no. S5193) was used, with 3.3 ng of cDNA from 3.3 ng of RNA and 400 nm of each primer. The expression of each gene was calculated by raising the efficiency of the different primer pairs to the negative cycle threshold value and then normalizing this to Receptor for Activated C Protein Kinase1 as the reference gene. For C. merolae, PCR was used to amplify sequences from cDNA using Taq DNA polymerase (Bioline). Reaction mixtures contained 125 nm of each primer, 2 mm MgCl2, and 5% (v/v) dimethyl sulfoxide. Amplification conditions on a TC-512 gradient instrument (Techne) were as follows: 94°C for 5 min, cycles of 94°C for 45 s, 63°C for 45 s, and 72°C for 2 min, and 72°C for 10 min. Cycle number was optimized (Supplemental Figure S4), and 35 and 23 cycles were chosen for METE and Elongation factor1a, respectively.
Western Blotting
Cells from 50 mL of midexponential phase culture were harvested by centrifugation (600g, 5 min), resuspended in 150 µL of extraction buffer (97.1 mm potassium phosphate buffer, pH 7.5, 5 mm dithiothreitol [DTT], and 2 mm EDTA, pH 8), and lysed using sonication (Biorupter; Diagenode). Protein concentration was quantified using a Bradford assay (Bradford, 1976), and 4 µg of protein was separated by SDS-PAGE on 12% (w/v) gels with a 5% (w/v) stacking gel, blotted onto a nitrocellulose membrane (Protran BA 83; Whatman), and labeled with METE antibodies (Schneider et al., 2008). Band intensity (integrated density) was quantified using ImageJ (Schneider et al., 2012), and relative METE levels were calculated (normalized to the quantity of loaded protein calculated from a Coomassie Brilliant Blue-stained gel).
Plasmid Construction
Plasmids were generated using Gibson assembly (Gibson et al., 2009). PCR was carried out using the iProof DNA polymerase (Bio-Rad) plus 3% (v/v) dimethyl sulfoxide with the primers listed in Supplemental Table S6. Transformation into E. coli, isolation of positive clones, and sequence confirmation were according to standard protocols.
Genetic Manipulation of C. reinhardtii and Reporter Gene Growth Assays
Transformation of a cell wall-less strain of C. reinhardtii (cw15, mt−) was carried out by electroporation via a procedure modified from Shimogawara et al. (1998). Briefly, 2-d-old cultures (1 × 106 cells mL−1 to 5 × 106 cells mL−1) were harvested via centrifugation (at 1,500g) and resuspended in Tris-acetate phosphate (TAP) medium + 60 mm Suc. Concentrated cells (300 µL) were combined with 3 µg of linear plasmid DNA per transformation in a chilled 0.4-mm-gap cuvette. Electroporation was performed with a Bio-Rad Gene Pulser Xcell set to exponential decay protocol, 800-V field strength, 25 µF, and no shunt resistance. A typical pulse time of 9.8 to 15 ms was obtained. Cells were subsequently transferred to 10 mL of TAP + 60 mm Suc and incubated at 20°C in low light for 16 to 18 h. For selection, cells were spread on TAP 1.5% (w/v) agar plates containing 10 µg mL−1 zeocin. Colonies appeared after 5 d and were further processed after 7 d.
To assess the effect of vitamin B12 on the various upstream sequences controlling the BLE reporter gene, growth assays were used to test resistance to zeocin in the presence of B12. For each transformed line of C. reinhardtii, three replicate precultures were set up on 24-well plates on nonselective TAP medium (without zeocin) either with B12 (1,000 ng L−1) or without. After 72 h, 15 µL of these precultures was inoculated into 50 mL of the appropriate medium for the plus or minus B12 condition, this time with 10 µg mL−1 zeocin. Growth was then monitored over time by measuring the optical density at 730 nm.
2D-DIGE
Proteins were extracted from frozen cell pellets of C. reinhardtii cw15 by a procedure modified from Gillet et al. (2006). Cells from 50 mL of culture, grown either with B12 (1 µg L−1) or with no addition, were resuspended in 500 μL of 50 mm Tris-Cl, pH 8.0, and 0.07% (w/v) DTT containing protease inhibitor cocktail (Roche no. 11697498001) and lysed by three cycles of freezing in liquid nitrogen and thawing at room temperature. After the removal of debris via centrifugation (45 min, 15,000g, 4°C), nucleic acids were removed by the addition of 2% (w/v) streptomycin sulfate and centrifugation for 45 min at 60,000g, 4°C. Proteins were precipitated with 10% (w/v) TCA and 80% (v/v) acetone at −20°C for 24 h and centrifuged (15 min, 17,000g, 4°C), washed once with acetone and once with 80% (v/v) acetone, dried in a SpeedVac centrifuge, and dissolved in 200 μL of 10 mm Tris-Cl, pH 8.0, 5 mm magnesium acetate, 8 m urea, and 2% (w/v) amidosulfobetaine-14. Protein concentration was determined with the DC Protein Assay kit (Bio-Rad) using bovine serum albumin as a standard.
For each of eight samples (four samples per condition), 50 μg of protein was labeled with 18 μm Cy5 dye (GE Healthcare no. RPK0275) and a standard, made of equal amounts of each sample, was labeled with Cy3 (no. RPK0273), according to the manufacturer’s instructions (for 30 min at 4°C). After quenching with 0.8 mm Lys for 10 min, individual Cy5-labeled samples were combined with the same amount of Cy3-labeled standard, and 1 volume of 4% (w/v) CHAPS, 8 m urea, 2% (w/v) DTT, and 2% (w/v) IPG Buffer pH 4-7 (GE Healthcare no. 17-6000-86) was added and the solution was incubated for 15 min at 4°C. Another 8 volumes of 4% (w/v) CHAPS, 8 m urea, 0.2% (w/v) DTT, and 1% (v/v) IPG Buffer/pharmalytes was added before isoelectric focusing was performed using a 13-cm Immobiline DryStrip pH 4-7 (GE Healthcare no. 17-6001-13). Proteins were separated on an Ettan IPGphor isoelectric focusing unit for 10 h at 20 V, 1 h at 500 V, 1 h at 1,000 V, and 5 h at 8,000 V and subsequently in the second dimension by SDS-PAGE using 12% (v/v) polyacrylamide. Cy3 and Cy5 spot maps were generated by scanning the gel with a Typhoon 9400 imager (GE Healthcare). Eight two-dimensional gels were run in total, loaded with equal amounts of sample and standard. Matching, quantification, and evaluation of spots were carried out using the software package DeCyder 5.02 (GE Healthcare). To calculate overall expression ratios for the proteins with several putative isoforms, the ratios of the individual isoforms were weighted by the average intensity of the Cy3-labeled standard spot.
For spot identification, gels were silver stained according to Carpentier et al. (2005). Excised gel pieces were destained with 30 mm K3Fe(CN)6/100 mm Na2S2O3, washed with water, and dried under vacuum. Gel pieces were incubated with 10 mm DTT in 50 mm NH4HCO3 for 1 h at 56°C, then with 5 mm iodoacetamide in 50 mm NH4HCO3 at room temperature in the dark for 45 min, before they were washed with 50 mm NH4HCO3 in 50% (v/v) CH3CN and dried under vacuum. Protein was digested with 12.5 ng μL−1 trypsin in 50 mm NH4HCO3 at 37°C overnight, and peptides were extracted with 50% (v/v) CH3CN in 5% (v/v) formic acid.
Liquid chromatography-MS/MS experiments were performed using the NanoLC-1D Plus (Eksigent Technologies) HPLC system and an LTQ Orbitrap mass spectrometer (ThermoFisher). Separation of peptides was performed by reverse-phase chromatography at a flow rate of 300 nL min−1 and an LC-Packings (Dionex) PepMap 100 column (C18; 75 μm i.d. × 150 mm, 3-μm particle size). Peptides were loaded onto a precolumn (Dionex Acclaim PepMap 100 C18; 5-μm particle size, 100 A, 300 μm i.d. × 5 mm) from the autosampler with 0.1% (v/v) formic acid for 5 min at a flow rate of 10 μL min−1. After this period, the 10-port valve was switched to allow elution of peptides from the precolumn onto the analytical column. Solvent A was water + 0.1% (v/v) formic acid and solvent B was acetonitrile + 0.1% (w/v) formic acid. The gradient employed was 5% to 50% (v/v) B in 40 min. The liquid chromatography eluent was sprayed into the mass spectrometer by means of a New Objective nanospray source. All mass-to-charge ratio values of eluting ions were measured in the Orbitrap mass analyzer, set at a resolution of 7,500. Peptide ions with charge states of 2+ and 3+ were then isolated and fragmented in the LTQ linear ion trap by collision-induced dissociation, and MS/MS spectra were acquired. Postrun, the data were processed using Bioworks Browser (version 3.3.1 SP1; ThermoFisher) and submitted to the Mascot Daemon 2.2.0 search algorithm (Matrix Science; Perkins et al., 1999) to search against the proteins predicted from assembly version 4.0 of the C. reinhardtii genome sequence (Merchant et al., 2007), using a fixed modification of carbamidomethylation and a variable modification of oxidation. A protein was considered identified if the highest Mascot score was above the background composed of the remaining hits (Supplemental Table S1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic diagram of the reporter constructs used in this study.
Supplemental Figure S2. B12 dose response of pMS4-1 and pMS4-2 to different concentrations of vitamin B12.
Supplemental Figure S3. RT-PCR reveals that METE expression is not repressed by B12 in the thermophilic red alga C. merolae.
Supplemental Figure S4. RT-PCR cycle number optimization for METE and Elongation factor1a expression in C. merolae.
Supplemental Figure S5. Investigating B12-responsive genes in C. reinhardtii by 2D-DIGE.
Supplemental Table S1. Identification of proteins from 2D-DIGE.
Supplemental Table S2. Expression ratio of identified spots from 2D-DIGE.
Supplemental Table S3. Characteristics of proteins involved in C1 metabolism in C. reinhardtii down-regulated by B12.
Supplemental Table S4. Strains, origins, and growth conditions for each alga grown during this study.
Supplemental Table S5. Primers and conditions used for RT-PCR.
Supplemental Table S6. PCR primers employed for the amplification of DNA fragments for reporter construct generation.
Supplementary Material
Acknowledgments
We thank Roger Sloboda (Department of Biological Sciences, Dartmouth College) for providing the METE antibody, and Dr. Elena Kazamia and Trinh Nguyen (University of Cambridge) and Dr. Glen Wheeler (Plymouth Marine Laboratory/Marine Biological Association of the United Kingdom) for helpful discussions.
Glossary
- UTR
untranslated region
- RT
reverse transcription
- qPCR
quantitative real-time PCR
- cDNA
complementary DNA
- 2D-DIGE
two-dimensional difference gel electrophoresis
- MS/MS
tandem mass spectrometry
- DTT
dithiothreitol
- TAP
Tris-acetate phosphate
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (grant no. BB/I013164/1), the Swiss National Science Foundation (grant nos. PBEZA–115703 and PA00P3–124169 to S.S.), and the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico, Brazil (to A.P.U.A.)
The online version of this article contains Web-only data.
References
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp 28–36 [PubMed] [Google Scholar]
- Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar TM, Bai J, Valas RE, Saito MA. (2012) Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci USA 109: E1762–E1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, McIlvin MR, Hoffman JM, Allen AE, Saito MA. (2013) Methionine synthase interreplacement in diatom cultures and communities: implications for the persistence of B12 use by eukaryotic phytoplankton. J Limnol Ocean 58: 1431–1450 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cai XY, Maxon ME, Redfield B, Glass R, Brot N, Weissbach H. (1989) Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc Natl Acad Sci USA 86: 4407–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci AF. (1970) The ecology of the plankton off La Jolla, California in the period April through September. II. Vitamin B12, thiamine and biotin. Bull Scripp Ins Ocean 17: 23–30 [Google Scholar]
- Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B. (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5: 2497–2507 [DOI] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. (2005) Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93 [DOI] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. (2007) Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA 104: 20770–20775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Warren MJ, Smith AG. (2006) Algae need their vitamins. Eukaryot Cell 5: 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubini A, Mus F, Seibert M, Grossman AR, Posewitz MC. (2009) Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J Biol Chem 284: 7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gillet S, Decottignies P, Chardonnet S, Le Maréchal P. (2006) Cadmium response and redoxin targets in Chlamydomonas reinhardtii: a proteomic approach. Photosynth Res 89: 201–211 [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ. (2012) Vitamins in the sea. Proc Natl Acad Sci USA 109: 13888–13889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, Norman C, Panzeca C, Taylor GT, Sanudo-Wilhelmy SA. (2007) Effect of B-vitamins (B-1, B-12) and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat Microb Ecol 49: 181–194 [Google Scholar]
- González JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. (1992) Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry 31: 6045–6056 [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. (2011) Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28: 2921–2933 [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Smith AG. (2013) Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet 29: 469–478 [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrázek J, Campbell A, Kaiser D. (2001) Characterizations of highly expressed genes of four fast-growing bacteria. J Bacteriol 183: 5025–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazamia E, Czesnick H, Nguyen TT, Croft MT, Sherwood E, Sasso S, Hodson SJ, Warren MJ, Smith AG. (2012) Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 14: 1466–1476 [DOI] [PubMed] [Google Scholar]
- Kurata A (1986) Blooms of Uroglena americana in relation to concentrations of B group vitamins. In J Kristiansen, RA Andersen, eds, Chrysophytes: Aspects and Problems. Cambridge University Press, Cambridge, UK, pp 185–196 [Google Scholar]
- Kurvari V, Qian F, Snell WJ. (1995) Increased transcript levels of a methionine synthase during adhesion-induced activation of Chlamydomonas reinhardtii gametes. Plant Mol Biol 29: 1235–1252 [DOI] [PubMed] [Google Scholar]
- Lilley KS, Friedman DB. (2004) All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics 1: 401–409 [DOI] [PubMed] [Google Scholar]
- Martin C, Butelli E, Petroni K, Tonelli C. (2011) How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Maxon ME, Redfield B, Cai XY, Shoeman R, Fujita K, Fisher W, Stauffer G, Weissbach H, Brot N. (1989) Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc Natl Acad Sci USA 86: 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner L, Whitfield C, Weissbach H. (1969) Effect of L-methionine and vitamin B12 on methionine biosynthesis in Escherichia coli. Arch Biochem Biophys 133: 413–419 [DOI] [PubMed] [Google Scholar]
- Moscatelli A, Scali M, Prescianotto-Baschong C, Ferro M, Garin J, Vignani R, Ciampolini F, Cresti M. (2005) A methionine synthase homolog is associated with secretory vesicles in tobacco pollen tubes. Planta 221: 776–789 [DOI] [PubMed] [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR. (2004) Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltean S, Banerjee R. (2003) Nutritional modulation of gene expression and homocysteine utilization by vitamin B12. J Biol Chem 278: 20778–20784 [DOI] [PubMed] [Google Scholar]
- Oltean S, Banerjee R. (2005) A B12-responsive internal ribosome entry site (IRES) element in human methionine synthase. J Biol Chem 280: 32662–32668 [DOI] [PubMed] [Google Scholar]
- Panzeca C, Beck AJ, Tovar-Sanchez A, Segovia-Zavala J, Taylor G, Gobler CJ. (2009) Distributions of dissolved vitamin B12 and Co in coastal and open-ocean environments. Estuar Coast Shelf Sci 85: 223–230 [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Plener L, Boistard P, González A, Boucher C, Genin S. (2012) Metabolic adaptation of Ralstonia solanacearum during plant infection: a methionine biosynthesis case study. PLoS ONE 7: e36877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix JD. (2013) Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA, Prokopenko MG, Berelson WM, Karl DM. (2012) Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA 109: 14041–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MJ, Ulland M, Sloboda RD. (2008) A protein methylation pathway in Chlamydomonas flagella is active during flagellar resorption. Mol Biol Cell 19: 4319–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M. (2006) RNA silencing in Chlamydomonas: mechanisms and tools. Curr Genet 49: 69–84 [DOI] [PubMed] [Google Scholar]
- Scott J, Weir D. (1994) Folate/vitamin B12 inter-relationships. Essays Biochem 28: 63–72 [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C. (2007) Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23–35 [DOI] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. (1993) Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YZ, Koch F, Gobler CJ. (2010) Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci USA 107: 20756–20761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unlü M, Morgan ME, Minden JS. (1997) Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18: 2071–2077 [DOI] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer GV. (1989a) Role of homocysteine in metR-mediated activation of the metE and metH genes in Salmonella typhimurium and Escherichia coli. J Bacteriol 171: 3277–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer GV. (1989b) Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J Bacteriol 171: 5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, Cypionka H, Daniel R, Drepper T, Gerdts G, et al. (2010) The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J 4: 61–77 [DOI] [PubMed] [Google Scholar]
- Warner DF, Savvi S, Mizrahi V, Dawes SS. (2007) A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J Bacteriol 189: 3655–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach H, Brot N. (1991) Regulation of methionine synthesis in Escherichia coli. Mol Microbiol 5: 1593–1597 [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956 [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Xie B, Bishop S, Stessman D, Wright D, Spalding MH, Halverson LJ. (2013) Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J 7: 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.