The Chlamydomonas reinhardtii pgr5 mutant shows photosynthetic and phenotypic traits that support the conservation of PGR5 in both redox-controlled cyclic electron flow and PSI photoprotection.
Abstract
The Chlamydomonas reinhardtii proton gradient regulation5 (Crpgr5) mutant shows phenotypic and functional traits similar to mutants in the Arabidopsis (Arabidopsis thaliana) ortholog, Atpgr5, providing strong evidence for conservation of PGR5-mediated cyclic electron flow (CEF). Comparing the Crpgr5 mutant with the wild type, we discriminate two pathways for CEF and determine their maximum electron flow rates. The PGR5/proton gradient regulation-like1 (PGRL1) ferredoxin (Fd) pathway, involved in recycling excess reductant to increase ATP synthesis, may be controlled by extreme photosystem I acceptor side limitation or ATP depletion. Here, we show that PGR5/PGRL1-Fd CEF functions in accordance with an ATP/redox control model. In the absence of Rubisco and PGR5, a sustained electron flow is maintained with molecular oxygen instead of carbon dioxide serving as the terminal electron acceptor. When photosynthetic control is decreased, compensatory alternative pathways can take the full load of linear electron flow. In the case of the ATP synthase pgr5 double mutant, a decrease in photosensitivity is observed compared with the single ATPase-less mutant that we assign to a decreased proton motive force. Altogether, our results suggest that PGR5/PGRL1-Fd CEF is most required under conditions when Fd becomes overreduced and photosystem I is subjected to photoinhibition. CEF is not a valve; it only recycles electrons, but in doing so, it generates a proton motive force that controls the rate of photosynthesis. The conditions where the PGR5 pathway is most required may vary in photosynthetic organisms like C. reinhardtii from anoxia to high light to limitations imposed at the level of carbon dioxide fixation.
Photosynthesis is a highly regulated process that integrates different electron transfer pathways to convert light energy into ATP and NADPH and balance this production of chemical energy with its use in anabolic metabolism. Linear electron flow accounts for the major flux of electrons from the primary electron donor water to PSII and intersystem carriers to reduce NADP+, the terminal acceptor associated with PSI. Electron transfer is coupled to proton transfer through reactions involving plastoquinones/plastoquinols that are dependent on the activity of the cytochrome b6f complex (cyt f); the protons are transferred from the stroma into the thylakoid lumen. The proton motive force generated is used for ATP synthesis by the ATP synthase. The NADPH and ATP produced in the light serve as the energy/reductant that drives the fixation of carbon dioxide (CO2) by Rubisco and the Calvin-Benson cycle and also supports other downstream metabolic reactions.
The Calvin-Benson cycle has a stoichiometric requirement of 3 ATP and 2 NADPH per CO2 molecule; this requirement is not fulfilled by linear electron flow, because it is slightly imbalanced in favor of NADPH production. Cyclic electron flow (CEF) pathways allow the cells to fulfill the energetic requirement for sustained CO2 fixation through recycling or reoxidation of NADPH and/or reduced ferredoxin (Fd) through the plastoquinone (PQ) pool, the cyt f, and PSI; this CEF pathway increases the transmembrane proton gradient that, in turn, allows for increased synthesis of ATP (Kramer et al., 2004). The enzymes involved in PQ reduction can vary according to the organism and the environmental conditions. The cyanobacteria Synechocystis spp. may have two direct and two indirect pathways for CEF (Jeanjean et al., 1999), whereas there are only two known pathways in vascular plants and green algae (Chlamydomonas reinhardtii): proton gradient regulation5 (PGR5) dependent and NADPH dehydrogenase (NDH) dependent. Vascular plants have a multisubunit type I NDH that is active in chloroplast thylakoids and also reduces plastoquinone in an Fd-dependent manner (Burrows et al., 1998; Kofer et al., 1998; Shikanai et al., 1998; Yamamoto et al., 2011), whereas in green algae, the type II NADPH dehydrogenase2, NDA2, has been implicated in electron recycling through NADPH (Jans et al., 2008). Arabidopsis (Arabidopsis thaliana) mutants devoid of both PGR5 and Chlororespiratory reduction pathways (pgr5 crr-1) can only sustain very poor photosynthetic growth (Munekage et al., 2004), illustrating the requirement of these pathways for efficient photosynthesis.
Other alternative electron transfer pathways fulfill roles for supplementary ATP synthesis and redox balancing and have been shown to function as photoprotective electron valves. Most of these alternative pathways use molecular oxygen (O2) as a terminal electron acceptor, and their prevalence is even more species dependent than CEF. The plastoquinol terminal oxidase (PTOX) is active in chlororespiration to balance dark chloroplast redox poise and can also work as a valve under specific conditions to oxidize plastoquinols while reducing oxygen when the PQ pool is overreduced (Bailey et al., 2008; Houille-Vernes et al., 2011). The Mehler reaction describes a water–water cycle in which electrons generated by the splitting of water at the donor side of PSII are used to reduce O2 at the acceptor side of PSI. This reaction produces superoxide radicals that can be detoxified through superoxide dismutase, which generates hydrogen peroxide, and ascorbate peroxidase, which converts the hydrogen peroxide to water and O2 (Mehler, 1951; Radmer and Kok, 1976; Badger et al., 2000). Chloroplast-reducing power can also be transferred to mitochondria through C3 or C4 carbon intermediates to fuel oxidative phosphorylation, which represents another O2 consuming reaction. This trafficking of reductant can be mediated by the exchange of malate/oxaloacetate (C4), where oxaloacetate is reduced to malate in the chloroplast through NADP-malate dehydrogenase, with the reverse reaction producing NADH in mitochondria; this pathway is referred to as the malate valve (Krömer and Scheibe, 1996). Export of reducing equivalents can also occur through the triose-phosphate translocator (Heldt and Rapley, 1970), which facilitates shuttling of the Calvin-Benson cycle intermediates, glyceraldehyde-3-P and 3-phosphoglycerate. Although also coupled to ATP, triose-phosphate translocator reactions are considered a shunt, because they consume more NADPH per ATP relative to that produced by linear electron flow. Plants and algae can also perform photorespiration in which O2 instead of CO2 is used by Rubisco to catalyze the formation of glycolate, which is shuttled to different cellular compartments, although this pathway is considered minor in green algae because of the intracellular accumulation of HCO3−/CO2 as a consequence of the carbon-concentrating mechanism (Badger et al., 2000).
The molecular identification of the PGR5 pathway was the result of a screen for mutants of Arabidopsis that exhibited a reduced flux of protons across the thylakoid membranes (Munekage et al., 2002). This pathway was shown to contribute to the production of an elevated thylakoid ΔpH (hence its name), which induces the thermal dissipation component (qE; E for energy dependent) of nonphotochemical quenching (NPQ). This mode of energy dissipation is proposed to be relevant when Calvin-Benson cycle activity is limiting (Heber and Walker 1992; Ruban et al., 1993; Niyogi et al., 1998; Li et al., 2000). PGR5 was shown to be an intermediate in the transfer of electrons from Fd to the PQ pool (Munekage et al., 2002); thus, it is considered the Fd-dependent pathway for CEF, which was proposed earlier by Arnon (Tagawa et al., 1963). Physiologically, the PGR5 pathway was proposed to function in limiting overreduction of electron carriers on the acceptor side of PSI, thus preventing PSI photoinhibition (Munekage et al., 2002). More recent work with Arabidopsis has shown that, under fluctuating light, PGR5 is essential for early developmental growth, because it functions in photoprotection of PSI (Suorsa et al., 2012). This study and previous studies (Avenson et al., 2005; Joliot and Johnson, 2011) suggest that the PGR5 pathway operates as a regulator of linear electron flow.
Proton gradient regulation-like1 (PGRL1) was shown to be coregulated with PGR5 in Arabidopsis and proposed to be in the same pathway (DalCorso et al., 2008). PGRL1 is an integral thylakoid membrane protein with both the N and C termini of the mature protein exposed to the stroma. Based on split ubiquitin assays, it has been shown to interact with PGR5, PSI subunits, Fd-NADPH reductase, and the cyt f; interestingly, PGR5 protein is absent in the pgrl1 mutant line (DalCorso et al., 2008). In Arabidopsis, PGR5 interacts with N- and C-terminal Cys residues of PGRL1, which are located on the stromal side of the thylakoid membranes (Hertle et al., 2013). In vitro, PGRL1 can reduce plastoquinone analogs when supplied with reduced Fd (Hertle et al., 2013). Reducing conditions can activate PGR5/PGRL1-Fd-dependent CEF, which might be mediated by an m-type thioredoxin and depend on a complex interplay of inhibition or activation of NDH and PGR5/PGRL1-Fd CEF pathways as well as regulation of the Calvin-Benson cycle enzymes (Courteille et al., 2013; Hertle et al., 2013).
In the green Chlorophyte alga, C. reinhardtii, supercomplex formation between PGRL1 and PSI-LHCI (for Light-Harvesting Complex)-LHCII-Fd-NADPH reductase-cytochrome b6f (Iwai et al., 2010; Takahashi et al., 2013) as well as the Ca2+ sensor protein and the protein Anaerobic Response1 (ANR1; Terashima et al., 2012) is promoted under anoxic conditions. Anoxia reduces the redox poise of the stroma, which has been shown to enhance CEF [measured in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU)] in the wild-type line but not pgrl1 mutant cells (Tolleter et al., 2011). Furthermore, pgrl1 knockdown lines exhibited hypersensitivity to iron deficiency, linking iron limitation to formation/remodeling of the supercomplex associated with CEF (Petroutsos et al., 2009). Here, it was also shown that conformational changes occur to the PGRL1 protein in relation to the redox state and the presence and absence of iron ions, suggesting that the protein binds iron.
Both the C. reinhardtii PGRL1 and PGR5 genes show transcript coaccumulation and coregulation under iron deficiency (Petroutsos et al., 2009) and in response to a number of environmental stimuli as revealed by open access transcriptomic data (http://genomes.mcdb.ucla.edu/cgi-bin/hgGateway). The pgrl1 knockout mutant was isolated in a forward screen based on its remarkable chlorophyll fluorescence phenotype (Tolleter et al., 2011). The fluorescence kinetics over a 2-min light exposure could be separated into two phases: a lack of transient quenching of PSII fluorescence and an increase in PSII yield (ϕPSII). As in Arabidopsis, a decrease in NPQ was associated with a decreased proton gradient, a consequence of the absence of PGR5/PGRL1-Fd–mediated CEF. Furthermore, this decrease in the proton gradient in the mutant resulted in an increased ϕPSII.
There has been uncertainty about the involvement of PGR5 in algal CEF (see comment in Leister and Shikanai, 2013) because of the lack of PGR5 protein in the C. reinhardtii PSIb6f supercomplexes when analyzed by either western blots and/or mass spectroscopy (Iwai et al., 2010; Terashima et al., 2012). Indeed, according to Hertle et al. (2013), in Arabidopsis, PGR5 is a low-abundance protein, approximately 7 times less abundant than its binding partner PGRL1, with PGRL1 being one-half as abundant as cyt f. A C. reinhardtii pgr5 mutant (the putative protein is orthologous to Arabidopsis PGR5) was isolated in a large screen for identifying mutants defective in their responses to high light (Dent et al., 2005). The PGR5 protein is also included in the GreenCut set of proteins derived from phylogenomic analysis, many of which are likely to be involved in chloroplast function and photosynthesis (Merchant et al., 2007; Grossman et al., 2010; Karpowicz et al., 2011; Heinnickel et al., 2013). Here, we report that the functional attributes of the C. reinhardtii pgr5 mutant are congruent with those attributes observed for the analogous mutant in vascular plants as well as those attributes of the Crpgrl1 mutant. Furthermore, we combine the pgr5 mutation with other defects to the photosynthetic apparatus to dissect the contribution of PGR5/PGRL1-Fd CEF to the regulation of photosynthesis in C. reinhardtii and explore the activities of alternative routes of electron flow in the absence of PGR5/PGRL1-Fd CEF.
RESULTS
C. reinhardtii PGR5 Participates in NPQ and CEF
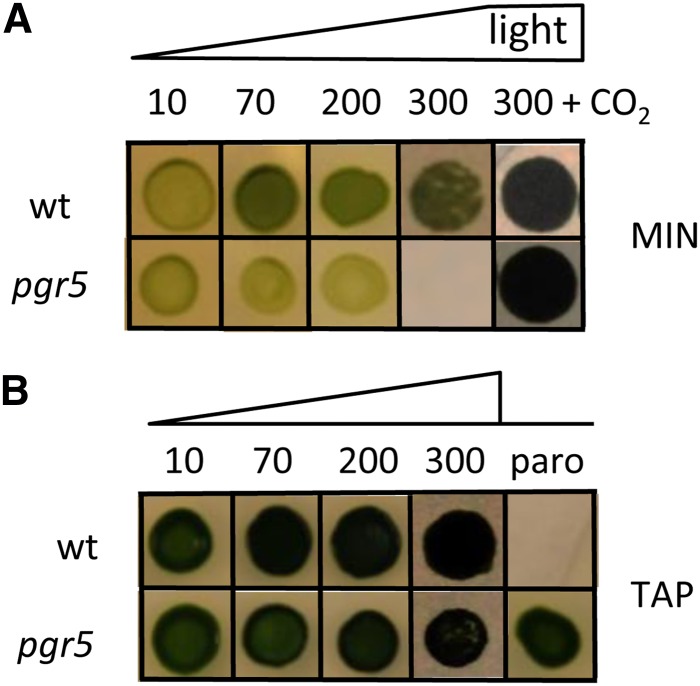
The pgr5 mutant examined in this study was generated by DNA insertional mutagenesis (Dent et al., 2005) with linearized plasmid encoding paromomycin resistance (Tran et al., 2012) and designated CAL028.01.15. The mutant contains an aminoglycoside 3'-phosphotransferase gene, APHVIII insertion, identified by PCR, that was 3,250 bp upstream of the putative PGR5 coding region (Cre05.g242400; 55% amino acid sequence identity with AtPGR5) and is on chromosome 5 in the most recent version of the C. reinhardtii genome, V5.3.1, and Phytozome 9.1. Furthermore, the insertion of the resistance gene caused a deletion that resulted in the loss of the PGR5 coding region, which was verified by PCR of the mutant genomic DNA (Supplemental Fig. S1A). Initial screening showed that the mutant is sensitive to metronidazole and high light on minimal medium, which we confirm here (Fig. 1).
Figure 1.
Comparisons of growth of pgr5 and wild-type (wt) line strains at various light intensities (as indicated in micromoles photons minute−2 second−1) on MIN (A) and acetate-containing medium (TAP; B). Phototrophic growth of pgr5 is impaired in moderate/high light, but growth can be restored under elevated CO2. The presence of the APHVIII insert in pgr5 makes it resistant to the antibiotic paromomycin (paro). [See online article for color version of this figure.]
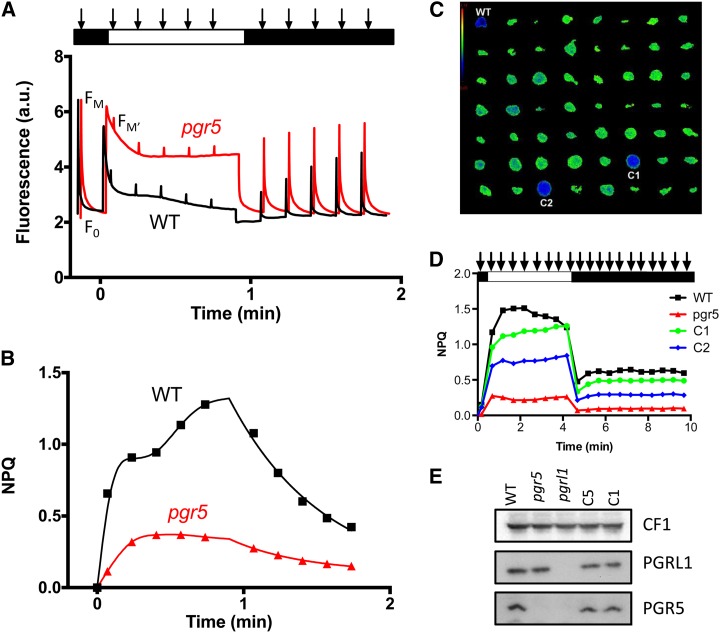
When C. reinhardtii cells were grown under phototrophic conditions (i.e. in minimal medium and >200 μmol photons m−2 s−1 light), wild-type cells exhibited the qE component of NPQ (Peers et al., 2009), which is shown in Figure 2. Figure 2A shows that the dark-adapted minimum fluorescence yield (F0) and the maximum fluorescence yield in the dark-adapted state (Fm) as measured by a saturating light pulse (Fig. 2A, first arrow) were the same in the wild-type line (Fig. 2A, black) and the pgr5 mutant (Fig. 2A, red). However, when the cells were exposed to 1,500 μmol photons m−2 s−1 of light during the analysis (white rectangle in Fig. 2A), the normal level of NPQ = (Fm − maximum fluorescence yield in the light-adapted state [Fm′])/Fm′ observed in the wild-type cells was severely compromised in the pgr5 mutant (compare the black and red curves in Fig. 2, A and B). The NPQ in the pgr5 mutant was reduced to approximately 30% of the wild-type line level. A similar finding was reported for the pgr5 mutant in Arabidopsis (Munekage et al., 2002) as well as the Atpgrl1 (DalCorso et al., 2008; Hertle et al., 2013) and Crpgrl1 (Tolleter et al., 2011) mutants. The fluorescence quenching elicited by high light quickly reversed in the dark in both the wild-type line and the pgr5 mutant, which is normally observed for the energy-dependent or pH-dependent qE component of NPQ.
Figure 2.
A, Chlorophyll fluorescence kinetics show that the C. reinhardtii pgr5 mutant shows diminished NPQ of chlorophyll fluorescence calculated in B from the data presented in A. The pgr5 mutant was grown in liquid MIN medium at 200 μmol photons m−2 s−1 light intensity. The minimum (F0) and maximum (Fm) dark fluorescence yields measured by a saturating flash (denoted by arrows) are the same in the wild-type (WT) line and pgr5 mutant; however, when exposed to very high light (1,500 μmol photons m−2 s−1), WT line cells undergo a fluorescence quenching that is severely compromised in the pgr5 mutant. This type of quenching quickly recovers in the WT line strain, which is evidenced by the dark recovery of fluorescence yield. C, Image of chlorophyll fluorescence from clones restreaked on solid minimum medium at 200 μmol photons m−2 s−1 of light calculated as specified in the text. The pgr5 clones show a higher fluorescence because of a lack of quenching (green spots) compared with WT line cells, and functionally complemented clones (blue spots) show a lower fluorescence under the same conditions. D, NPQ analysis for cultures preacclimated to 200 μmol photons m−2 s−1. Two of the complemented strains, C1 and C2, are compared with the impaired NPQ in pgr5 and normal NPQ in the WT line. E, Detection of PGR5 and PGRL1 proteins in thylakoids of the WT line, pgr5, pgrl1, and pgr5 complemented lines, C5, and C1 grown in TAP medium at <10 μmol photons m−2 s−1 light. [See online article for color version of this figure.]
To verify that this altered NPQ phenotype was a consequence of the absence of PGR5, genetic backcrosses with the wild-type line and rescue of the mutant phenotype with the wild-type PGR5 gene were performed. Genetic and phenotypic analyses showed that antibiotic resistance and disruption of PGR5 with the drug resistance cassette (based on PCR amplifications) cosegregated with the aberrant fluorescence phenotype. For consistency and ease of manipulation, genetic analyses were done on acetate-containing medium under low light, conditions that are often used to maintain nonphotosynthetic or light-sensitive mutants (see ΔrbcL and ΔATPase below). Under such conditions, C. reinhardtii cells do not show significant NPQ (Peers et al., 2009); however, chlorophyll fluorescence kinetics were still very different for pgr5 and the wild-type line. The chlorophyll fluorescence kinetics of pgr5 grown in low light in acetate-containing medium were similar to the kinetics reported for pgrl1 (Tolleter et al., 2011), showing a lack of a transient rise in fluorescence after 30 s of illumination (Supplemental Fig. S1B). The pgr5 fluorescence phenotype, which is seen in Supplemental Figure S1B, was always observable regardless of whether the cells were grown in the dark, in the light on acetate, or under photoautotrophic conditions, which suggests that the PGR5 pathway is constitutively active.
Rescue of the NPQ fluorescence phenotype (qE component) by introduction of the wild-type PGR5 gene into the pgr5 mutant was used to screen for complementation of the mutant phenotype. Mutant cells were cotransformed with the PGR5 coding region under the control of its own promoter and a spectinomycin resistance cassette. Spectinomycin-resistant transformants were selected and then screened for a restored qE component after spotting the transformants on minimal solid medium, growing the cells at 200 μmol photons m−2 s−1, and then, after 1 h of acclimation in the dark, monitoring chlorophyll fluorescence quenching using a video imaging camera (Johnson et al., 2009). Figure 2C shows the ratio between two images Fm′ (at t = 1 min) over Fm. The pgr5 clones show elevated fluorescence resulting from their defect in qE (Fig. 2C, green spots). In contrast, the wild-type line and complemented strains show lower fluorescence under the same conditions (Fig. 2C, blue spots). Of 700 resistant clones screened, 10 clones showed resistance to spectinomycin and paramomycin as well as restored qE. The intact PGR5 gene could also be amplified using genomic DNA templates isolated from these strains. Although the number of rescued strains among the transformants seems low, it does not reflect cotransformation efficiency (which is usually 2%–5%) but rather, the functional complementation efficiency. A detailed NPQ analysis for two of the complemented strains is shown in Figure 2D. In these strains, the impaired NPQ phenotype of pgr5 is rescued, with C1 showing essentially full rescue and C2 showing partial rescue. Additionally, the complemented strains C1 and C5 were tested for accumulation of PGR5 protein by western-blot analysis using the AtPGR5 antibody. A protein of the expected size (approximately 9 kD) was identified in the thylakoids of the wild-type line and the two complemented lines but not the original pgr5 mutant or the Crpgrl1 mutant (Fig. 2E), the latter dependence for coaccumulation having already been observed in Arabidopsis (DalCorso et al., 2008). Interestingly, the pgr5 mutant grown under these conditions accumulated wild-type levels of the PGRL1 protein, like the two complemented lines analyzed.
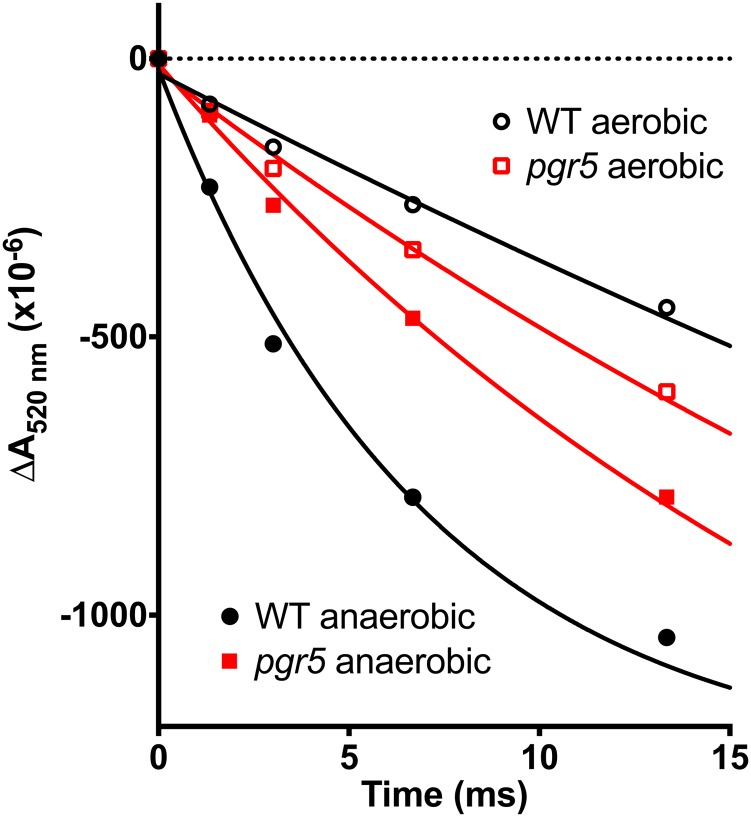
A diminished qE associated with pgr5 or pgrl1 mutations was previously reported (Munekage et al., 2002; Joliot and Johnson, 2011; Tolleter et al., 2011; Leister and Shikanai, 2013). It results from a decreased proton gradient in the light, itself caused by the lack of PGR5/PGRL1-Fd CEF. Similar to the Crpgrl1 study (Tolleter et al., 2011) but for a more detailed analysis and methodological description (Alric, 2014), we measured the rates of CEF in the absence of active PSII. Wild-type and pgr5 mutant cells were grown in acetate-containing medium in low light (<10 μmol photons m−2 s−1) and treated with DCMU and hydroxylamine to inhibit PSII. Under such conditions, only PSI is active, and any sustained electron flow in the light is attributable to CEF around PSI, the membrane potential being exclusively generated by CEF. The electrochromic shift (ECS) of carotenoids, which is induced by changes in the membrane potential, was measured at 520 nm. After an exposure of 5 s to 1,000 μmol photons m−2 s−1 of light, a steady-state signal was attained that reflects equilibrium between the generation of a proton motive force by CEF and the dissipation of this proton motive force through the activity of the ATP synthase. This steady-state level is arbitrarily shifted to zero in Figure 3 (dotted line). After the light is extinguished, the membrane potential decays in the dark, which is shown in Figure 3, at a rate that is equal to that of CEF in the light. The flow rate determined from the dark relaxation kinetic is given in charges (electrons or protons) transferred per second per PSI reaction center, with the amplitude of one electron per PSI determined by the absorbance change induced after a single turnover saturating flash (one charge separation); in this instance, the value is approximately 3 × 10−3. In aerobic conditions, in the presence of DCMU, the rates of CEF were very similar in the wild-type line and pgr5 cells at 9.5 ± 3.3 s−1 and 11.5 ± 4.8 s−1 (Table I), which supports the suggestion that the NADPH-mediated CEF is the dominant pathway for recycling reducing equivalents under these conditions (for review, see Alric, 2010). Under anaerobic conditions, there was a much greater difference in the CEF between the wild-type line (60.6 ± 9.2 s−1) and pgr5 mutant (16.3 ± 6.3 s−1) cells. As a verification, using the primary electron donor of PSI, measurements of P700 similar to those reported in the work by Takahashi et al. (2013), we have checked that, in such anaerobic conditions, the slower CEF does not stem from a more pronounced acceptor side limitation in the pgr5 mutant with respect to the control strain.
Figure 3.
Measurement of CEF rates after the ECS of carotenoids at 520 nm. Cells were grown in TAP and low light and treated with 10 μm DCMU and 1 mm hydroxylamine to inhibit PSII. Samples were adapted to aerobic (black and red symbols) or anaerobic (white symbols) conditions and preilluminated for 5 s with a saturating orange light delivering 1,000 μmol photons m−2 s−1. Flow rates were calculated after the cessation of illumination for more than three biological replicates and reported in Table I. One electron per PSI was calibrated with the ECS induced by a single turnover saturating flash. WT, Wild type. [See online article for color version of this figure.]
Table I. Rates of CEF measured in the wild-type line and pgr5 strains placed under aerobic or anaerobic conditions and treated with 10 μm DCMU and 1 mm hydroxylamine (typical experiments are shown in Fig. 3).
Additional details are in “Materials and Methods.” Means ± sd (n = 3) for biological replicates.
| Conditions | Wild-Type Line | pgr5 |
|---|---|---|
|
e− s−1 PSI−1 |
||
| Aerobic | 9.5 ± 3.3 | 11.5 ± 4.8 |
| Anaerobic | 60.6 ± 9.2 | 16.3 ± 6.3 |
These results support the hypothesis that the redox state of the cells controls CEF, which was previously suggested: when stromal redox carriers are reduced, which is the case under light anaerobic conditions, Fd-mediated CEF is strongly activated (Kessler, 1973; Alric, 2010; Alric et al., 2010; Terashima et al., 2012; Takahashi et al., 2013). It also shows that, like PGRL1 (Petroutsos et al., 2009; Tolleter et al., 2011), PGR5 is involved in CEF around PSI in C. reinhardtii. This analysis also provides an estimate of the maximum capacity of pathways other than the PGR5/PGRL1-Fd pathway that include the NDH pathway, which is 15 to 20 electrons (e−) s−1 PSI−1 (measured in anaerobic pgr5 cells), as well as an estimate of the additional contribution of the PGR5/PGRL1-Fd pathway, which is 40 to 45 e− s−1 PSI−1, with a total of approximately 60 e− s−1 PSI−1 in the anaerobic wild-type line cells.
PGR5 Protects PSI under Conditions Where Photosynthesis Is Acceptor Side Limited
Figure 1A shows that an absence of PGR5 results in reduced photosynthetic growth and ultimately, light sensitivity when the cells are grown on minimal medium in air. When the atmosphere is enriched with 2% CO2 or the medium is supplemented with acetate, the photosensitivity of pgr5 is relieved, which is shown in Figure 1B; growth of the mutant and wild-type lines is similar at a light intensity of 300 μmol photons m−2 s−1.
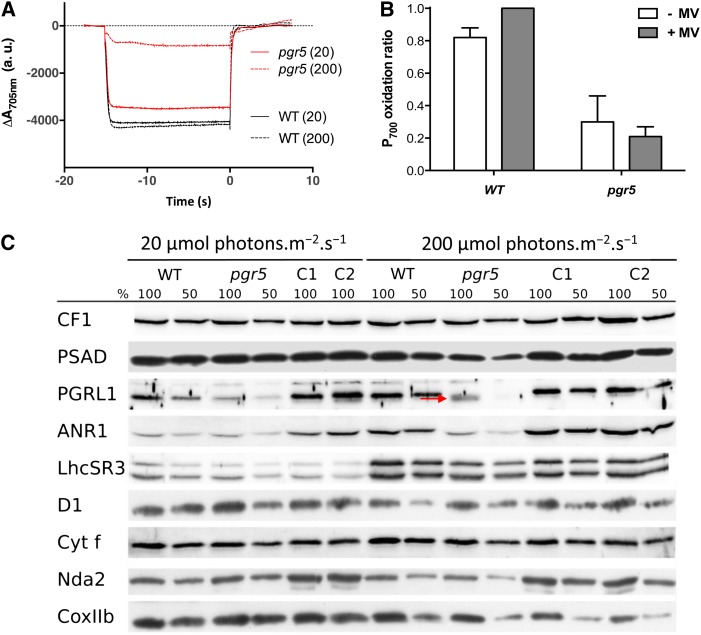
To determine if pgr5 photosensitivity is a consequence of reduced electron flow through PSI, the mutant and wild-type cells were grown in Tris-acetate phosphate (TAP), and then, they were resuspended and incubated for 24 h in liquid minimal medium at light intensities of 20 and 200 μmol photons m−2 s−1. In the presence of DCMU, the fraction of active PSI was measured at 705 nm as the relative amount of P700 (primary electron donor of PSI) that can be oxidized to P700+. At low-light intensities, the amount of oxidizable P700 was almost equal in the wild-type line and the pgr5 mutant strain (Fig. 4A), with a rereduction rate of 6.56 (±1.45) e−1 s−1 (Supplemental Fig. S2C), which is typical for CEF in such conditions (Alric et al., 2010). However, as also shown in Figure 4A, the quantity of oxidizable P700 was severely reduced in the pgr5 mutant compared with the wild-type line after growth in moderate/high light. In contrast to findings for Arabidopsis (Munekage et al., 2002), there was little change in the signal for P700+ in the pgr5 mutant, even after the addition of methylviologen, an efficient PSI electron acceptor (Fig. 4B). These results suggest that PSI photochemistry is severely reduced, not only because of a limitation of electron transfer at the acceptor side of PSI caused by the low CO2 levels in air but also, because the PSI reaction center is being photoinhibited.
Figure 4.
PSI is photoinhibited in pgr5 cells adapted to phototrophic conditions at light intensities above 20 μmol photons m−2 s−1. Cells were acclimated to low-light conditions (TAP medium at 20 μmol photons m−2 s−1 illumination) before incubation in MIN medium for 24 h at given light intensities measuring P700 oxidoreduction kinetics and protein accumulation. Under aerobic conditions, P700 can be oxidized in the presence of DCMU. Using an actinic light intensity of 3,300 μmol photons m−2 s−1 gives access to the full amount of oxidizable P700. A, P700 kinetics for the wild-type (WT) line and pgr5 at two light intensities: 20 and 200 μmol photons m−2 s−1 grown cells. B, P700 oxidation ratio (at a light intensity of 3,300 μmol photons m−2 s−1) for high-light-grown cells in the presence or absence of 1 mm methylviologen (MV) cannot restore the oxidizable portion of P700 shown to be lost in A. Histograms show mean values ± sd (n = 3). C, PSAD is reduced by 50% in 200 μmol photons m−2 s−1-adapted pgr5 compared with the WT line. Western-blot analysis on extracted whole proteins runs on denaturing SDS-PAGE. WT line and pgr5 whole-cell extracts were loaded per chlorophyll basis (100% = 2.5 μg). Filters were incubated with antibodies against CF1, PSI subunit (PSAD), CEF protein (PGRL1), supercomplex protein (ANR1), the qE-related protein (LHCSR3), PSII subunit (D1), cyt f, NDH (Nda2), and mitochondrial protein (COXIIb). [See online article for color version of this figure.]
To assess if the exposure of pgr5 to phototrophic conditions and/or high light had a specific effect on PSI or if other functions were also affected by the light treatment, we have done the western blots shown in Figure 4C. Antibodies against subunits of the major photosynthetic complexes were used for PSI (PSAD), PSII (PsbA D1), chloroplast ATPase (CF1), and cyt f. The mitochondrial cytochrome oxidase COXIIb was also probed (alternative oxidase1 [AOX1] shown in Fig. 6). To check whether the deficiency in PGR5 impacts other alternative pathways, we blotted PGRL1, ANR1, LHCSR3, and Nda2 (PTOX2 shown in Fig. 6). When grown under 20 μmol photons m−2 s−1 of light, pgr5 cells accumulated the same amount of proteins as the wild type, except for PGRL1, which was decreased by 50%. This situation contrasts with that observed in the dark, where pgr5 accumulates wild-type levels of PGRL1 (Fig. 2E). At 200 μmol photons m−2 s−1, PGRL1 amounts were further decreased in pgr5, and we noticed that the PGRL1 band was shifted downward (denoted here by a red arrow in Fig. 4), indicating a faster migration on SDS-PAGE. A similar observation was reported previously by Petroutsos et al. (2009) and attributed to a change in the redox state of PGRL1, the possible reduction of which in high-light conditions is dependent on PGR5 (Hertle et al., 2013). Similar to the result shown for dark conditions (Fig. 2E), the pgr5-complemented strain C1 (and here, C2) showed a restored accumulation of PGRL1 in both low-light and high-light conditions, further substantiating the functional link between PGR5 and PGRL1.
Figure 6.
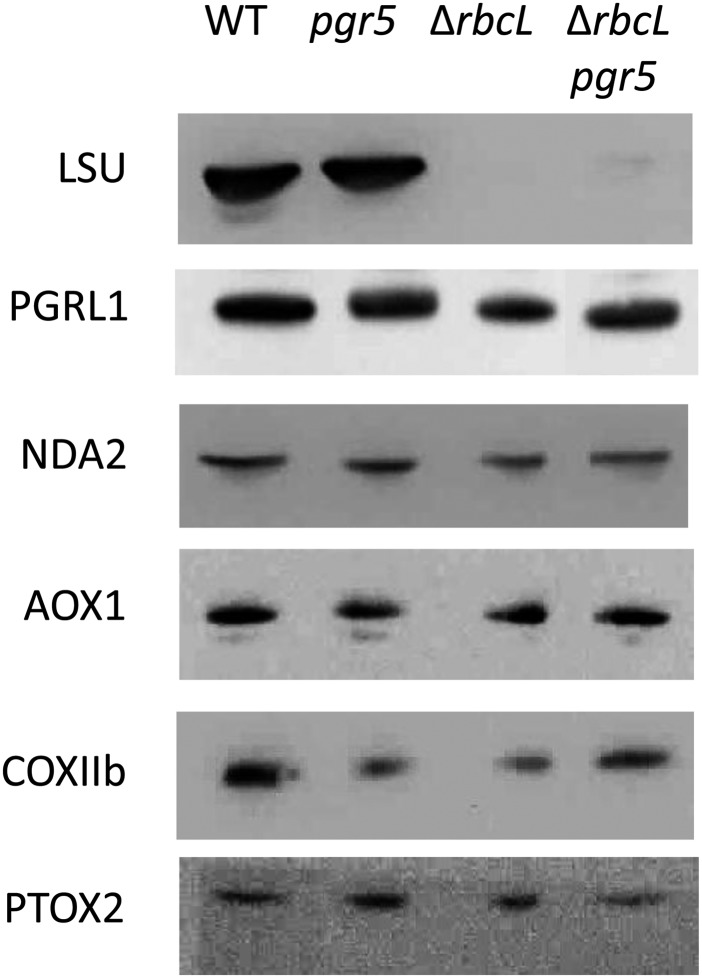
Western-blot analysis on whole-cell proteins grown in TAP medium at <10 μmol photons m−2 s−1 light shows that ΔrbcL and ΔrbcL pgr5 mutants do not accumulate Rubisco protein (Large Subunit of Rubisco [LSU]) and that no significant differences in protein accumulation can be observed for enzymes involved in the alternative pathways (PGRL1 and NDA2) and O2-consuming pathways (mitochondrial, AOX1 and COXIIb; chloroplastic, PTOX2) contemplated in Figures 5 and 7. WT, Wild type.
PGRL1 was not the only protein affected by high-light treatment. Although most of the proteins stayed almost unchanged (CF1, cyt f, NDA2, and COXIIb), thereby providing accurate loading controls, an accumulation of LHCSR3 on high-light treatment was observed, which was already reported for the wild type in the work by Peers et al. (2009). This light-induced accumulation of LHCSR3 was also observed here in pgr5, although perhaps to a smaller extent, showing that the NPQ defect in pgr5 (Fig. 2B) is not caused by a lack of the protein responsible for qE but rather, a smaller proton motive force as shown in Figure 3. Consistent with the results obtained from Figure 4, A and B (the relative amount of photoactive PSI is smaller in pgr5 after exposure to high light), we show here in Figure 4C that PSAD was decreased to less than 50% of wild-type levels, whereas PSII (D1) seemed unaffected or perhaps, even more stable in pgr5. Interestingly, the ANR1 protein, found in the anaerobic supercomplex associating PSI and cytochrome b6f (Iwai et al., 2010; Terashima et al., 2012), was decreased in pgr5. It shows a possible role for PGR5 in not only PSI accumulation but also, stabilization of the PSI-b6f supercomplex proteins, PGRL1 and ANR1.
Effects of a Lack of PGR5 on Acceptor Side Limitations
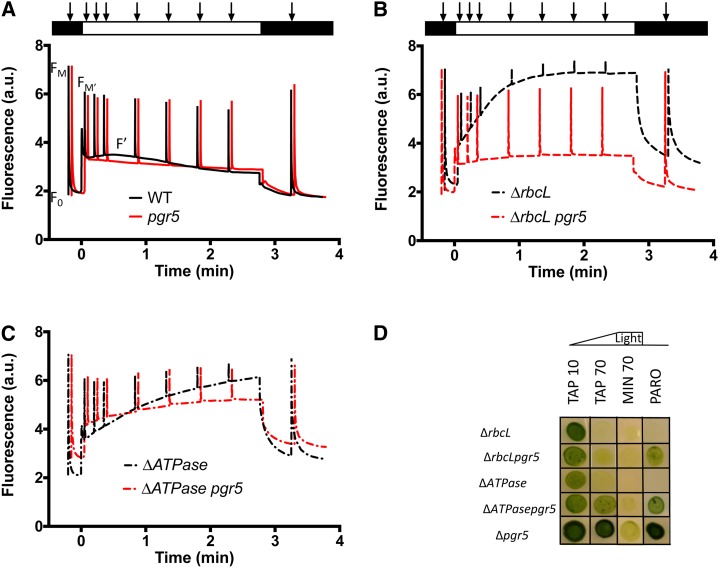
Taking advantage of the plasticity of green algae and their ability to remain green and grow on a reduced carbon source, even when a major photosynthetic complex is missing, we studied the functional consequences of combining the pgr5 mutation with lesions that eliminate the chloroplast ATP synthase (ΔATPase) or the Rubisco (ΔrbcL). The electron transport rates were measured using chlorophyll fluorescence kinetics for the various strains grown on acetate-supplemented medium. In the ΔrbcL and ΔATPase mutants, the fluorescence (F′) increased steadily throughout the 3-min illumination period at 170 μmol photons m−2 s−1. Saturating pulses induce Fm′, which is only slightly different than F′ for the ΔrbcL or ΔATPase mutants, showing that they are severely limited in electron flow compared with wild-type line cells (compare black curves in Fig. 5, A to C). When these mutations were combined with the pgr5 mutation, the decrease in photochemical yield of PSII, ϕPSII = (Fm′ − F′)/Fm′, is reduced, and the chlorophyll fluorescence transients more closely resemble the transients of the pgr5 strain (Fig. 5, A–C, red curves). This finding was especially apparent for the ΔrbcL pgr5 double mutant, where with respect to the fluorescence phenotype, pgr5 could be considered epistatic to rbcL. The effect of the pgr5 mutation on ΔrbcL was surprising to us, because we previously attributed the rise in fluorescence in Rubisco-less strains to PSI acceptor side limitations (Johnson, 2011). If this result were the case, the combination of the Rubisco mutant with that of pgr5 would have been expected to exacerbate rather than alleviate the acceptor side limitation, unless an alternative electron sink was present in the mutant. Therefore, we determined if any compensatory alternative electron transfer pathway (including PGRL1, NDA2, COXIIb, AOX1, and PTOX) was up-regulated in ΔrbcL pgr5. No significant differences were observed at the protein level, except the absence of Rubisco in ΔrbcL lines (Fig. 6). We note again that the cells were cultivated in acetate and low light, like those cells in Figure 2, and therefore, PGRL1 accumulation was unaffected in the absence of PGR5 here.
Figure 5.
Fluorescence kinetics of wild-type (WT) line and pgr5 (A), ΔrbcL and ΔrbcL pgr5 (B), and ΔATPase and ΔATPase pgr5 (C). Cells were grown in low light and TAP liquid media and analyzed with a JTS-10. The program monitored a number of variables to follow photosynthetic yield: an initial saturating flash (5,000 μmol photons m−2 s−1) gives access to the dark-adapted photosynthetic yield followed by a 3-min period of actinic light (170 μmol photons m−2 s−1) with saturating flashes to probe the yield of PSII, a return to darkness, and a final saturating flash to monitor PSII centers that recovered. D, Growth tests on TAP and MIN solid media for single and double mutants at light intensities shown (in micromoles photons minute−2 second−1). [See online article for color version of this figure.]
As expected from the absence of ATPase or Rubisco, both the single and double mutants were nonphototrophic, which is shown in Figure 5D (western-blot analysis confirmed the lack of ATPase protein in the ΔAtpase mutants; Supplemental Fig. S3). Combining the pgr5 mutation with a mutant devoid of Rubisco did not reduce the photosensitivity observed for the ΔrbcL strain; however, the pgr5 mutation alleviated photosensitivity of the ΔATPase mutant at light intensities of 70 μmol photons m−2 s−1 (Fig. 5D).
O2 Substitutes for CO2 as a Terminal Electron Acceptor
Comparing the ΔrbcL chlorophyll fluorescence kinetics with the kinetics of ΔrbcL pgr5, it seems that the rise in steady-state fluorescence in the former is caused by a gradual slowing down of linear electron transfer that depends on CEF. With the additional loss of PGR5 function (ΔrbcL pgr5), the restoration of PSII yield (nearly as high as in wild-type line cells at the light intensity tested) in the absence of CO2 fixation must reflect the recruitment of another electron acceptor than CO2.
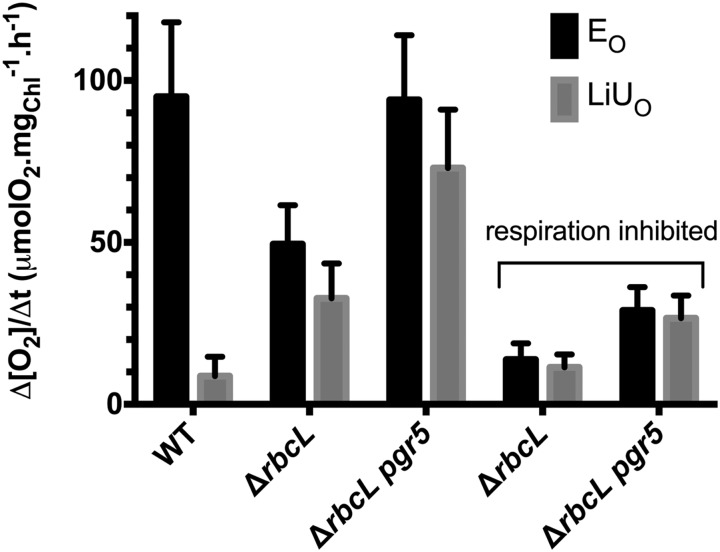
Figure 7 shows O2 exchange rates measured for the ΔrbcL and ΔrbcL pgr5 mutants compared with wild-type line cells grown in acetate containing medium under low-light conditions; typical raw data from gas exchange measurement are shown in Supplemental Figure S4. Gas exchange measurements using membrane inlet mass spectrometry (MIMS; Cournac et al., 2002) allow for isotopic discrimination of different O2 species. The PSII-dependent oxygen evolution from water (EO) is monitored at a mass-to-charge ratio (m/e = 32 [unlabeled 16O2]). The dark respiration rate (UO) and the light-induced uptake (LiUO) are measured at m/e = 36 (labeled 18O2) added to the sample before the measurement. Over a 5-min period of illumination, CO2 is the major electron acceptor in wild-type line cells, leading to a large net O2 evolution (Net = EO − LiUO). In the single ΔrbcL and double ΔrbcL pgr5 mutants, there is a significant light-induced O2 uptake (LiUO), which corresponds to nearly 70% of the O2 evolution (EO) and likely compensates for the absence of CO2 fixation in ΔrbcL strains. The single ΔrbcL mutant has an EO of around one-half of that observed for wild-type line cells, whereas the EO in the ΔrbcL pgr5 double mutant was fully restored. These results suggest a more active alternative electron flow in the double mutant than the single mutant. To determine if the alternative electron flow in the mutants was a consequence of a stimulated mitochondrial activity in the light, the cells were treated with the respiratory inhibitors myxothiazol (20 μM) and salicylhydroxamic acid (200 μM), inhibitors of the mitochondrial cytochrome bc1 complex and AOX, respectively. Although the dark respiration UO was similar in the wild-type line and the mutants, addition of mitochondrial inhibitors decreased UO by a factor of more than three. In the light, inhibitor addition resulted in a 70% reduction in both EO and LiUO. It shows that, in the mutants, the shuttling of reducing power to mitochondrial respiration is a prominent pathway, whereas it is not in the wild type, where electrons extracted from water are directed to CO2 fixation (a more detailed analysis of O2 evolution and uptake in the wild-type strain in the presence or absence of respiratory inhibitors is in the work by Peltier and Thibault, 1985). The maximum flow rate supported by this shuttle can be calculated as untreated LiUO − inhibited LiUO, which is approximately 46 μmol O2 mg Chl−1 h−1 for the double mutant. The remaining light-induced O2 uptake in the double mutant after the addition of inhibitors had a greater magnitude than in the single mutant. This finding suggests that O2 reduction reactions or Mehler-type reactions are stimulated in the absence of CEF. The maximum rate of Mehler-type O2 uptake is here around 26 μmol O2 mg Chl−1 h−1 for the double mutant, which is in agreement with previous estimates (Peltier and Thibault, 1985; Badger et al., 2000).
Figure 7.
Gas exchange measurements using labeled O2 to discriminate respiratory and photosynthetic fluxes in the wild-type (WT) line and ΔrbcL and ΔrbcL pgr5. The y axis shows the rate of EO or LiUO corrected for UO normalized to the chlorophyll content of the cells. The x axis shows the different strains tested. Respiratory inhibitors (20 μm myxothiazol and 200 μm SHAM) were added to measure the proportion of O2 exchange attributable to light-induced mitochondrial respiration in the ΔrbcL and ΔrbcL pgr5 strains. The data shown are the mean of n ≥ 3 replicates, and ses are shown for these replicates.
To determine if O2 uptake/evolution experiments and fluorescence experiments are consistent, gas exchange measurements were performed with cells that were either dark adapted or exposed to light for 5 min before making measurements. These conditions reflect the situation for the mutants during the prolonged light exposure used to monitor the fluorescence kinetics shown in Figure 5. When cells were dark adapted first, the ΔrbcL strain had an O2 evolution two times greater than that observed after the 5-min light exposure (Supplemental Fig. S4, A and B). This finding for the ΔrbcL strain correlates with the changes observed in the chlorophyll fluorescence analysis (Fig. 5). In stark contrast to the findings for the single mutant, the ΔrbcL pgr5 double mutant has sustained O2 evolution, and light induced O2 uptake regardless of whether the cells were preilluminated (Supplemental Fig. S4, C and D), which again correlates well with the fluorescence analysis.
In the Rubisco-less mutant, mrl1, a low but nonnull level of net O2 evolution was measured with a Clark electrode (Johnson et al., 2010). A Clark electrode does not discriminate between evolution and uptake rates and measures the net exchange rate (Net = EO − LiUO). Interestingly, in the gas exchange experiments shown here, LiUO did not quite reach the EO level. It does not appear very clearly from Figure 7, where the error bars reflect the variations between independent experiments rather than the real difference between EO and LiUO measured simultaneously for each different sample. Such a difference between EO and LiUO was reproduced for each of the experiments that we have done using different batches of algae (Supplemental Fig. S4 shows a representative experiment). For each sample, EO and LiUO yielded a residual net O2 evolution of ∼20% of EO (i.e. in the same range as that previously measured with a Clark electrode; Johnson et al., 2010). Interestingly, this net evolution was eliminated by the addition of inhibitors of mitochondrial electron transport, suggesting that a respiratory-dependent but non- O2-consuming pathway can occur downstream of photosynthesis in C. reinhardtii. This flow rate has a maximum of 21 μmol O2 equivalents mg Chl−1 h−1 in the double mutant.
DISCUSSION
We have shown here that the C. reinhardtii pgr5 mutant has phenotypic traits that are virtually identical to the traits described for pgrl1 (Petroutsos et al., 2009; Tolleter et al., 2011). The pgr5 mutant exhibited a defect in NPQ with a modified fluorescence kinetic that is similar to that described for pgrl1. As in other studies (Tolleter et al., 2011; Terashima et al., 2012), we show that CEF is most active under conditions when stromal carriers are reduced and that this finding reflects the activity of PGR5/PGRL1-Fd CEF. Highly reducing conditions were obtained here in the absence of oxygen. Anoxia inhibits respiration and likely increases ADP and inorganic phosphate (Pi) concentrations, therefore maintaining high membrane conductivity through ATPase, a situation favorable to CEF (see below). Our experiments (Fig. 3) have allowed us to estimate maximum CEF rates mediated by PGR5/PGRL1-Fd (40–45 e− s−1 PSI−1) and pathways other than PGR5, such as NDH (15–20 e− s−1 PSI−1), when the former is abolished in the pgr5 mutant. The rates of PGR5/PGRL1-Fd CEF that we have measured under anaerobic conditions are in agreement with recent reports (Tolleter et al., 2011; Terashima et al., 2012; Takahashi et al., 2013). In the general case (i.e. under aerobic conditions), when DCMU is absent, linear electron flow steadily produces NADPH and reduces Fd at the acceptor side of PSI. The PGR5/PGRL1-Fd pathway is then expected to contribute significantly to CEF, which is shown here by the differences in chlorophyll fluorescence kinetics obtained with and without PGR5 (Fig. 5, A–C). Under low-CO2 conditions or in the absence of CO2 fixation, the redox pressure would become more severe, and the role of PGR5 would seem more critical. An analysis of ECS in noninhibited conditions would require the deconvolution of the relative contributions of linear and cyclic to the proton motive force, which has been done in plants (Avenson et al., 2005). Here, we proposed a more straightforward diagnostic, assessing the maximal capacity for CEF, although our method does not apply to steady-state conditions, where CEF never represents 100% of the total electron flow.
Although most of our conclusions concerning the function of CrPGR5 are congruent with the conclusions drawn from studies of AtPGR5, there is one difference with respect to PSI photoinhibition. Strong photoinhibition of PSI occurs when the Crpgr5 mutants are grown in photoautotrophic conditions and exposed to light intensities of 200 μmol photons m−2 s−1. This photoinhibition results in a significant decrease in PSI, PGRL1, and ANR polypeptides (Fig. 4) and cannot be reversed by the addition of an artificial electron acceptor (methylviologen). This result is in contrast with that reported for mature Atpgr5 plants, where PSI photoinhibition could be relieved by the addition of methylviologen (Munekage et al., 2002). Interestingly, the phenotype that we observe here is similar to that of developing Atpgr5 plants grown under fluctuating light conditions; under such conditions, there was PSI photoinhibition and a loss of PSI subunits (Suorsa et al., 2012). We also observed that, under the high-light growth conditions, where the amount of PSI is reduced relative to other photosynthetic complexes, the reduction kinetics of P700 in Crpgr5 are accelerated (Supplemental Fig. S2, B and C). In these photoinhibited conditions, there is less PSI for the same stable amount of Nda2 and b6f (Fig. 4C), and therefore, more electrons are now funneling through the remaining PSI centers. Because PQ reduction is limiting for CEF (and not PSI), decreasing PSI levels will not limit CEF; on the contrary, it will increase the flow rate through the remaining PSI that we use here to measure CEF. We propose that CEF normalized against the chlorophyll content or any other complex in the photosynthetic chain would not seem to be accelerated under these conditions. This finding substantiates a simple enzymatic model for CEF (Maxwell and Biggins, 1976; Alric et al., 2010).
To further investigate the consequences of acceptor side limitations in the pgr5 mutant, we constructed double mutants that would magnify acceptor side limitation; we combined pgr5 with mutations affecting the chloroplast ATPase and Rubisco that stop the Calvin-Benson cycle. The pgr5 mutation slightly relieved the light sensitivity of the ΔAtpase mutant but was unable to alleviate light sensitivity of ΔrbcL (Fig. 5). This result is because the origins of the photosensitivity are different: in ΔrbcL, photosensitivity has been attributed, in large part, to the production of reactive O2 species at the acceptor side of PSI (Johnson et al., 2010; Johnson, 2011), whereas photosensitivity in ΔAtpase is caused by multiple phenomena. The ATPase mutant is probably both acceptor and donor side limited, with effects on PSII stability caused by an overacidification of the thylakoid lumen in both C. reinhardtii and plants (Majeran et al., 2001; Rott et al., 2011). The large ΔpH built up in the light in the absence of ATPase should slow down electron transfer through the cyt f and lead to photoinhibition. Such an exaggerated photosynthetic control in ΔAtpase could be released by the pgr5 mutation. In agreement with this view, we show here that the pgr5 mutation relieves the photosensitivity of the ΔAtpase strain. As observed in Figure 5D, ΔAtpase pgr5 grows at 70 μmol photons m−2 s−1 of light, a light intensity at which the ΔAtpase single mutant dies. In contrast, the pgr5 lesion does not alleviate the photosensitive phenotype of ΔrbcL; both single ΔrbcL and double ΔrbcL pgr5 mutants tolerate only very weak light ≤10 μmol photons m−2 s−1.
Such a difference in the growth phenotypes of the double mutants shows that, although ATP and NADPH are common substrates for the Calvin-Benson cycle, blocking photosynthesis at the level of Rubisco or the chloroplast ATPase is not strictly equivalent. Hence, we shall treat the two double mutants separately and try to elucidate the precise alterations in molecular mechanisms that may explain the phenotypic differences between the mutant strains.
The study of Arabidopsis pgr5 mutant under steady-state illumination revealed that the marked difference in qE observed in pgr5 compared with the wild type was largely attributable to a decreased proton motive force. Such a decrease in proton motive force in pgr5 was mainly caused by acceptor side limitations of linear electron flow (>2-fold) rather than the lack of the comparatively smaller contribution (approximately 13%) of CEF (Avenson et al., 2005). Because of an ATP deficiency in Atpgr5, a buildup of stromal [Pi] above its Km at the ATPase would maintain a high ATPase conductivity, accounting for a low proton gradient and a low qE (Avenson et al., 2005). The phenotype of Atpgr5 is, therefore, a combined effect of stromal overreduction and aberrant modification of ATPase conductivity decreasing qE capacity (Shikanai, 2010). It makes the study of our C. reinhardtii single ΔATPase and double ΔATPase pgr5 mutant strains interesting, because the presence or absence of PGR5 can be studied here with a low membrane conductivity (absence of ATPase). Unfortunately, qE analysis is precluded in C. reinhardtii ΔATPase, because the strain is photosensitive and LHCSR3, required for NPQ, accumulates only in high light (Peers et al., 2009; Fig. 4C). It explains why only a little chlorophyll fluorescence quenching (of Fm′) is observed in Figure 5C. Nonetheless, the analysis of linear electron flow in Figure 5C, which is indicated by the photochemical yield of PSII ϕPSII = (Fm′ − F′)/Fm′, remains informative. Contrary to Arabidopsis, where ϕPSII is smaller in pgr5 than in the wild type under high light (Munekage et al., 2002, 2008; Avenson et al., 2005), in C. reinhardtii, it is almost unaffected, even greater (Fig. 5A; Tolleter et al., 2011). In the absence of ATPase, ϕPSII is diminished, and the double mutant ΔATPase pgr5 shows a greater ϕPSII than the single mutant ΔATPase (Fig. 5C). It parallels some earlier observations. In isolated chloroplasts, a slowing down of linear electron flow under acceptor-limited conditions was attributed to proton gradient backpressure generated by CEF (Slovacek and Hind, 1977). In tobacco (Nicotiana spp.) knockdowns of chloroplast ATPase (Rott et al., 2011; Yamori et al., 2011), a decrease in linear electron flow revealed the negative feedback of an excess proton motive force on cytochrome b6f activity. In C. reinhardtii, an overacidification of the lumen in ΔATPase would result in an exaggerated photosynthetic control and be alleviated in ΔATPase pgr5. Therefore, we conclude that, in accordance with several other reports (Avenson et al., 2005; Rott et al., 2011), membrane conductivity for protons and counterions is going to affect not only the proton motive force but also, as a consequence, linear electron flow. Whether the modulation of membrane conductivity is determined only by ATPase efflux or other parameters, whether ATPase efflux is mainly controlled by the availability of ADP and Pi, or if ATPase accumulation also plays a role in the regulation of linear electron flow remain open questions. Here, in contrast with some observations in Arabidopsis (Suorsa et al., 2012) but in accordance with another study (Avenson et al., 2005), no difference in ATPase accumulation was observed in the C. reinhardtii pgr5 mutant grown under high-light phototrophic conditions (Fig. 4C).
In the absence of Calvin-Benson cycle activity, whereas the photosynthetic yield of ΔrbcL became increasingly reduced over an illumination when measured by chlorophyll fluorescence kinetics (Fig. 5B), the double mutant ΔrbcL pgr5 had a PSII yield restored to wild-type levels. In other words, ΔrbcL pgr5 shows a sustained electron flow that is steadily arrested in the single ΔrbcL mutant, although in both cases, the Calvin-Benson cycle is inactive and little net oxygen evolution can be measured (Johnson et al., 2010). Here, we show, by using O2 labeling techniques, that this small net O2 evolution previously reported for Rubisco-less mutants (Johnson et al., 2010) was, in fact, hiding larger fluxes of O2 uptake and evolution. What is truly surprising is that electron transfer to O2 could be as rapid and sustained as to CO2. Our results suggest that a disruption of the photosynthetic control, associated with the loss of PGR5, increased linear electron flow. The resulting redox pressure buildup in the stroma opened, in turn, a respiratory overflow valve (Figs. 5 and 7). An increased light-induced respiration was also described for the PGRL1 knockdown and knockout mutants (Petroutsos et al., 2009; Tolleter et al., 2011). Possible scenarios involve metabolic shuttles between the chloroplast and the mitochondrion. Oxaloacetate and 3-phosphoglycerate reduction rates have been shown to increase in the absence of CEF in isolated chloroplasts under acceptor-limiting conditions (Slovacek and Hind, 1977). Similar in vitro enzymatic assays showed that the malate valve could operate as rapidly as the major (linear) photosynthetic flow rate (Fridlyand et al., 1998). The malate valve was suggested to occur in plants under stress conditions and shown to have an elevated flow rate in the absence of CEF (Backhausen et al., 2000). Thus, in vivo alternative shuttling of reductants to the mitochondria can carry the full load of linear electron flow and totally substitute for CO2 fixation. The finding that linear electron flow can be entirely redirected to O2 but is usually directed to CO2 fixation under normal photosynthetic conditions is in line with the idea of a hierarchy of alternative pathways (Backhausen et al., 2000). In the wild type, this hierarchy would limit the leakage of possibly harmful electrons to other pathways to conserve electron flow to CO2 fixation.
CONCLUSION
Our data provide clear evidence that PGR5 is involved in the recycling of excess reductants to supplement ATP synthesis and that this process is vital for protection of PSI. In the presence of acceptor side and ATP limitations and the absence of PGR5, a sustained flow of electrons can be maintained in the absence of CO2 fixation. Alternative pathways to O2, involving mitochondrial respiration, can compensate for these limitations to a greater (ΔrbcL pgr5) or lesser (ΔATPase pgr5) extent. It reflects a connection between PGR5/PGRL1-Fd–mediated CEF, the ATP budget of the chloroplast, and the Calvin-Benson cycle at the level of regulating photosynthetic electron transfer. As clearly observed in Figure 6, PGR5/PGRL1-Fd CEF does not represent a valve, because it only recycles electrons without being a sink: in acceptor side-limited conditions, an increase in chlorophyll fluorescence is accordingly observed when PGR5/PGRL1-Fd CEF is active. The control of CEF is likely sensitive to redox conditions and factors, such as thioredoxin m (Courteille et al., 2013; Hertle et al., 2013) and/or the chloroplast calcium status and the calcium sensor Ca2+ sensor protein (Terashima et al., 2012). Regulation of shuttles, such as the malate dehydrogenase (Wolosiuk et al., 1977), represents a promising next step to unraveling the interwoven and possibly hierarchical network of pathways that regulates energy partitioning and photoprotection in photosynthetic organisms.
MATERIALS AND METHODS
Cell Cultures
Chlamydomonas reinhardtii, wild-type line, mutant, and complemented strains were grown in 500-mL flasks and agitated on a gyratory shaker (120 rpm) at 25°C under continuous light of various intensities (5–200 μmol photons m−2 s−1). Medium containing TAP or lacking acetate (MIN) was used for heterotrophic or phototrophic growth, respectively. The pgr5 mutant (strain CAL028.01.15) was generated by DNA insertional mutagenesis (Dent et al., 2005) with linearized pBC1 plasmid encoding paromomycin resistance (Tran et al., 2012). Crosses were performed with the 137c wild-type line strain using a standard protocol, and tetrads were dissected as described in the work by Harris (1989). Mutants were backcrossed two times, and four progeny from 20 tetrads for the second generation were retained for additional studies. The mutant devoid of Rubisco, ΔrbcL, was obtained by deleting the chloroplast rbcL gene as previously described (Johnson et al., 2010). It was used for crosses and gave the ΔrbcL pgr5 double mutant. Growth tests were conducted by placing 25-µL drops of liquid cultures at a density of 1 × 105 cells mL−1 under 10, 70, 150, or 300 μmol photons m−2 s−1 illumination onto solid medium. The ΔATPase and ΔATPase pgr5 mutants were generated by chloroplast transformation as described below.
Transformation
Complementation
A 2,040-bp sequence of PGR5 comprising 500 bp upstream of the start site, the 5′ and 3′ untranslated regions (UTRs), and the entire coding region were amplified from wild-type genomic DNA and used to complement the pgr5 mutant. Amplification of the fragment used a forward primer (i) and reverse primer (viii; presented in Supplemental Table S1 with the positions of the primers shown in Supplemental Fig. S1A) using Phusion high-fidelity DNA polymerase (Finnzyme) as per the manufacturer’s guidelines. The amplified fragment was cloned into pTOPO-PCR II blunt zero (Invitrogen), and the vector with the PGR5 insert was linearized before transformation. A spectinomycin cassette, in which the coding sequence of the drug resistance marker gene was driven by the hsp70a-RbcS promoter (Meslet-Cladiére and Vallon, 2011) in vector pALM32 and recoded for C. reinhardtii nuclear gene expression, was linearized with KpnI resolved by electrophoresis on agarose gels and excised from the gel. This linearized, purified cassette was cotransformed with DNA encoding the spectinomycin resistance gene; C. reinhardtii transformants were selected for spectinomycin resistance. The recipient strain, pgr5 2b24 (derived from a second backcross with the 137c wild-type strain), was transformed by electroporation (Shimogawara et al., 1998) in the presence of 2 µg of PGR5-containing plasmid DNA and 2 µg of pALM32 cassette. Selection was performed under 10 µE m−2 s−1 light at 25°C on TAP solid medium supplemented with 100 µg/mL spectinomycin (Sigma). Clones were then screened as described above for complementation of the NPQ phenotype using a laboratory-built chlorophyll fluorescence imaging system (Johnson et al., 2009).
Chloroplastic Knockout Mutant ΔATPase
A recycling 5′ psaA-driven aadA selectable cassette was constructed by digesting the 483-bp repeat-recycling cassette (Fischer et al., 1996) by NruI and NcoI to remove the atpA 5′ UTR upstream of the aadA coding sequence. The resulting 5,140-bp fragment was then ligated with the 242-bp fragment containing the psaA promoter and 5′ UTR recovered by digesting the 5′psaA-aadA-3′rbcL plasmid (Wostrikoff et al., 2004) with EcoRV and NcoI to create the pExc-aAKR cassette. To replace the atpI coding sequence by the Exc-aAKR cassette for selection of transformed clones based on spectinomycin resistance, a DNA fragment containing the atpI 5′ UTR fused to the 3′ UTR but lacking the coding sequence was created by a two-step PCR procedure (Higuchi, 1990): two pairs of primers (atpI-dr_DelF1/atpI-dr_R1 and atpI-dr_F1/atpI-dr_DelR1) allowed the amplification from the template plasmid p70 (http://chlamycollection.org/plasmid/p-70-cpdna-ecori-17-4-9-kb/) of two partially overlapping fragments that were mixed and used as templates in a third PCR with the external primers atpI-dr_F1 and atpI-dr_R1 (primer sequences are in Supplemental Table S1). The final amplicon contained the SacI and XhoI restriction sites, introduced in the sequence of the primers (underlined or bolded in Supplemental Table S1), at the junction between the atpI 5′ and 3′ UTRs. It was digested by HpaI and SnaBI and ligated into the p70 vector digested by the same enzymes to create plasmid pΔcodI. This latter plasmid was digested with SacI and XhoI and ligated with the 2,504-bp fragments obtained by digesting pExc-aAKR by the same enzymes to yield plasmid pΔatpI. This construction was then introduced in the chloroplast genome of C. reinhardtii by biolistic transformation as described by Boulouis et al. (2011), and transformed clones were recovered on TAP plates supplemented with spectinomycin (100 µg mL−1). The transformants were then individually streaked onto 500 μg mL−1 spectinomycin to generate homoplasmic strains.
PCR and Reverse Transcription PCR
A rapid total DNA extraction method was performed with Chelex 100 (Sigma) adapted from the work by Werner and Mergenhagen (1998). PCR was performed using the Taq PCR Master Mix Kit (Qiagen) as per the manufacturer’s protocol with the primers shown in Supplemental Table S1; annealing of the primers was at 57°C. PCR products were separated on 1.5% (w/v) agarose gels.
As shown in Supplemental Figure S1B, total RNA was isolated using the hot phenol method, treated with 5 units of DNAse at room temperature for 1 h, and further purified using the RNeasy MinElute Kit (Qiagen). RNA was quantified by Nanodrop (Thermoscientific), and complementary DNA (cDNA) was synthesized from that RNA using the standard SuperScript III protocol (Invitrogen). cDNA was diluted 10 times and amplified by PCR. The transcript encoding the housekeeping polypeptide Chlamydomonas β subunit-Like Polypeptide was used as a positive control; amplification of the transcript was for 30 cycles (Pootakham et al., 2010). To isolate the PGR5 cDNA, total cDNA was added undiluted into the reaction mixture and then amplified for 45 cycles. DNA separation was achieved on 1.5% (w/v) agarose gels.
Protein Analysis
Proteins were extracted from total cells and separated under denaturating conditions on 10% or 13% (v/v) PAGE Bis-Tris SDS-MOPS minigels (Invitrogen). Proteins were loaded based on chlorophyll content at a concentration of 2.5 or 1 μg mL−1 chlorophyll (Figs. 4 and 6; Supplemental Fig. S3). Proteins were transferred onto nitrocellulose filters (BioTrace NT; Pall Corporation) using a semidry apparatus (BIORAD). Primary antibodies were sourced from Michael Hippler (PSAD, LHCSR3, PGRL1, and ANR1; Naumann et al., 2005, 2007; Petroutsos et al., 2009; Terashima et al., 2012), Gilles Peltier (PGRL1; Tolleter et al., 2011), Pierre Cardol (NDA2; Jans et al., 2008), Agrisera (LSU, COXIIb, Psba D1, CF1, and AOX1), and Francis-Andre Wollman (PTOX2 and cyt f; Vallon et al., 1991; Houille-Vernes et al., 2011) and decorated by incubation in Tris-buffered saline. Secondary antibody was anti-rabbit (Invitrogen). Horseradish peroxidase chemiluminescent substrate was used to reveal the antibody signal using the GBOX imaging system (Syngene). For the western-blot analysis shown in Figure 2E, thylakoids were isolated according to the work by Chua and Bennoun (1975) and loaded on 16.5% (v/v) acrylamide Schägger and von Jagow Tris-Tricine gels (Schägger and von Jagow, 1987). Thylakoids were loaded at 10 μg mL−1 chlorophyll for decoration by CF1 and PGR5 antibodies and 2 μg mL−1 chlorophyll for PGRL1 antibody. The PGR5 antibody was made against an Arabidopsis (Arabidopsis thaliana) peptide and sourced from Toshiharu Shikanai (Munekage et al., 2002).
Chlorophyll Fluorescence and Spectroscopic Analysis
The chlorophyll fluorescence imaging system was described previously (Johnson et al., 2009). Fluorescence and absorbance (P700 and ECS) kinetics were performed using the JTS-10 spectrophotometer (BioLogic) as previously described (Joliot and Joliot, 2002). For fluorescence measurements, two parameters were calculated: for NPQ, NPQ = (Fm − Fm′)/Fm′, and for the photochemical yield of PSII, ϕPSII = (Fm′ − F′)/Fm′. For ECS measurements, samples were treated as follows. Cells were grown in TAP media at <10 μmol photons m−2 s−1 and harvested at exponential phase. Centrifugation for 3 min in falcon tubes at 2,000 rpm in swinging rotor centrifuge was followed by 10-fold concentration by resuspension in Ficoll 20% (w/v) HEPES-KOH (pH 7.2), leaving 2 mL of TAP supernatant (chlorophyll concentrations are measured and adjusted at this point so that different samples have equal chlorophyll). Resuspended cultures were left in the darkness in 50-mL open Erlenmeyer flasks, well-agitated, and poisoned with 10 μm DCMU and 1 mm hydroxylamine just before the experiment; 1.5 mL of culture is was added to a cuvette and covered with mineral oil until anoxia was reached (on average, 15 min). Single turnover flashes were delivered to normalize the ECS signal from different samples against total PSI and monitor when anaerobic conditions were reached (ECS decay rate). For P700 measurements, cultures were adapted to phototrophic conditions at either 20 or 200 μmol photons m−2 s−1. Cultures were centrifuged, concentrated, resuspended in Ficoll HEPES-KOH (pH 7.2), and left to dark adapt as described for the ECS measurements.
MIMS Analysis of O2 Exchange
A description of the protocols for MIMS and the methods used for analyzing MIMS data were previously described (Cournac et al., 2002). For O2 exchange experiments, cultures were grown in TAP at <10 μmol photons m−2 s−1 illumination. Cells were centrifuged and resuspended in fresh medium to a concentration of 30 μg mL−1 chlorophyll and either left in the dark for a minimum of 30 min or briefly preilluminated at 120 μmol photons m−2 s−1 in an INFORS incubator before performing the experiments; 1.5 mL culture was added to the MIMS cuvette. For experiments testing the impact of mitochondrial inhibitors on photosynthetic activities, the cultures were made of 20 μm myxothiazol and/or 200 μm SHAM (Sigma) and agitated in the dark for 10 min before initiating experiments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular and genetic characterization of the PGR5 mutation.
Supplemental Figure S2. PSI photoinhibition gives elevated rates of P700 rereduction.
Supplemental Figure S3. Western-blot analysis of ATPase and ATPase pgr5 mutants.
Supplemental Figure S4. MIMS raw data for oxygen uptake and evolution rates.
Supplemental Table S1. Primers used for PCR amplifications in this article.
Supplementary Material
Acknowledgments
We thank Gilles Peltier for critical reading of this manuscript and support during the completion of this work.
Glossary
- ANR1
anaerobic response1
- AOX1
alternative oxidase1
- cDNA
complementary DNA
- CEF
cyclic electron flow
- CF1
chloroplast ATPase
- COXIIb
cytochrome oxidase subunit IIb
- cyt f
cytochrome b6f complex
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- ECS
electrochromic shift
- EO
PSII-dependent oxygen evolution from water
- Fd
ferredoxin
- F′
fluorescence
- Fm
maximum fluorescence yield in the dark-adapted state
- Fm′
maximum fluorescence yield in the light-adapted state
- LiUO
light-induced uptake
- MIMS
membrane inlet mass spectrometry
- MIN
medium lacking acetate
- NDA2
type II NADPH dehydrogenase2
- NDH
NADPH dehydrogenase
- NPQ
nonphotochemical quenching
- PGR5
proton gradient regulation5
- PGRL1
proton gradient regulation-like1
- Pi
inorganic phosphate
- PQ
plastoquinone
- PTOX
plastoquinol terminal oxidase
- qE
thermal dissipation component
- TAP
Tris-acetate phosphate
- UO
dark respiration rate
- UTR
untranslated region
- O2
molecular oxygen
- e−
electrons
Footnotes
This work was supported by the Centre National de la Recherche Scientifique, Agence Nationale pour la Recherche (ALGOMICS and ALGOH2 projects); the National Science Foundation (award no. MCB0951094 to A.R.G.); the HélioBiotec platform funded by the European Union (European Regional Development Fund); the Région Provence Alpes Côte d’Azur; the French Ministry of Research; the Commissariat à l’Energie Atomique et aux Energies Alternatives; the U.S. Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division (Field Work Proposal no. 449B; isolation and initial characterization of the pgr5 mutant); the Centre National de la Recherche Scientifique, the Université Pierre et Marie Curie, Paris 06, the Unité Mixte de Recherche 7141 (to S.-I.O.); the Agence Nationale pour la Recherche (contract no. BLAN–NT09_451610 Chloro/Mito Control by Epistasy of Synthesis to S.-I.O.); the Region Ile de France (to L.H.-V.); the Centre National de la Recherche Scientifique and the Initiative d'Excellence program from the French state (grant no. DYNAMO ANR–11–LABX–0011–01 to F.R.); the Deutsche Forschungsgemeinschaft (grant no. HI739/9.1 to M.H.); the Gordon and Betty Moore Foundation (grant no. GBMF3070 to K.K.N.); and Brigitte Berthelemot (private funds). K.K.N. is an investigator of the Howard Hughes Medical Institute.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Alric J. (2010) Cyclic electron flow around photosystem I in unicellular green algae. Photosynth Res 106: 47–56 [DOI] [PubMed] [Google Scholar]
- Alric J. (2014) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii: II. Involvement of the PGR5-PGRL1 pathway under anaerobic conditions. Biochim Biophys Acta 1837: 825–834 [DOI] [PubMed] [Google Scholar]
- Alric J, Lavergne J, Rappaport F. (2010) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii (I) aerobic conditions. Biochim Biophys Acta 1797: 44–51 [DOI] [PubMed] [Google Scholar]
- Avenson TJ, Cruz JA, Kanazawa A, Kramer DM. (2005) Regulating the proton budget of higher plant photosynthesis. Proc Natl Acad Sci USA 102: 9709–9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhausen JE, Kitzmann C, Horton P, Scheibe R. (2000) Electron acceptors in isolated intact spinach chloroplasts act hierarchically to prevent over-reduction and competition for electrons. Photosynth Res 64: 1–13 [DOI] [PubMed] [Google Scholar]
- Badger MR, von Caemmerer S, Ruuska S, Nakano H. (2000) Electron flow to oxygen in higher plants and algae: rates and control of direct photoreduction (Mehler reaction) and rubisco oxygenase. Philos Trans R Soc Lond B Biol Sci 355: 1433–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Melis A, Mackey KR, Cardol P, Finazzi G, van Dijken G, Berg GM, Arrigo K, Shrager J, Grossman A. (2008) Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim Biophys Acta 1777: 269–276 [DOI] [PubMed] [Google Scholar]
- Boulouis A, Raynaud C, Bujaldon S, Aznar A, Wollman FA, Choquet Y. (2011) The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ. (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Bennoun P. (1975) Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA 72: 2175–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournac L, Mus F, Bernard L, Guedeney G, Vignais P, Peltier G. (2002) Limiting steps of hydrogen production in Chlamydomonas reinhardtii and Synechocystis PCC 6803 as analysed by light-induced gas exchange transients. Int J Hydrogen Energy 27: 1229–1237 [Google Scholar]
- Courteille A, Vesa S, Sanz-Barrio R, Cazalé AC, Becuwe-Linka N, Farran I, Havaux M, Rey P, Rumeau D. (2013) Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol 161: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D. (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Stampacchia O, Redding K, Rochaix JD. (1996) Selectable marker recycling in the chloroplast. Mol Gen Genet 251: 373–380 [DOI] [PubMed] [Google Scholar]
- Fridlyand LE, Backhausen JE, Scheibe R. (1998) Flux control of the malate valve in leaf cells. Arch Biochem Biophys 349: 290–298 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Karpowicz SJ, Heinnickel M, Dewez D, Hamel B, Dent R, Niyogi KK, Johnson X, Alric J, Wollman FA, et al. (2010) Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth Res 106: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Heber U, Walker D. (1992) Concerning a dual function of coupled cyclic electron transport in leaves. Plant Physiol 100: 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinnickel ML, Alric J, Wittkopp T, Yang W, Catalanotti C, Dent R, Niyogi KK, Wollman FA, Grossman AR. (2013) Novel thylakoid membrane GreenCut protein CPLD38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J Biol Chem 288: 7024–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldt HW, Rapley L. (1970) Specific transport of inorganic phosphate, 3-phosphoglycerate and dihydroxyacetonephosphate, and of dicarboxylates across the inner membrane of spinach chloroplasts. FEBS Lett 10: 143–148 [DOI] [PubMed] [Google Scholar]
- Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D. (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Higuchi R. (1990) Recombinant PCR. In PCR Protocols: A Guide to Methods and Application. In Innis M.A., Geffond D.H., Sninsky J.J., White T.J., eds, Academic Press, London, pp 177–183 [Google Scholar]
- Houille-Vernes L, Rappaport F, Wollman FA, Alric J, Johnson X. (2011) Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc Natl Acad Sci USA 108: 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J. (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Jans F, Mignolet E, Houyoux PA, Cardol P, Ghysels B, Cuiné S, Cournac L, Peltier G, Remacle C, Franck F. (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA 105: 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanjean R, van Thor JJ, Havaux M, Joset F, Matthijs HCP. (1999) Identification of plastoquinone–cytochrome/b6f reductase pathways in direct or indirect photosystem I driven cyclic electron flow in Synechocystis PCC 6803. In Peshek G.A., Loffelhardt W., Schmetterer G., eds, The Phototrophic Prokaryotes. Kluwer/Plenum, New York, pp 251–258 [Google Scholar]
- Johnson X. (2011) Manipulating RuBisCO accumulation in the green alga, Chlamydomonas reinhardtii. Plant Mol Biol 76: 397–405 [DOI] [PubMed] [Google Scholar]
- Johnson X, Vandystadt G, Bujaldon S, Wollman FA, Dubois R, Roussel P, Alric J, Béal D. (2009) A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth Res 102: 85–93 [DOI] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. (2010) MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Johnson GN. (2011) Regulation of cyclic and linear electron flow in higher plants. Proc Natl Acad Sci USA 108: 13317–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Joliot A. (2002) Cyclic electron transfer in plant leaf. Proc Natl Acad Sci USA 99: 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E. (1973) Effect of anaerobiosis on photosynthetic reactions and nitrogen metabolism of algae with and without hydrogenase. Arch Mikrobiol 93: 91–100 [DOI] [PubMed] [Google Scholar]
- Kofer W, Koop HU, Wanner G, Steinmüller K. (1998) Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol Gen Genet 258: 166–173 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Avenson TJ, Edwards GE. (2004) Dynamic flexibility in the light reactions of photosynthesis governed by both electron and proton transfer reactions. Trends Plant Sci 9: 349–357 [DOI] [PubMed] [Google Scholar]
- Krömer S, Scheibe R. (1996) Function of the chloroplastic malate valve for respiration during photosynthesis. Biochem Soc Trans 24: 761–766 [DOI] [PubMed] [Google Scholar]
- Leister D, Shikanai T. (2013) Complexities and protein complexes in the antimycin A-sensitive pathway of cyclic electron flow in plants. Front Plant Sci 4: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Majeran W, Olive J, Drapier D, Vallon O, Wollman FA. (2001) The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol 126: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PC, Biggins J. (1976) Role of cyclic electron transport in photosynthesis as measured by the photoinduced turnover of P700 in vivo. Biochemistry 15: 3975–3981 [DOI] [PubMed] [Google Scholar]
- Mehler AH. (1951) Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch Biochem Biophys 33: 65–77 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslet-Cladière L, Vallon O. (2011) Novel shuttle markers for nuclear transformation of the green alga Chlamydomonas reinhardtii. Eukaryot Cell 10: 1670–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T. (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Munekage YN, Genty B, Peltier G. (2008) Effect of PGR5 impairment on photosynthesis and growth in Arabidopsis thaliana. Plant Cell Physiol 49: 1688–1698 [DOI] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M. (2005) N-terminal processing of Lhca3 Is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O. (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peltier G, Thibault P. (1985) O2 uptake in the light in Chlamydomonas: evidence for persistent mitochondrial respiration. Plant Physiol 79: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D, Terauchi AM, Busch A, Hirschmann I, Merchant SS, Finazzi G, Hippler M. (2009) PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J Biol Chem 284: 32770–32781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W, Gonzalez-Ballester D, Grossman AR. (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmer RJ, Kok B. (1976) Photoreduction of O2 Primes and Replaces CO2 Assimilation. Plant Physiol 58: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott M, Martins NF, Thiele W, Lein W, Bock R, Kramer DM, Schöttler MA. (2011) ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell 23: 304–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV, Young AJ, Horton P. (1993) Induction of nonphotochemical energy dissipation and absorbance changes in leaves (evidence for changes in the state of the light-harvesting system of photosystem II in vivo). Plant Physiol 102: 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2010) Regulation of photosynthetic electron transport. In Rebeiz CABC, Bohnert HJ, Daniell H, Hoober JK, Lichtenhaler HK, Portis AR, Tripathy BC, eds, The Chloroplast: Basics and Applications. Advances in Photosynthesis and Respiration. Springer, Dordrecht, The Netherlands, pp 347–361 [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A. (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovacek RE, Hind G. (1977) Influence of antimycin a and uncouplers on anaerobic photosynthesis in isolated chloroplasts. Plant Physiol 60: 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M, Järvi S, Grieco M, Nurmi M, Pietrzykowska M, Rantala M, Kangasjärvi S, Paakkarinen V, Tikkanen M, Jansson S, et al. (2012) PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 24: 2934–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa K, Tsujimoto HY, Arnon DI. (1963) Role of chloroplast ferredoxin in the energy conversion process of photosynthesis. Proc Natl Acad Sci USA 49: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Clowez S, Wollman FA, Vallon O, Rappaport F. (2013) Cyclic electron flow is redox-controlled but independent of state transition. Nat Commun 4: 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M, Petroutsos D, Hüdig M, Tolstygina I, Trompelt K, Gäbelein P, Fufezan C, Kudla J, Weinl S, Finazzi G, et al. (2012) Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc Natl Acad Sci USA 109: 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Sharifi MN, Poddar S, Dent RM, Niyogi KK. (2012) Intragenic enhancers and suppressors of phytoene desaturase mutations in Chlamydomonas reinhardtii. PLoS ONE 7: e42196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O, Bulte L, Dainese P, Olive J, Bassi R, Wollman FA. (1991) Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc Natl Acad Sci USA 88: 8262–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner R, Mergenhagen D. (1998) Mating type determination of Chlamydomonas reinhardtii by PCR. Plant Mol Biol Rep 16: 295–299 [Google Scholar]
- Wolosiuk RA, Buchanan BB, Crawford NA. (1977) Regulation of Nadp-malate dehydrogenase by light-actuated ferredoxin-thioredoxin system of chloroplasts. FEBS Lett 81: 253–258 [Google Scholar]
- Wostrikoff K, Girard-Bascou J, Wollman FA, Choquet Y. (2004) Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J 23: 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Peng L, Fukao Y, Shikanai T. (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Takahashi S, Makino A, Price GD, Badger MR, von Caemmerer S. (2011) The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol 155: 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.