The redox state of the plastoquinone pool of Synechocystis sp. strain PCC 6803 is regulated between narrow limits, in contrast to the more dynamic chlorophyll a fluorescence signal.
Abstract
A method is presented for rapid extraction of the total plastoquinone (PQ) pool from Synechocystis sp. strain PCC 6803 cells that preserves the in vivo plastoquinol (PQH2) to -PQ ratio. Cells were rapidly transferred into ice-cold organic solvent for instantaneous extraction of the cellular PQ plus PQH2 content. After high-performance liquid chromatography fractionation of the organic phase extract, the PQH2 content was quantitatively determined via its fluorescence emission at 330 nm. The in-cell PQH2-PQ ratio then followed from comparison of the PQH2 signal in samples as collected and in an identical sample after complete reduction with sodium borohydride. Prior to PQH2 extraction, cells from steady-state chemostat cultures were exposed to a wide range of physiological conditions, including high/low availability of inorganic carbon, and various actinic illumination conditions. Well-characterized electron-transfer inhibitors were used to generate a reduced or an oxidized PQ pool for reference. The in vivo redox state of the PQ pool was correlated with the results of pulse-amplitude modulation-based chlorophyll a fluorescence emission measurements, oxygen exchange rates, and 77 K fluorescence emission spectra. Our results show that the redox state of the PQ pool of Synechocystis sp. strain PCC 6803 is subject to strict homeostatic control (i.e. regulated between narrow limits), in contrast to the more dynamic chlorophyll a fluorescence signal.
The photosynthetic apparatus of oxygenic phototrophs consists of two types of photosynthetic reaction centers: PSII and PSI. Both photosystems are connected in series, with electrons flowing from PSII toward PSI through an intermediate electron transfer chain, which comprises the so-called plastoquinone (PQ) pool, plastocyanin and/or cytochrome c553, and the cytochrome b6f complex. The redox potential of the PQ pool is clamped by the relative rates of electron release into and uptake from this pool. Within the PSII complex, electrons are extracted from water at the lumenal side of the thylakoid membrane and transferred to the primary accepting quinone (QA) at the stromal side. The electron is subsequently transferred to a PQ molecule in the secondary accepting quinone (QB) of PSII. The intermediate QB semiquinone, which is formed accordingly, is stable in the QB site for several seconds (Diner et al., 1991; Mitchell, 1993) and subsequently can be reduced to plastoquinol (PQH2). The midpoint potential of QA reduction is approximately −100 mV (Krieger-Liszkay and Rutherford, 1998; Allakhverdiev et al., 2011), whereas the corresponding midpoint potential of the QB semiquinone is close to zero (Nicholls and Ferguson, 2013). PQH2 equilibrates with the PQ pool in the thylakoid membranes, which has a size that is approximately 1 order of magnitude larger than the number of PSII reaction centers (Melis and Brown, 1980; Aoki and Katoh, 1983).
PQ is a lipophilic, membrane-bound electron carrier, with a midpoint potential of +80 mV (Okayama, 1976), that can accept two electrons and two protons to form PQH2 (Rich and Bendall, 1980). PQH2 can donate both electrons to the cytochrome b6f complex, one to low-potential cytochrome b6, by which reduced high-potential cytochrome b6 is formed, and one to the cytochrome f moiety on the lumenal side of the thylakoid membrane, where the two protons are released. High-potential cytochrome b6 then donates an electron back to PQ on the stromal side of the membrane, rendering a semiquinone in the PQ-binding pocket on the cytoplasmic face of the b6f complex ready as an acceptor of another electron from PSII, and reduced cytochrome f feeds an electron to a water-soluble electron carrier (i.e. either plastocyanin or cytochrome c553) for subsequent transfer to the reaction center of PSI or to cytochrome c oxidase, respectively (Rich et al., 1991; Geerts et al., 1994; Schubert et al., 1995; Paumann et al., 2004; Mulkidjanian, 2010).
Electron transfer through the cytochrome b6f complex proceeds according to the Q-cycle mechanism (Rich et al., 1991). As a result, maximally two protons from the stroma are released into the lumen per electron transferred. This electrochemical proton gradient can be used for the synthesis of ATP by the ATP synthase complex (Walker, 1998). In PSI, another transthylakoid membrane charge separation process is energized by light. Electron transfer within the PSI complex involves iron-sulfur clusters and quinones and leads to the reduction of ferredoxin, the reduced form of which serves as the electron donor for NADPH by the ferredoxin:NADP+ oxidoreductase enzyme (van Thor et al., 1999). The ATP and NADPH generated this way are used for CO2 fixation in a mutual stoichiometry that is close to the stoichiometry at which these two energy-rich compounds are formed at the thylakoid membrane. Normally, this ratio is ATP:NADPH = 3:2 (Behrenfeld et al., 2008).
Photosynthetic and respiratory electron transport in cyanobacteria share a single PQ pool (Aoki and Katoh, 1983; Aoki et al., 1983; Matthijs et al., 1984; Scherer, 1990). Respiratory electron transfer provides cells the ability to form ATP in the dark, but this ability is not limited to those conditions. Transfer of electrons into the PQ pool is the result of the joint activity of PSII, respiratory dehydrogenases [in particular those specific for NAD(P)H and succinate], and cyclic electron transport around PSI (Mi et al., 1995; Cooley et al., 2000; Howitt et al., 2001;Yeremenko et al., 2005), whereas oxidation of PQH2 is catalyzed by the PQH2 oxidase, the cytochrome b6f complex, the respiratory cytochrome c oxidase (Nicholls et al., 1992; Pils and Schmetterer, 2001; Berry et al., 2002), and possibly plasma terminal oxidase (Peltier et al., 2010). Multiples of these partial reactions can proceed simultaneously, including respiratory electron transfer during illumination (Schubert et al., 1995), which includes oxygen uptake through a Mehler-like reaction (Helman et al., 2005; Allahverdiyeva et al., 2013).
Because of its central location between the two photosystems, the redox state of the PQ pool has been identified as an important parameter that can signal photosynthetic imbalances (Mullineaux and Allen, 1990; Allen, 1995; Ma et al., 2010; Allen et al., 2011). Yet, an accurate estimation of the in vivo redox state of this pool has not been reported in cyanobacteria so far. Instead, the redox state of the PQ pool is widely assumed to be reflected in, or related to, the intensity of the chlorophyll a fluorescence emissions (Prasil et al., 1996; Yang et al., 2001; Gotoh et al., 2010; Houyoux et al., 2011). Imbalance in electron transport through the two photosystems may lead to a loss of excitation energy and, hence, to a loss of chlorophyll a fluorescence emission (Schreiber et al., 1986). Therefore, patterns of chlorophyll a fluorescence (pulse-amplitude modulated [PAM] fluorimetry; Baker, 2008) have widely been adopted for the analysis of (un)balanced photosynthetic electron transfer and, by inference, for indirect recording of the redox state of the PQ pool. However, the multitude of electron transfer pathways in the thylakoid membranes of cyanobacteria (see above) makes it much more complex to explain PAM signals in these organisms than in chloroplasts (Campbell et al., 1998). Additional regulatory mechanisms of nonphotochemical quenching, via the xanthophyll cycle in chloroplasts (Demmig-Adams et al., 2012) and the orange carotenoid protein (Kirilovsky and Kerfeld, 2012) in cyanobacteria, and energy redistribution via state transitions (Allen, 1995; Van Thor et al., 1998) complicate such comparisons even further.
Several years ago, an HPLC-based technique was developed for the detection of the redox state of PQH2 in isolated thylakoids (Kruk and Karpinski, 2006), but these results have neither been related to physiological conditions nor to the results of chlorophyll a fluorescence measurements. In this report, we describe an adaptation of this method with elements of a method for estimation of the redox state of the ubiquinone pool in Escherichia coli (Bekker et al., 2007). This modified method allows for reliable measurements of the redox state of the PQ pool of Synechocystis sp. strain PCC 6803 under physiologically relevant conditions. The method uses rapid cell lysis in an organic solvent to arrest all physiological processes, followed by extraction and identification of PQH2 by HPLC separation with fluorescence detection. Next, we manipulated the redox state of the PQ pool with various redox-active agents, with inhibitors of photosynthetic electron flow, and by illumination with light specific for either PSII or PSI. The measured redox state of the PQ pool was then related to the chlorophyll a fluorescence signal and 77 K fluorescence emission spectra of cell samples taken in parallel and to oxygen-exchange rates measured separately. These experiments reveal that, despite highly fluctuating conditions of photosynthetic and respiratory electron flow, a remarkably stable redox state of the PQ pool is maintained. This homeostatically regulated redox state correlates poorly in many of the conditions tested with the more dynamic signal of chlorophyll a fluorescence emission, as measured with PAM fluorimetry. The latter signal only reflects the redox state of QA and not that of the PQ pool.
RESULTS
Method Development for PQ Pool Extraction and Quantitative Estimation
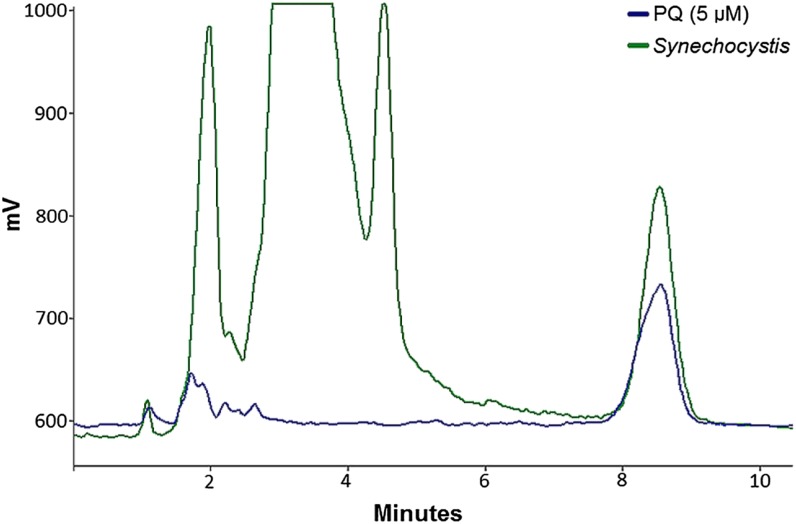
Established protocols for the quantitative estimation of the amount of quinone and quinol present in extracts of rapidly quenched intact cells proved not directly applicable to cyanobacteria. In principle, both PQ and PQH2 can be detected with HPLC through absorbance (255 nm) and fluorescence (290-nm excitation, 330-nm emission) measurements, respectively. However, for technical reasons, the maximal volume in rapid sampling had to be restricted to 2 mL. The amount of PQ that can be maximally extracted from such a sample (approximately 1 nmol) gives too low an absorbance signal in our detection system for meaningful quantitation. Therefore, we switched to the more sensitive detection of fluorescence emission from PQH2 for the analysis of the extracts. Separation of PQH2 from the other components in the extracts was done by HPLC; the peak eluting at 8.5 min (Fig. 1) was identified as PQH2 via comparison with a pure standard. For estimation of the in vivo PQ-PQH2 ratio, quadruplicate samples were quenched immediately during the experiment, and at the end of each experiment an additional quadruplicate sample was taken and reduced with NaBH4 before rapid extraction. For complete reduction of the PQ pool, we observed that an amount of 2.5 mg NaBH4 µg−1 chlorophyll a is optimal (Supplemental Fig. S1). The total size of the PQ pool does not change significantly over the course of a typical experiment with a maximal duration of 1 h (data not shown). The in vivo PQH2 content was determined in the immediately quenched sample, and the total PQ pool size in vivo was determined in the fully reduced sample. The in vivo redox state of the PQ pool is then expressed as [PQH2]/([PQ] + [PQH2]) × 100%.
Figure 1.
HPLC trace of a fully reduced 5 µm PQH2 standard (blue) and a fully reduced PQH2-containing extract from Synechocystis sp. strain PCC 6803 (green). Samples were reduced with NaBH4. The peak in the extract eluting at 8.5-min coelutes with the PQH2 standard. Fluorescence excitation/emission was at 290/330 nm. The cutoff at 1,000 mV is the upper detection limit of the system. [See online article for color version of this figure.]
The relatively low midpoint potential of the PQ/PQH2 couple (+80 mV) made it necessary to protect PQH2 against autooxidation. We found that the rate of oxidation of PQH2 in methanol is so fast that methanol alone is unsuitable for PQH2 extraction (Table I). In contrast, petroleum ether (PE) proved to prevent the oxidation of PQH2, and when PE was used as a 1:1 (v/v) mixture with methanol, PQH2 autooxidation in a 5-min period was negligible. Therefore, this latter mixture was selected as an appropriate solvent mixture for rapid extraction of PQH2. The extraction efficiency was tested at a chlorophyll a concentration of 3 mg L−1 by repeating the PQ extraction steps four times and determining the PQ compound of each fraction. Under these conditions, we found that the first and second fractions contained approximately 80% and 20% of all PQ extracted, respectively, whereas the third and fourth fractions contained around 1.5% and less than 0.5%, respectively, (data not shown); from this, we concluded that two extraction steps are sufficient. After extraction, the combined PE phases (see “Materials and Methods”) were immediately dried in a flow of N2 and stored at –20°C in 100 µL of hexanol in an HPLC vial until processing. We observed that both for storage at low temperature and subsequent analysis by HPLC, hexanol as a solvent showed the lowest autooxidation rates (Table I). Shorter chain alcohols (ethanol, propanol, and n-butanol) show higher rates, and longer chains (octanol and decanol) do not significantly lower it. Alkanes and PE generate peaks in the HPLC scans that distort the PQ peak (data not shown). Although hexanol was the most suitable solvent, it did not completely prevent autooxidation at room temperature. For technical reasons, HPLC had to be performed at room temperature, and at this temperature, PQH2 in hexanol has an autooxidation rate of approximately 5% per hour during the first 3 h (Supplemental Fig. S2). Therefore, we limited each HPLC run to a maximal run time of 2.5 h. The data obtained were then corrected for the time the sample spent at room temperature in the autosampler prior to HPLC analysis.
Table I. Half-life of PQH2 (due to autooxidation) in different solvents at various temperatures.
Oxidation was measured over a time course of 5 min at 4°C, 8 h at 21°C, and 24 h at −20°C and −80°C. Data are from a typical experiment.
| Solvent | Half-Life h | Temperature °C |
|---|---|---|
| Methanol | 0.04 | 4 |
| 1:1 Methanol:PE (40°C–60°C) | ∞ | 4 |
| Ethanol | 2.89 | 21 |
| Hexanol | 9 | 21 |
| Hexanol | 1,565 | −20 |
| Hexanol | 376 | −80 |
| Dry | 84 | −20 |
| Dry | 232 | −80 |
The Redox State of the PQ Pool in Growing Cells
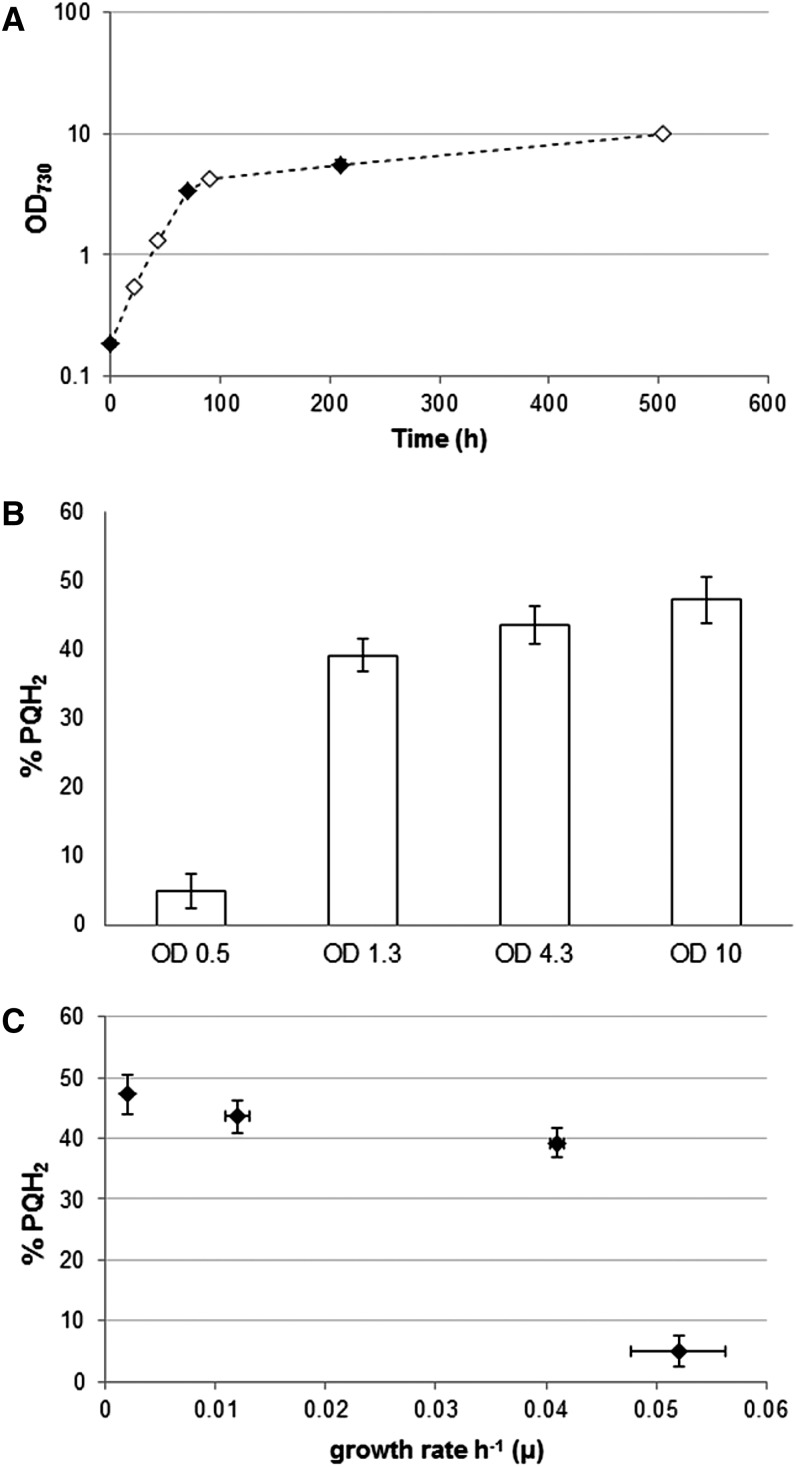
To determine the effect of redox manipulations through changing physiological conditions on the in vivo redox state of the PQ pool, we tested both actively growing and stationary phase cells. We found that non-light-limited, fast-growing Synechocystis sp. strain PCC 6803 cells have an oxidized PQ pool while stationary phase cells have a rather reduced PQ pool (Fig. 2). In between, during lower exponential growth rates caused by light limitation, we consider a growth phase with an intermediate redox state of the PQ pool around optical density at 730 nm (OD730) = 0.8. To be able to monitor both the reduction and oxidation of the redox state of the PQ pool, this latter growth phase was selected for further experiments.
Figure 2.
Growth of Synechocystis sp. strain PCC 6803 in batch culture in BG-11 medium with 25 mm NaHCO3 (A) and the corresponding PQ redox states plotted against OD730 (B) and growth rate per hour (µ; C) at the four time points indicated with white diamonds in A. Error bars indicate sd of biological duplicates.
Redox-Active Reagents and Inhibitors of Photosynthetic Electron Flow
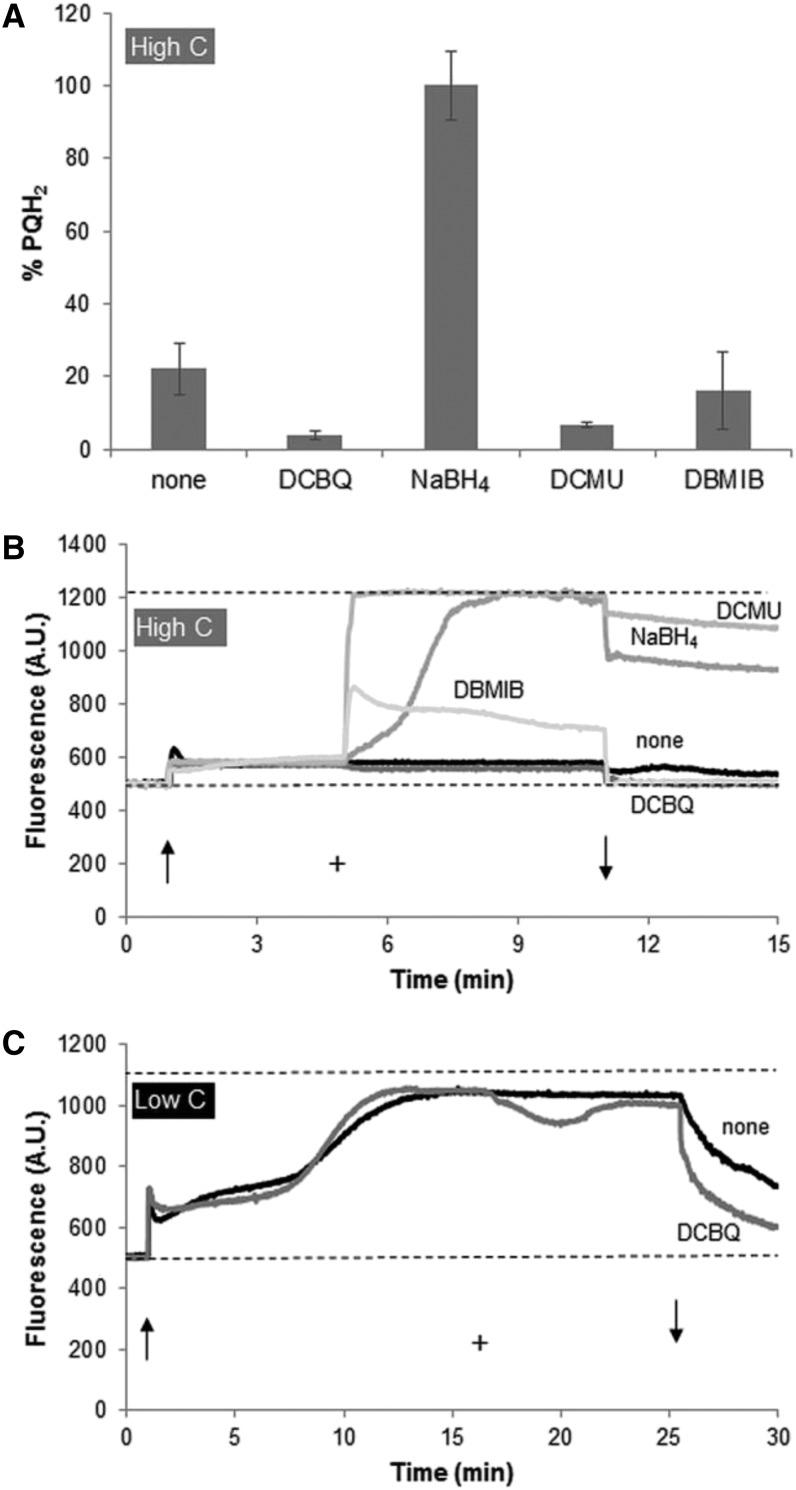
The quinone analog 2,6-dichloro-p-benzoquinone (DCBQ), in combination with 1 mm Fe3+, can take all electrons from PQH2 and completely oxidize the pool to PQ; conversely, NaBH4 is a strong reducing agent that can convert all PQ into PQH2. The use of both chemicals permits one to mark the fully oxidized and the fully reduced states of the PQ pool, respectively. Setting of those conditions was confirmed through the PQ extraction procedure (Fig. 3A). Figure 3B shows that the addition of NaBH4 raises chlorophyll a fluorescence to the maximum PSII fluorescence in the dark-adapted state (Fm) level, whereas DCBQ does not affect the chlorophyll a fluorescence signal. Since the steady-state fluorescence signal is close to the initial (minimum) PSII fluorescence in the dark-adapted state (F0) in cells incubated in high carbon and moderate red light intensities (655 nm, 60 µmol photons m−2 s−1), the DCBQ experiment was repeated with cells incubated in low-carbon conditions and 100 µmol photons m−2 s−1 red light (Fig. 3C). This figure shows that, under these latter conditions, DCBQ does lower the chlorophyll a fluorescence signal but it does not approach F0, as one would expect with a fully oxidized PQ pool, provided the fluorescence signal reflects the redox state of the PQ pool.
Figure 3.
PQ redox state (A) and chlorophyll a fluorescence recordings (B and C) demonstrating the response of Synechocystis sp. strain PCC 6803 cells to the addition of a range of different redox-active substances. PQ samples were taken 5 min after the addition of each chemical. Arrows indicate light on (↑) and light off (↓), and + indicates the addition of the chemical. A and B, Experiments conducted in BG-11 with 50 mm NaHCO3 and 60 µmol photons m−2 s−1 655-nm light (high-carbon conditions). C, Experiments conducted in BG-11 with 0.5 mm NaHCO3 and 100 µmol photons m−2 s−1 655-nm light (low-carbon conditions). Final concentrations were 10 µm DCBQ + 1 mm K3Fe(CN6), 2 mg mL−1 NaBH4, 20 µm DCMU, and 0.5 µm DBMIB. PQ redox state data in A are averages of three independent experiments, and error bars indicate sd. Chlorophyll a fluorescence data in B and C are from typical experiments. AU, Arbitrary units.
The photosynthetic electron transfer inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) has clear effects on the PAM signal (Fig. 3B). DCMU blocks the QA-to-QB electron transfer in PSII, which, in the light, causes complete reduction of QA and yields the maximal fluorescence signal. DCMU prevents electrons from PSII from flowing into the PQ pool and thus would leave the PQ pool fully oxidized, provided that the efflux of electrons (e.g. via PSI and/or the respiratory oxidases) continues. Although oxidation of the PQ pool was observed in the presence of DCMU, some PQH2 still remained (Fig. 3A). 2,5-Dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) prevents the outflow of electrons from the PQ pool by blocking the PQH2-binding site on the lumenal side of the thylakoid membrane of the cytochrome b6f complex (Qo). With an active PSII, this should cause strong reduction of the PQ pool, which, in turn, would be expected to cause an increase in the chlorophyll a fluorescence signal. Addition of DBMIB does cause a strong rise in chlorophyll a fluorescence, followed by a slow drop and a stabilization of the signal at a level that is about twice as high as without the addition (Fig. 3B). Interestingly, addition of DBMIB does not reduce the PQ pool; if anything, a small oxidation can be observed (Fig. 3A). This experiment was repeated under very low oxygen conditions (N2 sparging in the presence of Glc and Glc oxidase) and in the presence of 5 mm d-isoascorbic acid to fully reduce DBMIB beforehand; these latter experiments yielded similar results (data not shown).
PQ Redox State in Phycobilisomes Only Light, and in Phycobilisomes Plus PSI Light
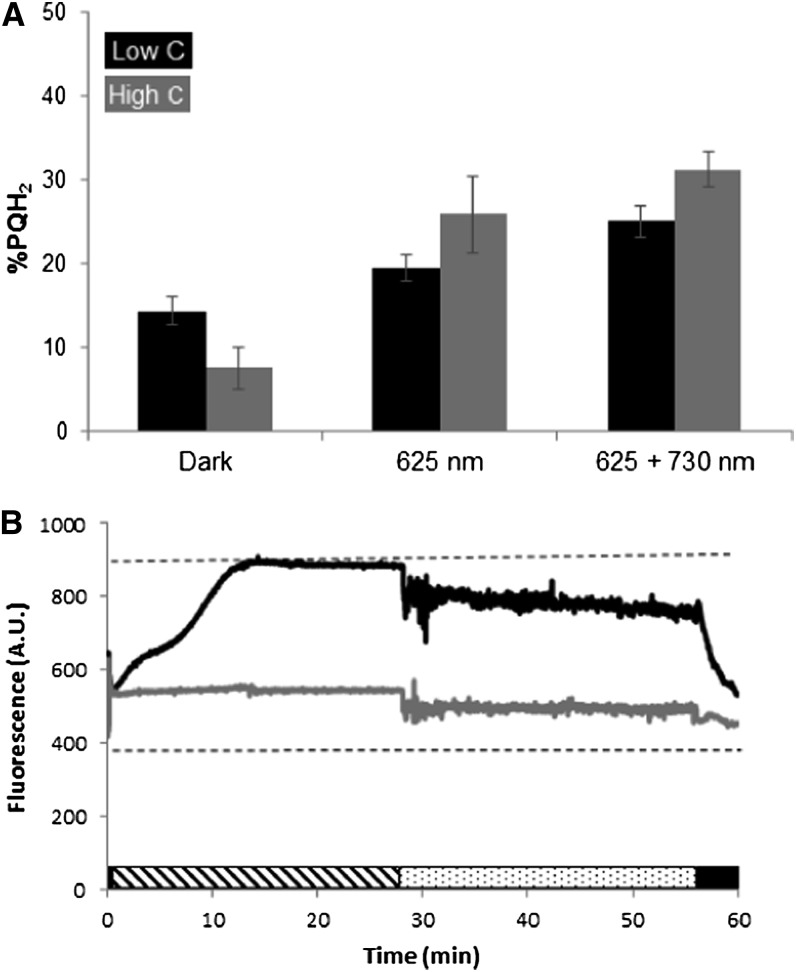
In order to achieve different redox states in the PQ pool under physiological conditions, we used different mixtures of actinic light, absorbed by the phycobilisomes (PBSs; 625 nm) and PSI (730 nm). Accordingly, an experiment was set up in which cultures with low or high carbon availability were illuminated with 100 µmol photons m−2 s−1 625-nm light. After 25 min, 25 µmol photons m−2 s−1 730-nm light was added to the 625-nm illumination. Samples for analysis of the PQ redox state and for 77 K fluorescence measurements were taken after 30 min in the dark and 10 min after the start of each of the illumination conditions. Figure 4A shows that the redox state of the PQ pool is somewhat more reduced in high-carbon than in low-carbon conditions after 10 min of 625-nm light, whereas in the dark, the redox pool of the low-carbon sample is more reduced. This should be compared with the massive difference in chlorophyll a fluorescence signals in 625-nm light (Fig. 4B). The small drop in chlorophyll a fluorescence and the increase in noise after the addition of 730-nm light is an artifact caused by scattering of the 730-nm actinic light into the PAM detector; therefore, no information could be extracted from the PAM signal in the presence of 730-nm illumination. The effect of 730-nm light on the redox state of the PQ pool, however, can be interpreted: it is more reduced under 625-nm illumination than in darkness and becomes even more reduced when 730-nm light is added (Fig. 4B). The latter observation is counterintuitive, since one would expect that PSI-specific light will oxidize the PQ pool.
Figure 4.
Redox state of the PQ pool and chlorophyll a fluorescence emission of Synechocystis sp. strain PCC 6803 cells under various illumination conditions. A, PQ redox states. Samples were taken after 30 min in the dark, after 10 min in 100 µmol photons m−2 s−1 625-nm light, and 10 min after the addition of 25 µmol photons m−2 s−1 730-nm light. Black bars, BG-11 medium with 0.5 mm NaHCO3; gray bars, BG-11 with 50 mm NaHCO3. Data shown are averages of biological duplicates with sd. B, Chlorophyll a fluorescence recordings. Striped bar, 100 µmol photons m−2 s−1 625-nm light; dotted bar, 100 µmol photons m−2 s−1 625-nm light with 25 µmol photons m−2 s−1 730-nm light; black bar, dark; dashed lines, F0 and Fm. AU, Arbitrary units.
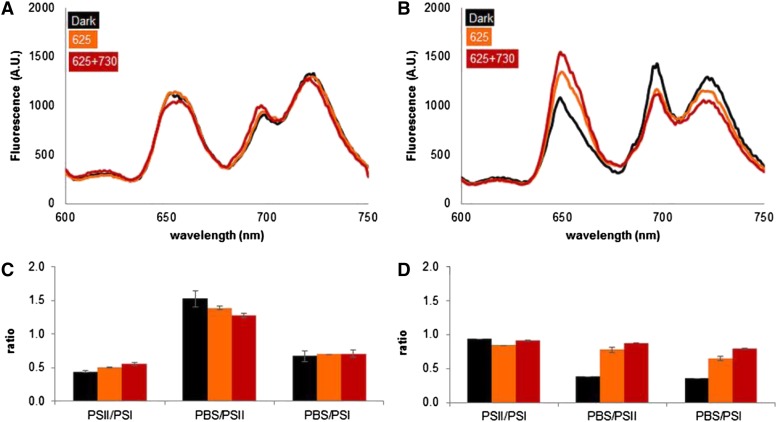
At 77 K, the photosynthetic pigments are locked in place, but they can still transfer their excitation energy to the photosystem they are bound to. By illuminating cell samples at 77 K with light specific for PBS excitation and recording the fluorescence spectra, we can get some insight into the level of coupling of the PBS to the photosystems. Fluorescence at 655 nm is emitted by phycocyanin, fluorescence at 685 nm indicates coupling of the PBS to PSII, and fluorescence at 720 nm indicates coupling to PSI. Figure 5 shows that the addition of 730-nm light triggers coupling of the PBS particularly to PSII in low-carbon medium (Fig. 5A), while with high carbon availability it triggers the release of PBS mainly from PSI (Fig. 5B).
Figure 5.
77 K fluorescence emission spectra, recorded with 590-nm excitation, of Synechocystis sp. strain PCC 6803 cells sampled under different conditions of carbon availability and illumination (A and B) and ratios of peak areas derived through skewed Gaussian deconvolution of these spectra (C and D). A and C, BG-11 with 0.5 mm NaHCO3. B and D, BG-11 with 50 mm NaHCO3. Black bars, samples taken after 30 min in the dark; orange bars, samples taken after 10 min in 100 µmol photons m−2 s−1 625-nm light; red bars, samples taken 10 min after the addition of 25 µmol photons m−2 s−1 730-nm light to a background of 100 µmol photons m−2 s−1 625-nm light. The data shown are averages of biological duplicates; error bars in C and D indicate sd. The peaks at 655, 685, and 720 nm are due to emission from phycocyanin, PSII, and PSI, respectively. AU, Arbitrary units. [See online article for color version of this figure.]
Oxygen Evolution from PSII and Selective Activation of PSI with 730-nm Light
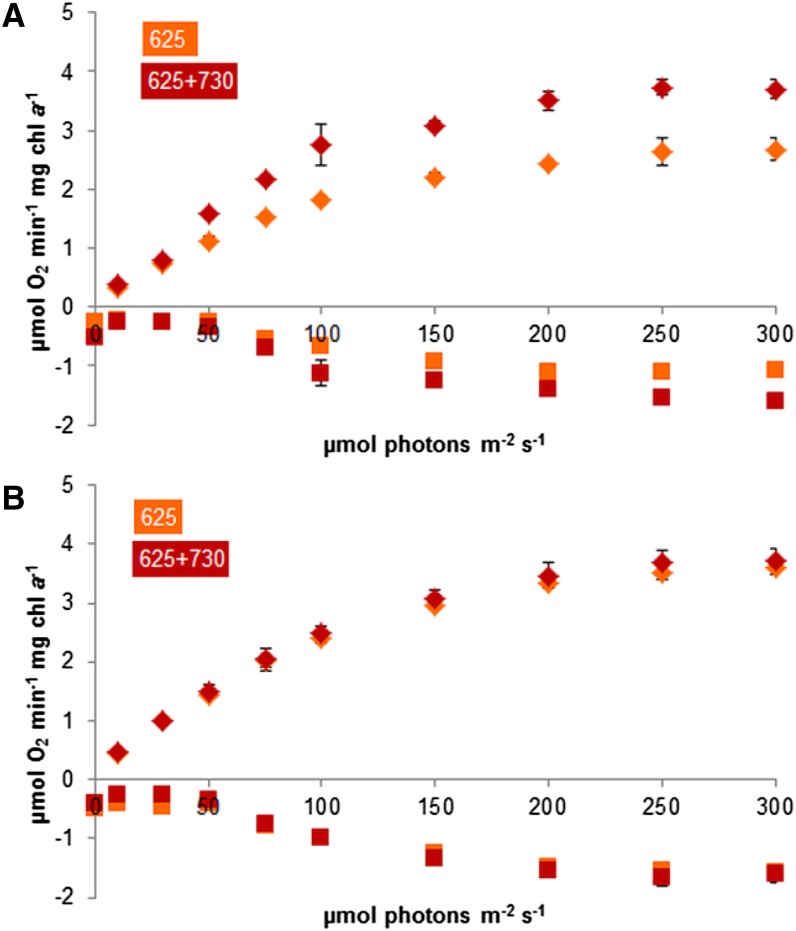
To further assess the effect of PBS- and PSI-specific illumination on the function of PSII, we measured oxygen evolution rates. The use of oxygen evolution as a proxy for PSII performance in whole cells requires an insight into oxygen uptake processes to discriminate overall oxygen exchange from net oxygen production at PSII. The membrane inlet mass spectroscopy (MIMS) technique permits this. By adding a small amount of 18O2 gas to the culture, we were able to detect oxygen uptake in the light; more details of this approach are presented in “Materials and Methods.” With an increasing 625-nm photon flux, the addition of 25 µmol photons m−2 s−1 730-nm light induces a large increase in the rate of oxygen evolution in cells with low carbon availability (Fig. 6), allowing the cells to evolve oxygen at a rate that is comparable to cells in conditions of carbon excess. Values for the maximal rate of oxygen evolution (Pmax) and the affinity for light (the initial tangent to the curve that relates the rate of oxygen evolution to the light intensity [α]) were determined from these data using Sigmaplot (Table II). Also, oxygen uptake in the light appears proportional to the amount of 625-nm light provided and exceeds respiration in the dark in all but the conditions with the lowest light intensities.
Figure 6.
Oxygen evolution rates of Synechocystis sp. strain PCC 6803 cultures in increasing intensities of 625-nm light, recorded with MIMS. 18O2 was added at the start of the experiment to monitor oxygen consumption in the light. Depicted are net photosynthesis (which is the sum of the measured oxygen production and the consumption [diamonds]) and respiration (squares). A, BG-11 with 0.5 mm NaHCO3. B, BG-11 with 50 mm NaHCO3. Orange symbols, 625-nm light only; red symbols, 625-nm light + 25 µmol photons m−2 s−1 730-nm light. Data shown are averages of biological duplicates with sd. [See online article for color version of this figure.]
Table II. α and Pmax values of net photosynthesis rates as depicted in Figure 6, calculated with Sigmaplot.
LC, BG-11 with 0.5 mm NaHCO3; HC, BG-11 with 50 mm NaHCO3; orange, 625-nm light; far-red, 625-nm light with the addition of 25 µmol photons m−2 s−1 730-nm light. α, Affinity for light; Pmax, maximal rate of oxygen evolution. Sigmaplot was used for calculations.
| Condition | α | ± | Pmax | ± | P |
|---|---|---|---|---|---|
| LC orange | 0.0228 | 0.0008 | 2.66 | 0.049 | <0.0001 |
| LC far-red | 0.0324 | 0.0012 | 3.75 | 0.078 | <0.0001 |
| HC orange | 0.0300 | 0.0009 | 3.6 | 0.060 | <0.0001 |
| HC far-red | 0.0308 | 0.0010 | 3.75 | 0.067 | <0.0001 |
DISCUSSION
In this study, we have developed an extraction method with which we can reproducibly assay the in vivo PQ redox state of Synechocystis sp. strain PCC 6803. Because the extinction coefficient of PQ is too small to detect this quinone in cell extracts by spectrophotometry, we measured its concentration indirectly by making use of the detection of fluorescence emission by PQH2. To determine the PQ redox state in a cell culture, two sets of samples were taken and subsequently differently processed in quadruplicate: one sample was the PQ/PQH2 extracted as is, and the other was first completely reduced with NaBH4 before extraction of PQH2. The difference in PQH2 content in the two types of sample preparations is equal to the in vivo PQ redox state. The PQ redox state of the cells is defined as the ratio of the in vivo amount of PQH2 over the total amount of PQ/quinol, expressed as a percentage. It should be noted that because of this approach, the method becomes relatively inaccurate when the PQ pool becomes very reduced (e.g. more than 90%). We observed, however, that the actual physiological reduction level of the PQ pool never exceeded 50% in our experiments. Hence, a substantial part of the PQ pool is always present as PQ. Also, very small changes in the redox state of the PQ pool cannot be monitored with our technique. In the permissible domain between 10% and 90%, the sd ranges from 5% to about 10%, and technical restraints (for more details, see “Materials and Methods”) limit the number of parallel samples that can be analyzed for a given condition. Our data show that we can accurately detect transitions in the redox state of the PQ pool from more reduced to more oxidized and vice versa in response to changes in the physiology of the Synechocystis sp. strain PCC 6803 cells. With the data we acquired, we can also estimate the size of the PQ pool and how this size changes with growth phase. We found that each cell contains around 1.2 fg of PQ and 27 fg of chlorophyll a and that these values are stable in the range of 5 × 107 to 3 × 108 cells mL−1 (i.e. in concomitantly measured OD730, values range from 0.5–3; Table III). This implies that the PQ content of the cells does not change significantly between the linear light-limited growth phase and the phase of growth in which carbon limitation presumably starts to contribute. Only in stationary phase do the PQ and chlorophyll a contents per cell go down (Table III).
Table III. Approximate amounts of chlorophyll a and PQ per cell at different cell densities.
n, Number of replicates the estimate was based on.
| No. of Cells | Chlorophyll a | PQ | n |
|---|---|---|---|
| fg cell−1 | |||
| 5 × 10−7 to 3 × 10−8 | 27 ± 2.7 | 1.2 ± 0.33 | 17 |
| 4 × 10−8 to 5 × 10−8 | 24 ± 0.1 | 0.8 ± 0.08 | 3 |
| 1 × 10−9 | 16 | 0.2 | 1 |
A basic assumption in this study has been that Synechocystis sp. strain PCC 6803 cells contain a single, homogenous, redox-equilibrated PQ pool. Nevertheless, some studies have shown that the distribution of protein components over the thylakoid membrane is nonhomogenous (Vermaas et al., 2008; Liu et al., 2012), and between the thylakoid and cytoplasmic membranes there are definitely differences in their abundance. This may cause differences in the activity of respiratory and photosynthetic electron flow in these two types of membranes (but see Scherer, 1990). However, there is currently no evidence for the existence of local proton gradients. A further complication is that the literature reports the existence in chloroplasts and cyanobacteria of active and inactive PQ pools, such that the inactive pool is located in small lipidic compartments called plastoglobuli. The inactive pool may comprise up to two-thirds of the total amount of PQ in chloroplasts (Kruk and Karpinski, 2006; Piller et al., 2012), and genes encoding plastoglobulin-related proteins have also been identified in Synechocystis sp. strain PCC 6803 (Cunningham et al., 2010).
Many studies in photosynthesis have used eukaryotic organisms (be they plants or [green] algae), in which oxidative phosphorylation (including respiratory electron transfer) and photosynthesis are separated into separate cellular organelles. In cyanobacteria, however, photosynthesis and respiration are intertwined and share PQ as a mobile electron carrier (Aoki and Katoh, 1983; Matthijs et al., 1984; Scherer, 1990; Mullineaux, 2014). From this, it follows that both photosynthetic and respiratory electron flow determine the PQ redox state in cyanobacteria. This difference between cyanobacteria and chloroplasts may be the underlying reason for the observed strong homeostatic control of the redox state of the PQ pool of Synechocystis sp. strain PCC 6803 under a wide range of physiological incubation conditions that include anaerobiosis and exposure to high light intensities (see “Results”; data not shown). The same broad spectrum of electron entry and exit pathways that is present in cyanobacteria (see the introduction) is not available in chloroplasts of green algae and plants, although both some PQ reduction and PQ oxidation systems, called chlororespiration, have been demonstrated in plant and microalgal chloroplasts (Heber and Walker, 1992; Corneille et al., 1998; Casano et al., 2000; Dijkman and Kroon, 2002; Jans et al., 2008; Miyake et al., 2009; Peltier et al., 2010; Houyoux et al., 2011). Nevertheless, the experimental procedure of PQ pool extraction that we present here can be applied in green algae as well, because the different types of quinones in mitochondria and chloroplasts permit their separate analysis with HPLC.
Our interest in the in vivo redox state of the PQ pool in cyanobacteria emerged from the relationship between the thylakoid redox state and several of the regulatory mechanisms that plants, microalgae, and cyanobacteria use to cope with dynamic changes in their environmental conditions, in particular light intensity. Quite a few regulatory mechanisms and underlying signal-transducing pathways have been attributed to the redox state of the PQ pool already (Fujita et al., 1987; Mullineaux and Allen, 1990; Escoubas et al., 1995; Oswald et al., 2001; Liu et al., 2012). With a strong homeostatic regulation and a highly stabilized PQ pool redox state, the proposed redox control of these regulatory processes may not be as straightforward to interpret as anticipated previously. Evidence acquired in this work includes the fact that the PSII inhibitor DCMU causes a maximal increase of chlorophyll a fluorescence by blocking QA-to-QB electron transfer, but this leads to only a partial oxidation of the PQ pool. This illustrates the active role of respiratory dehydrogenases and cyclic electron flow around PSI (Mi et al., 1995; Howitt et al., 2001; Yeremenko et al., 2005) in the supply of electrons to the PQ pool. DBMIB prevents the access of PQH2 to the Qo pocket of the cytochrome b6f complex (Roberts and Kramer, 2001). With PSII active, this should lead to an increase in the degree of reduction of the PQ pool. However, this study shows that the addition of DBMIB does not lead to reduction of the PQ pool (Fig. 3A). The binding of DBMIB shows only moderate affinity (Nanba and Katoh, 1984; Rich et al., 1991; Trebst, 2007), and competition for the PQH2 binding site depends on the redox state of DBMIB, which can be modulated by the cells (Rich et al., 1991). As the respiratory oxidases are insensitive to DBMIB, this allows electrons to exit the PQ pool even in the presence of this inhibitor (Roberts et al., 2004). It has been reported that DBMIB can cause oxidation of the PQ pool and that it can stimulate oxygen uptake (Nanba and Katoh, 1984). Also, oxidized DBMIB could function as an electron acceptor or a quencher of fluorescence (Berry et al., 2002). So the experiment was repeated with DBMIB in the presence of 5 mm d-isoascorbate to fully reduce DBMIB prior to the experiment and in cultures that were continuously sparged with nitrogen and to which Glc and Glc oxidase were added. Even under these conditions, DBMIB addition did not cause a reduction of the PQ pool (data not shown).
Previous studies of a functional relationship between the redox state of the PQ pool and regulatory processes in the cells often relied on the use of inhibitors of photosynthetic electron transport, such as DCMU and DBMIB. As demonstrated in this work, these agents do not exactly have the predicted effect on the redox state of the PQ pool in Synechocystis sp. strain PCC 6803, which supports the notion that some of the regulatory processes are controlled via the sensing of other components, like the occupancy in the Qo or QA site (Zito et al., 1999; Mao et al., 2002; Ma et al., 2010), and not by the redox state of the PQ pool itself. However, the method used in this work cannot monitor the PQ redox state continuously; samples are taken manually and require immediate processing, making one sample per minute the maximal time resolution. So it is possible that the addition of inhibitors or changes in the illumination conditions may result in rapid, but transient, changes in the redox state to which the cell may respond.
Among the processes for which the signal transduction route urgently awaits clarification, and for which redox regulation has been implied, are the state transitions that regulate the distribution of photon energy over the two photosystems (Van Thor et al., 1998; Joshua and Mullineaux, 2004; Mullineaux, 2008; Dong et al., 2009; Kondo et al., 2009).
The impact of the redox-active chemicals DCBQ (redox midpoint potential under conditions with a pH of 7 equals +315 mV) and NaBH4 (redox midpoint potential under standard conditions equals −1.24 V), which have an absolute effect on the redox state of the PQ pool, is also reflected in the chlorophyll a fluorescence signal (Fig. 3, B and C). NaBH4 is a reducing agent, which should completely reduce the PQ pool, leaving no electron acceptors available for PSII. In this respect, it is not surprising that the addition of NaBH4 results in a maximal chlorophyll a fluorescence signal, which is essentially similar to the level to which DCMU raises this fluorescence, consistent with the expectation that NaBH4 will also fully reduce QA. When the light is switched off in the presence of NaBH4, there is a steep drop in fluorescence, which goes against the idea that NaBH4 will fully reduce QA. However, the chlorophyll a fluorescence signal in cyanobacteria is, in part, distorted by fluorescence from unbound PBS (Campbell et al., 1998). This could, in part, explain the sudden drop in fluorescence. In contrast, the addition of DCBQ (in the presence of Fe3+) takes all the electrons out of the PQ pool (Shevela and Messinger, 2012) but only slightly lowers the chlorophyll a fluorescence signal, and only when this signal is high to start with. In low-carbon medium, there is an imbalance between ATP supply and electron acceptor availability. Generally, lack of CO2 is accompanied by an increase in the QA reduction level (and, therefore, a strong increase in the chlorophyll a fluorescence signal; see above) but also a higher resistance to photoinhibition (Sane et al., 2003). Under conditions of high excitation pressure, there is increased cyclic electron flow around PSII via cytochrome b559 (Buser et al., 1992). It has also been shown that in low-carbon conditions, the flavodiiron proteins Flv2 and Flv4 help protect PSII by accepting electrons from the reaction center (Zhang et al., 2012; Hakkila et al., 2013). Transfer of electrons to cytochrome b559 is much slower than transfer to the PQ pool, which could lead to the accumulation of reduced QA, which would explain the high fluorescence level. DCBQ is a general quinone analog, and it would certainly be possible for DCBQ to accept electrons from cytochrome b559 or even from QA or QB directly. This would alleviate the back pressure on PSII and explain the drop in chlorophyll a fluorescence in low-carbon conditions. Hence, while chlorophyll a fluorescence measurements report on the redox state of PQ in the QA site of PSII, the redox state of this component may differ from the redox state of the PQ pool.
The direct comparison between the redox state of the PQ pool and the intensity of the chlorophyll a fluorescence signal under a range of physiological conditions further demonstrates the poor correlation between these two parameters (Fig. 4). To further illustrate their lack of direct correlation, a typical discrepancy between the two is shown in 625-nm light (that selectively excites the PBS), in which the PQ pool is somewhat more reduced in high-carbon conditions than it is in low-carbon conditions, while the chlorophyll a fluorescence signal is much lower in high-carbon conditions (Fig. 4). This discrepancy is most likely caused by an up-regulation of cyclic electron flow around PSII, which lowers the electron transfer to the PQ pool and further supports our interpretation that the redox states of QA and of the PQ pool are not directly correlated to one another. In the redox midpoint potentials on the acceptor side of PSII, a clear gradient is observed: QA/QA− ∼ −100 mV (Krieger-Liszkay and Rutherford, 1998; Allakhverdiev et al., 2011), QB/QB− ∼ 0 mV (Nicholls and Ferguson, 2013), and the PQH2/PQ pool = +80 mV (Okayama, 1976). Hence, it is understandable that, if for some reason the kinetics of electron transfer in the initial part of the Z-scheme are impaired, the correlation between the redox states of QA, QB, and the PQ pool is lost.
Further studies on the effect of additional PSI light on this correlation were hampered by the limitations of our equipment to measure chlorophyll a fluorescence. With a chlorophyll a extract in 80% (v/v) acetone, with added milk powder to introduce light scatter, we confirmed that the initial drop that we observed in the PAM signal (Fig. 4B) after the addition of 730-nm light is an artifact in the form of an offset of the PAM measuring system (data not shown).
We expected the PQ pool to be more oxidized in the presence of PSI-specific illumination because this light should enable PSI to oxidize the PQ pool at a higher rate. However, this was not observed (Fig. 4A). The dynamically variable connection of the PBS antenna to either PSII or PSI, or to neither of these two, was considered as a possible cause. Therefore, we analyzed fluorescence excitation and emission spectra recorded at 77 K. These confirm that PSI-specific light triggers a state 1 transition (Fig. 5). Especially in low-carbon conditions, the light-to-dark state transition in Synechocystis sp. strain PCC 6803 is small (Fig. 5) compared with the corresponding transition in other species such as Synechococcus sp. strain PCC 7942 (Campbell et al., 1998). Species in which state transitions, and therefore energy redistribution, are more pronounced may also display stronger variations in the PQ pool redox state. Mullineaux and Allen (1990) have proposed that state transitions in cyanobacteria are triggered by changes in the redox state of the PQ pool, or of a closely associated electron carrier, but others have ascribed this trigger function to the PQ in the Qo site of the cytochrome b6f complex (Vener et al., 1997; Mao et al., 2002). Here, we see that cells that are in state 1 have a more reduced PQ pool, rather than a more oxidized pool, than cells that are in state 2. That is, regardless of whether redox-related triggers sense the redox state of PQ, or the occupation of the Qo and/or the QA site, this sensing mechanisms cannot respond to the steady-state redox level of the PQ pool.
Conditions such as low carbon availability and high light intensities induce many protective mechanisms to limit the amount of electrons liberated from water, such as nonphotochemical quenching of PSII (Finazzi et al., 2006), state transitions (Mullineaux and Allen, 1990), cyclic electron flow around both photosystems (Prasil et al., 1996; Miyake et al., 2005), and energy-quenching mechanisms such as those facilitated by the orange carotenoid protein (Kirilovsky and Kerfeld, 2012) and the flavodiiron proteins Flv2 and Flv4 (Zhang et al., 2012; Hakkila et al., 2013). This means that, despite the high excitation pressure in low-carbon conditions, the cells in high-carbon conditions are at a greater risk of overreduction of the PQ pool if PBS binding to PSII is increased. This may explain the difference in the mechanism underlying the state transitions: the state 1, observed in high-carbon conditions, is achieved mainly by the uncoupling of PBS from PSI, while the state 1, observed in low-carbon conditions, is based on the coupling of PBS to PSII (Fig. 5, C and D; for an overview, see Supplemental Fig. S3). Further studies with sodium fluoride (a phosphatase inhibitor; Mullineaux, 1993) or the use of mutants impaired in state transitions may help to disclose in greater detail this versatility in relative PBS binding to the two photosystems in Synechocystis sp. strain PCC 6803.
Although the occurrence of a state 1 transition may not be convincing from the 77 K data alone, the oxygen evolution experiments (Fig. 6) show the strong contribution from low-intensity 730-nm light. While carbon limitation represses oxygen evolution from PSII, the addition of low-intensity 730-nm light completely abolishes this effect, lifting the oxygen evolution (rate) up to the same level as in carbon-replete conditions. The addition of 730-nm light will accelerate PSI activity and may increase the rate of cyclic electron flow around PSI. Via increased ATP synthesis, this will lower the NADPH-ATP ratio and make extra ATP available for active HCO3− uptake (Nishimura et al., 2008). The increased PSI activity and, possibly, higher availability of CO2 for carbon fixation will increase the turnover rate of PSII, in combination with the increased PBS coupling, induced by the state 1 transition, explaining the benefit of additional PSI light to cultures in low-carbon conditions and possibly under ATP stress in general. Finally, the oxygen uptake rates of Synechocystis sp. strain PCC 6803 in the light are higher than the respiration rates in the dark. An often-made assumption holds that the rate of oxygen uptake in the light will never exceed the corresponding dark respiration (Smetacek and Passow, 1990), although there have been previous reports that this assumption does not always hold (Kana, 1993; Claquin et al., 2004; Allahverdiyeva et al., 2013). To maintain a fairly oxidized PQ pool redox state, it may be necessary for the cell to dissipate quite a lot of redox energy (i.e. transfer electrons to oxygen) to prevent the overreduction of the electron transfer system. By using respiratory enzymes for this, the cell can still use the free energy of these electrons for the production of ATP. Moreover, Helman et al. (2005) have shown that up to 40% of the electrons extracted from water by PSII are directly transferred back to oxygen (to reform water) via a flavoprotein-catalyzed Mehler-like reaction. Allahverdiyeva et al. (2013) further stress the importance of this reaction in cyanobacteria. In this way, a customized combination of linear and cyclic electron flow and respiration allows the cell to balance its ATP and NADPH supplies according to the needs dictated by its physiology and its environment.
The largest variation in the redox state of the PQ pool observed in this study was in the different stages of growth (Fig. 2). With a nonlimiting supply of light and nutrients, photosynthesis can run at its highest capacity (and, indeed, cellular growth is exponential) during the first 1 or 2 d. This very rapid electron flow through the Z-scheme apparently leads to a highly oxidized PQ pool. When light and/or nutrients start to become limiting, the flow through the system will slow down and the homeostatic regulatory mechanisms will kick in.
It appears that, for cyanobacteria, the ability to homeostatically regulate the redox state of its PQ pool is more important than preserving maximum amounts of free energy in the form of NADPH and ATP. Since cyanobacteria thrive in open water columns, in which mixing can suddenly expose them to high light intensities and/or nutrient limitation, this mode of regulation may very well be an important survival strategy.
MATERIALS AND METHODS
Strains and Culture Conditions
Synechocystis sp. strain PCC 6803 was grown in a continuous culture photobioreactor (Huisman et al., 2002) with a volume of 1.8 L and a light penetration of 5 cm at a temperature of 30°C. Growth was in continuous red light-emitting diode (LED) light (650 nm, 60 µmol photons m−2 s−1 incident light) in BG-11 mineral medium (Rippka et al., 1979) complemented with 15 mm Na2CO3. Mixing was established with a stream of sparged air enriched with 2% (v/v) CO2 at a rate of 30 L h−1. The dilution rate was set to 0.015 h−1, and a light-limited steady state with a final culture density of 8 × 107 cells mL−1, an OD730 of 0.8, and chlorophyll a at 2 mg L−1 was reached.
For experiments in which the PQ pool redox state was manipulated, aliquots of the culture (between 50 and 300 mL) were taken and centrifuged (5 min, 1,500g), washed once in BG-11 complemented with 0.5 or 50 mm NaHCO3 (referred to as low- or high-carbon medium, respectively), and resuspended in the same medium to a chlorophyll a concentration of 2 mg L−1.
To study the PQ pool redox state during growth, batch cultures were inoculated at an OD730 of 0.1 in BG-11 medium complemented with 25 mm NaHCO3. The cells were grown at 30°C in a shaking incubator at 200 rpm under 30 µmol photons m−2 s−1 plant-specific fluorescent light (Sylvania Gro-Lux).
Sampling under Varying Light Conditions
Synechocystis sp. strain PCC 6803 cultures in low- or high-carbon medium with a chlorophyll a concentration of 2 mg L−1 were placed in a 300-mL flat panel culture vessel with a light path of 3 cm. The vessel was placed between two LED light sources to ensure a constant light climate inside the vessel. The monitoring optical fiber of the PAM was placed against the side of the vessel, perpendicular to the light sources. The vessel was equipped with a rapid sampler, used for PQ redox state determination, as well as a long needle connected to a 1-mL syringe for sampling for 77 K fluorescence emission spectroscopy (for details, see below). At the start of the experiments, the cultures were dark adapted for 30 min, after which samples for 77 K spectroscopy, PQ redox state analysis, and optical density measurements were taken, and the F0 and Fm of the PAM signal were determined with a saturating pulse generated by the LED lamps (for details, see below). The cultures were then exposed to 625-nm (PBS) light, which preferentially excites PSII, at an intensity of 100 µmol photons m−2 s−1, and after 10 min, samples for 77 K spectroscopy and for PQ pool extraction were taken. After 25 min in 625-nm light only, 25 µmol photons m−2 s−1 730-nm LED light for excitation of PSI was added to the 625-nm light, and after another 10 min, 77 K and PQ samples were collected again. The PAM signal was monitored continuously.
Chemical Agents Used for Modulation of the Redox State of the PQ Pool
The effect of the addition of various chemicals was tested in aliquots of cultures incubated in the light, and samples were taken 5 min after each addition. Final concentrations were as follows: 2 mg mL−1 NaBH4 (Sigma), 10 µm DCBQ (Kodak) with the addition of 1 mm K3Fe(CN)6 to allow the reoxidation of DCBQ, 20 µm DCMU (Sigma), and 0.5 µm DBMIB (Sigma).
PQH2 Detection by HPLC
By use of a rapid sampling device (Lange et al., 2001), a 2-mL cell culture was rapidly sampled (within 0.5 s) directly into the extraction agent, which consisted of a 12-mL ice-cold (4°C) 1:1 (v/v) mixture of methanol and PE (boiling point range 40°C–60°C). This ensured not only reproducible and quantitative sampling but, most importantly, freezing of the in vivo redox state of the cells. Then, the sample was thoroughly mixed in capped glass tubes (vortex) for 1 min. The mixture was immediately centrifuged (900g, 1 min, 4°C), and the upper PE phase was transferred to an N2-flushed glass tube. To the remaining lower phase, 3 mL of PE was added for a second extraction, and the mixing and centrifugation steps were repeated. The collected PE upper phases were combined, and the PE was evaporated to dryness under a flow of N2 at room temperature. The dried extract was resuspended in 100 µL of hexanol and stored at −20°C until analysis by HPLC, using a Pharmacia LKB gradient pump 2249 system. The instrument was equipped with a fluorescence detector (Agilent 1260 infinity FLD) and a reverse-phase Lichrosorb (Chrompack) 10 RP 18 column (4.6-mm i.d., 250-mm length). The column was equilibrated with pure methanol, which was also used as the mobile phase. The flow rate was set at 2 mL min−1. Fluorescence excitation/emission were at 290/330 nm. Methanol (Sigma), hexanol (Sigma), and PE (Biosolve) were of analytical grade. The presence of PQH2 was confirmed with a PQ-9 standard kindly provided to us by Dr. Jersey Kruk. PQ was reduced with NaBH4 prior to HPLC analysis.
PQ Reduction
In order to determine the redox state of the PQ pool, the total amount of PQ (i.e. the sum of PQH2 and PQ in the sample) for each condition was determined by fully reducing 2 mL of the cell culture at the end of each experiment with 5 mg mL−1 NaBH4, 1 min before rapid extraction, and subsequent HPLC analysis. The redox state of the PQ pool was then determined from the difference between the area of the peaks obtained from physiologically reduced (A) cells and fully (i.e. NaBH4) reduced (AM) cells: A/AM. In this approach, we assume, due to rapid disproportionation, that any plastosemiquinone formed in the non-protein-bound PQ pool will instantaneously be converted into a combination of PQ and PQH2. The redox state of the PQ pool is presented as percentage reduced (i.e. PQH2) of the total PQ pool.
Fluorescence Measurements
Measurements of PSII fluorescence were performed with a PAM-100/103 instrument (Walz). F0 and Fm were determined directly on the sample vessel after a dark incubation of 30 min using the light source of the photobioreactor switched on at maximum intensity (6,000 µmol photons m−2 s−1). Steady-state fluorescence was recorded under a range of different illumination conditions.
77 K Fluorescence Analysis
For 77 K fluorescence analysis, samples were taken from different illumination conditions, diluted four times in ice-cold medium with glycerol (final concentration, 30% [v/v]), and immediately frozen in liquid nitrogen. The samples were analyzed in an OLIS 500 spectrofluorimeter equipped with a Dewar cell. PBS-specific excitation light was used at 590 nm, and fluorescence emission spectra were recorded between 600 and 750 nm, a wavelength domain in which the PBS (655 nm), PSII (685 nm), and PSI (720) show well-separated emission peaks. Skewed Gaussian deconvolution was performed on the different peaks in order to assay the degree of coupling of the PBS to PSII and PSI.
MIMS Measurements
MIMS measurements were performed using the HPR-40 system (Hiden Analytical) in a 10-mL air-tight cuvette (a modified DW3 cuvette from Hansatech Instruments) containing a Synechocystis sp. strain PCC 6803 culture in low- or high-carbon medium with a density of 2 mg mL−1 chlorophyll a. The high-vacuum membrane inlet sensor of the mass spectrometry analyzer was placed in the liquid culture. A thin medical-grade silicon tube serving as a membrane secured the continuous passage of small amounts of gasses from the liquid phase into the sensor tube of the mass spectrometer. Prior to the experiment, the sample was dark adapted for 30 min and then briefly (±10 s) sparged with N2 to reduce the prevalent oxygen concentration to about 20% of the value in air-equilibrated incubation buffer, with the aim to prevent oxygen saturation during the experiment. After sparging, the cuvette was closed, and 1 µL L−1 18O2 (95%–98% pure; Cambridge Isotope Laboratories) was added in the head space, which, while stirring, equilibrated with the liquid. An up-sloping mass spectrometer signal denoted the dissolving 18O2 until a plateau was reached. When the desired concentration was reached (10%–15% of the total oxygen concentration), the chamber was sealed after removing the 18O2 bubble. In order to minimize noise, the signals were normalized to argon, as suggested by Kana et al. (1994). For more information on the calculation procedure, see Bañares-España et al. (2013). The cultures were subjected to increasing 625-nm light intensities, ranging from 10 to 300 µmol photons m−2 s−1, in steps of 20 µmol photons m−2 s−1 below 100 µmol photons m−2 s−1 and steps of 50 µmol photons m−2 s−1 above 100 µmol photons m−2 s−1, aimed to excite PSII via its attached PBS. This illumination was combined with (or without) the addition of 25 µmol photons m−2 s−1 730-nm light, which typically only excites PSI. The lowest light intensity at the start of the experiment was on for a period of 10 min, to secure light adaptation, and all subsequent light intensities were kept on for 3 min. After the incubation in the light, dark respiration was monitored for 10 min.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Reduction of PQ with different quantities of NaBH4.
Supplemental Figure S2. Oxidation of PQH2 in hexanol over time.
Supplemental Figure S3. Cartoon of PBS binding under different light and carbon conditions.
Supplementary Material
Acknowledgments
We thank Jersey Kruk for providing us with a pure standard of PQ-9 and Ivo van Stokkum for help in the analysis of the 77 K fluorescence emission data.
Glossary
- PQ
plastoquinone
- PQH2
plastoquinol
- QA
primary accepting quinone
- QB
secondary accepting quinone
- PAM
pulse-amplitude modulated
- PE
petroleum ether
- OD730
optical density at 730 nm
- DCBQ
2,6-dichloro-p-benzoquinone
- Fm
maximum PSII fluorescence in the dark-adapted state
- F0
initial (minimum) PSII fluorescence in the dark-adapted state
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- PBS
phycobilisomes
- MIMS
membrane inlet mass spectroscopy
- LED
light-emitting diode
Footnotes
This work was supported by the Dutch Ministry of Economic Affairs, Agriculture, and Innovation (research program BioSolar Cells).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Allahverdiyeva Y, Mustila H, Ermakova M, Bersanini L, Richaud P, Ajlani G, Battchikova N, Cournac L, Aro EM. (2013) Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc Natl Acad Sci USA 110: 4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Tsuchiya T, Watabe K, Kojima A, Los DA, Tomo T, Klimov VV, Mimuro M. (2011) Redox potentials of primary electron acceptor quinone molecule (QA)- and conserved energetics of photosystem II in cyanobacteria with chlorophyll a and chlorophyll d. Proc Natl Acad Sci USA 108: 8054–8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. (1995) Thylakoid protein phosphorylation, state 1-state 2 transitions, and photosystem stoichiometry adjustment: redox control at multiple levels of gene expression. Physiol Plant 93: 196–205 [Google Scholar]
- Allen JF, Santabarbara S, Allen CA, Puthiyaveetil S. (2011) Discrete redox signaling pathways regulate photosynthetic light-harvesting and chloroplast gene transcription. PLoS ONE 6: e26372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M, Hirano M, Takahashi Y, Katoh S. (1983) Contents of cytochromes, quinones and reaction centers of photosystems I and II in a cyanobacterium Synechococcus sp. Plant Cell Physiol 24: 517–525 [Google Scholar]
- Aoki M, Katoh S. (1983) Size of the plastoquinone pool functioning in photosynthetic and respiratory electron transport of Synechococcus sp. Plant Cell Physiol 24: 1379–1386 [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Bañares-España E, Kromkamp JC, López-Rodas V, Costas E, Flores-Moya A. (2013) Photoacclimation of cultured strains of the cyanobacterium Microcystis aeruginosa to high-light and low-light conditions. FEMS Microbiol Ecol 83: 700–710 [DOI] [PubMed] [Google Scholar]
- Behrenfeld MJ, Halsey KH, Milligan AJ. (2008) Evolved physiological responses of phytoplankton to their integrated growth environment. Philos Trans R Soc Lond B Biol Sci 363: 2687–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, Kramer G, Hartog AF, Wagner MJ, de Koster CG, Hellingwerf KJ, de Mattos MJ. (2007) Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153: 1974–1980 [DOI] [PubMed] [Google Scholar]
- Berry S, Schneider D, Vermaas WFJ, Rögner M. (2002) Electron transport routes in whole cells of Synechocystis sp. strain PCC 6803: the role of the cytochrome bd-type oxidase. Biochemistry 41: 3422–3429 [DOI] [PubMed] [Google Scholar]
- Buser CA, Diner BA, Brudvig GW. (1992) Photooxidation of cytochrome b559 in oxygen-evolving photosystem II. Biochemistry 31: 11449–11459 [DOI] [PubMed] [Google Scholar]
- Campbell D, Hurry V, Clarke AK, Gustafsson P, Öquist G. (1998) Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol Mol Biol Rev 62: 667–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casano LM, Zapata JM, Martín M, Sabater B. (2000) Chlororespiration and poising of cyclic electron transport: plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J Biol Chem 275: 942–948 [DOI] [PubMed] [Google Scholar]
- Claquin P, Kromkamp JC, Martin-Jezequel V. (2004) Relationship between photosynthetic metabolism and cell cycle in a synchronized culture of the marine alga Cylindrotheca fusiformis (Bacillariophyceae). Eur J Phycol 39: 33–41 [Google Scholar]
- Cooley JW, Howitt CA, Vermaas WF. (2000) Succinate:quinol oxidoreductases in the cyanobacterium Synechocystis sp. strain PCC 6803: presence and function in metabolism and electron transport. J Bacteriol 182: 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneille S, Cournac L, Guedeney G, Havaux M, Peltier G. (1998) Reduction of the plastoquinone pool by exogenous NADH and NADPH in higher plant chloroplasts: characterization of a NAD(P)H-plastoquinone oxidoreductase activity. Biochim Biophys Acta 1363: 59–69 [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Jr, Tice AB, Pham C, Gantt E. (2010) Inactivation of genes encoding plastoglobuli-like proteins in Synechocystis sp. PCC 6803 leads to a light-sensitive phenotype. J Bacteriol 192: 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Cohu CM, Muller O, Adams WW., III (2012) Modulation of photosynthetic energy conversion efficiency in nature: from seconds to seasons. Photosynth Res 113: 75–88 [DOI] [PubMed] [Google Scholar]
- Dijkman NA, Kroon BMA. (2002) Indications for chlororespiration in relation to light regime in the marine diatom Thalassiosira weissflogii. J Photochem Photobiol B 66: 179–187 [DOI] [PubMed] [Google Scholar]
- Diner BA, Petrouleas V, Wendoloski JJ. (1991) The iron-quinone electron-acceptor complex of photosystem II. Physiol Plant 81: 423–436 [Google Scholar]
- Dong CX, Tang AH, Zhao JD, Mullineaux CW, Shen GZ, Bryant DA. (2009) CpcD is necessary for efficient energy transfer from phycobilisomes to photosystem I and helps to prevent photoinhibition in the cyanobacterium Synechococcus sp PCC 7002. Biochim Biophys Acta 1787: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, LaRoche J, Falkowski PG. (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92: 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G, Johnson GN, Dall’Osto L, Zito F, Bonente G, Bassi R, Wollman FA. (2006) Nonphotochemical quenching of chlorophyll fluorescence in Chlamydomonas reinhardtii. Biochemistry 45: 1490–1498 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Murakami A, Ohki K. (1987) Regulation of photosystem composition in the cyanobacterial photosynthetic system: the regulation occurs in response to the redox state of the electron pool located between the two photosystems. Plant Cell Physiol 28: 283–292 [Google Scholar]
- Geerts D, Schubert H, de Vrieze G, Borrias M, Matthijs HCP, Weisbeek PJ. (1994) Expression of Anabaena PCC 7937 plastocyanin in Synechococcus PCC 7942 enhances photosynthetic electron transfer and alters the electron distribution between photosystem I and cytochrome-c oxidase. J Biol Chem 269: 28068–28075 [PubMed] [Google Scholar]
- Gotoh E, Matsumoto M, Ogawa K, Kobayashi Y, Tsuyama M. (2010) A qualitative analysis of the regulation of cyclic electron flow around photosystem I from the post-illumination chlorophyll fluorescence transient in Arabidopsis: a new platform for the in vivo investigation of the chloroplast redox state. Photosynth Res 103: 111–123 [DOI] [PubMed] [Google Scholar]
- Hakkila K, Antal T, Gunnelius L, Kurkela J, Matthijs HCP, Tyystjärvi E, Tyystjärvi T. (2013) Group 2 sigma factor mutant ΔsigCDE of the cyanobacterium Synechocystis sp. PCC 6803 reveals functionality of both carotenoids and flavodiiron proteins in photoprotection of photosystem II. Plant Cell Physiol 54: 1780–1790 [DOI] [PubMed] [Google Scholar]
- Heber U, Walker D. (1992) Concerning a dual function of coupled cyclic electron transport in leaves. Plant Physiol 100: 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman Y, Barkan E, Eisenstadt D, Luz B, Kaplan A. (2005) Fractionation of the three stable oxygen isotopes by oxygen-producing and oxygen-consuming reactions in photosynthetic organisms. Plant Physiol 138: 2292–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houyoux PA, Ghysels B, Lecler R, Franck F. (2011) Interplay between non-photochemical plastoquinone reduction and re-oxidation in pre-illuminated Chlamydomonas reinhardtii: a chlorophyll fluorescence study. Photosynth Res 110: 13–24 [DOI] [PubMed] [Google Scholar]
- Howitt CA, Cooley JW, Wiskich JT, Vermaas WFJ. (2001) A strain of Synechocystis sp. PCC 6803 without photosynthetic oxygen evolution and respiratory oxygen consumption: implications for the study of cyclic photosynthetic electron transport. Planta 214: 46–56 [DOI] [PubMed] [Google Scholar]
- Huisman J, Matthijs HCP, Visser PM, Balke H, Sigon CAM, Passarge J, Weissing FJ, Mur LR. (2002) Principles of the light-limited chemostat: theory and ecological applications. Antonie van Leeuwenhoek 81: 117–133 [DOI] [PubMed] [Google Scholar]
- Jans F, Mignolet E, Houyoux PA, Cardol P, Ghysels B, Cuiné S, Cournac L, Peltier G, Remacle C, Franck F. (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA 105: 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua S, Mullineaux CW. (2004) Phycobilisome diffusion is required for light-state transitions in cyanobacteria. Plant Physiol 135: 2112–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana TM. (1993) Rapid oxygen cycling in Trichodesmium thiebautii. Limnol Oceanogr 38: 18–24 [Google Scholar]
- Kana TM, Darkangelo C, Hunt MD, Oldham JB, Bennett GE, Cornwell JC. (1994) Membrane inlet mass spectrometer for rapid high-precision determination of N2, O2 and Ar in environmental water samples. Anal Chem 66: 4166–4170 [Google Scholar]
- Kirilovsky D, Kerfeld CA. (2012) The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim Biophys Acta 1817: 158–166 [DOI] [PubMed] [Google Scholar]
- Kondo K, Mullineaux CW, Ikeuchi M. (2009) Distinct roles of CpcG1-phycobilisome and CpcG2-phycobilisome in state transitions in a cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 99: 217–225 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Rutherford AW. (1998) Influence of herbicide binding on the redox potential of the quinone acceptor in photosystem II: relevance to photodamage and phytotoxicity. Biochemistry 37: 17339–17344 [DOI] [PubMed] [Google Scholar]
- Kruk J, Karpinski S. (2006) An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochim Biophys Acta 1757: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Lange HC, Eman M, van Zuijlen G, Visser D, van Dam JC, Frank J, de Mattos MJ, Heijnen JJ. (2001) Improved rapid sampling for in vivo kinetics of intracellular metabolites in Saccharomyces cerevisiae. Biotechnol Bioeng 75: 406–415 [DOI] [PubMed] [Google Scholar]
- Liu LN, Bryan SJ, Huang F, Yu J, Nixon PJ, Rich PR, Mullineaux CW. (2012) Control of electron transport routes through redox-regulated redistribution of respiratory complexes. Proc Natl Acad Sci USA 109: 11431–11436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Mi H, Shen Y. (2010) Influence of the redox state of QA on phycobilisome mobility in the cyanobacterium Synechocystis sp. strain PCC6803. J Lumin 130: 1169–1173 [Google Scholar]
- Mao HB, Li GF, Ruan X, Wu QY, Gong YD, Zhang XF, Zhao NM. (2002) The redox state of plastoquinone pool regulates state transitions via cytochrome b6f complex in Synechocystis sp. PCC 6803. FEBS Lett 519: 82–86 [DOI] [PubMed] [Google Scholar]
- Matthijs HCP, Ludérus EME, Löffler HJM, Scholts MJC, Kraayenhof R. (1984) Energy metabolism in the cyanobacterium Plectonema boryanum: participation of the photosynthetic electron transfer chain in the dark respiration of NADPH and NADH. Biochim Biophys Acta 766: 29–37 [Google Scholar]
- Melis A, Brown JS. (1980) Stoichiometry of system I and system II reaction centers and of plastoquinone in different photosynthetic membranes. Proc Natl Acad Sci USA 77: 4712–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Endo T, Ogawa T, Asada K. (1995) Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 36: 661–668 [Google Scholar]
- Mitchell P. (1993) David Keilin’s respiratory chain concept and its chemiosmotic consequences. In Nobel Lectures in Chemistry, 1971–1980. World Scientific Publishing, Singapore, pp 295–330 [Google Scholar]
- Miyake C, Amako K, Shiraishi N, Sugimoto T. (2009) Acclimation of tobacco leaves to high light intensity drives the plastoquinone oxidation system: relationship among the fraction of open PSII centers, non-photochemical quenching of Chl fluorescence and the maximum quantum yield of PSII in the dark. Plant Cell Physiol 50: 730–743 [DOI] [PubMed] [Google Scholar]
- Miyake C, Miyata M, Shinzaki Y, Tomizawa K. (2005) CO2 response of cyclic electron flow around PSI (CEF-PSI) in tobacco leaves: relative electron fluxes through PSI and PSII determine the magnitude of non-photochemical quenching (NPQ) of Chl fluorescence. Plant Cell Physiol 46: 629–637 [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY. (2010) Activated Q-cycle as a common mechanism for cytochrome bc1 and cytochrome b6f complexes. Biochim Biophys Acta 1797: 1858–1868 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. (1993) Inhibition by phosphate of light-state transitions in cyanobacterial cells. Photosynth Res 38: 135–140 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. (2008) Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth Res 95: 175–182 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. (2014) Electron transport and light-harvesting switches in cyanobacteria. Front Plant Sci 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW, Allen JF. (1990) State 1-state 2 transitions in the cyanobacterium Synechococcus 6301 are controlled by the redox state of electron carriers between photosystems I and II. Photosynth Res 23: 297–311 [DOI] [PubMed] [Google Scholar]
- Nanba M, Katoh S. (1984) Effects of dibromothymoquinone on oxidation-reduction reactions and the midpoint potential of the Rieske iron-sulfur center in photosynthetic electron transport of Synechococcus sp. Biochim Biophys Acta 767: 396–403 [Google Scholar]
- Nicholls DG, Ferguson SJ (2013) Bioenergetics 4. Academic Press, Amsterdam [Google Scholar]
- Nicholls P, Obinger C, Niederhauser H, Peschek GA. (1992) Cytochrome oxidase in Anacystis nidulans: stoichiometries and possible functions in the cytoplasmic and thylakoid membranes. Biochim Biophys Acta 1098: 184–190 [Google Scholar]
- Nishimura T, Takahashi Y, Yamaguchi O, Suzuki H, Maeda SI, Omata T. (2008) Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol Microbiol 68: 98–109 [DOI] [PubMed] [Google Scholar]
- Okayama S. (1976) Redox potential of plastoquinone A in spinach chloroplasts. Biochim Biophys Acta 440: 331–336 [DOI] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. (2001) Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA 98: 2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumann M, Bernroitner M, Lubura B, Peer M, Jakopitsch C, Furtmüller PG, Peschek GA, Obinger C. (2004) Kinetics of electron transfer between plastocyanin and the soluble CuA domain of cyanobacterial cytochrome c oxidase. FEMS Microbiol Lett 239: 301–307 [DOI] [PubMed] [Google Scholar]
- Peltier G, Tolleter D, Billon E, Cournac L. (2010) Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res 106: 19–31 [DOI] [PubMed] [Google Scholar]
- Piller LE, Abraham M, Dörmann P, Kessler F, Besagni C. (2012) Plastid lipid droplets at the crossroads of prenylquinone metabolism. J Exp Bot 63: 1609–1618 [DOI] [PubMed] [Google Scholar]
- Pils D, Schmetterer G. (2001) Characterization of three bioenergetically active respiratory terminal oxidases in the cyanobacterium Synechocystis sp. strain PCC 6803. FEMS Microbiol Lett 203: 217–222 [DOI] [PubMed] [Google Scholar]
- Prasil O, Kolber Z, Berry JA, Falkowski PG. (1996) Cyclic electron flow around photosystem II in vivo. Photosynth Res 48: 395–410 [DOI] [PubMed] [Google Scholar]
- Rich PR, Bendall DS. (1980) The redox potentials of the b-type cytochromes of higher plant chloroplasts. Biochim Biophys Acta 591: 153–161 [DOI] [PubMed] [Google Scholar]
- Rich PR, Madgwick SA, Moss DA. (1991) The interactions of duroquinol, DBMIB and NQNO with the chloroplast cytochrome b6f complex. Biochim Biophys Acta 1058: 312–328 [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Roberts AG, Bowman MK, Kramer DM. (2004) The inhibitor DBMIB provides insight into the functional architecture of the Qo site in the cytochrome b6f complex. Biochemistry 43: 7707–7716 [DOI] [PubMed] [Google Scholar]
- Roberts AG, Kramer DM. (2001) Inhibitor “double occupancy” in the Q0 pocket of the chloroplast cytochrome b6f complex. Biochemistry 40: 13407–13411 [DOI] [PubMed] [Google Scholar]
- Sane PV, Ivanov AG, Hurry V, Huner NPA, Öquist G. (2003) Changes in the redox potential of primary and secondary electron-accepting quinones in photosystem II confer increased resistance to photoinhibition in low-temperature-acclimated Arabidopsis. Plant Physiol 132: 2144–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S. (1990) Do photosynthetic and respiratory electron transport chains share redox proteins? Trends Biochem Sci 15: 458–462 [DOI] [PubMed] [Google Scholar]
- Schreiber U, Schliwa U, Bilger W. (1986) Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res 10: 51–62 [DOI] [PubMed] [Google Scholar]
- Schubert H, Matthijs HCP, Mur LR, Schiewer U. (1995) Blooming of cyanobacteria in turbulent water with steep light gradients: the effect of intermittent light and dark periods on the oxygen evolution capacity of Synechocystis sp. PCC 6803. FEMS Microbiol Ecol 18: 237–245 [Google Scholar]
- Shevela D, Messinger J. (2012) Probing the turnover efficiency of photosystem II membrane fragments with different electron acceptors. Biochim Biophys Acta 1817: 1208–1212 [DOI] [PubMed] [Google Scholar]
- Smetacek V, Passow U. (1990) Spring bloom initiation and Sverdrup’s critical-depth model. Limnol Oceanogr 35: 228–234 [Google Scholar]
- Trebst A. (2007) Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth Res 92: 217–224 [DOI] [PubMed] [Google Scholar]
- van Thor JJ, Geerlings TH, Matthijs HCP, Hellingwerf KJ. (1999) Kinetic evidence for the PsaE-dependent transient ternary complex photosystem I/ferredoxin/ferredoxin:NADP(+) reductase in a cyanobacterium. Biochemistry 38: 12735–12746 [DOI] [PubMed] [Google Scholar]
- Van Thor JJ, Mullineaux CW, Matthijs HCP, Hellingwerf KJ. (1998) Light harvesting and state transitions in cyanobacteria. Bot Acta 111: 430–443 [Google Scholar]
- Vener AV, van Kan PJ, Rich PR, Ohad I, Andersson B. (1997) Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc Natl Acad Sci USA 94: 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas WFJ, Timlin JA, Jones HDT, Sinclair MB, Nieman LT, Hamad SW, Melgaard DK, Haaland DM. (2008) In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proc Natl Acad Sci USA 105: 4050–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE. (1998) ATP synthesis by rotary catalysis (Nobel Lecture). Angewandte Chemie International Edition 37: 2308–2319 [DOI] [PubMed] [Google Scholar]
- Yang DH, Andersson B, Aro EM, Ohad I. (2001) The redox state of the plastoquinone pool controls the level of the light-harvesting chlorophyll a/b binding protein complex II (LHC II) during photoacclimation. Photosynth Res 68: 163–174 [DOI] [PubMed] [Google Scholar]
- Yeremenko N, Jeanjean R, Prommeenate P, Krasikov V, Nixon PJ, Vermaas WFJ, Havaux M, Matthijs HCP. (2005) Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 46: 1433–1436 [DOI] [PubMed] [Google Scholar]
- Zhang P, Eisenhut M, Brandt AM, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM. (2012) Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA. (1999) The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J 18: 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.