Maize and Setaria viridis have independently recruited syntenic orthologs into the C4 pathway, and transcript abundance in the mesophyll and bundle sheath cells of these species is highly convergent.
Abstract
Leaves of almost all C4 lineages separate the reactions of photosynthesis into the mesophyll (M) and bundle sheath (BS). The extent to which messenger RNA profiles of M and BS cells from independent C4 lineages resemble each other is not known. To address this, we conducted deep sequencing of RNA isolated from the M and BS of Setaria viridis and compared these data with publicly available information from maize (Zea mays). This revealed a high correlation (r = 0.89) between the relative abundance of transcripts encoding proteins of the core C4 pathway in M and BS cells in these species, indicating significant convergence in transcript accumulation in these evolutionarily independent C4 lineages. We also found that the vast majority of genes encoding proteins of the C4 cycle in S. viridis are syntenic to homologs used by maize. In both lineages, 122 and 212 homologous transcription factors were preferentially expressed in the M and BS, respectively. Sixteen shared regulators of chloroplast biogenesis were identified, 14 of which were syntenic homologs in maize and S. viridis. In sorghum (Sorghum bicolor), a third C4 grass, we found that 82% of these trans-factors were also differentially expressed in either M or BS cells. Taken together, these data provide, to our knowledge, the first quantification of convergence in transcript abundance in the M and BS cells from independent lineages of C4 grasses. Furthermore, the repeated recruitment of syntenic homologs from large gene families strongly implies that parallel evolution of both structural genes and trans-factors underpins the polyphyletic evolution of this highly complex trait in the monocotyledons.
C4 species represent many of the world’s most productive crops (Edwards et al., 2010), and in the tropics and subtropics, the C4 pathway allows increased productivity compared with ancestral C3 photosynthesis. The increased productivity of C4 plants is due to their ability to concentrate CO2 around Rubisco (Hatch et al., 1967), and in the majority of C4 species, this is achieved through spatial compartmentation of the photosynthetic apparatus into mesophyll (M) and bundle sheath (BS) cells (Langdale, 2011). Compared with ancestral C3 plants, the leaf anatomy of C4 species is altered such that the BS and M compartments are increased and decreased in size, respectively. Despite this complexity, C4 plants are now documented in more than 60 independent lineages of angiosperms (Sage et al., 2011).
In all C4 lineages, carbonic anhydrase (CA) catalyzes the conversion of CO2 to HCO3− in M cells. Subsequently, fixation of HCO3− by phosphoenolpyruvate carboxylase (PEPC) allows C4 acids to accumulate in the M. In combination with the drawdown of C4 acids caused by decarboxylation and subsequent reactions of the C4 cycle, this drives the diffusion of C4 acids from the M to the adjacent BS cells. This increase in CO2 concentration minimizes the oxygenation reaction of Rubisco and, therefore, reduces photorespiration. At least three C4 acid decarboxylases have been recruited in different C4 lineages to release CO2 around Rubisco in BS cells: NAD-dependent malic enzyme, NADP-dependent malic enzyme (NADP-ME), and phosphoenolpyruvate carboxykinase (PCK). To complete the C4 cycle, phosphoenolpyruvate is regenerated by pyruvate,orthophosphate dikinase (PPDK) in M chloroplasts.
For the two-celled C4 cycle to operate, enzymes and transporters must be specifically localized in either M or BS cells. This preferential accumulation of proteins is underpinned by transcriptional and posttranscriptional regulation of gene expression as well as posttranslational modification (Hibberd and Covshoff, 2010). While there is considerable diversity in the mechanisms responsible for cell-specific gene expression in C4 leaves, and modeling predicts that the evolution of C4 photosynthesis has occurred via distinct routes in various taxa (Williams et al., 2013), it is now clear that there are also examples where the same mechanism has been used by independent C4 lineages to generate expression in either M or BS cells. For example, the accumulation of NAD-dependent malic enzyme in Cleome gynandra and NADP-ME in maize (Zea mays) is mediated by a conserved element found within the coding sequence of these genes (Brown et al., 2011). Because this sequence element is present in orthologous genes of C3 Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), the most parsimonious explanation is that these elements have repeatedly been coopted from an unknown ancestral function into generating BS specificity in C4 leaves. Furthermore, the same chromatin marks have been documented on PEPC and NADP-ME genes in M and BS cells of both maize and Setaria italica (Heimann et al., 2013), showing that separate C4 lineages regulate the expression of genes in either the M or BS cells via the same mechanisms in cis. However, the extent to which the expression of other genes and pathways in these cells resemble each other is not clear, nor is it known whether the same mechanisms in trans have been coopted by multiple C4 lineages.
Deep sequencing now allows an unbiased analysis of complete mRNA populations. This approach has been used to investigate transcript abundance in M and BS cells from maize (Li et al., 2010b; Chang et al., 2012) and led to estimates that in the M between 53 and 78, while in the BS between 102 and 214, transcription factors accumulated preferentially. However, maize represents only one lineage of C4 plants. We sought to determine the patterns of gene expression in M and BS cells that underpin the photosynthetic reactions in a mature photosynthetic leaf of an independent lineage of C4 plant and to define the extent to which they overlap with those in maize. For this, we chose Setaria viridis, a weedy relative of domesticated S. italica. S. viridis has an annotated genome sequence and is increasingly being used as a model C4 grass (Bennetzen et al., 2012; Zhang et al., 2012). While S. viridis represents an independent origin of C4 photosynthesis from maize (Brutnell et al., 2010), both species use NADP-ME as the primary C4 acid decarboxylase in the BS cells.
In this study, we first defined the mRNA profiles of M and BS cells from S. viridis. We combined this information with publicly available databases to examine the extent to which patterns of transcript abundance are convergent in S. viridis and maize. We quantify convergence at the mRNA level in M and BS cells of these independent C4 lineages. This includes structural genes that are known to be required for the C4 pathway to operate but also trans-factors that previously have not been implicated in the function of M or BS cells.
RESULTS
Rapid Isolation of RNA and Protein from M Cells and BS Strands of S. viridis
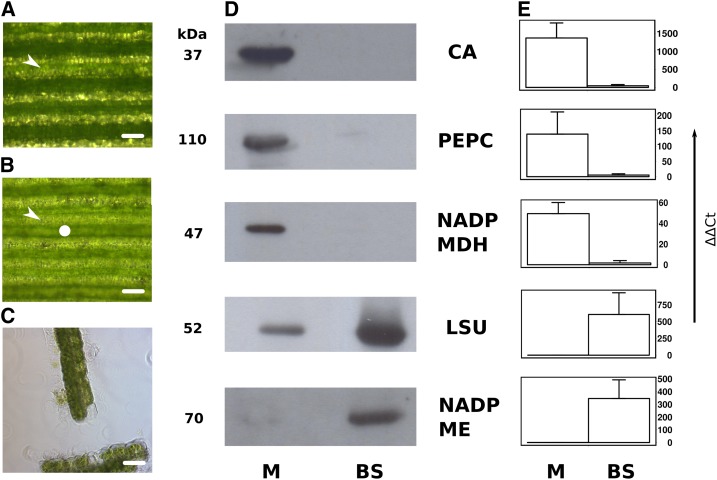
To enable the quantification of transcript abundance in M and BS cells from fully photosynthetic leaves of S. viridis, we used leaf rolling to extract M cell contents (Covshoff et al., 2013) followed by mechanical isolation of BS strands (Markelz et al., 2003). Leaf rolling reduced the chlorophyll content of S. viridis leaves (Fig. 1, A and B). After mechanical blending of rolled leaves, BS strands surrounding the veins were visible and very few M cells remained (Fig. 1C). Separation of soluble protein from these M and BS extracts followed by immunoblotting indicated that proteins characteristic of the C4 cycle were partitioned as expected between these two cell types. For example, CA, PEPC, and NADP-dependent malate dehydrogenase (NADP-MDH) were enriched in M cell extracts, while NADP-ME and the large subunit (LSU) of Rubisco (Fig. 1D) were enriched in the BS. As has been observed previously for maize (Majeran et al., 2005), LSU was detectable in M cells of S. viridis, although at much lower abundance than in the BS. We extracted RNA from analogous M and BS samples. Electropherograms confirmed that this RNA was good quality (RNA integrity number ≥ 7.4 and ratio of 28S ribosomal RNA to 18S ribosomal RNA ≥ 1; Supplemental Table S1), and quantitative PCR was carried out to determine the abundance of transcripts encoding proteins characteristic of the C4 cycle (Fig. 1E).
Figure 1.
Extraction of protein and RNA from M and BS cells of S. viridis. A, Representative image of a leaf prior to rolling, with alternating bands of M (arrowhead) and BS cells. B, After leaf rolling, the chloroplasts within the BS cells (circle) are visible. C, Representative image of BS preparation after blending. D, Immunoblotting demonstrates that CA, PEPC, and NADP-MDH proteins were abundant in M samples but not detectable in BS strands. In contrast, Rubisco LSU and NADP-ME were preferentially localized in BS strands. The molecular mass of each protein is annotated to the left of each blot. E, Quantitative PCR for CA, PEPC, NADP-MDH, RbcS, and NADP-ME indicated preferential transcript accumulation in the same cell type as each protein. Bars = 200 µm (A and B) and 4 µm (C).
Deep Sequencing of Transcripts from M and BS Cells of S. viridis
To quantify transcript abundance in the M and BS of S. viridis, we undertook deep sequencing of triplicate M rolled samples and BS strands. Approximately 200 million 91-bp paired-end reads were obtained (Table I), and after cleaning, 97.8% aligned to the genome. Of the 27,045 genes for which transcripts were detected, 9,680 were differentially expressed (adjusted P ≤ 0.05) between M and BS cells (Supplemental File S1). Based on homology to known maize C4 proteins (Li et al., 2010b; Chang et al., 2012) and strong cell-specific expression, we identified putative S. viridis C4 gene families, from which 31 genes were selected for further analysis (Table II; Supplemental File S2).
Table I. Summary of sequencing, read processing, mapping, and differential expression analysis.
| Sequence Metric | Value |
|---|---|
| Read length | 91 bp |
| Read type | Paired |
| Replicates | 3 |
| Reads before cleaning | 202,048,285 |
| Reads after cleaning | 183,508,816 |
| Average cleaned reads per library | 30,584,803 |
| Reads aligned | 179,478,816 |
| Read alignment percentage | 97.8% |
| Average reads aligned per library | 29,913,136 |
| Detected genes | 27,045 |
| Differentially expressed genes | 9,680 |
Table II. Abundance of transcripts encoding enzymes required for the C4 pathway in M or BS cells of S. viridis.
Each gene recruited into the C4 pathway in S. viridis is highly expressed and preferentially generates transcripts in either M or BS cells. Proteins are listed in groups according to whether they function in the core C4 pathway, metabolite transport, the Calvin-Benson cycle, or photorespiration. Library-normalized read counts, fold enrichment in each cell type, and Benjamini-Hochberg procedure adjusted P values are shown.
| Gene | Identifier | Role | Cell Type | M | BS | Fold Change, BS/M | Log2 Fold Change, BS/M | Adjusted P |
|---|---|---|---|---|---|---|---|---|
| CA | Si003882m.g | Core pathway | M | 453,421.09 | 12,315.93 | 0.03 | −5.20 | 5.23E-093 |
| PEPC | Si005789m.g | Core pathway | M | 1,413,131.82 | 47,109.98 | 0.03 | −4.91 | 7.31E-060 |
| NADP-MDH | Si013632m.g | Core pathway | M | 108,641.70 | 4,588.13 | 0.04 | −4.57 | 5.21E-076 |
| PPDK | Si021174m.g | Core pathway | M | 1,169,325.81 | 57,155.39 | 0.05 | −4.35 | 5.61E-071 |
| AK | Si014186m.g | Core pathway | M | 99,257.32 | 12,079.26 | 0.12 | −3.04 | 1.91E-039 |
| ASP-AT | Si017156m.g | Core pathway | M | 74,263.60 | 10,050.17 | 0.14 | −2.89 | 4.34E-036 |
| PPDK-RP | Si032116m.g | Core pathway | M | 17,359.84 | 7,810.30 | 0.45 | −1.15 | 3.20E-007 |
| NADP-ME | Si000645m.g | Core pathway | BS | 4,155.43 | 613,075.40 | 147.54 | 7.20 | 7.82E-084 |
| RBCS | Si023465m.g | Core pathway | BS | 2,717.93 | 290,283.67 | 106.80 | 6.74 | 6.79E-103 |
| PCK | Si034404m.g | Core pathway | BS | 240.11 | 3,170.30 | 13.20 | 3.72 | 3.18E-051 |
| RBCACT | Si026402m.g | Core pathway | BS | 6,245.84 | 77,396.56 | 12.39 | 3.63 | 3.21E-021 |
| OMT1 | Si024403m.g | Transport | M | 32,208.32 | 1,289.06 | 0.04 | −4.64 | 2.17E-077 |
| MEP3b | Si000451m.g | Transport | M | 63,801.82 | 10,077.02 | 0.16 | −2.66 | 2.68E-029 |
| PPT | Si013874m.g | Transport | M | 64,440.52 | 17,850.85 | 0.28 | −1.85 | 3.81E-016 |
| DIT1 | Si029415m.g | Transport | M | 3,703.36 | 2,349.68 | 0.63 | −0.66 | 9.64E-003 |
| DCT2 | Si035016m.g | Transport | BS | 650.50 | 137,421.82 | 211.25 | 7.72 | 3.68E-157 |
| MEP3a | Si024315m.g | Transport | BS | 675.13 | 11,279.22 | 16.71 | 4.06 | 1.93E-011 |
| TPI | Si030636m.g | Calvin cycle | M | 52,985.29 | 3,153.93 | 0.06 | −4.07 | 1.39E-063 |
| GAPDH | Si035707m.g | Calvin cycle | M | 230,052.78 | 124,502.17 | 0.54 | −0.89 | 1.41E-004 |
| PRK | Si017390m.g | Calvin cycle | BS | 1,348.39 | 210,577.18 | 156.17 | 7.29 | 2.86E-065 |
| FBA | Si010312m.g | Calvin cycle | BS | 4,611.67 | 616,786.63 | 133.74 | 7.06 | 6.1E-088 |
| SBP | Si001775m.g | Calvin cycle | BS | 1,174.47 | 134,922.00 | 114.88 | 6.84 | 1.20E-059 |
| RPI | Si030780m.g | Calvin cycle | BS | 436.32 | 24,891.21 | 57.05 | 5.83 | 2.01E-088 |
| RPE | Si036994m.g | Calvin cycle | BS | 1,528.85 | 70,559.89 | 46.15 | 5.53 | 5.86E-035 |
| FBP | Si035941m.g | Calvin cycle | BS | 3,018.51 | 56,039.35 | 18.57 | 4.21 | 2.46E-032 |
| TKL | Si005927m.g | Calvin cycle | BS | 24,364.05 | 131,028.52 | 5.38 | 2.43 | 8.46E-010 |
| PGK | Si021917m.g | Calvin cycle | BS | 59,178.50 | 87,873.79 | 1.48 | 0.57 | 3.05E-002 |
| GDC | Si000068m.g | Photorespiration | BS | 388.71 | 74,572.76 | 191.85 | 7.58 | 1.48E-103 |
| GCH | Si011200m.g | Photorespiration | BS | 65.68 | 9,987.76 | 152.06 | 7.25 | 3.05E-136 |
| SHMT | Si035240m.g | Photorespiration | BS | 3,133.86 | 60,501.53 | 19.31 | 4.27 | 7.07E-067 |
| GOX | Si040072m.g | Photorespiration | BS | 10,487.89 | 23,729.01 | 2.26 | 1.18 | 4.71E-007 |
Core C4 transcripts encoding CA, PEPC, NADP-MDH, and PPDK were all at least 20-fold more abundant in the M than the BS, while transcripts encoding NADP-ME, PCK, Rubisco activase, and the small subunit of Rubisco were at least 12-fold more abundant in BS cells compared with the M (Table II). Consistent with photosynthesis requiring significant amounts of C4 cycle proteins, for 23 of the 31 C4 genes, transcript abundance was very high in either the M or BS (transcripts per million [TPM] ≥ 1,000; Supplemental File S2). We conclude that the M rolled samples and BS strands showed patterns of transcript abundance consistent with the C4 pathway. We next determined the extent to which S. viridis and maize, which represent independent lineages of C4 grasses, show convergent patterns of transcript abundance in M and BS cells.
Convergence in C4 Transcript Abundance in Two Independent Grass Lineages
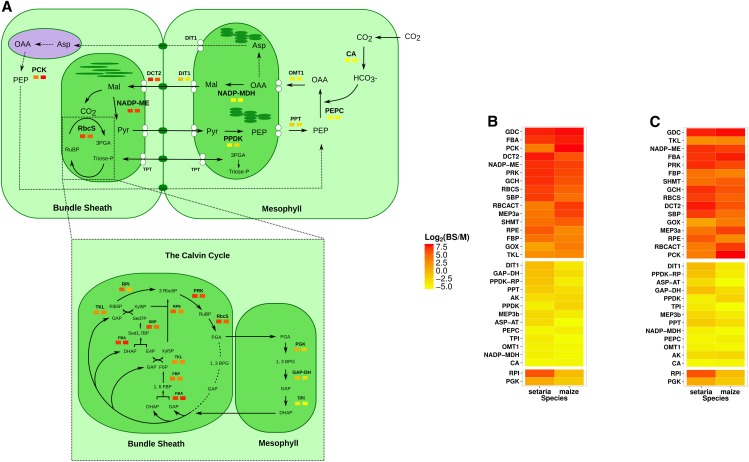
We compared the S. viridis M and BS mRNA data sets with analogous publicly available data from maize (Li et al., 2010b; Chang et al., 2012). Due to higher similarity in both sampling and sequencing procedures, unless explicitly stated, all comparisons were made with data from Chang et al. (2012). Transcripts encoding proteins known to be involved in the C4 cycle showed very similar compartmentalization between M and BS cells of these two C4 grass lineages (Fig. 2A). This included enzymes of the core C4 pathway, transporters that allow the flux of metabolites across organelle membranes, and proteins of the Calvin-Benson cycle, which are known to be compartmentalized between the two cell types (Fig. 2A). Notably, in addition to the accumulation of transcripts encoding the A subunit of glyceraldehyde phosphate dehydrogenase (GAPDH; Si010261m.g) in the BS of S. viridis, we also detected Si035707m.g encoding the B subunit of GAPDH in the M. It is thought that the BS-specific isoform of GAPDH forms the GAPDH-Chloroplast Protein12 (CP12)-phosphoribulokinase (PRK) supercomplex (Majeran et al., 2005) that regulates PRK activity (Howard et al., 2011). Consistent with this and with maize RNA-seq data (Li et al., 2010b; Chang et al., 2012), we also found that in S. viridis, CP12 (Si003343m.g) was BS specific.
Figure 2.
Abundance of transcripts encoding proteins of the C4 cycle in M and BS cells of S. viridis and maize. A, Summary of transcript quantification in M and BS cells; components of the Calvin-Benson cycle are shown at bottom. Transcripts that are more abundant in the M are colored yellow, while those that are more abundant in the BS are colored red (the scale is shown in the heat map to the right). For each component of the C4 cycle, quantifications for S. viridis and maize are shown on the left and right, respectively. B and C, Log2 fold change of transcript abundance in BS and M cells for all C4 genes sorted by mean enrichment (high to low) in S. viridis and maize (B) or convergence between the two species (C). The top and middle sections represent transcripts that in both species were preferential to BS and M cells, respectively, while the bottom section represents transcripts that showed divergent patterns between the two species. Abbreviations not defined in the text are as follows: AK, adenylate kinase; ASP-AT, Asp aminotransferase; DCT2, dicarboxylate transporter; DIT1, dicarboxylate transporter; FBP, Fru-1,6-bisphosphatase; GCH, Gly cleavage H-protein; GOX, glycolate oxidase; MEP3, putative protein/pyruvate symporter; PPDK-RP, pyruvate,orthophosphate dikinase regulatory protein; PPT, phosphoenolpyruvate/phosphate translocator; RBCACT, Rubisco activase; RbcS, Rubisco small subunit; RPE, ribulose-phosphate3 epimerase; SBP, sedoheptulose-1,7-bisphosphatase; SHMT, Ser hydroxymethyltransferase; TLK, trans-ketolase; TPT, triose phosphate/phosphate antiporter.
Each pair of homologs recruited into the C4 pathway in S. viridis and maize was ranked in terms of mean fold change in transcript abundance within M or BS cells (Fig. 2B; Supplemental Table S2) but also by the extent to which transcript abundances were convergent (Fig. 2C; Supplemental Table S3). In the BS, transcripts encoding glycine decarboxylase (GDC) and fructose bisphosphate aldolase (FBA) were highly enriched (Supplemental Table S2) and highly convergent in their cell specificity (Fig. 2C; Supplemental Table S3). However, while transcripts encoding PCK were highly enriched in the BS, they were far less convergent, and TKL transcripts were less abundant in BS cells but highly convergent (Fig. 2, B and C). CA and NADP-MDH transcripts were the most enriched in the M of both species (Fig. 2B). It was noticeable that OMT1 (for 2-oxoglutarate/malate transporter) transcripts were highly enriched in M to very similar extents in both maize and S. viridis (Fig. 2, B and C).
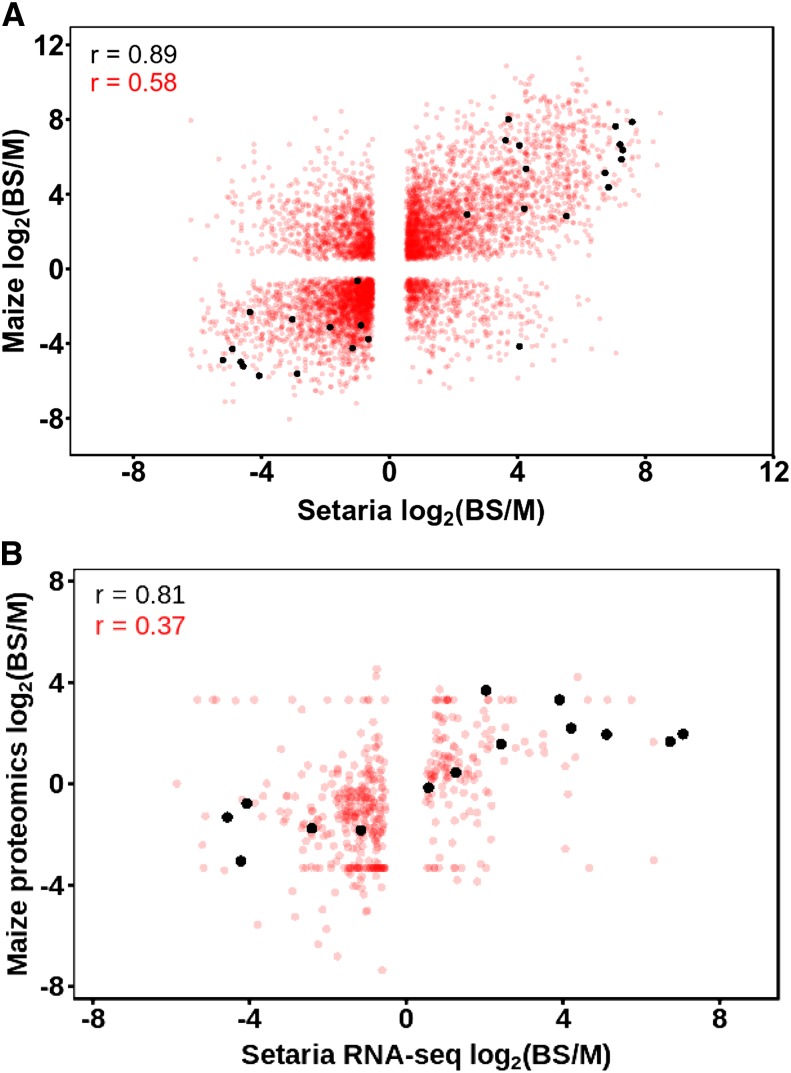
To quantify the convergence in patterns of gene expression between S. viridis and maize, we compared the enrichment in BS versus M cells for pairs of homologous genes in the two species (Fig. 3A). For all differentially expressed genes in M and BS cells, the Pearson’s correlation coefficient (r) was 0.58, while for genes important for the C4 cycle, the correlation coefficient was 0.89. These data indicate a high degree of convergence in the relative abundance of transcripts encoding C4 cycle proteins between M and BS cells of these species. We also investigated the correlation between S. viridis RNA-seq and maize chloroplast proteomic data (Fig. 3B) and found that C4 genes were more highly correlated than the background. This finding extends previous analysis indicating that the abundance of transcripts encoding components of the core C4 cycle in M and BS cells of maize was highly correlated (r = 0.95) with their cognate proteins (Li et al., 2010b).
Figure 3.
Convergence in the abundance of transcripts and proteins between S. viridis and maize. A, Relationship between the abundance of transcripts in BS and M cells of S. viridis and maize. B, Relationship between the abundance of transcripts in BS and M cells of S. viridis and chloroplast proteins in maize defined by best BLASTP hits (from Friso et al., 2010). All differentially expressed genes are represented in red, while C4 genes are in black. Pearson’s correlation coefficients (r) are shown.
Two genes encoding proteins of the Calvin-Benson cycle showed opposite patterns of expression in S. viridis and maize (Fig. 2, B and C). First, in S. viridis, RIBOSE-5P-ISOMERASE (RPI) transcripts were strongly preferential to the BS and highly expressed, and this is consistent with the maize RPI protein preferentially accumulating in the BS (Majeran et al., 2005; Friso et al., 2010). However, transcripts from the only strongly expressed (TPM ≥ 100) maize RPI gene (GRMZM5G874903) were weakly M specific. An independent study using laser-capture microdissection to isolate RNA also detected RPI transcripts in M cells (Li et al., 2010b), so it would appear likely that in maize strong posttranscriptional or translational control leads to the accumulation of RPI in the BS. Second, while in maize, both RNA-seq (Fig. 2D) and proteomics (Majeran et al., 2005; Friso et al., 2010) indicated enrichment of phosphoglycerate kinase (PGK) in the M, in S. viridis, a chloroplast-targeted PGK (Si021917m.g) was highly expressed but weakly preferential to the BS (Table II; Fig. 2, B and C). As the presence of PGK, GAPDH, and triose phosphate isomerase (TPI) in M cells of C4 leaves allows balancing of reducing equivalents between M and BS (Majeran et al., 2005), we propose that PGK is under strong posttranscriptional or posttranslational control in M cells of S. viridis.
Although C4 plants have been classified into three subtypes depending on the major C4 acid decarboxylase that they use to provide CO2 to Rubisco, it is clear that mixed and flexible decarboxylase systems are common (Furbank, 2011). In maize, although NADP-ME is considered the primary C4 acid decarboxylase, a simultaneous decarboxylation reaction mediated by PCK (Wingler et al., 1999; Leegood and Walker, 2003; Furbank, 2011) also takes place. Of the two PCK genes in maize, transcripts derived from GRMZM2G001696 (TPM = 11,202) are much more abundant than those from GRMZM5G870932 (TPM = 68). There is only one PCK gene in S. viridis (Si034404m.g), and although it is syntenic to the strongly expressed maize isoform, its expression was relatively low (BS TPM = 26). Therefore, we conclude that S. viridis likely operates a minimal PCK decarboxylase pathway.
The light-dependent reactions of photosynthesis can also be compartmented between M and BS cells in C4 leaves. For example, in NADP-ME subtypes, linear and cyclic electron transport occur preferentially in the M and BS, respectively (Takabayashi et al., 2005). This is because PSII is enriched in the M, while PSI and the NAD(P)H dehydrogenase (NDH) complex are enriched in the BS (Majeran et al., 2005; Friso et al., 2010; Li et al., 2010b). Consistent with these maize proteomics data, but in contrast to RNA-seq (Chang et al., 2012), in S. viridis transcripts encoding components of PSII were more abundant in M cells but transcripts encoding components of PSI or proteins allowing cyclic electron transport were more abundant in the BS (Table III). We suggest that differences between the S. viridis and maize RNA-seq data sets are because the leaf rolling approach is faster than the protoplasting that has previously been used to release M cells (Chang et al., 2012), although it is also possible that differences in growth conditions are responsible.
Table III. Number of genes encoding multiprotein complexes required for the light-dependent reactions of photosynthesis for which differential transcript abundance was detected in M or BS cells of maize and S. viridis.
Many proteins used in the C4 pathway are coopted from multigene families in C3 species (Aubry et al., 2011). For proteins that become more abundant in C4 compared with C3 leaves, these multigene families provide evolution with a rich resource for natural selection to up-regulate a gene. To investigate the extent to which the same members of multigene families have been recruited into the C4 pathway in maize and S. viridis, we searched a database of syntenic orthologs within the grasses for genes of the core C4 cycle, the Calvin-Benson cycle, and associated transporters (Schnable et al., 2012). All 10 of the genes defined as enzymes of the core C4 cycle were syntenic orthologs, while three of the six metabolite transporters were syntenic (Supplemental Table S4). These data imply that for many of the key enzymes that are up-regulated in M or BS cells of these C4 grasses, a specific member of each gene family is repeatedly recruited. As almost all of these genes belong to sizeable gene families (Supplemental Table S4), these data indicate strong selective pressure to recruit particular isoforms into C4 photosynthesis.
Global Comparisons of the M and BS Transcriptomes from S. viridis and Maize
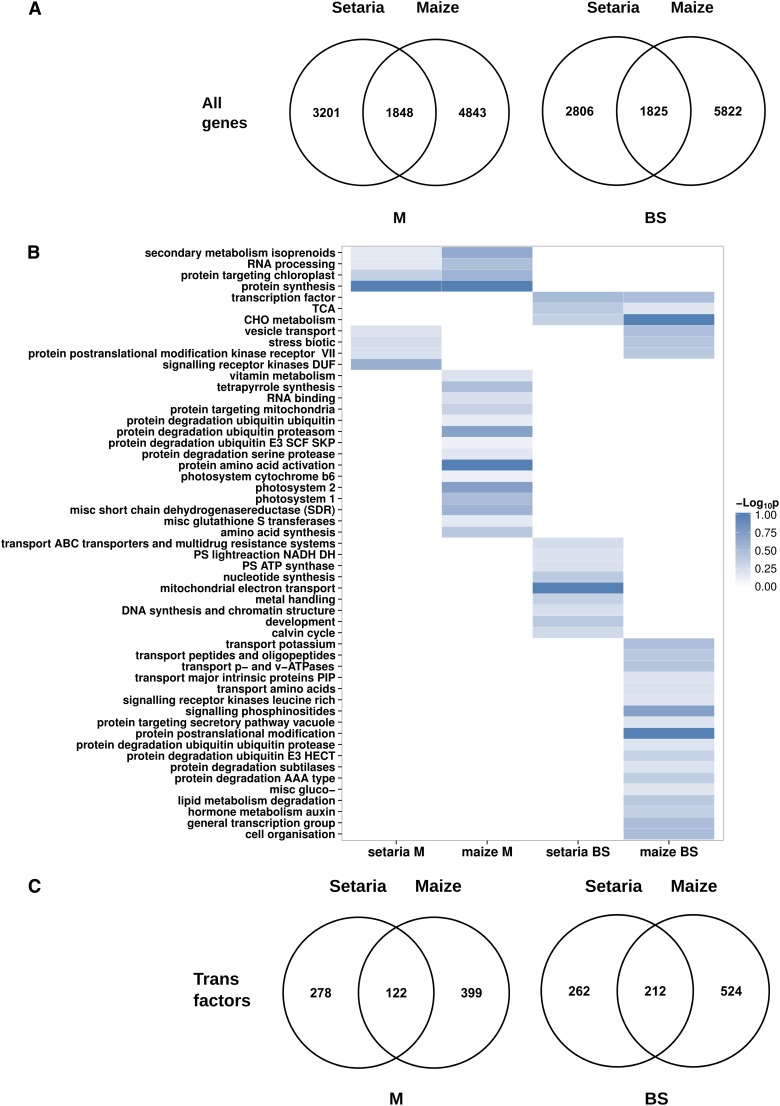
To investigate the extent to which global patterns of transcript abundance were similar in M and BS cells of S. viridis and maize, we used protein alignments to link differentially expressed genes in S. viridis to homologs in maize. This resulted in the annotation of 92% of genes whose transcripts accumulated differentially between M and BS cells of S. viridis with maize homologs. Of the 9,680 transcripts that accumulated differentially in S. viridis, 5,049 were preferential to the M while 4,631 were preferential to the BS. In maize, 14,338 transcripts accumulated differentially between the two cell types, with 6,691 being more abundant in the M and 7,647 up-regulated in the BS. The highly duplicated and complex nature of the maize genome (Schnable et al., 2009) likely contributes to higher numbers in this species. Of the transcripts that were more abundant in the M and BS of S. viridis, 1,848 and 1,825, respectively, shared homologs that were M or BS specific in maize (Fig. 4A; Supplemental File S3). Therefore, we detected a higher degree of similarity in M and BS mRNA profiles between maize and S. viridis than that estimated by two separate studies in maize (Li et al., 2010b; Chang et al., 2012).
Figure 4.
Global convergence in transcript abundance within M and BS cells of S. viridis and maize. A, Venn diagrams show the number of homologous genes from S. viridis and maize that were differentially expressed (FDR = 5%) in the two cell types. B, Functionally enriched gene categories in S. viridis and maize, defined by Fisher’s exact test (FDR = 10%). C, Venn diagrams showing the extent to which transcripts encoding homologous transcription factors accumulate in either M or BS cells of S. viridis and maize (FDR = 5%).
We investigated the functional enrichment of Gene Ontology terms within the M and BS cells of both maize and S. viridis. In maize, of 201 categories, 44 were functionally enriched (false discovery rate [FDR] = 10%), while in S. viridis, of 197 gene categories, 20 were enriched (FDR = 10%; Fig. 4B; Supplemental File S4). Although we detected many differences between the two species (Fig. 4), we also found convergence in a small number of functional categories. Ten categories were enriched in both species, and seven of these were enriched within the same cell type (Table IV). In the M, we detected convergent enrichment of the secondary metabolism of isoprenoids, protein targeting to the chloroplast, protein synthesis, and RNA processing categories (Fig. 4B). In the BS, the tricarboxylic acid cycle, transcription factor, and carbohydrate metabolism categories were overrepresented in both species (Fig. 4B).
Table IV. Statistical enrichment of MapMan categories in either M or BS cells of S. viridis and maize.
Within each category, the number of genes that showed differential transcript abundance in each cell type is listed, along with the Benjamini-Hochberg procedure adjusted P values.
| Category | S. viridis | Maize | Enrichment | ||||
|---|---|---|---|---|---|---|---|
| M | BS | Adjusted P | M | BS | Adjusted P | ||
| Protein synthesis | 119 | 37 | 7.03E-8 | 443 | 41 | 2.23E-099 | M |
| Protein targeting to chloroplast | 19 | 1 | 0.002 | 33 | 5 | 7.26E-006 | M |
| RNA processing | 46 | 20 | 0.07 | 119 | 68 | 3.59E-005 | M |
| Secondary metabolism isoprenoids | 30 | 10 | 0.07 | 47 | 11 | 5.60E-007 | M |
| Carbohydrate metabolism | 47 | 77 | 0.026 | 43 | 125 | 2.68E-006 | BS |
| Tricarboxylic acid cycle | 11 | 32 | 0.01 | 16 | 37 | 0.09 | BS |
| Transcription factors | 400 | 474 | 0.004 | 521 | 736 | 0.002 | BS |
Our data also imply compartmentation of protein-degrading enzymes between the two cell types. In both species, the metalloproteases category was enriched in the M (Table V). In maize, Ser proteases were enriched in the M, while subtilases and AAA-type proteases were more abundant in the BS (Fig. 4B). Although not statistically significant, we also note that categories defined as the Calvin-Benson cycle, development, fatty acid lipid degradation, ATP-binding cassette transporters, and amino acid transporters were more represented in the BS, while lipid metabolism, PSII, and RNA binding were up-regulated in the M of both species (Table V). In summary, the analysis of functional categories indicated shared patterns in transcript abundance within M and BS of two C4 monocot lineages, but we also note a significant amount of functional diversification (Fig. 4B). It is not clear to what extent the similarities in transcript abundance in these cells is associated with their ancestral roles in C3 species or specializations associated with the evolution of the C4 pathway.
Table V. Enrichment of MapMan categories in either M or BS cells of S. viridis and maize.
These categories missed the statistical cutoff but were clearly enriched in both species. The number of genes in each category is shown.
| Category | S. viridis | Maize | Enrichment | ||
|---|---|---|---|---|---|
| M | BS | M | BS | ||
| Calvin-Benson cycle | 9 | 25 | 12 | 20 | BS |
| Development | 55 | 92 | 83 | 107 | BS |
| Lipid metabolism degradation | 19 | 32 | 13 | 42 | BS |
| Transport ATP-binding cassette | 19 | 38 | 24 | 46 | BS |
| Transport amino acids | 23 | 32 | 14 | 36 | BS |
| Lipid metabolism fatty acids | 29 | 20 | 33 | 24 | M |
| PSII | 22 | 8 | 37 | 4 | M |
| Protein degradation metalloprotease | 11 | 4 | 22 | 12 | M |
| RNA binding | 38 | 16 | 79 | 50 | M |
Transcription Factors Underpinning M and BS Gene Expression in Maize, S. viridis, and Sorghum
We identified transcription factors using the MapMan (Thimm et al., 2004) and PlantTFDB (Riaño-Pachón et al., 2007) databases. Transcripts encoding 400 transcription factors were more abundant in the M than the BS of S. viridis, and 122 of these had a direct homolog that accumulated in the M of maize. In addition, of 474 transcription factors whose transcripts were more abundant in the BS compared with M cells of S. viridis, 212 had a BS-specific homolog in maize (Fig. 4C).
Within these transcriptional regulators, we assessed those with known roles in the regulation of either nucleus- or chloroplast-encoded photosynthesis genes to identify the extent to which they have been recruited into cell-specific roles in independent lineages of C4 plants. This included GOLDEN-LIKE1 (GLK1) and GLK2 transcripts that preferentially accumulated in the M and BS, respectively (Table VI). Many of the genes induced by GLK1 and GLK2 in Arabidopsis (Waters et al., 2009) were differentially expressed between the M and BS of S. viridis (Supplemental Table S5). In both S. viridis and maize, transcripts encoding two sigma factors, SIG2 (Si026193m.g and GRMZM2G143392) and SIG3 (Si021619m.g and GRMZM5G830932), were enriched in M and BS cells, respectively (Fig. 5).
Table VI. Abundance of transcripts encoding transcriptional regulators and RNA-binding proteins implicated in chloroplast function in M and BS cells of S. viridis and maize.
Gene identifiers for the homologs in S. viridis and maize are provided. In two cases, a gene duplication appears to have occurred in S. viridis. Library-normalized read counts, fold enrichment in each cell type, and Benjamini-Hochberg procedure adjusted P values are shown.
| Transcript | S. viridis Identifier | Maize Identifier | Role | Cell Type | M | BS | Fold Change, BS/M | Log2 Fold Change, BS/M | Adjusted P |
|---|---|---|---|---|---|---|---|---|---|
| Transcriptional regulators | |||||||||
| Unnamed | Si026826m.g | GRMZM2G177895 | Transcription factor | M | 3,339.75 | 95.63 | 0.03 | −5.13 | 8.39E-83 |
| Si011021m.g | M | 2,479.71 | 155.53 | 0.06 | −3.99 | 1.41E-58 | |||
| IDD5 | Si016736m.g | GRMZM2G129261 | Transcription factor | M | 2,933.07 | 115.56 | 0.04 | −4.66 | 3.3E-74 |
| Si009633m.g | M | 3,833.30 | 171.80 | 0.04 | −4.48 | 2.95E-69 | |||
| IDD5 | Si029540m.g | GRMZM2G042666 | Transcription factor | M | 2,677.52 | 97.22 | 0.04 | −4.78 | 5.33E-74 |
| IDD5 | Si013546m.g | GRMZM2G179677 | Transcription factor | M | 2,117.55 | 79.71 | 0.04 | −4.73 | 3.54E-33 |
| GLK1 | Si006400m.g | GRMZM2G026833 | Transcription factor | M | 3,123.41 | 150.79 | 0.05 | −4.37 | 1.14E-65 |
| PTAC12 | Si000722m.g | GRMZM5G897926 | Transcription factor | M | 368.73 | 115.97 | 0.31 | −1.67 | 3.29E-02 |
| SIG2 | Si026193m.g | GRMZM2G143392 | Transcription factor | M | 2,714.03 | 871.80 | 0.32 | −1.64 | 2.89E-07 |
| Unnamed | Si034188m.g | GRMZM2G089696 | Transcription factor | M | 2,311.88 | 1,106.47 | 0.48 | −1.06 | 7.18E-6 |
| TCP4 | Si004293m.g | GRMZM2G148022 | Transcription factor | BS | 22.38 | 413.95 | 18.5 | 4.21 | 1.61E-41 |
| Unnamed | Si002496m.g | GRMZM2G166946 | Transcription factor | BS | 364.60 | 1,527.56 | 4.19 | 2.07 | 4.45E-18 |
| GLK2 | Si001336m.g | GRMZM2G087804 | Transcription factor | BS | 2,891.51 | 8,242.37 | 2.85 | 1.51 | 8.94E-11 |
| MFP1 | Si000389m.g | GRMZM2G142413 | Transcription factor | BS | 768.46 | 1,800.07 | 2.34 | 1.23 | 3.72E-07 |
| RNA-binding proteins | |||||||||
| Unnamed | Si010801m.g | GRMZM2G090271 | RNA binding | M | 3,999.80 | 840.82 | 0.21 | −2.25 | 2.18E-13 |
| ORRM1 | Si017445m.g | GRMZM5G899787 | RNA binding | M | 601.31 | 236.49 | 0.39 | −1.35 | 8.87E-08 |
| Unnamed | Si036195m.g | GRMZM2G016084 | RNA binding | M | 13,032.89 | 5,137.39 | 0.39 | −1.34 | 3.16E-09 |
| CRB | Si022373m.g | GRMZM2G165655 | RNA binding | BS | 1,079.18 | 12,874.32 | 11.93 | 3.58 | 3.62E-49 |
Figure 5.
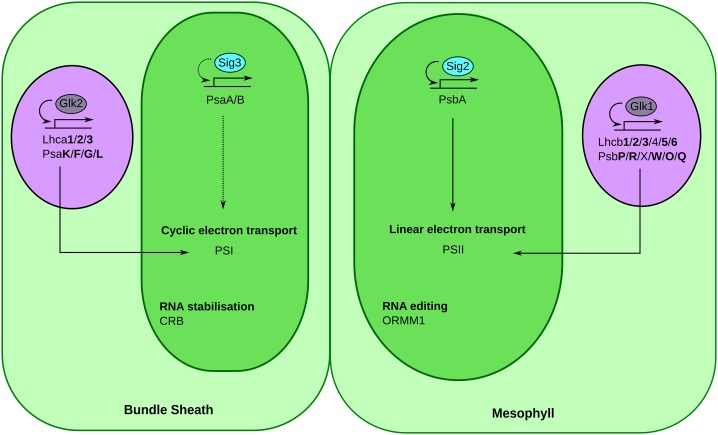
Model for the regulation of photosynthesis gene expression in M and BS cell chloroplasts of maize and S. viridis. Genes previously implicated in the regulation of photosynthesis genes in the nucleus (GLK1 and GLK2) and chloroplast (SIG2) are depicted with solid arrows. The proposed regulation by SIG3 of PsaA/B genes is shown with dashed black arrows. Purple and dark green ovals represent the nucleus and chloroplast, respectively. Genes highlighted in boldface encode components of the light-harvesting complexes known to be differentially expressed between M and BS cells of maize (Li et al., 2010b).
To identify regulatory factors involved in C4 chloroplast organization, we selected genes that were differentially expressed between M and BS cells of S. viridis and maize that were associated with an Arabidopsis Gene Ontology term for plastid organization defined by coexpression analysis as well as physical and genetic interactions (GO:0009657; Obayashi et al., 2011). This identified 183 transcripts that encode proteins involved in PSII assembly, maintenance, and repair as well as others involved in the Calvin-Benson cycle, photorespiration, and cyclic electron transport (Supplemental File S5). A relatively small number of transcriptional regulators were present in this data set. Because of two apparent gene duplications within S. viridis, we identified 14 genes from S. viridis that were homologous to 12 loci from maize (Table VI). This included GLK1, GLK2, and SIG2 but also pTAC12, a regulator of plastid transcription (Gao et al., 2011). In S. viridis, four homologs of Indeterminate domain5 (IDD5), an uncharacterized transcription factor in Arabidopsis, were highly expressed and M specific, as were two uncharacterized SMAD/Forkhead associated domain (FHA) transcriptional regulators (Si026826m.g and Si011021m.g). Both the IDD5 and SMAD/FHA regulators were predicted by TargetP to be chloroplast localized (Emanuelsson et al., 2000). Therefore, we propose that these proteins, along with GLK1 and GLK2 contribute to cell-specific C4 plastid differentiation in S. viridis and maize.
Gene expression in the plastid is also under posttranscriptional control (del Campo, 2009). Within the plastid organization Gene Ontology term, four RNA-binding proteins known or predicted to be chloroplast targeted were identified (Table VI). While two are currently unnamed, the others correspond to Organelle RNA recognition motif protein1 (ORRM1; Sun et al., 2013) and chloroplast RNA binding (CRB; Qi et al., 2012). CRB (Si022373m.g) transcripts preferentially accumulated in BS cells, and the protein is known to bind and stabilize chloroplast transcripts ribulose bisphosphate carboxylase large subunit (RbcL), as well as components of PSI (PsaA/B) and PSII (PsbC/D; Qi et al., 2012).
Many of these transcription factors whose mRNAs accumulated preferentially in either M or BS cells of S. viridis and maize belong to large gene families. To investigate the extent to which evolution coopted genes derived from a common ancestor independently into C4 photosynthesis, we used synteny to determine orthology. Remarkably, this showed that the vast majority of these transcriptional and posttranscriptional regulators in S. viridis and maize were syntenic (Supplemental Table S6). We then used this information to define syntenic orthologs to these regulators in a third C4 grass, sorghum (Sorghum bicolor), and using quantitative reverse transcription-PCR investigated the extent to which each was preferentially expressed in M or BS cells of this species. Of the 11 syntenic orthologs in sorghum, all but two were preferentially expressed in the same cell type as in maize and S. viridis (Supplemental Table S7). Overall, these data imply that different lineages of C4 grass have repeatedly recruited the same trans-factors during evolution.
DISCUSSION
Quantifying C4 Transcript Convergence between S. viridis and Maize
C4 photosynthesis is thought to have evolved independently in at least 62 lineages of angiosperms (Sage et al., 2011), and in almost all cases this requires modifications to gene expression in M and BS cells (Hibberd and Covshoff, 2010). Twenty-six of these lineages are found in the monocotyledons, with a large cluster being restricted to the Panicoideae (Sage et al., 2011). However, to date, patterns of transcript abundance in M and BS cells have only been investigated in the maize lineage (Li et al., 2010b; Chang et al., 2012). Using deep sequencing, we aimed to initiate an understanding of the extent to which the mRNA profiles of M and BS cells are the same in separate C4 grass lineages. While genes recruited into the C4 pathway showed a very high degree of convergence (Pearson’s r = 0.89) in terms of relative transcript abundance in M or BS cells, for all genes with direct homologs in the two species, the correlation was relatively low (r = 0.58). Of thirty-one C4 genes, only two were expressed in opposite cell types in maize and S. viridis. We were also able to quantify the degree to which the abundance of particular transcripts converged in M and BS cells, and this indicated that GDC and CA were the most convergent in BS and M cells, respectively. This is presumably because strong selection pressure leads to very similar levels of gene expression in both cell types of these two species. The extent to which this conservation is ubiquitous in C4 plants will require this type of analysis in many more lineages. There was also a strong correlation between the abundance of transcripts encoding C4 cycle proteins in S. viridis and the abundance of those proteins in maize (r = 0.81). This implies that these cell-specific patterns of transcript accumulation make an important contribution to the compartmentation of C4 cycle proteins in both species.
Quantifying the Convergence in M and BS Transcriptomes of C4 Grasses
We estimate that of the genes for which transcripts were compartmentalized between M and BS of S. viridis, 37% and 39%, respectively, shared this distribution with a direct homolog in maize. This may represent an upper estimate of cell-specific convergence in independent C4 lineages, as these species are both panicoid monocotyledons and belong to the NADP-ME subtype (Brutnell et al., 2010; Sage et al., 2011). Previous work has shown that the M of maize plays important roles in protein synthesis, chloroplast protein targeting, secondary metabolism, and RNA processing, while the BS is critical for transport and carbon metabolism (Majeran et al., 2005; Friso et al., 2010; Li et al., 2010b; Chang et al., 2012). Many gene categories showed divergent patterns of transcript abundance in S. viridis and maize, but transcripts encoding proteins required for chloroplast targeting, isoprenoid metabolism, RNA processing, and protein synthesis were up-regulated in the M of both maize and S. viridis. Increased representation of transcripts encoding components of the protein synthesis machinery in M cells was largely associated with structural components of the chloroplast ribosomes, which may facilitate the synthesis of chloroplast-encoded components of PSII with high turnover rates (Majeran et al., 2005). The chloroplast-targeting term was associated with an up-regulation of translocon components in the M, which is surprising given that the majority of the Calvin-Benson cycle is found in the BS in C4 leaves. It is possible that photosynthesis proteins in the M have faster turnover rates, so increased import is required. Overrepresentation of genes encoding proteins required for isoprenoid synthesis in the M may be related to the presence of PSII in the M. For example, PIGMENT DEFECTIVE EMBRYO181, an enzyme involved in the synthesis of xanthophyll pigments (Josse et al., 2000), was preferentially expressed in M cells. Transcripts derived from 18 genes encoding RNA-binding proteins targeted to the chloroplast were more abundant in the M, while only two were more abundant in the BS. These genes include members of the RNA recognition motif (RRM)/RNA binding domain/Ribonucleoprotein family involved in RNA stabilization, editing, and splicing (Sun et al., 2013).
Previous analyses indicated that transcripts encoding enzymes associated with proteolysis were overrepresented in the BS of maize (Li et al., 2010b; Chang et al., 2012). Our finding that the tricarboxylic acid cycle and transcription factor categories were overrepresented in both species, therefore, extends our understanding of the C4 BS. We also report compartmentation of transcripts encoding specific classes of protein-degrading enzymes between the two cell types, with Ser and metalloproteases enriched in the M while subtilases and AAA-type proteases were enriched in the BS. This specialization in the protein degradation machinery between the two cell types may be important for posttranslational control of C4 proteins (Meierhoff and Westhoff, 1993; Roth et al., 1996; Brutnell et al., 1999).
Regulators Underlying Patterns of Convergence in C4 M and BS Cells
In maize, the GOLDEN2 (G2) protein regulates the accumulation of photosynthesis proteins in the BS (Roth et al., 1996). This role in photosynthesis gene expression of BS cells of C4 plants appears to represent a spatially more confined version of its function in all photosynthetic cells of C3 leaves (Waters et al., 2009). For example, in C3 Arabidopsis, GLK1 and GLK2 redundantly regulate nucleus-encoded photosystem genes, including components of PSI (PSAD-O), PSII (PSBO-Z), as well as their respective light harvesting complexes (LHCA1-5 and LHCB1-6; Waters et al., 2009), and the pale-leaf phenotype of glk mutants in maize and rice suggests that this function is maintained in the monocotyledons (Langdale and Kidner, 1994; Wang et al., 2013). The fact that transcripts homologous to GLK1 and GLK2 in S. viridis accumulate in the M and BS, respectively, indicates that they likely perform analogous roles to GLK and G2 in maize.
The GLK family of transcription factors is the only one with a confirmed role in maintaining C4 photosynthesis. Our data indicate that 122 and 212 additional trans-factors were preferentially expressed in the M and BS, respectively, of maize and S. viridis. We also note that of these trans-factors, 8% and 2% were annotated as fulfilling roles in chloroplast function in the M and BS, respectively. Our analysis of S. viridis and maize, therefore, identifies a small number of regulators whose transcripts preferentially accumulate in either M or BS cells of two independent C4 grass lineages. Compared with C3 species, it appears that for two of these trans-factors, their targets may have diverged. For example, in Arabidopsis, SIG3 regulates PsbN (Zghidi et al., 2007), and while SIG3 transcripts in S. viridis and maize were both BS specific, we did not detect any cell specificity of PsbN transcripts. Furthermore, ORRM1 is involved in editing transcripts encoding the NDH complex of Arabidopsis (Sun et al., 2013). While transcripts encoding the NDH complex accumulated in the BS of S. viridis and maize, ORRM1 transcripts were more abundant in M of all three species, implying that its targets may have altered.
In contrast, of transcripts from S. viridis and maize that accumulate in the same cell types as their known targets in other species, one was a transcription factor while two were involved in posttranscriptional regulation. Nucleus-encoded sigma factors control chloroplast-encoded genes, including components of the photosystems (Tsunoyama et al., 2004; Noordally et al., 2013; Puthiyaveetil et al., 2013). SIG2 is thought to regulate PsbA in C3 Arabidopsis (Woodson et al., 2013), and since both transcripts derived from the SIG2 and PsbA genes are enriched in the M of S. viridis and maize, we infer that SIG2 drives the enrichment of PsbA in both species.
ORGANELLE TRANSCRIPT PROCESSING82 (OTP82) and CHLOROPLAST RNA BINDING PROTEIN (CRB) likely play roles in RNA binding in the BS of maize and S. viridis. In Arabidopsis, OTP82 edits NDH-B (Hammani et al., 2009), so the accumulation of NDH transcripts and the OTP82 homolog (Si006246m.g) in the BS of maize and S. viridis is consistent with this function. CRB stabilizes rbcL transcripts in C3 Arabidopsis (Qi et al., 2012), and CRB transcripts accumulate in the BS of maize and S. viridis where rbcL transcripts are abundant. The fact that transcripts encoding three of the four RNA-binding proteins implicated in plastid gene regulation were M preferential reflects a trend in S. viridis and maize (Li et al., 2010b; Chang et al., 2012) for increased accumulation of mRNAs encoding RNA-binding and RNA-processing proteins in M cells.
Recruitment of Syntenic Orthologs in C4 Grasses
Phylogenetic reconstructions have led to the inference that specific members of multigene families have repeatedly been coopted into the C4 cycle and, therefore, that parallel evolution underlies their recruitment into the C4 pathway (Christin et al., 2013). Using information on the relative abundance of transcripts in M and BS cells, which is a hallmark of C4 photosynthesis, as well as synteny (Schnable et al., 2012), we show that a high proportion of genes recruited into the C4 pathway are syntenic. For example, all 10 structural genes of the C4 cycle and half of the metabolite transporters that are up-regulated in either M or BS cells of maize and S. viridis are syntenic. Our analysis supports the proposals of Christin et al. (2013), but we also find that syntenic homologs from the OMT1 and Rubisco Activase gene families have been recruited into C4 photosynthesis. We excluded genes encoding Ala aminotransferase and pyrophosphorylase from our analysis because the former is not associated with the NADP-ME pathway used by maize and S. viridis (Furbank et al., 2011) and the latter was not differentially expressed between M and BS cells. As genes are recruited into the C4 cycle they are up-regulated, but their expression is also restricted to M or BS cells (Hibberd and Covshoff, 2010). The extent to which parallel evolution underlies both of these alterations in gene expression (Christin et al., 2013) may differ for each gene. The ancestral localization of each protein in M and BS cells of C3 species will need to be determined to provide insight into this phenomenon. The high proportion of syntenic orthologs that are recruited into the C4 cycle is remarkable and indicates that specific members of multigene families are more likely to be coopted into the C4 pathway than others. The simplest explanation for repeated recruitment of syntenic orthologs is presumably that they are part of existing gene regulatory networks in C3 species that are altered in the same way in C4 leaves. It is also possible that the ancestral characteristics of these specific isoforms are more appropriate for a role in C4 photosynthesis than others (Christin et al., 2013).
Notably, in addition to these structural genes, we also detect strong cell-specific expression of transcriptional regulators that are both homologous and syntenic in the maize, Setaria species, and sorghum genomes. The fact that some of these transcription factors belong to families that contain more than 10 genes makes this result compelling. The repeated recruitment of GLK genes from redundant and constitutive expression in C3 leaves (Waters et al., 2009) into cell-specific functions in C4 plants indicates parallel evolution of trans-factors. Analysis of expression patterns in the leaves of ancestral C3 species will be required to confirm whether additional trans-factors have undergone parallel evolution as they are recruited into cell-specific roles in C4 plants. If C4 species have repeatedly used homologous transcription factors to underpin the patterns of gene expression required for the C4 pathway, comparative analysis of multiple C4 and C3 lineages provides an alternative approach to mutant screens and reverse genetics to identify key regulators of this highly complex trait.
CONCLUSION
We report highly convergent patterns of transcript abundance in independent lineages of C4 grasses. The data strongly implicate the recruitment of homologous trans-factors into cell-specific roles in independent groups of C4 plants and also provide, to our knowledge, the first quantitative insight into the extent of convergence of transcript accumulation in M and BS cells of C4 leaves. Specific members of large gene families have been repeatedly recruited into M or BS roles in the C4 leaf. It will be interesting to determine the extent to which other C4 plants have recruited syntenic orthologs into the pathway and converged on very similar levels of transcript compartmentation between M and BS cells.
MATERIALS AND METHODS
Plant Growth, M and BS Separation, and RNA and Protein Isolation
Setaria viridis was grown in a mixture of 3:1 medium compost:fine vermiculite in a growth chamber. The light cycle was set at 12 h of light and 12 h of dark, with a photon flux density of 200 µmol photons m−1 s−1, relative humidity at 75%, and temperature of 23°C. Seeds were placed directly into soil, and after germination, plants were watered one to two times per week. Seventeen days after sowing, plants were watered, and 8 h into the photoperiod, M cell contents and BS cells were isolated from third leaves. The top and bottom 0.5 cm of these 8-cm leaves were discarded and the midrib removed to generate two leaf segments that were subsequently divided into two sections for rolling. Leaf rolling was performed after Covshoff et al. (2013) with the following modification: A glass rod was rolled twice over the surface of each leaf to release the M cell contents. These were then rapidly collected with a pipette filled with mirVana lysis/binding buffer (Ambion).
To isolate BS strands, leaves were cut into 2-mm2 segments placed in isolation buffer (0.33 m sorbitol, 0.3 m NaCl, 0.01 m NaCl, 0.01 m EGTA, 0.01 m dithiothreitol, 0.2 m Tris, pH 9.0, and 5 mm diethylthiophosphoryl chloride), and then pulsed for 10 s three times in a Waring blender on low speed. The suspension was then filtered through a 60-µm mesh, and blending buffer (0.35 m sorbitol, 5 mm EDTA, 0.05 m Tris, pH 8, and 0.1% [v/v] β-mercaptoethanol) was used to return the BS material back into the blender. Homogenization at maximum speed for 1 min followed by filtering was repeated three times. Purified BS cells were placed on a paper towel stack to remove excess moisture and then snap frozen in liquid nitrogen prior to RNA isolation. BS tissue was ground to a fine powder in liquid nitrogen and resuspended in 1 mL of lysis/binding buffer from the mirVana microRNA isolation kit (Ambion).
RNA was eluted in nuclease-free water, and quantity and quality were assessed using the 2100 Bioanalyzer (Agilent Technologies). M and BS proteins were extracted using the same mechanical methods used for RNA extraction, but 20 mm Na2PO4, pH 7.5, plus protease inhibitors (Roche) were used for resuspension. Samples were centrifuged at 12,000g for 10 min prior to the supernatant being removed and then snap frozen in liquid nitrogen. Three replicate samples were generated for sequencing, each of which was derived from 10 plants to provide sufficient RNA for analysis.
Soluble protein (5.5 µg per lane) was separated by SDS-PAGE (12% [v/v] polyacrylamide) and transferred to a 0.2-µm nitrocellulose membrane (Bio-Rad). After transfer, the membrane was placed in 5% (w/v) milk powder in wash buffer (0.33 m NaCl, 0.02 m Tris, and 0.3% [v/v] Tween 20) overnight at 4°C. This was followed by a 1-h incubation with primary antibody (1:1,000 dilution, rabbit anti-CA, anti-PEPC, anti-NADP-MDH, anti-NADP-ME, and anti-LSU of Rubisco) followed by washes and then a 1-h incubation in secondary antibody (anti-rabbit IgG peroxidase, 1:5,000; Sigma) followed by further washes. Antibodies were gifts from R.C. Leegood and J.C. Gray. Western Lightning chemiluminescent substrate (Perkin-Elmer) was applied to the film to allow visualization. Growth conditions as well as cell and RNA extractions from sorghum (Sorghum bicolor) were as described previously (Covshoff et al., 2013).
Quantitative PCR, Deep Sequencing, and Analysis of Gene Expression
For quantitative PCR, 400 ng of RNA was treated with RNase-free DNase (Promega) in 10 µL at 37°C for 30 min. The reaction was stopped with 1 µL of RQ1 DNase Stop solution at 65°C for 10 min. Reverse transcription was performed with SuperScript II according to the manufacturer’s protocol (Invitrogen). Each reaction was diluted 15-fold upon completion. Quantitative PCR was performed using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) with 4 µL of complementary DNA and 4 µm primers in each reaction. Relative expression was normalized based on an RNA spike (Agilent) and primer sequences provided in Supplemental File S6.
While there were two comparable maize (Zea mays) data sets available, we compared our data with those of Chang et al. (2012), as they were generated using experimental and sequencing procedures similar to those used in this study. For example, in this study and Chang et al. (2012), whole-leaf extractions were performed and sequencing was performed in triplicate with high depth and long paired-end reads. In contrast, Li et al. (2010b) used only leaf tips and sequenced two biological replicates with single-end reads.
RNA-seq libraries were prepared from 1 µg of total RNA (TruSeq RNA sample preparation version 2 guide; Illumina). Six libraries (three from each cell type) were sequenced by synthesis with TruSeq version 3 chemistry using one lane of the HiSeq 2000 to generate approximately 202 million 91-bp paired-end reads. Reads for Chang et al. (2012) and Li et al. (2010b) were obtained from the Short Read Archive. Reads were quality trimmed, and adapters were removed using Trimmomatic (Lohse et al., 2012). The latest versions of the genomes for Setaria italica (version 2.0.18) and maize (version 3.18) were used from Ensembl Plants (http://plants.ensembl.org/) with corresponding annotations. Reads were aligned with TopHat2 (default settings, set to two mismatches; Kim et al., 2013), and alignments were then counted to exons with HT-SEQ (Anders, 2011) with mode set to union. Read counts were used as input for DESEQ (Anders and Huber, 2010) for differential expression analysis. Multiple testing correction was by the Benjamini-Hochberg procedure with FDR set to 5%. Counts from HT-SEQ were TPM normalized following the method of Li et al. (2010b). Raw and normalized data are given in Supplemental File S7.
Annotation of genes with homologs was performed using alignments of S. italica (version 2.0.18) and maize (version 3.18) protein sequences obtained from Ensembl Plants (http://plants.ensembl.org/) using the usearch program (Edgar, 2010). Setaria species and maize C4 gene families were identified using the Ensembl Plants Biomart service (Kinsella et al., 2011) and target peptides designated by WoLF PSORT (Horton et al., 2007). Genes were annotated using MapMan mappings (Thimm et al., 2004), except for transcription factors that were supplemented with annotations using PlantTFDB (Riaño-Pachón et al., 2007). Functional enrichment was tested using Fisher’s exact test in R with Benjamini-Hochberg multiple testing correction with FDR set to 10%.
Sequence data from this article can be found in the European Nucleotide Archive under project accession PRJEB5074.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Summary of bioanalyzer traces of RNA samples from mesophyll and bundle sheath of S. viridis.
Supplemental Table S2. Mean fold change values for transcripts encoding C4 proteins.
Supplemental Table S3. Convergence in abundance of transcripts encoding components of the C4 cycle in S. viridis and maize.
Supplemental Table S4. Syntenic orthologs of C4 genes from maize and Setaria spp.
Supplemental Table S5. Differentially expressed S. viridis genes homologous to genes induced by expression of GLK1 or GLK2 in Arabidopsis (Arabidopsis thaliana).
Supplemental Table S6. Transcriptional regulators and RNA-binding proteins implicated in chloroplast function in mesophyll and bundle sheath cells of S. viridis and maize.
Supplemental Table S7. Transcripts encoding chloroplast regulators are preferentially abundant in either mesophyll or bundle sheath cells of sorghum.
Supplemental File S1. Transcript abundance in M and BS cells of Setaria spp. and maize.
Supplemental File S2. C4 related genes in Setaria spp. and maize.
Supplemental File S3. Gene homologs differentially expressed in M and BS cells of Setaria spp. and maize.
Supplemental File S4. Overrepresented Gene Ontology terms in M and BS cells of Setaria spp. and maize.
Supplemental File S5. Transcripts related to chloroplast processes.
Supplemental File S6. Primer sequences.
Supplemental File S7. Raw and normalized data.
Supplementary Material
Glossary
- M
mesophyll
- BS
bundle sheath
- CA
carbonic anhydrase
- PEPC
phosphoenolpyruvate carboxylase
- NADP-ME
NADP-dependent malic enzyme
- PCK
phosphoenolpyruvate carboxykinase
- PPDK
pyruvate,orthophosphate dikinase
- NADP-MDH
NADP-dependent malate dehydrogenase
- LSU
large subunit
- TPM
transcripts per million
- GAPDH
glyceraldehyde phosphate dehydrogenase
- PRK
phosphoribulokinase
- GDC
glycine decarboxylase
- FBA
fructose bisphosphate aldolase
- FDR
false discovery rate
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (studentships to C.R.J. and H.W.) and the Millennium Seed Bank (Ph.D. studentship to R.D.S.-U.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Anders S (2011) HTSeq: analysing high-throughput sequencing data with Python. http://www-huber.embl.de/users/anders/HTSeq/ (April 7, 2014) [DOI] [PMC free article] [PubMed]
- Anders S, Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM. (2011) The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J Exp Bot 62: 3049–3059 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, et al. (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30: 555–561 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. (2011) Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331: 1436–1439 [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Sawers RJ, Mant A, Langdale JA. (1999) BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu XG, Kellogg E, Van Eck J. (2010) Setaria viridis: a model for C4 photosynthesis. Plant Cell 22: 2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Liu WY, Shih ACC, Shen MN, Lu CH, Lu MYJ, Yang HW, Wang TY, Chen SC, Chen SM, et al. (2012) Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol 160: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP. (2013) Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol Evol 5: 2174–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covshoff S, Furbank RT, Leegood RC, Hibberd JM. (2013) Leaf rolling allows quantification of mRNA abundance in mesophyll cells of sorghum. J Exp Bot 64: 807–813 [DOI] [PubMed] [Google Scholar]
- del Campo EM. (2009) Post-transcriptional control of chloroplast gene expression. Gene Regul Syst Bio 3: 31–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CA, Smith SA, Bond WJ, Christin PA, Cousins AB, Duvall MR, Fox DL, Freckleton RP, et al. (2010) The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328: 587–591 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT. (2011) Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J Exp Bot 62: 3103–3108 [DOI] [PubMed] [Google Scholar]
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN. (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD, Slack CR, Johnson HS. (1967) Further studies on a new pathway of photosynthetic carbon dioxide fixation in sugar-cane and its occurrence in other plant species. Biochem J 102: 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann L, Horst I, Perduns R, Dreesen B, Offermann S, Peterhansel C. (2013) A common histone modification code on C4 genes in maize and its conservation in sorghum and Setaria italica. Plant Physiol 162: 456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. (2010) The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol 61: 181–207 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TP, Lloyd JC, Raines CA. (2011) Inter-species variation in the oligomeric states of the higher plant Calvin cycle enzymes glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase. J Exp Bot 62: 3799–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse EM, Simkin AJ, Gaffé J, Labouré AM, Kuntz M, Carol P. (2000) A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011: bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. (2011) C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23: 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Kidner CK. (1994) Bundle sheath defective, a mutation that disrupts cellular differentiation in maize leaves. Development 120: 673–681 [Google Scholar]
- Leegood RC, Walker RP. (2003) Regulation and roles of phosphoenolpyruvate carboxykinase in plants. Arch Biochem Biophys 414: 204–210 [DOI] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. (2010a) RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010b) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–W627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markelz NH, Costich DE, Brutnell TP. (2003) Photomorphogenic responses in maize seedling development. Plant Physiol 133: 1578–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Westhoff P. (1993) Differential biogenesis of photosystem II in mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: the non-stoichiometric abundance of the subunits of photosystem II in the bundle-sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 191: 23–33 [Google Scholar]
- Noordally ZB, Ishii K, Atkins KA, Wetherill SJ, Kusakina J, Walton EJ, Kato M, Azuma M, Tanaka K, Hanaoka M, et al. (2013) Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339: 1316–1319 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. (2011) ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Allen JF. (2013) Evolutionary rewiring: a modified prokaryotic gene-regulatory pathway in chloroplasts. Philos Trans R Soc Lond B Biol Sci 368: 20120260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Armbruster U, Schmitz-Linneweber C, Delannoy E, de Longevialle AF, Rühle T, Small I, Jahns P, Leister D. (2012) Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J Exp Bot 63: 1251–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth R, Hall LN, Brutnell TP, Langdale JA. (1996) Bundle sheath defective2, a mutation that disrupts the coordinated development of bundle sheath and mesophyll cells in the maize leaf. Plant Cell 8: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF, Christin PA, Edwards EJ. (2011) The C4 plant lineages of planet Earth. J Exp Bot 62: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M, Lyons E. (2012) Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol Evol 4: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sun T, Germain A, Giloteaux L, Hammani K, Barkan A, Hanson MR, Bentolila S. (2013) An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc Natl Acad Sci USA 110: E1169–E1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi A, Kishine M, Asada K, Endo T, Sato F. (2005) Differential use of two cyclic electron flows around photosystem I for driving CO2-concentration mechanism in C4 photosynthesis. Proc Natl Acad Sci USA 102: 16898–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. (2004) Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc Natl Acad Sci USA 101: 3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Fouracre J, Kelly S, Karki S, Gowik U, Aubry S, Shaw MK, Westhoff P, Slamet-Loedin IH, Quick WP, et al. (2013) Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta 237: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Johnston IG, Covshoff S, Hibberd JM. (2013) Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2: e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen ZH, Leegood RC. (1999) Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol 120: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghidi W, Merendino L, Cottet A, Mache R, Lerbs-Mache S. (2007) Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Res 35: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, et al. (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30: 549–554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.