A pepper receptor-like cytoplasmic kinase and Xanthomonas effector complex promote cell death in a phosphorylation-dependent manner.
Abstract
Xanthomonas campestris pv vesicatoria type III effector protein, AvrBsT, triggers hypersensitive cell death in pepper (Capsicum annuum). Here, we have identified the pepper SGT1 (for suppressor of the G2 allele of skp1) as a host interactor of AvrBsT and also the pepper PIK1 (for receptor-like cytoplasmic kinase1). PIK1 specifically phosphorylates SGT1 and AvrBsT in vitro. AvrBsT specifically binds to the CHORD-containing protein and SGT1 domain of SGT1, resulting in the inhibition of PIK1-mediated SGT1 phosphorylation and subsequent nuclear transport of the SGT1-PIK1 complex. Liquid chromatography-tandem mass spectrometry of the proteolytic peptides of SGT1 identified the residues serine-98 and serine-279 of SGT1 as the major PIK1-mediated phosphorylation sites. Site-directed mutagenesis of SGT1 revealed that the identified SGT1 phosphorylation sites are responsible for the activation of AvrBsT-triggered cell death in planta. SGT1 forms a heterotrimeric complex with both AvrBsT and PIK1 exclusively in the cytoplasm. Agrobacterium tumefaciens-mediated coexpression of SGT1 and PIK1 with avrBsT promotes avrBsT-triggered cell death in Nicotiana benthamiana, dependent on PIK1. Virus-induced silencing of SGT1 and/or PIK1 compromises avrBsT-triggered cell death, hydrogen peroxide production, defense gene induction, and salicylic acid accumulation, leading to the enhanced bacterial pathogen growth in pepper. Together, these results suggest that SGT1 interacts with PIK1 and the bacterial effector protein AvrBsT and promotes the hypersensitive cell death associated with PIK1-mediated phosphorylation in plants.
Gram-negative phytopathogenic bacteria inject effector proteins into the host cell via the type III secretion system to subvert host basal defense or pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI; Boller and Felix, 2009; Boller and He, 2009; Wilton et al., 2010). Once delivered, effector proteins are targeted to various subcellular locations (Nomura et al., 2005). In effector-triggered immunity (ETI; Jones and Dangl, 2006), pathogen effector proteins are recognized by specific resistance (R) proteins in the host cells. Upon recognition, some effector proteins cause programmed cell death, also known as the hypersensitive response (HR), which is characterized by the rapid death of plant cells at the site of pathogen invasion (Greenberg and Yao, 2004; Lam, 2004). In plants, the molecular events that lead to HR during ETI include the accumulation of salicylic acid (SA), reactive oxygen species (ROS), and nitrogen oxide, the activation of mitogen-activated protein kinase cascades, changes in intracellular calcium levels, and the transcriptional reprogramming and synthesis of antimicrobial compounds (Coll et al., 2011). These cellular responses in ETI are highly accelerated and amplified compared with those in PTI, suggesting that quantitative rather than qualitative differences account for HR cell death induction (Jones and Dangl, 2006).
Xanthomonas campestris pv vesicatoria (Xcv), which causes bacterial spot disease of pepper (Capsicum annuum) and tomato (Solanum lycopersicum), secretes approximately 25 type III effector proteins into the host cell (Bonas et al., 1991; White et al., 2009). Among the Xcv effector proteins, AvrBsT contains a putative YopJ (Yersinia outer protein J)-like Ser/Thr acetyltransferase domain (Mukherjee et al., 2006). However, the exact enzymatic activity of AvrBsT is not fully understood. AvrBsT induces cell death in pepper but suppresses defense responses in tomato (Kim et al., 2010). AvrBsT also suppresses host immunity in most Arabidopsis (Arabidopsis thaliana) ecotypes, with the exception of ecotype Pitztal (Cunnac et al., 2007). Ecotype Pitztal lacks the SUPPRESSOR OF AVRBST-ELICITED RESISTANCE1 phospholipase activity required for the suppression of phosphatidic acid accumulation (Cunnac et al., 2007; Kirik and Mudgett, 2009). AvrBsT acetylates Arabidopsis ACETYLATED INTERACTING PROTEIN1 (ACIP1), a protein that associates with microtubules and is required for immunity (Cheong et al., 2014). AvrBsT-dependent acetylation in planta alters ACIP1’s defense function, which is linked to the activation of ETI. In pepper, AvrBsT was shown to suppress AvrBs1-specific ETI (Szczesny et al., 2010). Importantly, transiently expressed AvrBsT in pepper and Nicotiana benthamiana induces rapid hypersensitive cell death and a strong defense response (Orth et al., 2000; Escolar et al., 2001; Kim et al., 2010). This response caused by AvrBsT is reminiscent of a typical R gene-mediated plant defense (Eitas et al., 2008; Eitas and Dangl, 2010). However, the cognate R protein for AvrBsT has not yet been identified.
Pathogen effector proteins are recognized by nucleotide-binding leucine-rich repeat (NLR) immune receptor proteins using structurally conserved Toll-interleukin receptor-like or coiled coil-nucleotide binding-Leu-rich repeat domains (Eitas and Dangl, 2010). Some periphery proteins have been identified as essential to ETI in Arabidopsis. These proteins function with a chaperone complex including HSP90 (for heat shock protein90), RAR1 (for required for Mla12 resistance), and SGT1 (for suppressor of the G2 allele of skp1; Shirasu, 2009; Zhang et al., 2010). SGT1 is a highly conserved eukaryotic protein involved in the regulation of cell division, SCF (for Skip1p/Cdc53p-Cullin F-box)-mediated ubiquitination of proteins, and the regulation of the cAMP pathway in Saccharomyces cerevisiae (Kitagawa et al., 1999). In S. cerevisiae and humans, SGT1 dimers are involved in chromosome segregation during the cell cycle, leading to SCF complex formation and ubiquitination (Kitagawa et al., 1999; Steensgaard et al., 2004). In S. cerevisiae, SGT1 dimerization is negatively regulated by protein kinase CK2-mediated phosphorylation at Ser-361 (Bansal et al., 2009). Notably, SGT1 has been shown to interact with Leu-rich repeat domains and to be essential for the maintenance of steady-state R protein levels (Leister et al., 2005; Azevedo et al., 2006). In pepper, SGT1 was found to be essential for normal development and growth as well as basal defense responses (Chung et al., 2006).
Protein kinases play a key role in signal transduction by phosphorylating target proteins involved in the plant defense response (Romeis, 2001; Asai et al., 2002; Kim and Hwang, 2011). Among pattern-recognition receptors in PTI, receptor-like kinases are required for the perception and integration of immune responses (Zipfel, 2008; Boller and Felix, 2009). Signals triggered by these pattern recognition receptors are transduced downstream by protein kinase cascades (Pitzschke et al., 2009). Pepper PATHOGEN-INDUCED PROTEIN KINASE1 (PIK1) was previously identified in a complementary DNA (cDNA) screen as up-regulated during Xcv infection (Kim and Hwang, 2011). PIK1 regulates the expression of SA-dependent genes and the generation of ROS, thus conferring resistance to pepper plants (Kim and Hwang, 2011). There is comprehensive evidence that plant pathogens evolve effector proteins that specifically target host kinases and downstream signaling components (Mittal et al., 2006; Zipfel and Rathjen, 2008; Gimenez-Ibanez et al., 2009), further highlighting the critical role of phosphorylation during the resistance response of the host plant.
In this study, we report the identification of the pepper SGT1 that interacts with AvrBsT and pepper PIK1. The Xcv effector, AvrBsT, interacts with SGT1 in vivo. Interestingly, SGT1 was also identified as a PIK1-interacting protein by yeast two-hybrid assays. More importantly, Kim and Hwang (2011) have identified PIK1 as essential for plant signaling of defense and cell death responses. Coimmunoprecipitation and multicolor bimolecular fluorescence complementation (BiFC) analyses (Waadt et al., 2008) revealed that SGT1 forms a heterotrimeric complex with both AvrBsT and PIK1 mainly in the cytoplasm. AvrBsT binds mainly to the CS (for CHORD-containing protein and SGT1) domain of SGT1, suppressing the PIK1-mediated phosphorylation of SGT1 and, subsequently, its nuclear transport. AvrBsT-triggered cell death is promoted by the expression of both SGT1 and PIK1 in plants. SGT1 is essential for AvrBsT recognition and cell death as well as the PIK1 recognition associated with phosphorylation. Silencing of SGT1 and/or PIK1 significantly compromises PTI as well as avrBsT-triggered cell death in pepper. Agrobacterium tumefaciens-mediated transient expression of both SGT1 and PIK1 enhances avrBsT-triggered cell death in N. benthamiana. We conclude that SGT1 specifically interacts with the type III effector AvrBsT and thus promotes the hypersensitive cell death associated with PIK1-mediated phosphorylation.
RESULTS
SGT1 Interacts with AvrBsT and PIK1
AvrBsT is a key avirulence determinant of Xcv in pepper plants (Kim et al., 2010). A yeast two-hybrid cDNA library of pepper leaves infected by the Xcv avirulent strain Bv5-4a was screened for AvrBsT-interacting proteins. As a result, SGT1 (GenBank accession no. JN252483) was identified (Supplemental Figs. S1 and S3A). The isolated SGT1 shared 77% amino acid sequence identity with SGT1b (Chung et al., 2006), previously identified in pepper as essential for growth, development, and basal defense responses (Supplemental Fig. S2). Interestingly, SGT1 was also identified as a PIK1-interacting protein by yeast two-hybrid assays (Supplemental Fig. S3B). PIK1 is a pepper receptor-like cytoplasmic protein kinase that is necessary for plant signaling of defense and cell death responses (Kim and Hwang, 2011).
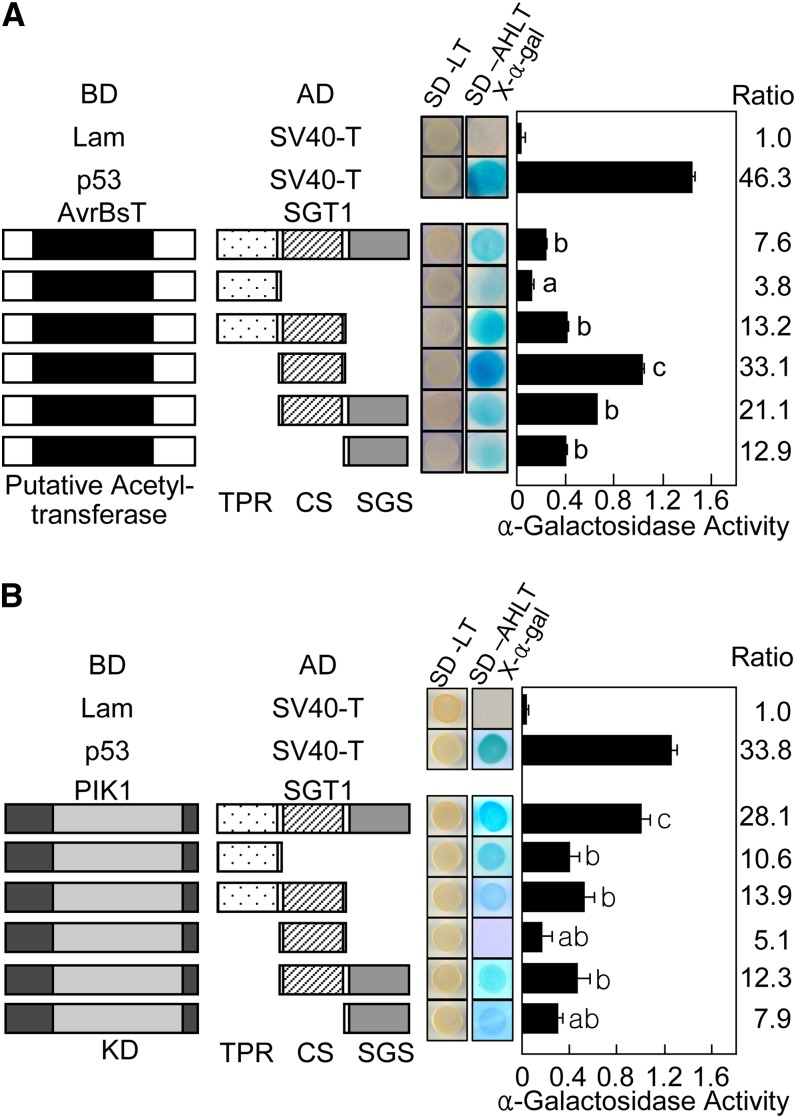
The SGT1 protein contains three distinct regions: tetratricopeptide repeats (TPRs), the CS domain, and a, SGT1-specific (SGS) domain (Supplemental Fig. S2). Full-length and deletion constructs of SGT1 were tested for interaction with both AvrBsT and PIK1 (Fig. 1). In yeast, AvrBsT most strongly interacted with the central CS domain region of SGT1; however, it interacted with the C-terminal SGS domain as strong as the full-length SGT1 but not the N-terminal TRR region (Fig. 1A). As a positive control, p53 strongly interacted with SV40-T in yeast cells. This yeast two-hybrid assay indicates that the CS domain of SGT1 is required for its interaction with AvrBsT. In contrast, all truncated SGT1 interacted poorly with PIK1, compared with the whole SGT1 protein (Fig. 1B).
Figure 1.
AvrBsT and PIK1 interact with SGT1 in yeast. A, Interactions between AvrBsT and SGT1 proteins in yeast two-hybrid assays. B, Interactions between PIK1 and SGT1 proteins in yeast two-hybrid assays. Plasmids containing fusions to the GAL4 DNA-binding domain and transcriptional activation domain are indicated by BD and AD, respectively. Lam-SV40-T and p53-SV40-T combinations were used as negative and positive controls, respectively. KD, Kinase domain; SD, synthetic dropout agar medium; SD-LT, SD minus Leu (L) and Trp (T); SD-ALTH-X-α-gal, SD minus adenine (A), Leu (L), Trp, (T), and His (H) with 5-bromo-4-chloro-3-indoyl-α-d-galactoside (X-α-gal). Different letters indicate statistically significant differences (lsd; P < 0.05). The results represent mean values ± sd from three independent experiments.
SGT1, AvrBsT, and PIK1 Form a Complex in Planta
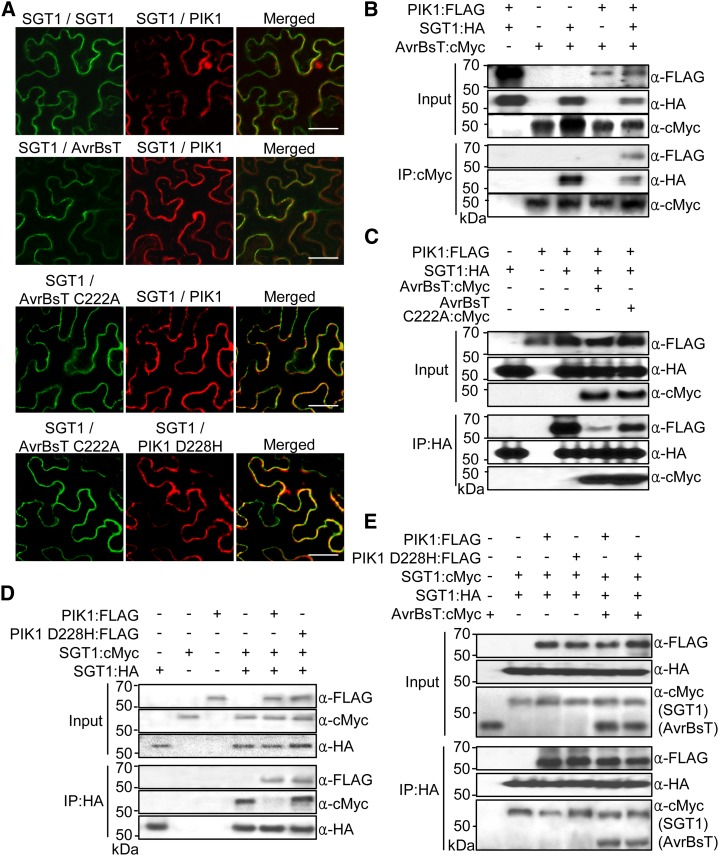
The multicolor BiFC assay (Waadt et al., 2008) was employed to visualize the simultaneous formation of SGT1-SGT1, SGT1-PIK1, SGT1-SGT1-PIK1, and AvrBsT-SGT1-PIK1 protein complexes in N. benthamiana leaves (Fig. 2). In the multicolor BiFC, we used N-terminal fragments of Venus (VYNE) and N-terminal (SCYNE) and C-terminal (SCYCE) fragments of SCFP3A fluorescent proteins (Nagai et al., 2002; Kremers et al., 2006; Waadt et al., 2008). Both SGT1 and AvrBsT have been reported to be localized to the cytoplasm and the nucleus (Noël et al., 2007; Szczesny et al., 2010). SGT1:GFP, PIK1:GFP, and PIK1D228H:GFP fusion proteins were localized to the cytoplasm and the nucleus (Fig. 2; Supplemental Fig. S4). In the PIK1 D228H mutant, a conserved protein kinase active site Asp (D) is replaced with His (H). Transient coexpression of VYNE:AvrBsT with SCYCE:SGT1 resulted in green fluorescence in the cytoplasm (Supplemental Fig. S3C, top row), indicating that AvrBsT interacts with SGT1 in the cytoplasm. When SCYCE:SGT1 and SCYNE:PIK1 were coexpressed, however, blue fluorescence was detected in the cytoplasm and the nucleus (Supplemental Fig. S3C, bottom row), indicating the presence of SGT1-PIK1 complexes. The multicolor BiFC assay revealed that the transient coexpression of SGT1:VYNE, SGT1:SCYCE, and SCYNE:PIK1 results in the formation of SGT1-SGT1 complexes in the cytoplasm and also SGT1-PIK1 complexes in both the cytoplasm and the nucleus (Fig. 2A). This suggests the simultaneous formation of SGT1-SGT1-PIK1 complexes specifically within the cytoplasm (Fig. 2A, top row, light yellow fluorescence in the merged image). Transient coexpression of VYNE:AvrBST, SGT1:SCYCE, and SCYNE:PIK1 resulted in the colocalization of AvrBsT-SGT1 and SGT1-PIK1 complexes in the cytoplasm (Fig. 2A, second row). The AvrBsT C222A mutant was able to interact with SGT1, suggesting that the functional enzymatic activity of AvrBsT, required for the HR (Orth et al., 2000), is not essential for the interaction with SGT1 (Fig. 2A, third row). The PIK1 D228H mutation abolished PIK1 kinase activity (Fig. 3B), but the interaction of PIK1 D228H with SGT1 was not greatly reduced in terms of intensity and frequency (Fig. 2A, fourth row). Immunoblot analyses show that all BiFC fusion proteins were stably synthesized in N. benthamiana leaves 30 h after agroinfiltration (Supplemental Fig. S5).
Figure 2.
AvrBsT, SGT1, and PIK1 form a complex in planta. A, Multicolor BiFC assay of interactions among AvrBsT, SGT1, and PIK1 in N. benthamiana leaves. The signals were visualized 30 h after agroinfiltration using a confocal laser scanning microscope. Fluorescence signals of VYNE/SCYCE (515 nm) and SCYNE/SCYCE (477 nm) channels were digitally colored in green and red, respectively. Bars = 50 µm. B, Coimmunoprecipitation and immunoblotting of AvrBsT and PIK1 with SGT1. The indicated proteins were transiently expressed in N. benthamiana leaves under the control of the 35S promoter. C, AvrBsT inhibits the binding of PIK1 to SGT1. D, SGT1 forms dimers in vivo, and phosphorylation by PIK1 negatively regulates dimer formation. E, AvrBsT inhibits the monomerization of SGT1. Protein extracts were incubated with anti-HA agarose beads, and coimmunoprecipitated proteins were analyzed by immunoblotting using the indicated antibodies.
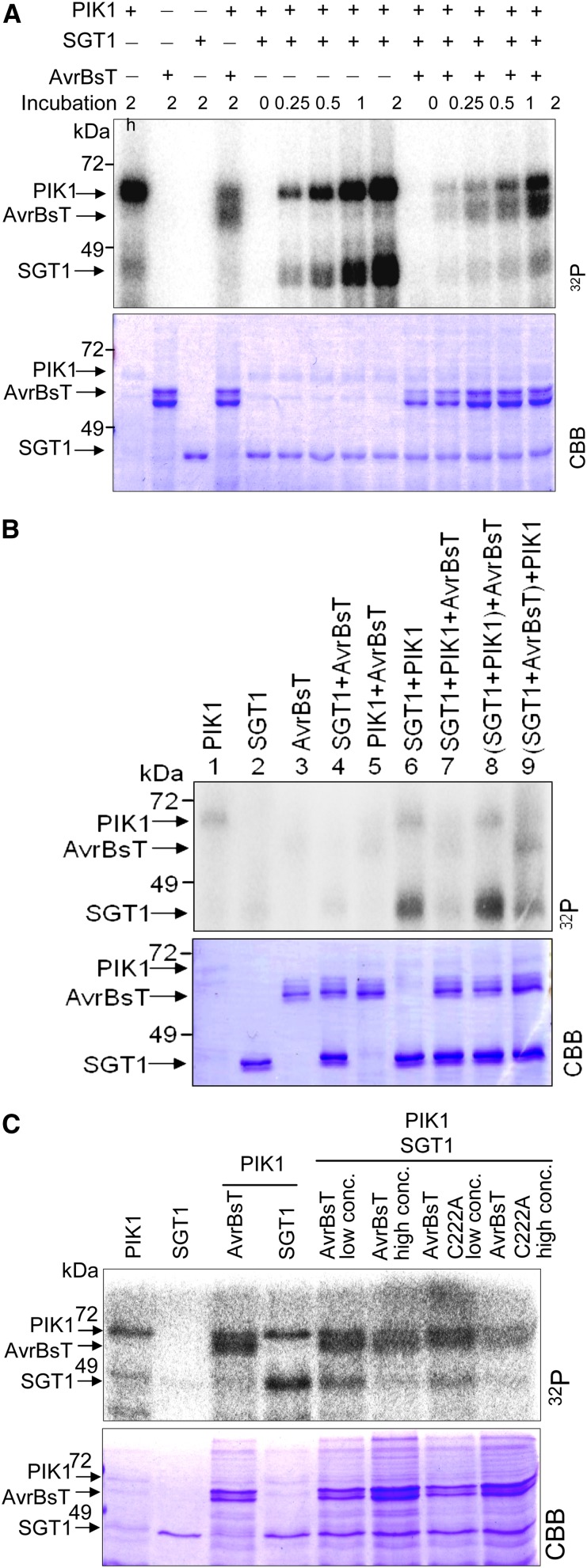
Figure 3.
SGT1 is phosphorylated by PIK1 in vitro. A, SGT1 is a specific target protein of PIK1 for phosphorylation. GST-tagged PIK1, His-tagged SGT1, MAL-tagged CaMBL1, MAL-tagged CaRING1, His-tagged CaPR4b, and MAL proteins were purified from E. coli and separated by SDS-PAGE. Proteins were incubated with [γ-32P]ATP, separated by SDS-PAGE, and visualized by autoradiography and Coomassie Brilliant Blue (CBB) staining. Arrows indicate His-tagged proteins separated on the Coomassie Brilliant Blue-stained gel. B, PIK1 mutant proteins with defective kinase activity do not phosphorylate SGT1. K130R and D228H mutants do not contain functional kinase activity. WT, Wild type. [See online article for color version of this figure.]
The formation of AvrBsT-SGT1-PIK1 complexes was analyzed by coimmunoprecipitation and immunoblotting. AvrBsT, SGT1, and PIK1 were transiently expressed in N. benthamiana, followed by their total protein extraction. The protein extracts were incubated with anti-cMyc-agarose, anti-hemagglutinin (HA)-agarose, or anti-FLAG-agarose, eluted, resolved by SDS-PAGE, and immunoblotted. Immunoblots show that both SGT1 and PIK1 were coimmunoprecipitated with AvrBsT (Fig. 2B). However, PIK1 was only detected in the presence of SGT1. We next investigated whether the residues of AvrBsT that are critical for cell death (Orth et al., 2000) affect the binding of SGT1 to PIK1 (Fig. 2C). PIK1 were coimmunoprecipitated and immunoblotted with SGT1 when both SGT1 and PIK1 were expressed without AvrBsT. The addition of AvrBsT strongly inhibited the coimmunoprecipitation of PIK1 with SGT1; however, the mutant AvrBsT C222A did not inhibit the binding of PIK1 with SGT1. SGT1 dimer formation was also confirmed by coimmunoprecipitation (Fig. 2D). SGT1:cMyc alone did not cross-react with anti-HA agarose beads, and SGT1:cMyc was only detected in the presence of SGT1:HA. SGT1 dimerization was disrupted when PIK1 was coexpressed, as shown by the weak band intensity of SGT1:cMyc (Fig. 2D). In contrast, the disruption of SGT1 dimerization was abolished when phospho-inactive PIK1 D228H was coexpressed with SGT1:cMyc and SGT1:HA. Coexpression of AvrBsT with SGT1:cMyc, SGT1:HA, and PIK1:FLAG increased the level of coimmunoprecipitated SGT1:cMyc (Fig. 2E) compared with the coexpression of SGT1:cMyc, SGT1:HA, and PIK1:FLAG without AvrBsT (Fig. 2D), suggesting that AvrBsT inhibits the disruption of SGT1 dimerization. Collectively, these results indicate that AvrBsT, SGT1, and PIK1 form a protein complex in the cytoplasm in planta.
PIK1 Specifically Phosphorylates SGT1 and AvrBsT
An in vitro kinase assay was used to investigate whether SGT1 is a specific phosphorylation substrate of PIK1 (Fig. 3). The recombinant PIK1 was autophosphorylated, and the resulting protein, visualized as a band of 69 kD, corresponded to the combined molecular mass of PIK1 and glutathione S-transferase (GST; Fig. 3A; Kim and Hwang, 2011). PIK1 specifically phosphorylated SGT1 but not mannose-binding lectin1 (MBL1; Hwang and Hwang, 2011), really interesting new gene1 (RING1; Lee et al., 2011), pathogenesis-related protein4b (PR4b; Hwang et al., 2014), or maltose-binding protein (MAL) proteins used as negative controls. Autophosphorylation and SGT1 phosphorylation did not occur when using the PIK1 K130R and D228H mutants. In the PIK1 K130R mutant, an ATP-binding site Lys (K) is substituted with Arg (R; Fig. 3B). Significantly, PIK1 did not interact with AvrBsT in yeast (data not shown), but PIK1 did phosphorylate AvrBsT in vitro (Fig. 4A).
Figure 4.
AvrBsT inhibits PIK1 autophosphorylation and phosphorylation of SGT1. A, Time courses of the inhibition of PIK1 and SGT1 phosphorylation by AvrBsT. B, Inhibitory effects of AvrBsT on PIK1 and SGT1 phosphorylation during their different in vitro interactions. Lanes 1 to 7, Each mixture was incubated with [γ-32P]ATP at 30°C for 1 h; lanes 8 and 9, after the first incubation of SGT1/PIK1 or SGT1/AvrBsT with [γ-32P]ATP at 30°C for 30 min, AvrBsT or PIK1 was added to the mixtures, respectively, followed by incubation for 30 min. C, Inhibition of the phosphorylation of SGT1 and PIK1 is independent of AvrBsT enzymatic activity. PIK1, AvrBsT, AvrBsT C222A, and SGT1 proteins were incubated with [γ-32P]ATP for 1 h. Proteins were separated by SDS-PAGE and visualized by autoradiography (top) and Coomassie Brilliant Blue (CBB) staining (bottom). AvrBsT low concentration was 10 μg, and AvrBsT high concentration was 20 μg. [See online article for color version of this figure.]
AvrBsT Inhibits SGT1 Phosphorylation as Well as PIK1 Autophosphorylation
A time course in vitro kinase assay was used to investigate whether AvrBsT binding to the SGT1-PIK1 complex affects phosphorylation. The assay revealed that both PIK1 autophosphorylation and SGT1 phosphorylation were reduced in the presence of AvrBsT (Fig. 4A). The inhibitory effects of AvrBsT on PIK1 and SGT1 phosphorylation were examined during their differential in vitro interactions (Fig. 4B). Preincubation of SGT1 with PIK1 resulted in strong SGT1 phosphorylation, regardless of whether AvrBsT was later added to the reaction mixture. PIK1 was autophosphorylated at similar levels during the SGT1 phosphorylation, indicating that SGT1 does not affect PIK1 kinase activity. However, preincubation of SGT1 with AvrBsT significantly reduced SGT1 phosphorylation by PIK1, although AvrBsT was apparently phosphorylated as an alternative. AvrBsT also reduced PIK1 autophosphorylation. Together, these results suggest that AvrBsT specifically binds to SGT1 in order to block SGT1 phosphorylation sites. This results in the inhibition of SGT1 phosphorylation as well as PIK1 autophosphorylation. However, how AvrBsT could inhibit PIK1 autophosphorylation remains to be determined.
To determine whether the putative YopJ-like Ser/Thr acetyltransferase activity of AvrBsT is responsible for the phosphorylation inhibition, an AvrBsT mutant was generated. In this mutant (designated C222A), the 222nd Cys (C) was substituted with Ala (A; Orth et al., 2000). The AvrBsT C222A mutant was able to inhibit SGT1 phosphorylation similar to the wild-type AvrBsT in a dosage-dependent manner (Fig. 4C). These results indicate that the phosphorylation inhibition by AvrBsT was not due to the putative enzymatic activity of AvrBsT. Instead, the inhibition results from AvrBsT binding to SGT1, causing a blockage on or around the SGT1 phosphorylation sites.
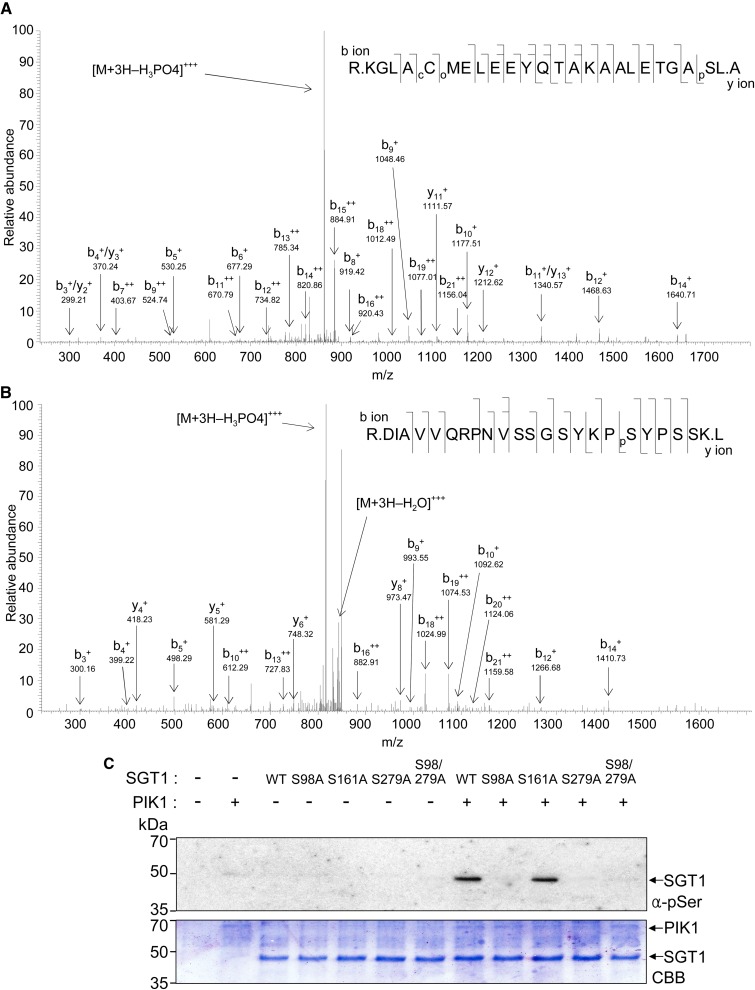
Mapping of SGT1 Phosphorylation Sites by Liquid Chromatography-Tandem Mass Spectrometry
To identify SGT1 phosphorylation sites, SGT1 protein was in vitro phosphorylated by PIK1. After SDS-PAGE of the in vitro-phosphorylated SGT1 protein, the SGT1 band was excised, digested with trypsin, and subjected to ion-trap liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. SGT1 phosphorylation sites were mapped by LC-MS/MS, and the two residues Ser-98 and Ser-279 were identified from the fragmentation spectra (Fig. 5, A and B). The identified Ser (S) sites were mutated to Ala (A) to deny phosphorylation and mimic dephosphorylation. Ser-161 was mutated to Ala to use as a negative control. Wild-type SGT1 and SGT1 S161A mutant proteins were phosphorylated by PIK1 in vitro (Fig. 5C). In contrast to the wild type and the S161A mutant, the mutants SGT1 S98A, S279A, and S98/279A were not phosphorylated by PIK1, suggesting that these residues, Ser-98 and Ser-297, can be putative PIK1-mediated phosphorylation sites. However, both mutations in SGT1 may also cause structural changes that hinder phosphorylation by PIK1.
Figure 5.
SGT1 is phosphorylated on Ser-98 and Ser-279 residues by PIK1. The in vitro-phosphorylated SGT1 protein was PAGE purified and in-gel digested with trypsin, and the resulting peptides were extracted and analyzed by LC-MS/MS using a capillary LC system directly connected to the LTQ linear ion-trap mass spectrometer. Each MS/MS spectrum is a collection of ions produced by collision-induced dissociation of the intact peptide. A and B, Amino acid sequences and electrospray ionization MS/MS spectra of tryptic peptides of SGT1. The predominant ion peaks of N-terminal fragments (b ions) and C-terminal fragments (y ions) are labeled accordingly, with the subscripts denoting their positions in the identified peptide and the + and ++ superscripts indicating singly and doubly protonated ions, respectively. Product ions eliciting neutral mass losses of H3PO4 and water (H2O) are also indicated. Identified phosphoseryl resides are denoted as pS. cC denotes carbamidomethylation of the Cys residue, and oM denotes oxidation of the Met residue. C, In vitro phosphorylation of the wild-type SGT1 (WT) and S98A, S161A, S279A, and S98A/S279A mutant proteins by PIK1. Phosphorylated proteins were detected using the anti-phosphoserine antibody (top). The SDS-PAGE gel was stained with Coomassie Brilliant Blue (CBB; bottom). [See online article for color version of this figure.]
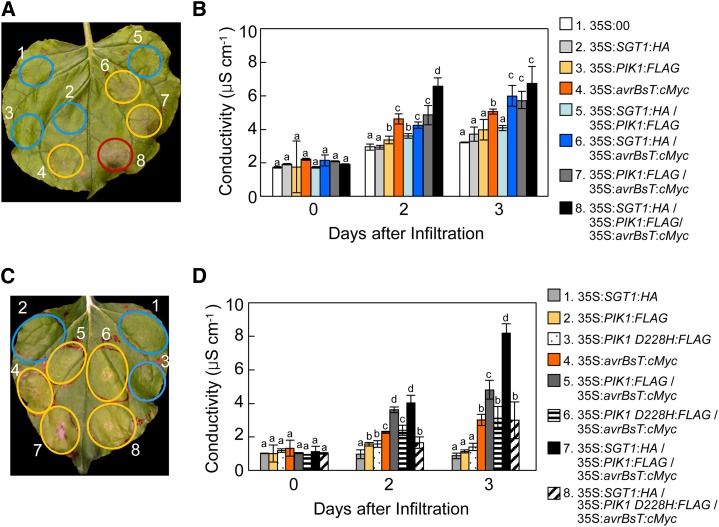
SGT1 and PIK1 Coexpression Enhances avrBsT-Triggered Cell Death
An A. tumefaciens-mediated transient expression experiment was conducted to investigate whether the coexpression of SGT1 and/or PIK1 affects the avrBsT-triggered cell death phenotype in N. benthamiana leaves (Fig. 6). To clearly observe the additive effect of the coexpression of SGT1 and/or PIK1, A. tumefaciens titer was set to optical density at 600 nm (OD600) = 0.05, at which AvrBsT induces full cell death at 5 d after agroinfiltration. The transient coexpression of SGT1 and PIK1 or the expression of either SGT1 or PIK1 did not induce any cell death (Fig. 6A). In contrast, the expression of avrBsT alone or in combination with SGT1 and/or PIK1 distinctly triggered a typical cell death response 3 and 5 d after agroinfiltration (Fig. 6A). However, transient expression of the avrBsT C222A mutant did not trigger hypersensitive cell death, as observed previously (Orth et al., 2000; Supplemental Fig. S6, A and B). Coexpression of avrBsT with either SGT1 or PIK1 produced a more severe necrotic cell death phenotype than avrBsT expression alone. The induction of the cell death response was intimately associated with increased electrolyte leakage from the leaf tissues (Fig. 6B). Transient expression of avrBsT with either SGT1 or PIK1 enhanced electrolyte leakage. AvrBsT coexpression with both SGT1 and PIK1 resulted in the greatest electrolyte leakage among the coexpression combinations. The PIK1 D228H mutant did not enhance avrBsT-triggered cell death as compared with wild-type PIK1 (Fig. 6, C and D). Immunoblot analyses show that the epitope-tagged SGT1, PIK1, PIK1 D228H, and AvrBsT proteins were transiently expressed in agroinfiltrated N. benthamiana leaves (Supplemental Fig. S7, A and B). Together, these data indicate that PIK1-mediated phosphorylation of AvrBsT results in the enhancement of AvrBsT-triggered HR.
Figure 6.
Transient coexpression of SGT1 and PIK1 with avrBsT enhances avrBsT-triggered hypersensitive cell death. A and C, Cell death phenotypes in N. benthamiana leaves 3 d after agroinfiltration (OD600 = 0.05). The infiltrated sites with no visible, partial, and full cell death phenotypes are circled in blue, yellow, and red, respectively. B and D, Quantification of electrolyte leakage from the N. benthamiana leaves after agroinfiltration (OD600 = 0.05). The data represent means ± sd from three independent experiments. Different letters indicate statistically significant differences (lsd; P < 0.05).
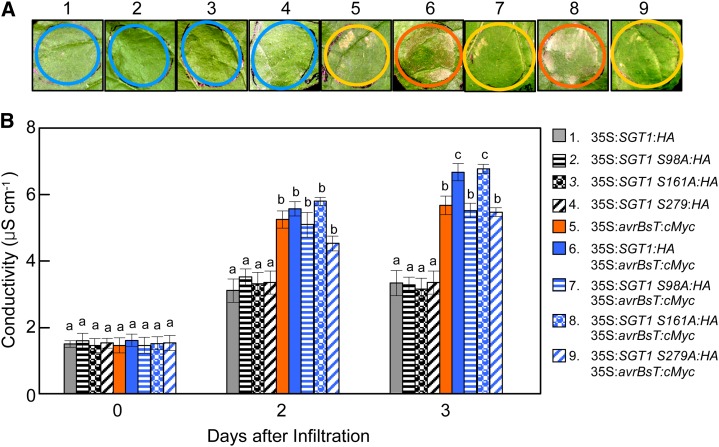
Coexpression of Phosphorylation-Defective SGT1 Mutants Does Not Enhance avrBsT-Triggered Cell Death
To investigate whether the identified SGT1 phosphorylation sites are involved in the activation of AvrBsT-triggered cell death, SGT1 and SGT1 phosphorylation-defective mutants were coexpressed with avrBsT by agroinfiltration into N. benthamiana leaves (Fig. 7). To clearly define the additive effect of the coexpression of SGT1 and SGT1 mutants, the A. tumefaciens titer was set to OD600 = 0.05, at which avrBsT expression induces full cell death 5 d after agroinfiltration. The transient expression of SGT1, SGT1 S98A, SGT1 S161A, or SGT1 S279A alone did not induce any cell death in N. benthamiana leaves (Fig. 7A). However, the expression of avrBsT alone or in combination with wild-type SGT1 distinctly triggered a typical cell death response 3 d after agroinfiltration. Coexpression of avrBsT with the SGT1 S161A mutant that was normally phosphorylated was effective enough to enhance cell death by avrBsT and SGT1 coexpression. In contrast, coexpression of the phosphorylation-defective mutants SGT1 S98A and SGT1 S279A with avrBsT did not enhance avrBsT-triggered cell death in N. benthamiana leaves (Fig. 7A). The induction of the cell death response (Fig. 7A) was consistent with the increased electrolyte leakage from the N. benthamiana leaf tissues (Fig. 7B). Transient expression of avrBsT with either SGT1 or SGT1 S161A distinctly enhanced electrolyte leakage from the leaf tissues. In contrast, the phosphorylation-defective SGT1 S98A and S279A mutants were not able to enhance avrBsT-triggered electrolyte leakage as compared with wild-type SGT1 (Fig. 7B). Immunoblot analyses show that the HA- or cMyc-tagged SGT1, SGT1 S98A, SGT1 S161A, SGT1 S279A, and AvrBsT proteins were transiently expressed in agroinfiltrated N. benthamiana leaves (Supplemental Fig. S8). Together, these results indicate that SGT1 phosphorylation is required for the enhancement of the AvrBsT-triggered hypersensitive cell death response.
Figure 7.
Transient coexpression of SGT1 phosphorylation-defective mutants with avrBsT does not enhance avrBsT-triggered hypersensitive cell death. A, Cell death phenotypes in N. benthamiana leaves 3 d after agroinfiltration (OD600 = 0.05). The infiltrated sites with no visible, partial, and severe cell death phenotypes are circled in blue, yellow, and orange, respectively. B, Quantification of electrolyte leakage from the N. benthamiana leaves after agroinfiltration (OD600 = 0.05). The data represent means ± sd from three independent experiments. Different letters indicate statistically significant differences (lsd; P < 0.05).
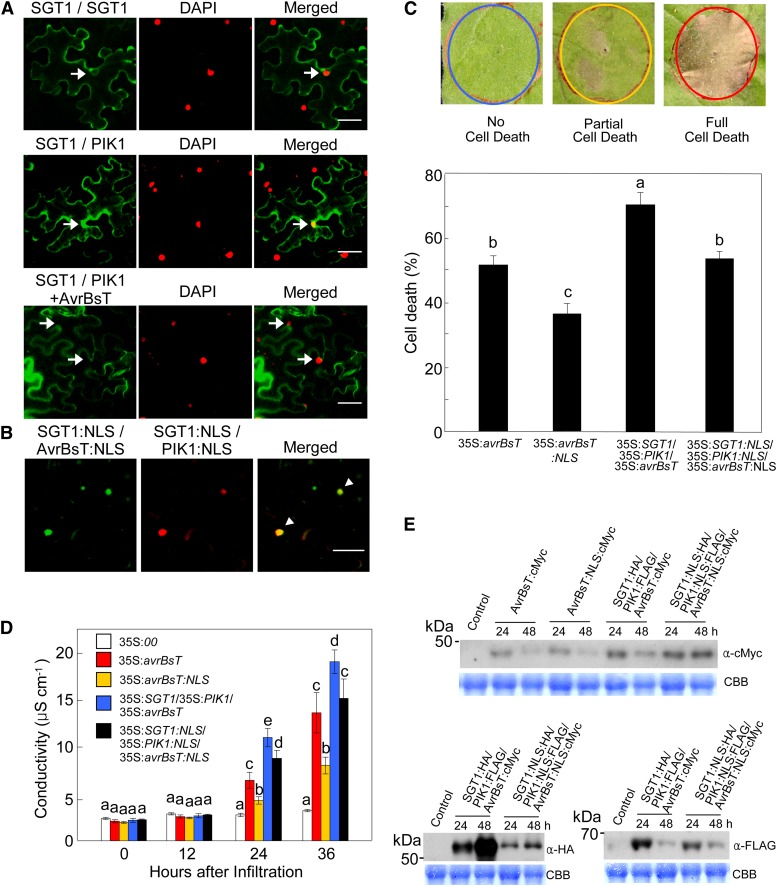
Nuclear Localization of SGT1/PIK1/AvrBsT Reduces HR Cell Death
The BiFC constructs were used to analyze the nuclear localization of SGT1/PIK1/AvrBsT. The nuclear localization of the SGT1-PIK1 complexes was confirmed by counterstaining the cell nuclei with 4′,6-diamidino-2-phenylindole (DAPI; Fig. 8A). VYNE/SCYCE signals from SGT1:VYNE/SGT1:SCYCE coexpression did not colocalize with DAPI signals, suggesting that SGT1 dimers remain in the cytoplasm (Fig. 8A). The BiFC assay and DAPI counterstaining data also indicate the cytoplasmic localization of the AvrBsT-SGT1-PIK1 complexes (Fig. 8A). To determine whether the localization of the SGT1-PIK1-AvrBsT complex affects cell death induction, a nuclear localization signal (NLS; Slootweg et al., 2010; Choi and Hwang, 2011; Choi et al., 2012) was added to the BiFC constructs. Nuclear localization of the protein complexes following the addition of the NLS was confirmed using confocal microscopy (Fig. 8B; Supplemental Fig. 9A). Immunoblot analyses show that all BiFC fusion proteins were stably synthesized in N. benthamiana leaves 30 h after agroinfiltration (Supplemental Fig. S10). When cytoplasmic AvrBsT was nucleus localized, full cell death phenotypes were reduced in N. benthamiana leaves (Fig. 8C; Supplemental Fig. S10B). Likewise, nuclear targeting of the SGT1-PIK1-AvrBsT complex resulted in lower full cell death phenotypes (53%) than did the cytoplasmic SGT1-PIK1-AvrBsT complexes (67%). At 24 and 36 h after agroinfiltration, transient expression of the cytoplasmic SGT1-PIK1-AvrBsT complex induced the greatest electrolyte leakage from N. benthamiana leaves, followed in decreasing order by the nuclear SGT1-PIK1-AvrBsT complex, cytoplasmic AvrBsT, and the AvrBsT NLS (Fig. 8D). Nucleus-targeted expression of avrBsT resulted in the lowest cell death and electrolyte leakage (Fig. 8, C and D). These results indicate that the cytoplasmic localization of either AvrBsT or the SGT1-PIK1-AvrBsT complex is required for the induction of the hypersensitive cell death response. The transient expression of these proteins or protein complexes was clearly confirmed by immunoblot analysis (Fig. 8E).
Figure 8.
Nuclear localization of the SGT1-PIK1-AvrBsT complex reduces cell death phenotypes. A, BiFC analysis and DAPI counterstaining for the detection of nuclear localization. 35S:SGT1:SPYNE, 35:SGT1:SPYCE, and 35S:PIK1:SPYCE constructs were used for BiFC analysis. The 35S:avrBsT construct was also used to coexpress AvrBsT with SGT1:SPYNE and PIK1:SPYCE (SGT1/PIK1 + AvrBsT). Samples were counterstained with DAPI to visualize nuclei. Arrows indicate the nuclei colocalized with BiFC signals. Bars = 50 µm. B, Multicolor BiFC assay of the altered localizations of AvrBsT, SGT1, and PIK1. The NLS sequence was added to 35S:VYNE:avrBsT, 35S:SGT1:SCYCE, and 35S:SCYNE:PIK1 constructs. Arrowheads indicate the nuclei colocalized with SGT1/PIK1/AvrBsT. Fluorescence signals of VYNE/SCYCE (515 nm) or SPYNE/SPYCE (527 nm) and SCYNE/SCYCE (477 nm) or DAPI (470 nm) were digitally colored in green and red, respectively. Bar = 50 µm. C, Cell death scores in SGT1, PIK1, and AvrBsT localizations in N. benthamiana leaves. Cell death levels were rated based on a 1 to 3 scale: 1, no cell death (less than 10%); 2, partial cell death (10%–80%); and 3, full cell death (80%–100%). D, Quantification of electrolyte leakage from N. benthamiana leaves after agroinfiltration. E, Transient expression of AvrBsT:cMyc, SGT1:HA, and PIK1:FLAG after agroinfiltration. Total soluble proteins were resolved on 10% (w/v) SDS-PAGE gels, followed by immunoblotting with anti-cMyc, anti-HA, and anti-FLAG antibodies. CBB, Coomassie Brilliant Blue. For C and D, data are means ± sd from three independent experiments. Different letters indicate statistically significant differences (lsd; P < 0.05).
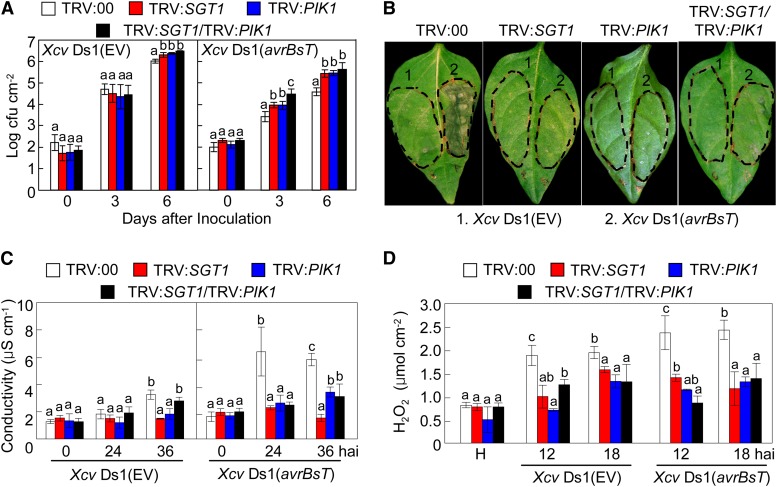
Silencing of SGT1 and PIK1 Compromises avrBsT-Triggered Cell Death
Virus-induced gene silencing was performed to investigate whether SGT1 and PIK1 are essential for avrBsT-triggered cell death in pepper (Fig. 9). Silencing of SGT1, PIK1, and SGT1/PIK1 conferred enhanced susceptibility to Xcv infection. Xcv virulent DS1 or avirulent DS1 (avrBsT) grew slightly better in the silenced plants than those in the empty vector control plants (Fig. 9A). These results suggest that SGT1 and PIK1 are necessary for resistance to Xcv infection. Notably, silencing of SGT1 and PIK1 compromised avrBsT-triggered cell death in pepper (Fig. 9B). No cell death phenotypes were observed in SGT1-, PIK1-, or SGT1/PIK1-silenced leaves. Infection by avirulent Xcv Ds1 (avrBsT) induced lower electrolyte leakage from SGT1-, PIK1-, or SGT1/PIK1-silenced leaves in comparison with empty vector leaves (Fig. 9C). This supports the hypothesis that transient expression of SGT1 or SGT1/PIK1 contributes to the avrBsT-triggered cell death. Additionally, infection by either Xcv Ds1 or Ds1 (avrBsT) did not significantly trigger hydrogen peroxide (H2O2) accumulation in the gene-silenced leaves (Fig. 9D; Supplemental Fig. S11). This indicates that AvrBsT, SGT1, or PIK1 acts as a general trigger for ROS burst.
Figure 9.
Silencing of SGT1, PIK1, and SGT1/PIK1 compromises the avrBsT-triggered hypersensitive cell death response. A, Bacterial growth in leaves of empty vector (EV) control (TRV:00) and silenced pepper plants infiltrated with Xcv (5 × 104 colony-forming units [cfu] mL−1). B, Cell death phenotypes developed on empty vector control (TRV:00) and silenced leaves 2 d after infiltration with Xcv (108 cfu mL−1). C, Quantification of electrolyte leakage from leaves infiltrated with Xcv (5 × 107 cfu mL−1). D, Quantification of H2O2 accumulation in leaves infiltrated with Xcv (5 × 107 cfu mL−1). Data are means ± sd from three independent experiments. Different letters indicate statistically significant differences (lsd; P < 0.05).
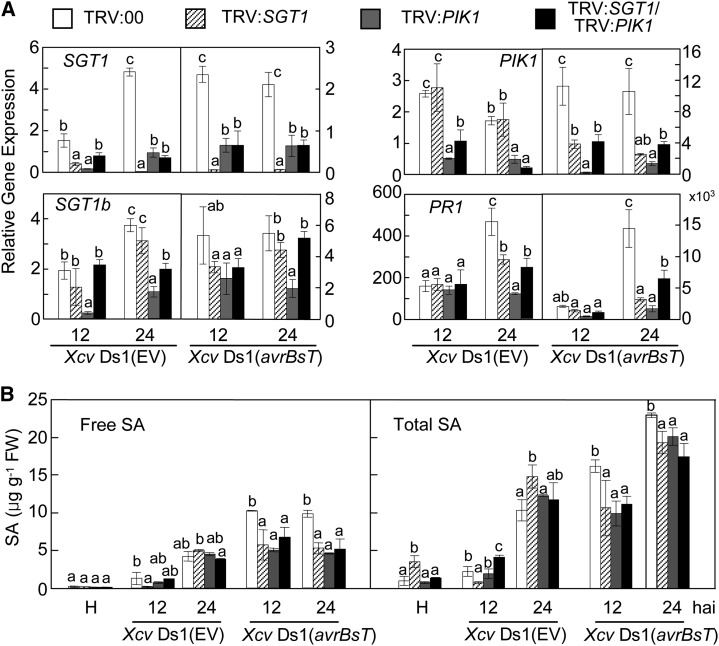
Transcripts of SGT1 and PIK1 were significantly reduced in the gene-silenced pepper leaves during Xcv infection (Fig. 10A), indicating that SGT1 and PIK1 were effectively silenced in pepper. The level of SGT1b transcripts was not significantly altered by SGT1 silencing. This indicates that the construct used for silencing SGT1 was SGT1 specific (Fig. 10A). Interestingly, PIK1 silencing compromised the expression of both SGT1 and SGT1b during Xcv Ds1 and Ds1 (avrBsT) infection. SGT1-silenced leaves also failed to accumulate the PIK1 transcript during Ds1 (avrBsT) infection. The defense-related genes pathogenesis-related protein1 (PR1; Kim and Hwang, 2000), defensin1 (DEF1; Do et al., 2004), and systemic acquired resistance gene8.2 (SAR8.2; Lee and Hwang, 2003; Fig. 10A; Supplemental Fig. S12A) were significantly down-regulated in silenced leaves during Xcv (avrBsT) infection. These results imply that the induction of these defense-related genes by SGT1 and PIK1 enhances avrBsT-triggered cell death in pepper.
Figure 10.
Silencing of SGT1, PIK1, and SGT1/PIK1 compromises defense-related gene expression and SA and JA accumulation. A, Quantitative real-time PCR analysis of the expression of SGT1, SGT1b, PIK1, and PR1 in pepper leaves infected by Xcv. The pepper 18S ribosomal RNA was used to normalize the mRNA abundance of the tested genes. B, Comparison of free SA and total SA (free SA plus Glc-conjugated SA) levels in leaves. Data are means ± sd from three independent experiments. Different letters indicate statistically significant differences (lsd; P < 0.05). EV, Empty vector; FW, fresh weight.
Previously, the accumulation of SA was shown to be impaired in PIK1-silenced plants during incompatible Xcv infections (Kim and Hwang, 2011). Consistent with the defense gene expression patterns, SGT1-, PIK1-, and SGT1/PIK1-silenced plants contained significantly lower SA levels when compared with the empty vector control plants 12 and 24 h after inoculation with Xcv Ds1 (avrBsT; Fig. 10B). These results indicate that SGT1 and PIK1 function in AvrBsT recognition upstream of SA. In contrast to the SA levels, the silencing of SGT1, PIK1, and SGT1/PIK1 did not significantly compromise the accumulation of jasmonic acid (JA) during Xcv infection, with the exception of lower JA levels exhibited by silenced leaves 24 h after inoculation with Xcv Ds1 (avrBsT; Supplemental Fig. S12B).
DISCUSSION
In this study, pepper SGT1 is demonstrated to interact with the Xcv type III effector, AvrBsT, to promote hypersensitive cell death associated with the receptor-like cytoplasmic PIK1-mediated phosphorylation. AvrBsT and PIK1 have been identified as SGT1-interacting proteins by yeast two-hybrid screening. AvrBsT triggers HR in pepper-Xcv interactions (Kim et al., 2010). SGT1 positively regulates PTI as well as AvrBsT-triggered cell death in pepper. PIK1 is suggested to be crucial for the plant signaling of defense and cell death responses (Kim and Hwang, 2011). PIK1 phosphorylates both AvrBsT and SGT1. AvrBsT binds to the CS domain of SGT1 and blocks the phosphorylation of SGT1 by PIK1 and the nuclear transport of SGT1. Silencing of SGT1 and/or PIK1 compromises PAMP-triggered immunity and avrBsT-triggered cell death in pepper.
SGT1, an essential eukaryotic protein, exhibits features of a cochaperone (Shirasu and Schulze-Lefert, 2003). Notably, SGT1 forms a chaperone complex with HSP90 to function as an immune sensor of the plant NLR immune receptor proteins (Shirasu, 2009). The residues Ser-98 and Ser-279 of SGT1 were identified as the major PIK1-mediated phosphorylation sites. Site-directed mutagenesis of SGT1 revealed that the identified SGT1 phosphorylation sites are required for the activation of AvrBsT-triggered cell death in N. benthamiana. Silencing of SGT1 in pepper leaves compromised the hypersensitive cell death response triggered by AvrBsT, suggesting that SGT1 is essential for the ETI. Drastically low expression of the defense-related genes PR1, DEF1, and SAR8.2 in SGT1-silenced plants supports the hypothesis that SGT1 functions as a positive regulator in the plant signaling of downstream defense responses.
Plant immune receptors have been suggested to integrate signals from multiple subcellular compartments to coordinate effective immune responses against pathogen attack (Heidrich et al., 2011; Bai et al., 2012). In vitro and in vivo phosphorylation assays revealed that SGT1 is specifically phosphorylated by PIK1. In yeast, SGT1 phosphorylation was shown to negatively regulate SGT1 dimerization (Bansal et al., 2009). The BiFC data suggest that PIK1/SGT1, but not SGT1 dimers, is localized to the nuclei of plant cells. These findings support the hypothesis that SGT1 phosphorylation positively regulates the monomerization of SGT1 and its nuclear localization. However, SGT1 and PIK1 do not contain any identifiable NLS peptides (Azevedo et al., 2002; Peart et al., 2002; Kim and Hwang, 2011). Thus, the nuclear localization of SGT1/PIK1 may depend upon interaction with other proteins, such as HSPs. In support of this, HSP70 has been demonstrated to interact with SGT1 and mediate its nuclear localization (Noël et al., 2007).
Of the three domains present in SGT1 (CS, TPRs, and SGS), the CS domain resembles an α-crystallin domain of the cochaperone HSP20 (Dubacq et al., 2002; Garcia-Ranea et al., 2002). RAR1 and HSP90, both required for the plant immune system, have been shown to interact with the CS domain of SGT1 (Azevedo et al., 2002; Takahashi et al., 2003). Consistent with these findings, AvrBsT binds to SGT1 at the CS domain, suggesting that the CS domain is essential for the interaction of SGT1 with AvrBsT.
It is possible that the binding of AvrBsT to SGT1 may induce a conformational change that triggers an alteration in the specific association between SGT1 and PIK1 in the cytoplasm (Botër et al., 2007; Shirasu, 2009). AvrBsT seems to reduce PIK1 autophosphorylation as well as SGT1 phosphorylation. This results in the inhibition of the nuclear localization of the SGT1-PIK1 complex. Therefore, the recognition of AvrBsT by the SGT1-PIK1 complex seems to occur in the cytoplasm. The forced nuclear localization of the SGT1-PIK1-AvrBsT complex by the addition of the NLS significantly suppressed cell death (Heidrich et al., 2011; Bai et al., 2012), supporting the cytoplasmic localization and recognition of AvrBsT. AvrBsT did not exhibit any kinase activity in isolation. PIK1 did not physically interact with AvrBsT in yeast cells; however, PIK1 may phosphorylate AvrBsT in the presence of SGT1. Phosphorylation of AvrBsT by PIK1 greatly increased in the presence of SGT1. Thus, AvrBsT is hypothesized to be activated in the host by phosphorylation via PIK1 kinase activity. Collectively, these results suggest that SGT1 is not only a substrate of PIK1 but also an interactor of AvrBsT. This ultimately leads to the PIK1-mediated phosphorylation of AvrBsT.
The catalytic triad of AvrBsT has been shown to be essential for HR cell death (Orth et al., 2000). However, the AvrBsT C222A mutant, incapable of triggering HR in plants, was able to bind SGT1 and inhibit SGT1 phosphorylation. Thus, the enzymatic activity of AvrBsT may be significant in triggering HR but not responsible for the inhibition of SGT1 phosphorylation. SGT1 silencing nearly abolished HR, suggesting that the not yet identified R or NLR protein responsible for AvrBsT recognition requires SGT1 for HR activation (Azevedo et al., 2006). AvrBsT seems to be phosphorylated by PIK1 only when SGT1 is present. Therefore, it is hypothesized that AvrBsT enters host cells via a type III secretion system and binds to SGT1, which then recruits PIK1 and is phosphorylated by PIK1. Consequently, the HR is triggered upon the recognition of the SGT1-PIK1-AvrBsT complex by host factors in the cytoplasm, where the enzymatic activity of AvrBsT plays a critical role.
Combining the data presented here, we propose a working model for the AvrBsT recognition of SGT1 to promote hypersensitive cell death associated with PIK1-mediated phosphorylation (Supplemental Fig. S13). The virulent Xcv Ds1 infection causes a rapid increase in PIK1 expression (Supplemental Fig. S13A). This may occur through a general pattern-recognition receptor signaling pathway, recognizing Xcv PAMPs (Jones and Dangl, 2006; Kim and Hwang, 2011). In turn, PIK1 may bind to SGT1 homodimers and phosphorylate SGT1, resulting in the positive regulation of SGT1 monomerization. The PIK1-SGT1 complex is likely to recruit other proteins, which may lead to its translocation into the nucleus (Noël et al., 2007). Inside the nucleus, the PIK1-SGT1 complex is proposed to up-regulate some defense-response genes, which may contribute to host basal defense (Kim and Hwang, 2011). However, once Xcv Ds1 successfully colonizes host plants, it is presumed that unknown type III effector proteins of Xcv Ds1 effectively suppress the host basal defense. In contrast, avirulent Xcv Ds1 (avrBsT) secretes a type III effector protein, AvrBsT, into host plant cells to bind SGT1 (Supplemental Fig. S13B). PIK1 binds to SGT1 and forms a transient AvrBsT-SGT1-SGT1-PIK1 complex. The AvrBsT-SGT1-SGT1-PIK1 complex is confined to the cytoplasm, possibly due to the negative regulation of SGT1 monomerization. PIK1 phosphorylates AvrBsT rather than SGT1 and dissociates from the AvrBsT-SGT1-SGT1-PIK1 complex, which may be transiently generated in the cytoplasm. The abnormal state of the AvrBsT-SGT1-SGT1 complex may activate unknown host R proteins in the cytoplasm, ultimately leading to the enhanced HR cell death in host plants. On the other hand, SGT1 may act as a scaffold for PIK1 as well as putative R proteins. Together, our results suggest that AvrBsT promotes hypersensitive cell death associated with PIK1-mediated phosphorylation by specifically interacting with SGT1.
MATERIALS AND METHODS
Plant Materials and Bacterial Inoculation
Pepper (Capsicum annuum ‘Nockwang’) and Nicotiana benthamiana were grown as described by Choi et al. (2012). The Xanthomonas campestris pv vesicatoria virulent Ds1 (empty vector) and avirulent Ds1 (avrBsT) strains and Agrobacterium tumefaciens strain GV3101 were used in this study (Kim et al., 2010). The Xcv strains were cultured overnight, harvested, and resuspended in 10 mm MgCl2 solution, followed by infiltration into pepper leaves. For transient coexpression of SGT1, PIK1, and avrBsT, A. tumefaciens cultures were grown overnight, resuspended in medium (10 mm MES, pH 5.7, 10 mm MgCl2, and 0.2 mm acetosyringone), and infiltrated into N. benthamiana leaves.
Yeast Two-Hybrid Assays
The yeast two-hybrid transformation and screening as well as quantitative α-galactosidase activity assays were performed according to the Matchmaker GAL4 Two-Hybrid System 3 (Clontech) protocol. The PCR-amplified full-length coding regions of avrBsT and PIK1 were cloned into the bait vector pGBKT7 (Supplemental Table S1). The yeast-two hybrid cDNA library was constructed in the GAL4 activation domain vector (pGADT7) using cDNAs prepared from pepper leaves infected with the Xcv avirulent strain Bv5-4a. Truncated SGT1 and PIK1 were also created by PCR amplification (Supplemental Table S1). A lithium acetate-mediated transformation method was used to introduce the prey cDNA library and the bait construct into yeast strain AH109 (Ito et al., 1983). Transformants were selected on interaction selection medium (adenine-His-Leu-Trp) supplemented with 5-bromo-4-chloro-3-indoyl-α-d-galactoside.
Multicolor BiFC
Multicolor BiFC analyses were conducted as described by Waadt et al. (2008). To generate the BiFC constructs, cDNAs encoding AvrBsT, SGT1, and PIK1 without termination codons were PCR amplified and subcloned into the binary vectors pVYNE(R), pSCYCE, and pSCYNE(R) under the control of the cauliflower mosaic virus 35S promoter (Supplemental Table S1). Oligonucleotides containing NLS and nuclear export signal sequences (Slootweg et al., 2010; Choi and Hwang, 2011) were inserted into the XhoI/KpnI site to create the NLS and nuclear export signal fusion constructs. In addition, SGT1 was subcloned into the pVYNE and pSPYNE vectors. PIK1 also was introduced into the pSPYCE vector. Pairwise combinations of pSCYCE::SGT1 with either pVYNE(R)::AvrBsT or pSCYNE(R)::PIK1 and triple combinations of pSCYCE::SGT1 with pVYNE::SGT1 and pSCYNE(R)::PIK1 or pVYNE::AvrBsT and pSCYNE(R)::PIK1 were coexpressed in N. benthamiana leaves by A. tumefaciens-mediated transient transformation.
For transient expression, the A. tumefaciens strain GV3101 carrying each of the AvrBsT, SGT1, and PIK1 BiFC constructs (OD600 = 0.5 each) was combined with the A. tumefaciens strain p19 (OD600 = 0.3) and used to infiltrate N. benthamiana leaves. The nuclei of the cells were counterstained with DAPI.
Virus-Induced Gene Silencing
The tobacco rattle virus (TRV)-based virus-induced gene silencing system was used for gene silencing in pepper (Liu et al., 2002; Lee et al., 2008; Hwang et al., 2011). To achieve SGT1-specific silencing, a 240-bp fragment with the lowest homology (no longer than 15 consecutive identical base pairs) to SGT1b was PCR amplified and cloned into pTRV2 to generate pTRV2:CaSGT1-240 (Supplemental Table S1). PIK1-silenced pepper plants were also generated as described previously (Kim and Hwang, 2011). A. tumefaciens GV3101 carrying pTRV1 and pTRV2:SGT1-240, pTRV2:PIK1, or pTRV2:SGT1-240/pTRV2:PIK1 was coinfiltrated into the fully expanded cotyledons of pepper plants.
RNA Isolation and Quantitative Reverse Transcription-PCR
Total RNA was isolated from pepper leaves inoculated with Xcv strains using Trizol reagent (Invitrogen). Two micrograms of RNA was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Enzynomics) and oligo(dT) primers. The reaction and real-time PCR were performed using iQ SYBR Green Supermix and iCycler iQ (Bio-Rad) with pepper gene-specific primers for SGT1, SGT1b, PIK1, BPR1, SAR82A, DEF1, and 18S ribosomal RNA (for the oligonucleotide sequences, see Supplemental Table S1). Relative expression levels were determined as described previously (Hwang and Hwang, 2011).
Immunoblot Analysis
Total soluble proteins were extracted from N. benthamiana leaves with 1 mL of denaturing buffer (50 mm Tris-HCl [pH 8.8], 4 m urea, 10 mm sodium phosphate [pH 7.8], 250 mm NaCl, 0.1% [v/v] Nonidet P-40, 1 mm EDTA, and 0.5% [w/v] SDS). For coimmunoprecipitation analyses, proteins were extracted in soluble buffer (50 mm Tris-HCl [pH 8.8], 50 mm NaCl, 10 mm EDTA, 0.1% [v/v] Triton X-100, and 2× protease inhibitor cocktail [Roche]). Proteins in the supernatant were resolved on 10% (w/v) SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (GE Healthcare Biosciences). Proteins tagged with HA, cMyc, or FLAG epitopes were detected using anti-HA-peroxidase, anti-cMyc-peroxidase, or anti-FLAG-peroxidase antibodies (Sigma), respectively. Phosphorylated proteins were detected using anti-phosphothreonine antibodies (Sigma).
Expression and Purification of Recombinant Proteins
GST-fused PIK1 and the two inactive mutants (K130R and D228H) were generated as described by Kim and Hwang (2011). GST-PIK1 and mutant proteins were induced by adding 0.3 mm isopropyl β-d-thiogalactopyranoside. Proteins were purified using a Glutathione Sepharose 4B column (GE Healthcare). The coding regions of avrBsT and SGT1 were PCR amplified and cloned into pGEX-5x-1 and pET22b, respectively (Supplemental Table S1). Escherichia coli strain BL21 (DE3) overexpressing GST-AvrBsT and SGT1-6xHis proteins was induced with 0.6 mm isopropyl β-d-thiogalactopyranoside and purified using Glutathione Sepharose 4B and nickel-nitrilotriacetic acid agarose (Qiagen) resins, respectively, according to the manufacturer’s instructions.
In Vitro Kinase Assays
Phosphorylation of SGT1 was verified by in vitro kinase assays performed as described by Kim and Hwang (2011). Purified GST-fused PIK1 was incubated in buffer (50 mm Tris-HCl [pH 7.0], 20 mm MnCl2, 40 μm ATP, and 1 mm 1,4-dithiothreitol) supplemented with 10 μCi of [γ-32P]ATP (3,000 Ci mmol−1) for 20 min at room temperature. The reactions were separated on a 12% (w/v) SDS-PAGE gel. Gels were dried and analyzed using an image plate reader (FLA-7000; Fujifilm).
To determine the effect of AvrBsT on SGT1 or PIK1 phosphorylation, purified AvrBsT was added to a PIK1/SGT1 reaction. To characterize the mechanism of phosphorylation inhibition by AvrBsT, SGT1 was preincubated with either PIK1 or AvrBsT in the reaction buffer for 30 min. AvrBsT or PIK1 was then added to the reaction, and incubation was continued for 1 h. The AvrBsT C222A mutant, incapable of inducing cell death (Orth et al., 2000), was also generated using the QuickChange site-directed mutagenesis kit (Stratagene) and pGEX-5x-1:AvrBsT as a template (Supplemental Table S1). AvrBsT wild-type and C222A proteins were purified and added to the PIK1/SGT1 reaction. Samples were separated by SDS-PAGE, followed by image plate analysis.
Measurement of SA and JA
SA and SA glycoside were extracted and quantified as described previously (Lee et al., 2011). JA was extracted and quantified as described previously (Hwang and Hwang, 2010).
Enzymatic In-Gel Digestion
In vitro-phosphorylated SGT1 protein was separated by SDS-PAGE. The excised gel pieces containing proteins were destained with 50% (v/v) acetonitrile containing 50 mm NH4HCO3 and vortexed until Coomassie Brilliant Blue was completely removed. These gel pieces were then dehydrated in 100% acetonitrile and vacuum dried for 20 min. For the digestion, gel pieces were reduced using 10 mm dithiothreitol in 50 mm NH4HCO3 for 45 min at 56°C, followed by alkylation by 55 mm iodoacetamide in 50 mm NH4HCO3 for 30 min in the dark. Finally, each of gel pieces was treated with 12.5 ng µL−1 sequencing-grade modified trypsin (Promega) in 50 mm NH4HCO3 buffer (pH 7.8) at 37°C overnight. Following digestion, tryptic peptides were extracted with 5% (v/v) formic acid in 50% (v/v) acetonitrile solution at room temperature for 20 min. The supernatants were collected and dried by SpeedVac. The samples were purified and concentrated in 0.1% (v/v) formic acid using C18 ZipTips (Millipore) before MS/MS analysis.
Nano-LC-Electrospray Ionization-MS/MS Analyses
The proteolytic peptides of SGT1 were loaded onto a fused silica microcapillary column (12 cm × 75 µm) packed with C18 reverse-phase resin (5 µm, 200 Å). LC separation was conducted under a linear gradient as follows: a 3% to 40% solvent B (0.1% [v/v] formic acid in acetonitrile) gradient (where solvent A is 0.1% [v/v] formic acid in water), with a flow rate of 250 nL min−1 for 60 min. The column was directly connected to the LTQ linear ion-trap mass spectrometer (Finnigan) equipped with a nano-electrospray ion source. The electrospray voltage was set at 1.95 kV, and the threshold for switching from mass spectrometry (MS) to MS/MS was 500. The normalized collision energy for MS/MS was 35% of the main radio frequency amplitude, and the duration of activation was 30 ms. All spectra were acquired in a data-dependent scan mode. Each full MS scan was followed by five MS/MS scans from the most intense to the fifth most intense peaks of the full MS scan. The repeat count of the peak for dynamic exclusion was one, and its repeat duration was 30 s.
The acquired LC-electrospray ionization-MS/MS fragment spectra were searched in the BioWorksBrowser (version Rev. 3.3.1 SP1; Thermo Fisher Scientific) with the SEQUEST search engines against the data in FASTA format generated from SGT1 in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The conditions for the search were trypsin as enzyme specificity, a permissible level for two missed cleavages, peptide tolerance of ±3 atomic mass units, a mass error of ±1 atomic mass units on fragment ions, and variable modifications of carbamidomethylation of Cys (+57 D), oxidation of Met (+16 D), and phosphorylation of Ser (+80 D) residues.
A. tumefaciens-Mediated Transient Expression
For A. tumefaciens-mediated transient expression of SGT1, PIK1, and avrBsT, these genes without the termination codons were PCR amplified and subcloned into the binary vectors pVYNE(R) (cMyc tagged), pSCYCE (HA tagged), and pSCYNE(R) (FLAG tagged) under the control of the cauliflower mosaic virus 35S promoter (Supplemental Table S1). Constructs were sequenced and transferred to A. tumefaciens strain GV3101 through electroporation. A. tumefaciens strain GV3101 harboring the avrBsT:cMyc, SGT1:HA, or PIK1:FLAG constructs were grown overnight in yeast extract peptone medium containing appropriate antibiotics. Cells were suspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone, pH 5.7). N. benthamiana leaves were infiltrated with A. tumefaciens cells (OD600 = 0.05).
Ion Leakage Assay
Ion leakage was measured as described previously (Hwang and Hwang, 2011; Lee et al., 2011). Leaf discs (0.5 cm in diameter) were washed and incubated in 10 mL of sterile double-distilled water for 3 h at room temperature. Ion conductivity of the leaf sample solutions was measured using a SensION 7 conductivity meter (Hach).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: pepper SGT1 (JN252483), SGT1b (AY899280), AvrBsT (GQ266402), PIK1 (GU295436), PR1 (AF053343), SAR82A (AF313766), DEF1 (AF442388), and 18S ribosomal RNA (EF564281); tobacco (Nicotiana tabacum) SGT1 (AAO85509); rice (Oryza sativa) Sgt1 (AAF18438); and Arabidopsis SGT1a (AAL33611) and SGT1b (AAL33612).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of SGT1 cDNA of pepper.
Supplemental Figure S2. Alignment of pepper SGT1 with SGT1 proteins of other plants.
Supplemental Figure S3. Yeast two-hybrid and BiFC assays of interactions between AvrBsT and SGT1 or SGT1 and PIK1.
Supplemental Figure S4. Subcellular localization of SGT1, PIK1, and PIK1 D228H.
Supplemental Figure S5. Immunoblot analyses of the expression of BiFC fusion proteins in Figure 2A.
Supplemental Figure S6. Transient expression of the avrBsT C222A mutant does not trigger hypersensitive cell death.
Supplemental Figure S7. Time-course analyses of the expression of 35S:SGT1:HA, 35S:PIK1:FLAG1, 35S:PIK1:FLAG1(D228H), and 35S:avrBsT:cMyc in N. benthamiana leaves by immunoblotting.
Supplemental Figure S8. Immunoblot analyses of the expression of AvrBsT and SGT1 mutant proteins in Figure 7.
Supplemental Figure S9. Neither nuclear nor cytoplasmic SGT1/PIK1 localization triggers HR without AvrBsT.
Supplemental Figure S10. Immunoblot analyses of the expression of BiFC fusion proteins in Figure 8, A and B.
Supplemental Figure S11. Visualization of H2O2 accumulation in empty vector control and SGT1-, PIK1-, and SGT1/PIK1-silenced pepper leaves by 3,3’-diaminobenzidine staining.
Supplemental Figure S12. Silencing of SGT1, PIK1, and SGT1/PIK1 compromises defense-related gene expression and JA accumulation.
Supplemental Figure S13. Proposed model of AvrBsT recognition of SGT1 to promote hypersensitive cell death associated with PIK1-mediated phosphorylation.
Supplemental Table S1. Oligonucleotides for plasmid constructs used in this study.
Supplementary Material
Glossary
- PTI
PAMP-triggered immunity
- ETI
effector-triggered immunity
- HR
hypersensitive response
- SA
salicylic acid
- ROS
reactive oxygen species
- PAMP
pathogen-associated molecular pattern
- Xcv
Xanthomonas campestris pv vesicatoria
- cDNA
complementary DNA
- BiFC
bimolecular fluorescence complementation
- HA
hemagglutinin
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- OD600
optical density at 600 nm
- DAPI
4′,6-diamidino-2-phenylindole
- NLS
nuclear localization signal
- H2O2
hydrogen peroxide
- JA
jasmonic acid
- MS
mass spectrometry
Footnotes
This work was supported by the Next Generation BioGreen21 Program (Plant Molecular Breeding Center, grant no. PJ008027), Rural Development Administration, Republic of Korea.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K. (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FLW, et al. (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal PK, Mishra A, High AA, Abdulle R, Kitagawa K. (2009) Sgt1 dimerization is negatively regulated by protein kinase CK2-mediated phosphorylation at Ser361. J Biol Chem 284: 18692–18698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U, Schulte R, Fenselau S, Minsavage GV, Staskawicz BJ, Stall RE. (1991) Isolation of a gene-cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol Plant Microbe Interact 4: 81–88 [Google Scholar]
- Botër M, Amigues B, Peart J, Breuer C, Kadota Y, Casais C, Moore G, Kleanthous C, Ochsenbein F, Shirasu K, et al. (2007) Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 19: 3791–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Kirik A, Kim JG, Frame K, Kirik V, Mudgett MB. (2014) AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog 10: e1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Hwang BK. (2011) Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23: 823–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Hwang IS, Hwang BK. (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Ryu CM, Oh SK, Kim RN, Park JM, Cho HS, Lee S, Moon JS, Park SH, Choi D. (2006) Suppression of pepper SGT1 and SKP1 causes severe retardation of plant growth and compromises basal resistance. Physiol Plant 126: 605–617 [Google Scholar]
- Coll NS, Epple P, Dangl JL. (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Wilson A, Nuwer J, Kirik A, Baranage G, Mudgett MB. (2007) A conserved carboxylesterase is a SUPPRESSOR OF AVRBST-ELICITED RESISTANCE in Arabidopsis. Plant Cell 19: 688–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do HM, Lee SC, Jung HW, Sohn KH, Hwang BK. (2004) Differential expression and in situ localization of a pepper defensin (CaDEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci 166: 1297–1305 [Google Scholar]
- Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C. (2002) Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell 1: 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Nimchuk ZL, Dangl JL. (2008) Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA 105: 6475–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar L, Van Den Ackerveken G, Pieplow S, Rossier O, Bonas U. (2001) Type III secretion and in planta recognition of the Xanthomonas avirulence proteins AvrBs1 and AvrBsT. Mol Plant Pathol 2: 287–296 [DOI] [PubMed] [Google Scholar]
- Garcia-Ranea JA, Mirey G, Camonis J, Valencia A. (2002) p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett 529: 162–167 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Yao N. (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6: 201–211 [DOI] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Hwang IS, An SH, Hwang BK. (2011) Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J 67: 749–762 [DOI] [PubMed] [Google Scholar]
- Hwang IS, Choi S, Kim NH, Kim DS, Hwang BK. (2014) Pathogenesis-related protein 4b interacts with leucine-rich repeat protein 1 to suppress PR4b-triggered cell death and defense response in pepper. Plant J 77: 521–533 [DOI] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK. (2010) The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol 152: 948–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK. (2011) The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol 155: 447–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim DS, Hwang BK. (2011) The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J 66: 642–655 [DOI] [PubMed] [Google Scholar]
- Kim NH, Choi HW, Hwang BK. (2010) Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol Plant Microbe Interact 23: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Hwang BK. (2000) Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol Plant 108: 51–60 [Google Scholar]
- Kirik A, Mudgett MB. (2009) SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci USA 106: 20532–20537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Kremers GJ, Goedhart J, van Munster EB, Gadella TW., Jr (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Lam E. (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Lee DH, Choi HW, Hwang BK. (2011) The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol 156: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Hwang BK. (2003) Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 216: 387–396 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang IS, Choi HW, Hwang BK. (2008) Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol 148: 1004–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Mittal R, Peak-Chew SY, McMahon HT. (2006) Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA 103: 18574–18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312: 1211–1214 [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE. (2007) Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Melotto M, He SY. (2005) Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol 8: 361–368 [DOI] [PubMed] [Google Scholar]
- Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. (2000) Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DAW, Xiao SY, Coleman MJ, et al. (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Romeis T. (2001) Protein kinases in the plant defence response. Curr Opin Plant Biol 4: 407–414 [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Schulze-Lefert P. (2003) Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci 8: 252–258 [DOI] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensgaard P, Garrè M, Muradore I, Transidico P, Nigg EA, Kitagawa K, Earnshaw WC, Faretta M, Musacchio A. (2004) Sgt1 is required for human kinetochore assembly. EMBO Rep 5: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, Bonas U. (2010) Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol 187: 1058–1074 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- White FF, Potnis N, Jones JB, Koebnik R. (2009) The type III effectors of Xanthomonas. Mol Plant Pathol 10: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. (2010) The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci USA 107: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH. (2010) Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol Cell 39: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20: 10–16 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Rathjen JP. (2008) Plant immunity: AvrPto targets the frontline. Curr Biol 18: R218–R220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.