Summary
How epigenetic information is transmitted from generation to generation remains largely unknown. Deletion of the C. elegans Histone H3 lysine 4 dimethyl (H3K4me2) demethylase spr-5 leads to inherited accumulation of the euchromatic H3K4me2 mark and progressive decline in fertility. Here we identified multiple chromatin-modifying factors, including novel H3K4me1/me2 and H3K9me3 methyltransferases, an H3K9me3 demethylase and an H3K9me reader, which either suppress or accelerate the progressive transgenerational phenotypes of spr-5 mutant worms. Our findings uncover a network of chromatin regulators that control the trans-generational flow of epigenetic information, and suggest that the balance between euchromatic H3K4 and heterochromatic H3K9 methylation regulates trans-generational effects on fertility.
Introduction
Most heritable information is transmitted by DNA, following Mendelian inheritance (Avery et al., 1944), but some traits such as longevity, fertility, disease susceptibility and obesity, can be inherited non-genetically in several model organisms (Daxinger and Whitelaw, 2012; Greer and Shi, 2012; Youngson and Whitelaw, 2008). The underlying molecular mechanisms of transgenerational epigenetic transmission remain unclear but chromatin changes may play a role.
Chromatin is composed of 146 base pairs of DNA wrapped around a histone octamer (2 copies each of histone H2A, H2B, H3 and H4). Both DNA and histones are modified, which impacts chromatin-templated processes. Among many histone modifications, lysine (K) methylation is of particular interest in the context of epigenetic inheritance as this modification is more stable, but can also be dynamically regulated. Histone methylation can be associated with either transcriptional activation or repression. For instance, histone H3K4 di- and trimethylation (H3K4me2/3) are associated with active or poised gene transcription (Bernstein et al., 2002; Pokholok et al., 2005; Santos-Rosa et al., 2002), while H3K9 di- and trimethylation (H3K9me2/3) are associated with transcriptional repression, gene silencing and heterochromatin (Bannister et al., 2001; Ebert et al., 2006; Li et al., 2007). Both H3K4 and H3K9 methylation events are regulated by multiple, site-specific methyltransferases and demethylases (Mosammaparast and Shi, 2010; Ruthenburg et al., 2007). When H3K4 is methylated, H3K9 is often demethylated and sometimes acetylated; likewise, when H3K9 is methylated, H3K4 is often unmethylated (Barski et al., 2007; Guenther et al., 2007; Heintzman et al., 2007; Mikkelsen et al., 2007; Wang et al., 2008). Antagonism between H3K4 and H3K9 methylation plays a critical role in dictating the boundaries between euchromatin and heterochromatin (Lan et al., 2007; Rudolph et al., 2007). However, the functional consequences of the crosstalk between methylation at H3K4 and H3K9 remain incompletely understood.
SPR-5, the C. elegans ortholog of the human H3K4me1/me2-specific demethylase LSD1, regulates transgenerational inheritance. C. elegans without spr-5 do not exhibit sterility initially, but successive generations lacking spr-5 display increasing infertility concomitant with global accumulation of H3K4me2 (Katz et al., 2009; Nottke et al., 2011). This progressive phenotype can be reversed by the addition of a single copy of spr-5. However, how this epigenetic memory is transmitted across generations is still unknown. To investigate the underlying molecular mechanism of these inherited epigenetic changes, we carried out targeted RNA interference (RNAi) screens to identify suppressors and enhancers of the progressive fertility phenotypes associated with loss of spr-5. Our findings not only uncovered a network of enzymes and reader proteins involved in regulating H3K4 and H3K9 methylation but also demonstrated that a functional interplay between H3K4 and H3K9 methylation plays a key role in regulating epigenetic inheritance in C. elegans.
Results
The Main RNAi Pathways Mediated by rde-1 and ergo-1 Are Not Involved in the Progressive Sterility of spr-5(by101) Mutant Worms
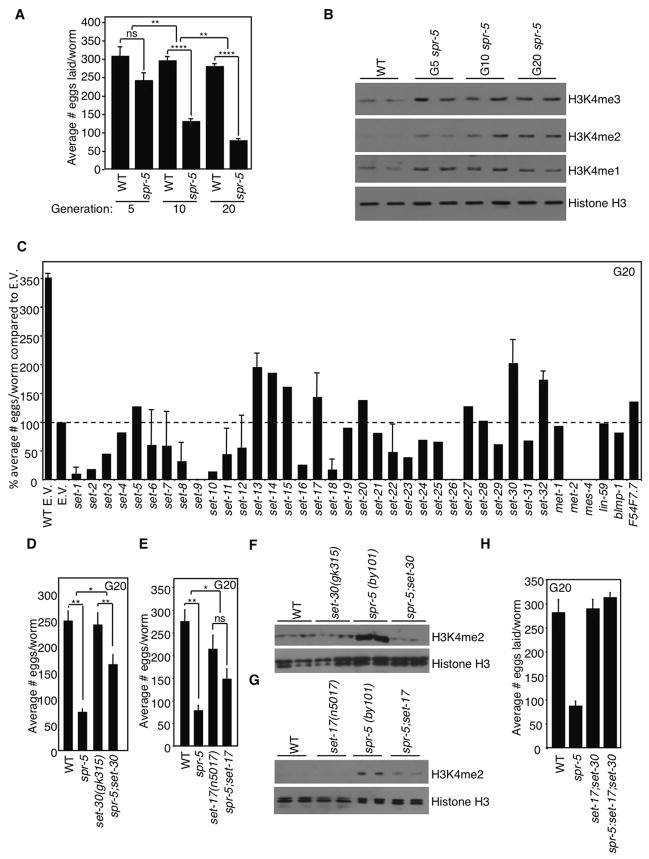
Genetic ablation of the H3K4me2 demethylase spr-5 in C. elegans leads to a progressive decrease in fertility and increase in H3K4me2 over generations (Katz et al., 2009; Nottke et al., 2011). We confirmed the progressive loss of fertility, assessed by counting laid eggs, in successive generations of worms using two genetically null, deletion strains of spr-5 (spr-5(by101) and spr-5(by134)) (Figures 1A and S1A), but for the remainder of the studies, we focused on the spr-5(by101) allele. We observed a generational accumulation of H3K4me2 in spr-5(by101) mutant worms (Figure 1B). In contrast, H3K4me1 and H3K4me3 levels, though elevated in spr-5(by101) mutant worms, did not change across generations.
Figure 1. set-17 and set-30 deletions suppress the progressive sterility of spr-5 mutant worms.
A) spr-5(by101) mutant worms display progressive fertility defects (bars represent mean +/− SEM for 4 experiments for generation 5, 15 experiments for generation 10, and 34 experiments for generation 20: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate) B) H3K4me2 increases across generations of spr-5(by101) mutant worms as assessed by whole worm western blots of L4 stage worms. H3K4me1 and H3K4me3 are higher in spr-5(by101) mutant worms but do not change across generations. Blots are representative of 4 independent experiments performed in duplicate. C) Number of eggs laid by spr-5(by101) mutant worms fed dsRNA of C. elegans potential methyltransferases or empty vector (E.V.) for 20 generations. D) spr-5;set-30 double mutants for 20 generations causes a partial suppression of decreased fertility capacity of spr-5(by101) mutant worms. This graph displays the mean +/− SEM of 4 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate. E) spr-5;set-17 double mutant worms have a partial suppression of the fertility defect of spr-5(by101) mutant worms at generation 20. This graph displays the mean +/− SEM of 4 independent experiments. F) spr-5(by101) mutant worms have increased H3K4me2 at generation 20 but spr-5;set-30 double mutants have normal H3K4me2 levels as assessed by whole worm western blots of L3 worms. G) spr-5;set-17 double mutants have lower H3K4me2 at generation 20 than spr-5(by101) mutants as assessed by whole worm western blots of L4 worms. h) spr-5;set-17;set-30 triple mutant worms have a complete suppression of the fertility defect of spr-5(by101) mutant worms at generation 20. This graph displays the mean +/− SEM of 3 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate. *: p<0.05, **: p<0.01, ****: p<0.0001
As RNAi inheritance has been implicated in transgenerational epigenetic inheritance in several species (Moazed, 2011), we first investigated whether RNAi pathways played a role in spr-5 induced epigenetic inheritance. The argonaute genes rde-1 and ergo-1 are largely required for exogenous and endogenous RNAi in C. elegans, respectively (Grishok et al., 2000; Yigit et al., 2006) although other argonautes in C. elegans could be required for specific RNA inheritance events (Conine et al., 2013). Worms carrying double mutations of spr-5(by101) with either rde-1 or ergo-1 laid the same number of eggs as spr-5(by101) at generation 10 (Figures S1B and S1C), suggesting that RNAi inheritance mediated by these argonautes does not play a role in the generational sterility inheritance of spr-5 mutants.
SET-17 and SET-30 suppress transgenerational phenotypes of the spr-5(by101) mutant worms
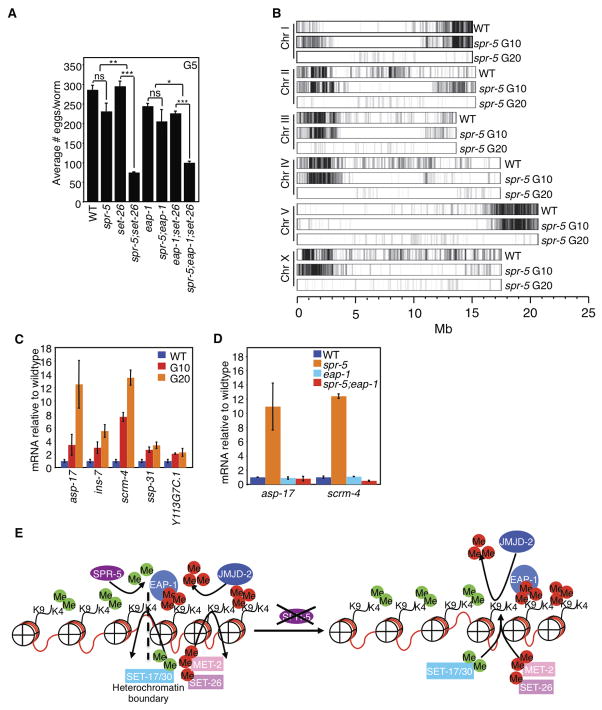
Because SPR-5 is an H3K4me1/me2 demethylase, we hypothesized that H3K4me2-specific methylases would act as suppressors. These enzymes are unknown in C. elegans, so we knocked down all genes containing predicted methyltransferase domains (Andersen and Horvitz, 2007; Herz et al., 2013) (Figure 1C). spr-5(by101) mutant worms fertility was assessed after being fed bacteria expressing dsRNA against 39 methyltransferase domain-containing genes for 20 generations. Knockdown of set-13, set-14, set-15, set-17, set-20, set-30, and set-32 all partially suppressed the progressive sterility of spr-5(by101) mutant worms. To rule out off-target effects, we crossed predicted null mutants of each of these genes with spr-5 mutants and examined the effects. We found that mutations of set-20 and set-32 had no effect on the fertility of spr-5(by101) mutant worms (Figures S2A and S2B), suggesting the RNAi suppression was due to off-target effects. A predicted genetic null mutation of set-25, which did not suppress the phenotype in our RNAi screen but is required for the maintenance of silencing triggered by piRNA in some instances (Ashe et al., 2012), also had no effect on spr-5(by101) fertility (Figure S2C). We failed to obtain progeny from spr-5;set-13 double mutants for reasons that are unclear (Figure S2D).
Importantly, maintaining either set-17 or set-30 as homozygous mutants for 20 generations significantly, albeit partially, suppressed spr-5(by101) transgenerational sterility (Figures 1D and 1E), confirming the initial RNAi screen results. Genetic ablation of either set-17 or set-30 also suppressed spr-5(by101) elevated H3K4me2 levels (Figures 1F and 1G). Furthermore, deletion of both set-17 and set-30 in spr-5(by101) mutants completely suppressed the transgenerational sterility (Figure 1H). The closest mammalian homologue of SET-17 is PRDM9, which has been suggested to mediate H3K4me2/me3 (Hayashi et al., 2005) (Full protein: 33.2% identity, SET domain: 46.67% identity). Collectively, these findings suggest that SET-17 and SET-30 are potential H3K4 methyltransferases.
SET-17 and SET-30 are H3K4me1/me2 methyltransferases
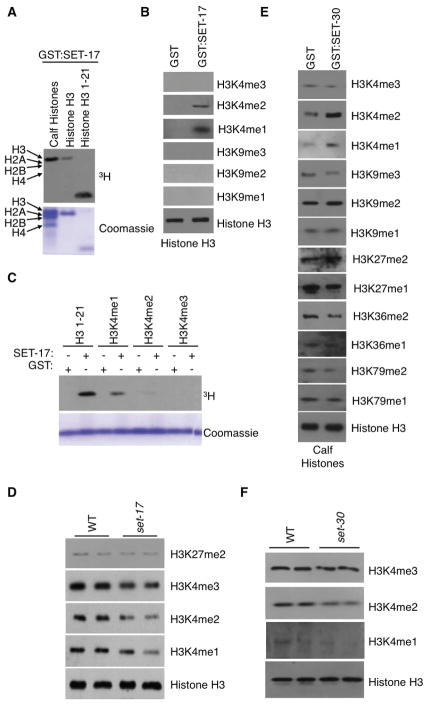
To determine whether SET-17 and SET-30 mediate H3K4 methylation, we performed in vitro radioactive methyltransferase assays using GST-tagged SET-17 and SET-30, expressed and purified from bacteria. SET-17 specifically methylated histone H3 of calf thymus histones, as well as unmodified recombinant histone H3 and an H3 peptide containing the first 21 amino acids (Figure 2A). Using histone methyl-specific antibodies, we found SET-17 mediated mono- and dimethylation of H3K4 in calf thymus histone or recombinant H3 (Figures 2B and S3A), while displaying no activities towards other lysine residues (Figures 2B, S3A, and data not shown). Furthermore we found that SET-17 methylated unmodified H3 peptide and to a lesser extent the H3K4me1 pre-methylated peptide but did not methylate the H3K4me2/me3 pre-methylated peptide (Figure 2C). In vivo, set-17 mutant worms displayed lower global levels of H3K4me but wildtype levels of H3K27me2 (Figure 2D). Together, these results suggest that SET-17 is an H3K4me1/me2 methyltransferase.
Figure 2. SET-17 and SET-30 are H3K4me1/me2 methyltransferases.
A) GST:SET-17 full length protein methylates histone H3 amino acids 1–21, histone H3, and only histone H3 of calf histones in vitro. B) GST:SET-17 full length protein methylates H3K4me1/me2 of Histone H3 as assessed by western blots of in vitro methylation assays performed on recombinant Histone H3. C) GST:SET:17 methylates H3K4me1 and H3K4me2 as assessed by radioactive methyltransferase assays of histone H3 amino acids 1–21 which are unmodified or premethylated on H3K4. D) set-17(n5017) mutant worms have lower H3K4 methylation as assessed by whole worm western blots of L4 worms. E) GST:SET-30 full length protein methylates H3K4me1/me2 as assessed by western blots of in vitro methylation assay performed on histones. F) set-30(gk315) mutant worms have lower H3K4 methylation as assessed by whole worm western blots of L1 worms.
Similar to SET-17, SET-30 preferentially mediates H3K4me1/me2 on calf thymus histones and 293T cell nucleosomes (Figures 2E and S3B). Unlike SET-17, SET-30 was unable to methylate recombinant histone substrates (data not shown). In vivo, early larval stage L1 and L2 (but not L3 and L4) set-30 mutant worms displayed lower H3K4me levels (Figures 1F, 2F, and data not shown), consistent with SET-30 being an H3K4 methyltransferase. Taken together, our results demonstrate that SET-17 and SET-30 mediate H3K4me1/me2 in vitro and in vivo and suggest that they may oppose the activity of the demethylase SPR-5. Combined deletion of set-17 and set-30 did not completely eliminate global H3K4 mono- and di-methylation (data not shown) suggesting the existence of additional H3K4 mono and di-methyltransferases.
Loss of SET-30, but not SET-17, reverts the progressive sterility of spr-5 mutant worms
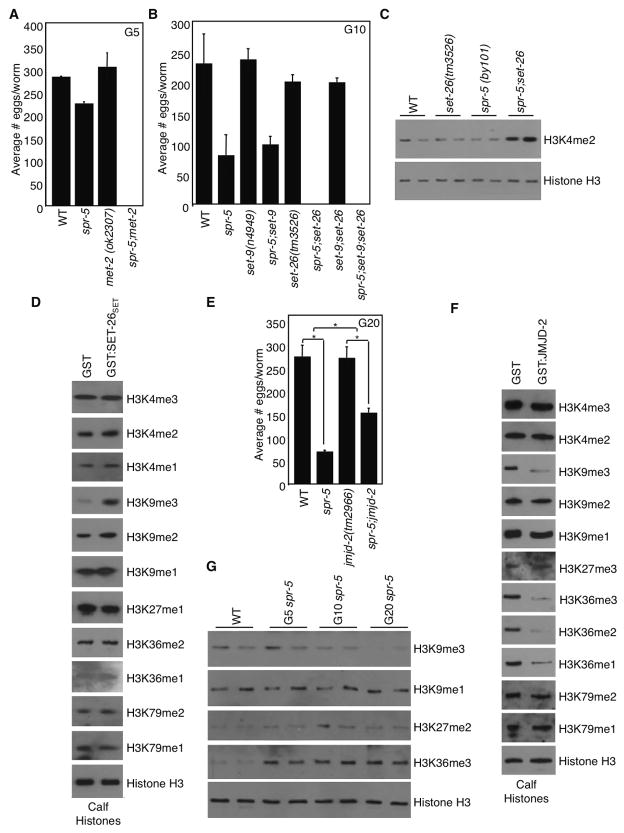
The above genetic suppression experiment involved simultaneous and persistent inhibition of SET-17 or SET-30 in spr-5 worms from generation zero. We wished to determine whether removal of set-17 or set-30 in later generation spr-5 mutants, which are already less fertile, is sufficient to revert the reproductive capacity. We therefore assessed spr-5(by101) mutants fertility after being maintained for 20 generations on empty vector control RNAi (E.V.) bacteria, then switched to set-17 or set-30 RNAi for an additional 5 generations. set-30, but not set-17, RNAi partially reverted the spr-5(by101) progressive sterility and increased H3K4me2 levels (Figures 3A and 3B). The reversion became evident after 2–3 generations on set-30 RNAi as spr-5(by101) mutant worms began to lay as many eggs as spr-5(by101) mutant worms fed set-30 RNAi for all generations (Figure 3C). These results suggest that while SET-17 may be required for initiating the transgenerational phenotypes, SET-30 might be important in both initiating and maintaining progressive sterility associated with the loss of SPR-5.
Figure 3. set-30 knockdown reverts the progressive phenotypes of spr-5 mutant worms.
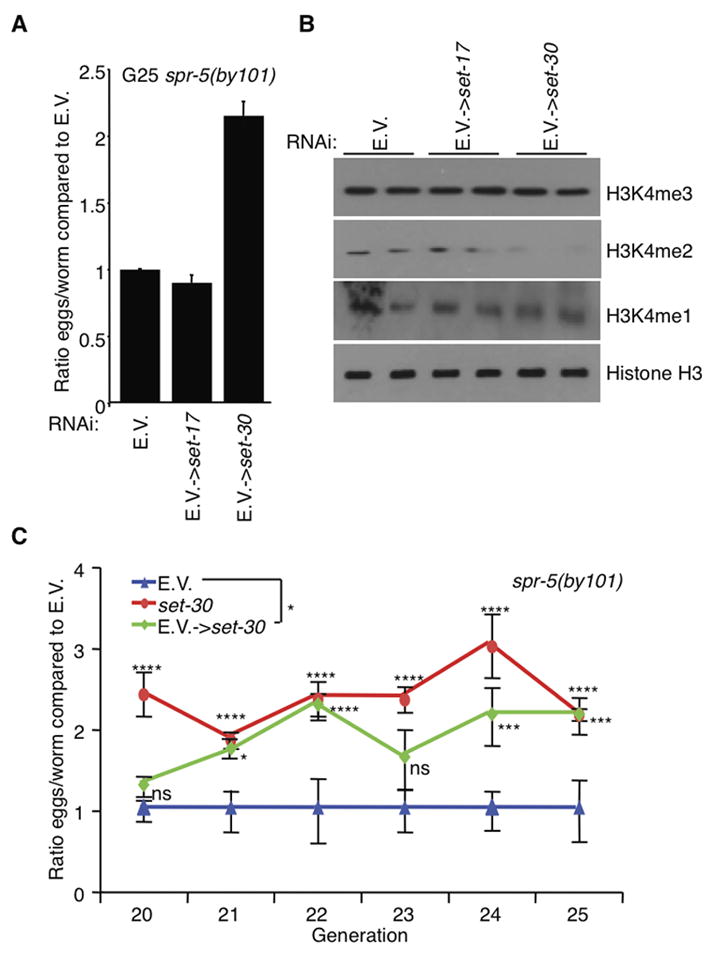
A) RNAi against set-30 but not set-17 for 5 generations partially reverted the fertility defect of spr-5(by101) mutant worms fed empty vector control RNAi (E.V.) for 20 generations prior. This graph represents the mean +/− SEM of 3 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate. B) spr-5(by101) mutant worms increased H3K4me2 at generation 25 is reverted by 5 generations of treatment with set-30 RNAi as assessed by whole worm western blots of L3 worms. C) The fertility defect of spr-5(by101) mutant worms fed empty vector RNAi bacteria (E.V.) for 20 generations and switched to set-30 RNAi suggests that set-30 knockdown for 2–3 generations causes the same degree of partial reversion of the fertility defect as spr-5(by101) mutant worms which had been fed dsRNA against set-30 for 22–23 generations. *: p<0.05, ***: p<0.001, ****: p<0.0001
Loss of the predicted H3K9 mono/dimethyltransferase MET-2 and the H3K9 trimethyltransferase SET-26 accelerate the progressive sterility and accumulation of H3K4me2 in spr-5 mutant worms
Our RNAi screen also identified genes whose knockdown accelerated the progressive sterility of spr-5 mutants (Figure 1C). Knockdown of set-9, set-26, met-2, and mes-4 had the strongest effect, rendering spr-5 mutants completely sterile by generation 2–13. mes-4 was previously identified as a sterility inducer after one generation in wild type worms (Capowski et al., 1991). met-2 mutants were previously reported to display a mortal germline phenotype after 18–28 generations (Andersen and Horvitz, 2007) while set-9 and set-26 have no reported fertility effects.
To confirm the RNAi result, we crossed the predicted null mutants, met-2(ok2307), met-2(n4256), set-9(n4949) and set-26(tm3526), with spr-5(by101) mutants. Crossing either met-2 mutant with spr-5 accelerated the progressive sterility such that spr-5;met-2 double mutants were completely sterile by generation 2 (Figure 4A and data not shown). Interestingly, mutation of set-26, but not set-9, accelerated the progressive sterility of spr-5 such that the spr-5;set-26 double mutants were completely sterile by generations 5 to 8 (Figure 4B). The reason that we identified set-9 as an enhancer in the RNAi screen was likely due to set-9 siRNA cross-inhibiting SET-26 expression, due to the high degree of sequence similarity between these two genes (97% sequence identity). Importantly, set-26(tm3526) mutation on its own did not cause a progressive decline in fertility (Figure 4B), suggesting that set-26 participates in fertility regulation specifically through genetic interactions with spr-5. Although set-26 worms did not show elevated levels of H3K4me2, spr-5;set-26 double mutants displayed significantly higher levels of H3K4me2 at generation 4 than spr-5 mutants (Figure 4C).
Figure 4. H3K9me regulation controls the spr-5(by101) progressive sterility.
A) spr-5;met-2 double mutants accelerates the progressive sterility of spr-5(by101) mutant worms after 5 generations. This graph represents the mean +/− SEM of 2 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate. B) spr-5;set-26 double mutants accelerates the progressive sterility of spr-5(by101) mutant worms after 10 generations. Graph is a representative experiment where each bar represents the mean +/− SEM for 3 replicates of 10 worms each. set-9(n4949) deletions’ effect on fertility has been tested 1 additional time. set-26(tm3526) deletions’ effect on fertility has been tested 5 additional times. C) spr-5;set-26 double mutants have significantly higher H3K4me2 at generation 4 as assessed by whole worm western blots of L3 worms. Representative blot of 4 independent experiments. D) GST:SET-26SET causes an increase in H3K9me2/me3 as assessed by western blots of in vitro methyltransferase assays of histones. E) spr-5;jmjd-2 double mutant worms have a suppression of the fertility defect of spr-5(by101) mutant worms at generation 20 (graph is the mean +/− SEM of 3 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate). *: p<0.05. F) GST:JMJD-2 causes a decrease in H3K9me3 and H3K36me as assessed by western blots of in vitro demethylase assays of histones. G) H3K9me3 decreases across generations of spr-5(by101) mutant worms as assessed by whole worm western blots of L4 stage worms. These blots are representative of 3 independent experiments performed in duplicate.
Interestingly, met-2, set-9 and set-26 are all predicted H3K9 methylases (Andersen and Horvitz, 2007; Bessler et al., 2010; Ni et al., 2011; Towbin et al., 2012). We performed in vitro radioactive methyltransferase assays using the catalytic SET-domain of SET-26 (SET-26SET) to identify it’s histone substrates. SET-26SET selectively methylated H3, but not H2A, H2B or H4 of 293T cell nucleosomes, but failed to methylate recombinant substrates (Figure S3C and data not shown). SET-26SET mediated H3K9me3 but not methylation of other H3 lysine residues, suggesting that SET-26 is an H3K9 trimethyltransferase (Figures 4D and S3C). met-2 mutants have lower H3K9me in embryos when assessed by mass spectrometry (Towbin et al., 2012) but have undetectable H3K9me2 and high levels of H3K9me3 in the adult germline as assessed by immunofluorescence (Bessler et al., 2010). Therefore MET-2 has been proposed to be an H3K9 mono and di-methyltransferase (Andersen and Horvitz, 2007; Bessler et al., 2010; Towbin et al., 2012) although its direct methyltransferase activity has not been biochemically demonstrated.
Loss of the H3K9me3 demethylase JMJD-2 suppresses the transgenerational fertility defects of spr-5(by101) mutant worms
If acceleration of the infertility of spr-5 mutants upon loss of MET-2 and SET-26 depends on their function as H3K9 methylases, the absence of an H3K9 demethylase should suppress this defect. Amongst the 11 demethylase candidates (Klose et al., 2006), we found that only mutation of jmjd-2(tm2966), which deletes the catalytic Jumonji C domain and should produce an enzymatically null protein, suppressed the spr-5 (by101) progressive fertility defect (Figures 4E and S2E–I). JMJD-2 is a putative H3K9me3/H3K36me3 demethylase based on its sequence homology with the mammalian JMJD2 family of demethylases (Black et al., 2010; Whetstine et al., 2006). Consistently, we found that JMJD-2 demethylated H3K9me3 and H3K36 methylation on calf histones, but not other H3 lysine residues (Figure 4F). Together, these results suggest that H3K9me3 regulates the transgenerational progressive sterility of spr-5(by101) mutant worms.
Indeed, we found that H3K9me3 levels in L4 spr-5(by101) mutants declined across generations (Figure 4G) while global H3K9me1 and H3K27me2 levels remained unchanged. Although global H3K36me3 was elevated in spr-5(by101) mutant worms, it did not change across generations (Figure 4G). Collectively, these findings suggest that the ability of JMJD-2 to regulate H3K9me3 is important and relevant for its effects on the spr-5 progressive sterility.
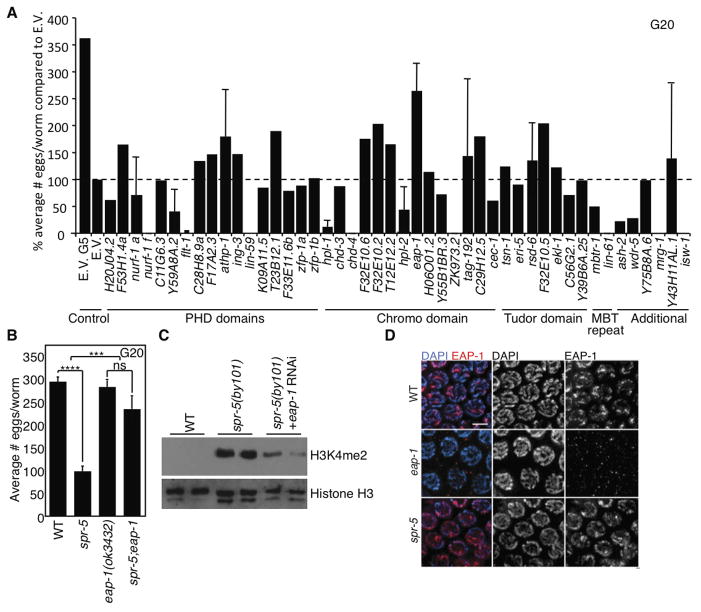
A chromodomain-containing gene, eap-1, suppresses transgenerational spr-5 phenotypes
To better understand how H3K9 methylation affects the transgenerational phenotypes of an H3K4me1/me2 demethylase mutant, we carried out an additional targeted fertility RNAi screen in spr-5(by101) mutants of 46 genes encoding potential histone methylation recognition modules (Taverna et al., 2007), including PHD, Chromo, MBT repeats, PWWP, or Tudor domains (Figure 5A). Knockdown of the chromodomain-containing gene cec-3 most potently suppressed the spr-5 transgenerational fertility defect. We therefore renamed this gene eap-1 (epigenetic memory antagonism protein 1). The eap-1 null mutant strain (ok3432) (confirmed by Western blot, Figure S5A), spr-5;eap-1 double mutants, and wild type worms laid the same numbers of eggs (Figure 5B). Knockdown or deletion of eap-1 in spr-5(by101) mutant worms also reduced the generational accumulation of H3K4me2 (Figure 5C and data not shown). However, deferred knockdown of eap-1 beginning at generation 20 failed to revert the transgenerational phenotypes (Figure S4B).
Figure 5. eap-1 deletion suppresses the progressive phenotypes of spr-5 mutant worms.
A) spr-5(by101) mutant worms fed dsRNA of C. elegans potential methyl binding genes for 20 generations’ effect on fertility as compared to E.V. treated spr-5(by101) mutant worms. B)spr-5;eap-1 double mutant worms have an almost complete suppression of the fertility defect of spr-5(by101) mutant worms at generation 20 (graph is the mean +/− SEM of 7 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate). ***: p<0.001, ****: p<0.0001. C) spr-5(by101) mutant worms display increased H3K4me2 at generation 20 which is suppressed by knockdown of eap-1 for 20 generations as assessed by western blots of whole worm lysates at the L3 stage. D) EAP-1 is expressed in every nucleus throughout the germline and localizes to chromatin as seen in immunofluorescence of mid-pachytene nuclei of dissected gonads from wild type, eap-1(ok3432), and spr-5(by101) mutants at generation 5.
Whole mount worm immunofluorescence revealed that EAP-1 was expressed in every cell in the embryo (Figure S5B). EAP-1 is predominantly expressed in the head region and the nuclei of the germline (Figures S5C and S5D) where H3K4me2 accumulates in spr-5 mutants (Nottke et al., 2011). A more detailed examination of EAP-1 expression in dissected gonads revealed that EAP-1 was expressed at all stages throughout the germline (Figures 5D and S5E–H).
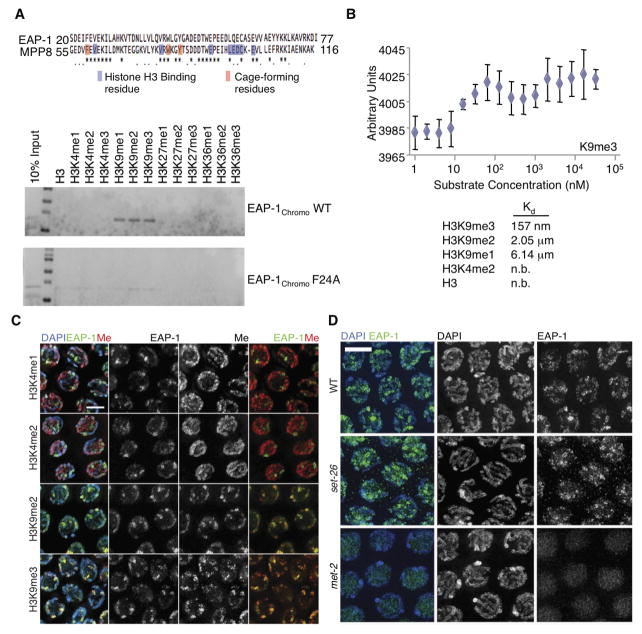
EAP-1 binds to methylated H3K9
The closest mammalian EAP-1 homologue is MPP8 (full length protein: 27.33% identity, chromodomain: 50% identity), which binds methylated H3K9 (Chang et al., 2011; Kokura et al., 2010). In vitro binding assays, using purified chromodomain (EAP-1chromo) or full length EAP-1 fused to GST, showed that EAP-1 selectively binds to H3K9 methylated peptides (Figures 6A and S6A). Using MPP8 as a guide, we identified F24, W45 and Y48 in EAP-1 as the predicted aromatic cage forming residues (Chang et al., 2011) (Figure 6A). Mutation of each of these sites to alanine eliminated binding of EAP-1 to H3K9 methylated peptides (Figures 6A, S6B, and data not shown). Binding assays using a histone peptide array harboring defined single and combinatorial modifications (Rothbart et al., 2012b) (Table S1), confirmed these findings (Figures S6C and D). In the same assay, we found that EAP-1 binding, like the chromodomain of MPP8 (Rothbart et al., 2012a), was inhibited by phosphorylation of threonine 6 and serine 10 (Figures S6C and D).
Figure 6. EAP-1 binds methylated H3K9.
A) EAP-1chromo binds to H3K9 methylated peptides in in vitro binding assays. Shown above is EAP-1 homology to MPP8 with conserved residues marked *. Mutation of any of the three cage-forming amino acids to alanine eliminate EAP-1chromo’s ability to bind to H3K9 methylated peptides (F24A is displayed). B) Microscale thermophoresis of EAP-1chromo and MLA histones shows that EAP-1 has highest binding affinity for H3K9me3 than H3K9me2 than H3K9me1 and no binding affinity for H3K4me2 or unmodified Histone H3. Binding affinity for H3K9me3 is displayed in the figure while other histone H3 affinities are displayed in Figure S6E. C) EAP-1 colocalizes with H3K9me2/me3 but not with H3K4me1/me2 as assessed by immunofluorescence of dissected gonads from wild type adult hermaphrodites. Pachytene nuclei are shown. Scale bar, 4 μm. D) EAP-1 no longer localizes to the chromatin when H3K9 methylation is reduced by mutation of the H3K9me1/me2 methyltransferase met-2. Pachytene nuclei are shown. Scale bar, 4 μm.
To assess the affinity of EAP-1 for differentially methylated H3K9, we performed Microscale Thermophoresis (MST) (Jerabek-Willemsen et al., 2011; Wienken et al., 2010) with MLA (Methyl Lysine Analog) histones. EAP-1 bound most tightly to H3K9me3 (Kd = 157 nM), followed by H3K9me2 (Kd = 2.05 μM) and H3K9me1 (Kd = 6.14 μM) (Figures 6B and S6E). In the same analysis, EAP-1 had no detectable affinity for unmodified histone H3 or H3K4me2 (Figures 6B and S6E). Consistently, we found that in dissected gonads of wild type worms, EAP-1 protein signal overlaps with those of H3K9me2/me3 but not H3K4me1/me2 (Figure 6C). Additionally, deletion of met-2, which reduces H3K9me1/me2 in adults (Bessler et al., 2010), reduced overall EAP-1 chromatin association (Figure 6D). However, deletion of set-26 had no overt impact on EAP-1 chromatin association globally, suggesting that SET-26 may play a locus-specific methylation role. Collectively, these results identify EAP-1 as an H3K9me reader.
To further examine the mechanistic interaction between EAP-1 and SET-26, we crossed eap-1(ok3432) mutants with set-26(tm3526) and spr-5(by101). We found that spr-5;eap-1;set-26 triple mutants laid a similar number of eggs as spr-5;set-26 double mutants (Figure 7A), suggesting that SET-26 is epistatic to EAP-1.
Figure 7. EAP-1 regulates transgenerational gene expression of spr-5 mutant worms.
A) spr-5;eap-1;set-26 triple mutant worms lay as many eggs as spr-5;set-26 double mutants at generation 5 suggesting that set-26 is epistatic to eap-1 (graph is the mean +/− SEM of 2 independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate). *: p<0.05, **: p<0.01, ***: p<0.001. B) EAP-1 binds to regions which are marked with H3K9me3 and decline across generations of spr-5(by101) mutant worms. Band intensity reflects EAP-1 binding. Darker regions reflect stronger binding affinity while whiter regions reflect weaker ones. Note that EAP-1 binding does occur in G20 worms but is weaker than in WT and spr-5 G10 mutant worms. C) EAP-1 bound target genes display increases in gene expression across spr-5(by101) generations. The results represent the mean +/− SD of 4 biological replicates of ~1000 young adult worms as compared to pan-actin expression. D) EAP-1 bound target genes do not increase in gene expression in generation 20 spr-5;eap-1 double mutant worms. The results presented correspond to the mean +/− SEM of 2 (scrm-4) or 4 (asp-17) independent biological experiments of replicates of ~1000 young adult worms as compared to pan-actin expression. E) Model for epigenetic inheritance of elevated H3K4me2.
spr-5 mutant worms lose EAP-1 binding across generations
We next investigated the genomic locations of EAP-1 binding by ChIP-seq experiments on whole worms in wildtype and spr-5 mutant backgrounds at generations 10 and 20 (Figure 7B). As a control for EAP-1 antibodies, we found no EAP-1 binding in eap-1(ok3432) null mutant worms (data not shown). EAP-1 binding was highest in genomic regions, which had previously been reported to have high H3K9me3 (Gu and Fire, 2010; Liu et al., 2011), consistent with EAP-1 being an H3K9me3 reader. In these regions, EAP-1 binding decreased across the generations (Figure 7B: G0–G10, G10–G20 for regions bound by EAP-1 in WT; p<2.2 × 10−16), similar to the decline of H3K9me3 seen in Western blots of whole worms (Figure 4G). In late generation spr-5 mutants, EAP-1 protein level was similar to wildtype worms (Figure S5A) and EAP-1 was still present on chromatin based on immunostaining (data not shown), but EAP-1’s binding near the chromosome ends showed a clear decrease (Figure 7B). Together, these results suggest that the decline in EAP-1 enrichment near the chromosome ends over generations may be the consequence of the global decline in H3K9me3 in spr-5 mutants.
Interestingly, the genes bound by EAP-1 in wildtype worms and in spr-5 mutants at generation 10 (Table S2) displayed a gene ontology (GO) enrichment for regulation of growth (p=0.00865346) and gamete generation (p=0.03873372). An examination of some of the genes in regions of high EAP-1 binding revealed that their expression increased as EAP-1 binding declined (Figure 7C) consistent with these regions becoming more euchromatic and accessible for transcription. The transgenerationally elevated expression of several of these genes was dependent on eap-1 as generation 20 spr-5;eap-1 double mutant worms had wildtype levels (Figure 7D).
Discussion
In this study, we identified novel H3K4me1/2 (SET-17 and SET-30) and H3K9me3 methylases (SET-26), as well as an H3K9me3 reader (EAP-1), that regulate transgenerational progressive decline of fertility associated with the persistent loss of the H3K4me1/me2 demethylase spr-5 in C. elegans. While H3K4me2 accumulates, H3K9me3 decreases across the generations of spr-5 mutants. Our ChIP-seq analysis of the genomic locations of EAP-1 in the spr-5 mutants suggests a model whereby progressive loss of EAP-1 chromatin association may be important for the trans-generational fertility phenotype associated with SPR-5 loss. Our findings lay the framework for a molecular model where the interplay between H3K4 versus H3K9 methylation impacts transgenerational epigenetic inheritance in C. elegans.
spr-5 mutant worms have reduced transgenerational fertility
A recent report (Alvares et al., 2013) suggested that spr-5(by134) mutant worms only displayed a transgenerational fertility defect at the elevated temperature of 25°C but not at 20°C. This result was contrary to the initial results reported by (Katz et al., 2009) as well as to our observations. The authors proposed that the transgenerational defect seen by Katz et al at 20°C was due to maintaining the spr-5(by101) strain as a heterozygous balanced strain or because of potential instability of the by101 Tc3 transposon insertion. We maintained our strains by crossing repeatedly with a wildtype strain, not as a heterozygous balanced strain, but still observed progressive fertility defects at 20°C (Figure 1A). We also observed a progressive fertility decline in the spr-5(by134) strain used by (Alvares et al., 2013) at 20°C (Figure S1A). The reduced fecundity of spr-5 mutant worms was also observed by a third independent group (Kim et al., 2012). Therefore, the discrepancy between the results of Alvares et al (2013) and those of us and other labs remains unexplained.
Suppression versus reversion of the trans-generational phenotypes
While knockdown of set-17, set-30, or eap-1 led to suppression of the progressive defects of spr-5 mutants, only deferred knockdown of set-30 reverted the phenotypes (Figures 3 and S4). These results suggest that the two H3K4 methyltransferases have both similar and distinct roles in regulating epigenetic inheritance. This difference could be due to differential expression across cell types, although in situ results suggest both genes are expressed in the germ cells (NEXTDB: http://nematode.lab.nig.ac.jp). Alternatively, their functions may be differentially regulated by existing modifications on the histone tails or they may target different genomic loci. Furthermore, their ability to regulate methylation states at H3K4 could be dictated by distinct protein partners. In mammalian cells, DNA methylation is regulated by the de novo methyltransferases DNMT3a/b and the maintenance methyltransferase DNMT1 (Bestor, 2000; Okano et al., 1999). Our findings suggest the possibility that, analogous to the mammalian DNA methyltransferases, SET-17 may be required only for maintaining H3K4me1/me2 levels, while SET-30 may be important for both maintaining and re-setting the H3K4me2 levels to that of the wild type worms.
Potential molecular mechanisms
spr-5 mutants display increased global H3K4me2 over generations. Is the altered H3K4me2 itself passed from generation to generation, or is the machinery that regulates H3K4 methylation inherited to allow the reacquisition and accumulation of H3K4me2? Recent mammalian cell studies argue for the latter. Specifically, the H3K4 methyltransferase MLL and the Polycomb group complexes, PRC1 and PRC2, are either maintained or re-established on chromatin through cell divisions (Blobel et al., 2009; Francis et al., 2009). According to this model, the enzymatic machinery responsible for establishing H3K4me2 states (such as SET-30) could be inherited at specific loci to reapply methyl marks upon DNA duplication.
What might be the molecular mechanisms that underlie the involvement of regulators of both H3K4 and H3K9 methylation in controlling the transgenerational phenotypes associated with the loss of the H3K4me2-specific demethylase SPR-5? We envision three different possibilities that are not mutually exclusive. First, upon loss of SPR-5, H3K4me2 may accumulate randomly, imparting a more open chromatin that is increasingly susceptible to chromatin damage. Indeed, SPR-5 deletion causes perturbation of meiotic DNA double-strand break repair (DSBR) and progressively increased germ cell apoptosis (Nottke et al., 2011). Additionally, PRDM9, the potential homologue of SET-17, has been implicated in mammals as a determinant of appropriate sites of meiotic recombination (Baudat et al., 2013). However, the spr-5 phenotypes are completely suppressed by adding back a single copy of spr-5, suggesting that the transgenerational phenotypes are not due to inherited accumulation of DNA damage. This is consistent with the previous finding that increased H3K9 methylation, rather than H3K4 methylation, correlates with increased mutation rates in human cancer cells (Schuster-Bockler and Lehner, 2012).
Alternatively, H3K4me2 may accumulate by spreading into nearby heterochromatic regions in the absence of SPR-5, thus changing heterochromatin-euchromatin boundaries, which can impact chromatin structure and gene expression. Consistent with this model, in Drosophila and S. pombe, the homologs of SPR-5 have been shown to play roles in euchromatin-heterochromatin boundary formation (Lan et al., 2007; Rudolph et al., 2007). This is also supported by the global narrowing of EAP-1 binding regions across generations in spr-5(by101) mutant worms (Figure 7B). This model, which we favor, predicts that the proteins identified in our screens would function in the same cells to regulate transgenerational inheritance. We therefore propose that in C. elegans, heterochromatic/euchromatin boundaries are maintained by coordinated actions of both the H3K4me1/me2 demethylase SPR-5 and H3K4me1/me2 methyltransferases SET-17 and SET-30 on one side of the equation, and the actions of the H3K9me binding protein EAP-1, the H3K9me3 demethylase JMJD-2, the H3K9me1/me2 methyltrasnfserase MET-2, and the H3K9me3 methyltransferase SET-26, on the other. Thus, loss of SPR-5 may enable the H3K4me2 mark to gradually encroach into the otherwise heterochromatic region (Figure 7E). Supporting this theory, a previous study reported that deletion of the predicted H3K9me1/me2 methyltransferase met-2 leads to a progressive fertility defect (Andersen and Horvitz, 2007). This suggests that altering either side of this balanced equation, the H3K4me1/me2 demethylase SPR-5 or the H3K9me1/me2 methyltransferase MET-2, will facilitate euchromatin spreading into heterochromatic regions. Although this second model favors the hypothesis that these proteins function in the same cells, our current data do not preclude the possibility that some of the proteins function in the soma as opposed to the germline to regulate the transgenerational phenotypes after the memory has been transmitted. This alternative scenario could help explain why SET-30 but not SET-17 deletion reverts the progressive fertility defects of spr-5(by101) mutant worms.
The third model, which could also explain the mis-regulation of specific genes involved in fertility regulation, involves SPR-5 impacting local gene expression independently of localized euchromatin expansion. In this scenario SPR-5 would affect gene expression at specific loci where it is recruited. A previous study reported a mis-regulation of spermatogenesis genes in spr-5 mutants (Katz et al., 2009). Similarly, EAP-1 bound genes had a significant enrichment of genes involved in reproduction. Whether these reproduction genes become mis-regulated through euchromatin expansion or are subject to localized SPR-5 recruitment remains to be determined.
In summary, our findings have revealed a molecular network that controls transgenerational inheritance in C. elegans, and raise the possibility that perturbation of the balance between histone H3K4 and H3K9 methylation regulation may impact epigenetic inheritance.
Experimental Procedures
Fertility assays
From day 3 to day 8 post-hatching, 10 worms were placed on NGM plates with OP50-1 in triplicate (30 worms total per condition). Worms were grown at 20°C. However, for initial RNAi screening only a single plate was used, but hits were repeated in triplicate. After 24 h, the adult worms were removed from each plate and placed on new plate. The numbers of eggs and hatched worms on the plate were counted. Statistical analyses of fertility were performed using two-way ANOVA tests with Bonferroni post-tests, or t-tests using mean and standard error values.
Methyltransferase assays
10 μg of glutathione S-transferase (GST)-purified SET-26SET, SET-30, or SET-17 were incubated with histone peptides (amino acids 1–21 of histone H3), recombinant histone H3 (NEB), histone octamers (Sigma), or nucleosomes purified from 293T cells in the presence of either 0.1 mM S-adenosyl-methionine (SAM) or 2 μCi [3H]SAM at 37°C for 2 hours in a methyltransferase reaction buffer (50 mM Tris-HCl pH 8.5, 20 mM KCl, 10 mM MgCl2, 10mM β-mercaptoethanol, 250 mM sucrose) as described (Rea et al., 2000). Reactions were subjected to SDS–PAGE and either autoradiography or western blot as described below.
Demethylase assays
2 μg of GST-purified JMJD-2 were incubated with histone octamers (Sigma) at 37°C for 4 hours in a demethylase reaction buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2, 1 mM α-mercaptoethanol, 2 mM ascorbic acid) as described (Whetstine et al., 2006). Reactions were subjected to SDS-PAGE and western blot as described below.
Microscale Thermophoresis
Fluorescence distribution measurements were taken of fluorescently labeled molecules inside a capillary upon laser irradiation. Temperature gradients were generated by an IR-Laser focused on the capillary. Binding affinities are calculating by measuring a temperature jump in the initial stage of irradiation, thermophoretic movement of the molecules within the gradient at later stages, or both (Jerabek-Willemsen et al., 2011; Wienken et al., 2010). At least 3 independent experiments were performed for each histone modification.
Gonad dissection, immunohistochemistry and analysis
Gonads from young adult hermaphrodites (24 hours post-L4) were dissected in M9 buffer (22 mM KH2PO4, 34 mM K2HPO4, 86 mM NaCl, 1 mM MgSO4) and fixed on slides with −20°C methanol for one minute. The remaining steps were carried out at room temperature. Slides were then fixed with 4% formaldehyde (4% formaldehyde in PBS with 80 mM HEPES (pH7.4), 0.8 mM EDTA, and 1.6 mM MgSO4) for 30 minutes. After a five minute wash in PBST, the slides were blocked in 0.5% BSA for one hour. Slides were incubated overnight with primary antibodies (αEAP-1, αH3K4me1 (CMA302),α H3K4me2 (CMA303),α H3K9me2 (CMA317), and αH3K9me3 (CMA318)) at a 1:100 dilution. Slides were then incubated with DAPI (Sigma, 1.7 μg/ml) and secondary antibodies from Jackson ImmunoResearch Laboratories (FITC αrabbit (111-095-144) and Cy3 αmouse (405309)at a 1:100 dilution for 2h.
Images were taken with a 100X objective combined with auxiliary magnification (1.6X) in 0.2 μm Z-stack intervals with an IX-70 microscope (Olympus) and cooled CCD camera (CH350; Roper Scientific) using the DeltaVision system (Applied Precision). Partial projections of half-nuclei are shown.
Additional information about worm strains, constructs, RNA interference, whole mount immunocytochemistry, genotyping, antibodies, western blotting, peptide binding assays, ChIPseq, and real-time analysis can be found in the Supplementary Information section
Supplementary Material
Highlights.
H3K4me2 increases and H3K9me3 decreases across L4 stage spr-5 mutant worms generations
Loss of H3K4 methyltransferases suppress transgenerational sterility of spr-5 mutants
H3K9me regulators control transgenerational sterility of the H3K4me2 demethylase spr-5
Acknowledgments
We thank T.K. Blackwell, J. Lieberman, and members of the Shi lab for discussions and critical reading of the manuscript. We thank A. Fire for discussions. We thank T. Stiernagle and the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), S. Mitani and the National Bioresource Project for the Experimental Animal “Nematode C. elegans”, and D. Moerman laboratories for C. elegans strains. E.L.G. was supported by T32-CAA009361, by a Helen Hay Whitney post-doctoral fellowship, and by a National Institute on Aging of the National Institute of Health (NIH) grant (K99AG043550). S.E.B.S. was supported by an individual NRSA postdoctoral fellowship (F32GM100515) from the NIH/NIGMS, E.B. was supported in part by an EMBO Fellowship, S.B.R. was supported by the UNC Lineberger Comprehensive Cancer Center Basic Sciences Training Program (T32CA09156) and an American Cancer Society post-doctoral fellowship (PF-13-085-01-DMC). Y.Z. and W.W. were supported in part by a National Institutes of Health (NIH) grant (GM096194). B.D.S. was supported in part by a National Institutes of Health (NIH) grant (GM068088). M.P.C. was supported by a National Institutes of Health (NIH) grant (GM072551), a John and Virginia Kaneb Fellowship, and a grant from the Charles E. W. Grinnell Fund. This work was supported by NIH grants to Y.S. (GM058012, CA118487, MH096066) and by an Ellison Foundation Senior Scholar Award to Y.S. YS is an American Cancer Society Research Professor.
Footnotes
Author Contributions
E.L.G. and Y.S. conceived and planned the study and wrote the paper. S.E.B-S. produced Figs 5D, 6C, and D, and S5E-H and was advised by M.P.C.. E.B. performed ChIP-seq experiments and helped with the analysis and produced Fig 7C. R.S. helped produce Figs 4D and S3D. Y.Z. performed ChIP-seq analysis and was advised by W.W.. S.B.R. produced Figs. S6C and D and was advised by B.D.S.. S.C. produced Fig 2D. D.A.C., S.C., and Q.J. helped produce and purify recombinant proteins. A.I.B. performed several Western blotting experiments and constructed several plasmids. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvares SM, Mayberry GA, Joyner EY, Lakowski B, Ahmed S. H3K4 demethylase activities repress proliferative and postmitotic aging. Aging cell. 2013 doi: 10.1111/acel.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development (Cambridge, England) 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery OT, Macleod CM, McCarty M. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types: Induction of Transformation by a Desoxyribonucleic Acid Fraction Isolated from Pneumococcus Type Iii. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nature reviews Genetics. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler JB, Andersen EC, Villeneuve AM. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS genetics. 2010;6:e1000830. doi: 10.1371/journal.pgen.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, Rinehart C, Quiton J, Walsh R, Smallwood A, et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Molecular cell. 2010;40:736–748. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Molecular cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Horton JR, Bedford MT, Zhang X, Cheng X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J Mol Biol. 2011;408:807–814. doi: 10.1016/j.jmb.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Jr, Yates JR, 3rd, Mello CC. Argonautes Promote Male Fertility and Provide a Paternal Memory of Germline Gene Expression in C. elegans. Cell. 2013;155:1532–1544. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nature reviews Genetics. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature reviews Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science (New York, NY. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Gu SG, Fire A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma. 2010;119:73–87. doi: 10.1007/s00412-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013 doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Govindan JA, Tu ZJ, Greenstein D. SACY-1 DEAD-Box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics. 2012;192:905–928. doi: 10.1534/genetics.112.143271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature reviews Genetics. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. The EMBO journal. 2010;29:3673–3687. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Molecular cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Mechanisms for the inheritance of chromatin States. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging cell. 2011 doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottke AC, Beese-Sims SE, Pantalena LF, Reinke V, Shi Y, Colaiacovo MP. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12805–12810. doi: 10.1073/pnas.1102298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nature structural & molecular biology. 2012a;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Strahl BD, Fuchs SM. Peptide microarrays to interrogate the “histone code”. Methods Enzymol. 2012b;512:107–135. doi: 10.1016/B978-0-12-391940-3.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Molecular cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.