Abstract
In immune responses, activated T cells migrate to B cell follicles and develop to T follicular helper (Tfh) cells, a new subset of CD4+ T cells specialized in providing help to B lymphocytes in the induction of germinal centers 1,2. Although Bcl6 has been shown to be essential in Tfh cell function, it may not regulate the initial migration of T cells 3 or the induction of Tfh program as exampled by C-X-C chemokine receptor type 5 (CXCR5) upregulation 4. Here, we show that Achaete-Scute homologue 2 (Ascl2), a basic helix-loop-helix (bHLH) transcription factor 5, is selectively upregulated in its expression in Tfh cells. Ectopic expression of Ascl2 upregulates CXCR5 but not Bcl6 and downregulates C-C chemokine receptor 7 (CCR7) expression in T cells in vitro and accelerates T cell migration to the follicles and Tfh cell development in vivo. Genome-wide analysis indicates that Ascl2 directly regulates Tfh-related genes while inhibits expression of Th1 and Th17 genes. Acute deletion of Ascl2 as well as blockade of its function with the Id3 protein in CD4+ T cells results in impaired Tfh cell development and the germinal center response. Conversely, mutation of Id3, known to cause antibody-mediated autoimmunity, greatly enhances Tfh cell generation. Thus, Ascl2 directly initiates Tfh cell development.
Development of Tfh cells is initiated by and dependent on their movement out of the T cell zone and into the B cell follicle. This migration process is regulated by upregulation of CXCR5 as well as downregulation of both CCR7 and P-selectin glycoprotein ligand 1 (PSGL1)2,6. Tfh cells have unique developmental regulation and Bcl6 was reported to be selectively expressed in Tfh cells7–9. However, although Bcl6 potentiates Tfh cell generation in vivo, recent data suggest that it may not regulate the CXCR5 upregulation by activated T cells or their migration to B cell follicles in vivo 3,4. Hence, the transcriptional mechanisms underlying initial Tfh cell commitment remain unclear 4.
Recently, we observed that a bHLH domain-containing transcription factor, Ascl2 was highly expressed in CXCR5hiBcl6hi in comparison with CXCR5−Bcl6− T cells 4. Interestingly, CXCR5+Bcl6lo T cells also exhibited upregulation of Ascl2 mRNA expression 4, suggesting that its upregulation may precede that of Bcl6. Moreover, Ascl2 gene locus was marked with active chromatin marker trimethylated histone H3 lysine 4 (H3K4me3) in Tfh and to a much less extent, Th2, but not other T cell subsets, while the other Tfh-regulating genes Bcl6 7–9, Maf 10, Batf 11,12 and Irf4 13 were uniformly associated with H3K4me3 in all T cell subsets (Extended Data Fig. 1a). To validate these results, we sorted three subpopulations of cells (CXCR5−RFP−, CXCR5+RFPlo and CXCR5hiRFPhi) from Bcl6-RFP reporter mice immunized with keyhole limpet hemocyanin (KLH)/complete Freund’s adjuvant (CFA) (Fig. 1a), and found that Ascl2 was highly expressed in Tfh cells at both mRNA and protein level (Fig. 1b and Extended Data Fig. 1b). Also, Ascl2 expression was closely correlated with that of CXCR5 (Fig. 1b) and higher in Tfh than that in other T cell subsets (Fig. 1c). In human T cells, expression of Ascl2 as well as CXCR5 and Bcl6 was found with human tonsil CXCR5hiPD1hi Tfh cell (Fig. 1d and e). Collectively, Ascl2 is highly expressed in Tfh cells and its expression may precede that of Bcl6.
Extended Data Figure 1. Ascl2 exhibits unique epigenetic regulation in Tfh cell, and its expression is dependent on Wnt signal.
a. Genome-wide histone modifications (H3K4me3, permissive marker; H3K27me3, suppressive marker) across Bcl6, Maf, Batf, Irf4 and Ascl2 loci in T cell subsets (in vivo- Tfh and non-Tfh data set were newly generated; the rest were derived from GEO database (GSE14254)28).
b. Flow cytometric analysis of Ascl2 expression in three populations of activated CD44+ CD4+ T cell in draining LNs from Bcl6-RFP mice: CXCR5hiBcl6hi (red), CXCR5+Bcl6lo (blue) and CXCR5−Bcl6− (black) cells.
c. Quantitative RT-PCR measurement of Ascl2, Bcl6, and Batf expression in Bcl6-RV-GFP, Batf-RV-GFP and control vector infected CD4+ T cells; WT and Stat5−/− naïve CD4+ T cells were cultured under Th0 condition, or together with IL-6, respectively. Ascl2, Bcl6, and Batf transcriptional expression were measured by qRT-PCR.
d. Quantitative RT-PCR measurement of Ascl2 in CD4+ T cells cultured under indicated conditions.
e. Quantitative RT-PCR measurement of CXCR5 and Bcl6 in control or TWS119 (1μM)- treated T cells.
All experiments were repeated at least three times with similar results. Bar graph displayed the relative level of mRNA as mean ± SD, n = 3 per group, *P<0.05, **P<0.01, two-tailed t-test, N.S, no significance.
Figure 1. Ascl2 is selectively expressed in both mouse and human Tfh cells.
a., Three populations of CXCR5hiBcl6-RFPhi (red), CXCR5+Bcl6-RFPlo (blue) and CXCR5−Bcl6-RFP− (black) cells were sorted from dLNs in Bcl6-RFP mice immunized with KLH emulsified in CFA subcutaneously. b. Ascl2, CXCR5 and Bcl6 transcriptional expression in sorted cells. c. Ascl2 mRNA expression among in vivo-generated Tfh, naïve, Th0, Th1, Th2, and Th17 cells by quantitative RT-PCR. d. Flow cytometric analysis of human tonsil CD4+ T cells by CXCR5 and PD-1 staining. e. The expression of Ascl2, CXCR5, Bcl6, and Gata3 mRNA in sorted cells. f. CXCR5 expression in CD4+ T cells activated by anti-CD3/anti-CD28 in the presence of TWS119 (1μM) (an inhibitor of glycogen synthase kinase-3beta (Gsk-3beta)) for three days. All experiments were repeated at least three times with similar results. (b, c, and e) bar graph displayed the relative level of mRNA as mean ± SD, n = 3 per group, *P<0.05, **P<0.01, two-tailed t-test.
Bcl6 and Batf are necessary in Tfh cell development 6,12, whereas Stat5 inhibits Tfh cell development 14,15. Overexpression of Bcl6 or Batf, or Stat5 deficiency, failed to increase Ascl2 expression (Extended Data Fig. 1c). None of the known stimuli including anti-CD3, anti-CD28, anti-ICOS, IL-6 and IL-21, nor their combination upregulated Ascl2 expression in T cells (Extended Data Fig. 1d). Ascl2 was previously shown as a target of canonical Wnt signaling in intestinal stem cell 5, and we found also that Ascl2 and CXCR5 but not Bcl6 expression in CD4+ T cells can be upregulated by TWS119 16 (Fig. 1f and Extended Data Fig. 1d–e) or other Wnt agonists (data not shown).
As a first step to examine the function of Ascl2 in Tfh cells, retroviral overexpression of Ascl2 was conducted in CD4+ T cells, leading to substantial induction of CXCR5 expression in over 30% of transduced cells, whereas Bcl6, Batf, or Maf in purified T cells did not (Fig. 2a and Extended Data Fig. 2a). Ascl2 overexpression increased Cxcr5 mRNA expression by ~60 folds (Fig. 2b), without affecting Bcl6, Prdm1, Batf, Sh2d1a, Cd40lg, Icos, Pdcd1, Btla, and Il21 expression (Fig. 2c). CXCR5 expression was equally induced by Ascl2 in wild-type (WT), Bcl6−/− and Batf−/− CD4+ T cells in vitro (Fig. 2d). Thus, our findings suggest that Ascl2 is unique in its ability to induce CXCR5 protein expression in CD4+ T cells in vitro. CCR7 and PSGL1 as well as CD25 (Il2ra) and CD122 (Il2rb) expression was downregulated in Ascl2-overexpressing T cells (Fig. 2e and Extended Data Fig. 2b), likely accounting for increased follicular homing ability and decreased IL-2 signaling in Tfh cells 14,15,17. In addition, Th1- and Th17-related signature genes were strongly suppressed by Ascl2 (Extended Data Fig. 2c).
Figure 2. Ascl2 expression induces the Tfh program.
a. Flow cytometry analysis of surface CXCR5 expression from empty-RV-GFP, Ascl2-RV-GFP, and Bcl6-RV-GFP retrovirus-infected T cells, respectively. b. Sorted GFP+ T cells were subject to the measurement of CXCR5 mRNA by quantitative RT-PCR. c. Measurement of genes expression including Cxcr5, Bcl6, Prdm1, Batf, Sh2d1a, Cd40lg, Icos, pdcd1, Btla, and Il21. d. Surface CXCR5 expression in Ascl2-RV-GFP-and vector virus-infected WT, Bcl6−/−, and Batf−/− T cells. e. CCR7, PSGL-1, CD25, and CD122 expression by flow cytometry analysis. (f–k) Ascl2-RV-GFP- or Empty-RV-GFP- transduced GFP+ OT-II cell were transferred into naïve TCRβ−/− mice subsequently immunized with NP-OVA/CFA. f. At day 2 and Day 6, flow cytometry analysis of donor cells with staining CXCR5 and Bcl6, n = 4. g. Quantification of CXCR5+ and CXCR5+Bcl6+ donor-derived T cells. h. GC B cells (GL-7hi Fashi) in recipient mice, n = 4. i. Quantification of GC B cells. j. At day 8, dLNs were collected and subject to histochemistry staining of GC center and donor T cells. Green, GFP; Red, PNA; Blue, anti-IgD; Scale bar, 100μm, dot graph represents donor cells in GC, displayed as mean ± SD, n = 10. k. Titers of NP-specific antibodies in serum from mice day 8 after immunization, n = 4. l. Distribution of Ascl2-RV-GFP- and vector-infected GFP+ OT-II donor cells in B220+ B cell follicles from dLNs in mice immunized with OVA/CFA for four days, scale bar, 100μm, dot graph represents distribution with a ratio of donor cells in B cell follicle versus T zone, displayed as mean ± SD. Empty-RV-GFP, n = 21; Ascl2-RV-GFP, n=15. All experiments were repeated at least three times with similar results. (b, c, g, i, and j–l) graph displayed as mean ± SD, two-tailed t-test, N.S, no significance.
Extended Data Figure 2. Ascl2 regulates a selective subset of Tfh-relevant genes.
a. Flow cytometry analysis of CXCR5 expression in CD4+ T cell transduced with vector control, Ascl2-RV-GFP, Bcl6-RV-GFP, Batf-RV-GFP and Maf-RV-GFP. Data are a representative of two independent experiments.
b. Transcriptional expression of Ccr7, Psgl1, Il2ra, and Il2rb in Ascl2-RV-GFP- or control Vector-infected T cells were measured by quantitative RT-PCR. Data are a representative of two independent experiments. Bar graph displayed the relative level of mRNA as mean ± SD, n = 3, two-tailed t-test.
c. Quantitative RT-PCR measurement of genes expression including Th1-related Tbx21 and Ifng, as well as Th17-related Rorc and Il17a. Data are a representative of three independent experiments. Bar graph displayed the relative level of mRNA as mean ± SD, n = 3, two-tailed t-test.
d. Bcl6-RV-GFP- or control viral vector-infected GFP+ OT-II cells were adoptively transferred into naïve congenic mice, respectively, followed with subcutaneous OVA/CFA immunization. At day 2 after immunization, flow cytometry analysis of donor-derived cells in dLNS with CXCR5 and PD1 staining. Data are a representative of two independent experiments, n = 3.
e. Quantification of donor-derived Tfh cells. Bar graph displayed as mean ± SD, n = 3, two-tailed t-test, N.S, no significance.
f. Vector-transduced GFP+CXCR5− CD4+ T (RV) cell, Ascl2-RV-GFP-infected GFP+CXCR5− (CXCR5−) and GFP+CXCR5+ (CXCR5+) CD4+T cell were sorted and subject to microarray assay.
g. Hierarchic clustering and principal component analysis (PCA) were applied on seven microarray dataset including RV, CXCR5−, CXCR5+ as well as Tfh and non-Tfh cells (derived from GSE40068 4).
h. The clusterd heatmap of ~350 genes from RV, CXCR5−, CXCR5+, Tfh, Non-Tfh cells. The color-coding applies to gene expression level (log2) with 0 as a median.
We then assayed the role of Ascl2 in vivo by transferring Ascl2-transduced OT-II cells into TCRβ−/− recipient mice. At day 2 post immunization with 4-Hydroxy-3-nitrophenyl (NP)- Ovalbumin (OVA)/CFA, neither CXCR5 nor Bcl6 expression were detectable in vector-transduced control group, whereas Ascl2 overexpression strongly increased CXCR5+Bcl6lo cells (Fig. 2f–g). In contrast, ectopic expression of Bcl6 did not promote Tfh generation at this time point (Extended Data Fig. 2d–e). At day 6 post immunization, Ascl2 overexpression induced higher percentage of CXCR5hiBcl6hi Tfh cells (Fig. 2f–g). Accordingly, germinal center (GC) B cells and the size of GC at day 8 in mice receiving Ascl2-transdued T cells were significantly increased (Fig. 2h–j); Anti-NP IgM, IgA, IgG1 as well as IgG3 titers were increased, while IgG2a and IgG2b was not affected (Fig. 2k), consistent with that IgG2a switching is primarily mediated by extrafollicular T cells 18.
We next assessed whether Ascl2 could promote T cell follicular homing in vivo. Ascl2-overexpressing OT-II cells were more preferentially accumulated in follicles (Fig. 2l), and even in GC (Fig. 2j) in comparison with control vector-infected T cells. Given that Bcl6 overexpression does not affect early Tfh generation (Extended Data Fig. 2d–e) and Tfh cell migration 3. Therefore, these observations collectively demonstrate that different from Bcl6, Ascl2 promotes T cell migration to the follicles and the initiation of Tfh cell development.
To investigate the mechanism of Ascl2-controlled Tfh cell generation, we performed microarray analysis, and found that the expression of 293 genes was changed over 2 folds by Ascl2 overexpression. Cross-referencing the current data set of Ascl2 vs. vector with our previous Tfh vs. non-Tfh data set 4 revealed that 85 of the 293 genes affected by Ascl2 were directly associated with Tfh cell differentiation: 22 genes were upregulated, and 63 genes were downregulated 4 (Fig. 3a–b and Supplementary Table 1). Chemokine receptors CXCR5 and CXCR4, a GC Tfh-related receptor 19, were at the top of the upregulated gene list, while Th1-related genes (Il12rb1, Tbx21, Ifng, and Gzmb) and Th17-related aryl hydrocarbon receptor (Ahr) gene were greatly suppressed by Ascl2 (Fig. 3a). When comparing Ascl2-RV-GFP-infected CXCR5+ T cells and CXCR5− T cells with in vivo-generated Tfh and non-Tfh cells 4, we found Ascl2-induced CXCR5+ T cells were more similar in gene expression to Tfh cells (Extended Data Fig. 2f–g), with ~350 genes commonly expressed in these cells (Extended Data Fig. 2h).
Figure 3. Ascl2-dependent transcriptional regulation of Tfh-related genes.
a. Scatterplot of the average signal of Tfh versus non-Tfh cell, and Ascl2-RV-GFP+ versus Empty-RV-GFP+ T cell gene expression microarray data. The green line indicates gene expression change by factor of 2; data normalized from two replicates are shown; n = 2. Genes with the most transcriptional changes are listed. b. Venn diagram of genes regulated by Ascl2 and Tfh-related genes. c. Distribution of Ascl2 ChIP-Seq peaks in Ascl2-overexpressed CD4+ T cells. d. Venn diagram of genes regulated by Ascl2 and Ascl2-bound genes. e. Ascl2 binding site is identical to E-box binding motif. f. Ascl2 binding peaks located at gene loci including Cxcr5, Cxcr4, Ccr7, Selplg1, Il2ra, Il2rb, Ifng, Tbx21, Il2, and Rorc. g. Comparison of mouse Cxcr5 genomic sequence to dog, human, and rat. Red regions denote CNS region. h. Ascl2 binding sites at Cxcr5 locus in in vivo-generated Tfh cells by ChIP-qPCR (primers information were listed in Supplementary Table II). Green indicates the CNS region. Data are a representative of two independent experiments.
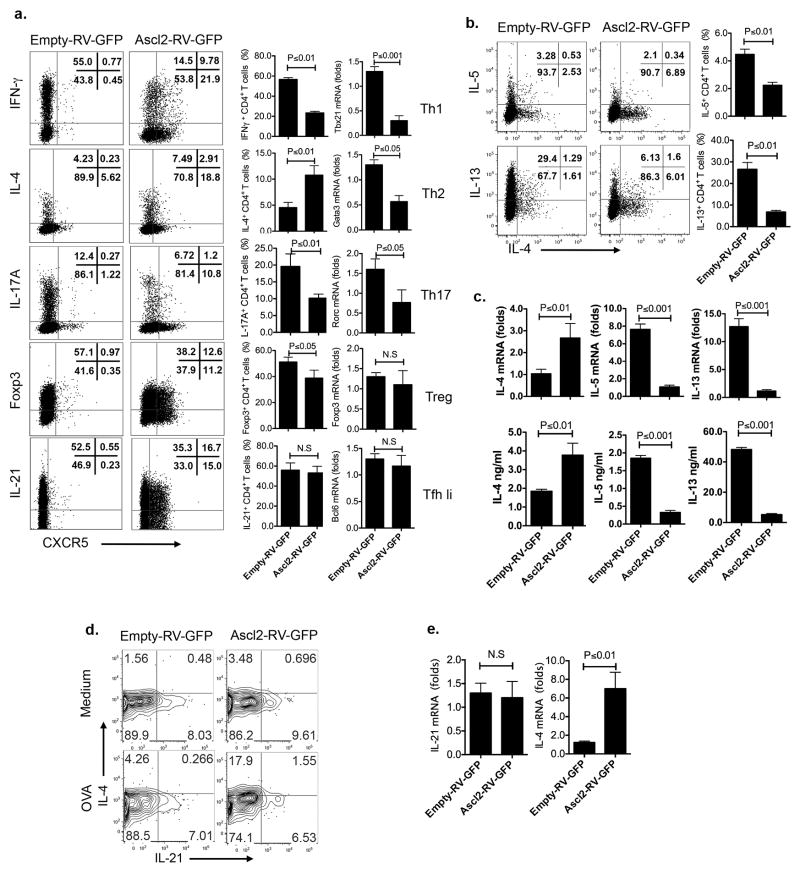
We further examined the effect of Ascl2 on Th1, Th2 and Th17 cell differentiation. As shown in Extended Data Fig. 3a, overexpression of Ascl2 suppressed both Th1 and Th17 differentiation and induced CXCR5 expression. Ascl2 had no effect on TGFβ-induced Foxp3 expression but induced CXCR5+ regulatory T (Treg) cells, suggesting that it may also be related with T follicular regulatory (Tfr) cell generation 20. Under Th2-polarized condition, Ascl2 enhanced IL-4 expression, while inhibited the expression of Gata3, IL-5 and IL-13 (Extended Data Fig. 3a–c), in agreement with recent studies that showed IL-4 but not Gata3, IL-5 or IL-13 expression in Tfh cells 21. Also, we observed that Ascl2 increased IL-4 but not IL-21 production in vivo (Extended Data Fig. 3d–e). Therefore, Ascl2 promotes Tfh gene expression and inhibits Th1-, Th2- and Th17-related gene expression.
Extended Data Figure 3. Regulation of Th cell differentiation by Ascl2.
a. Naïve CD4+ T cells from C57BL6 mice were activated under neutral condition and infected with Ascl2-RV-GFP or control vector (empty-RV-GFP) virus, followed with continuous culture under Th1, Th2, Th17, iTreg, and Tfh-like condition for 3–4 days, respectively. Quantification of signature genes by intracellular staining and real-time RT-PCR.
(b–c). Ascl2-RV-GFP- or control vector- transduced T cells were cultured under Th2 condition for 4 days.
b. After re-stimulation with PMA and ionomycin for 5h, Th2-related genes including IL-4, IL-5, and IL-13 expression were measured by flow cytometric analysis. c. GFP+ T cells were sorted and re-stimulated by plate-bound anti-CD3, transcriptional expression of IL-4, IL-5, and IL-13 were measured by quantitative RT-PCR; cytokines in supernatants of re-stimulation were subject to ELISA analysis.
(d–e). Ascl2-RV-GFP- or control vector- transduced OT-II cells were adoptively transferred into naïve congenic mice, respectively, followed with subcutaneous OVA/CFA immunization for seven days. d. After restimulation with OVA, flow cytometry analysis of donor-derived cells from dLNs with intracellular IL-4 and IL-21 staining. e. GFP+ donor-derived T cells were sorted from dLNs, re-stimulated with anti-CD3, and subject to quantitative RT-PCR measurement of IL-21 and IL-4 mRNA expression. All data are a representative of two independent experiments. (a–c, and e) bar graph displayed as mean ± SD, n = 3, two-tailed t-test, N.S, no significance.
We next assessed Ascl2 target genes by chromatin immunoprecipitation (ChIP) coupled with high throughput sequencing (ChIP-Seq). The analysis revealed a total of 10028 Ascl2-binding peaks, among which 41% and 36% were enriched in intronic and intergenic regions, respectively (Fig. 3c). Only 20% of Ascl2 binding sites were located at the promoter regions (Fig. 3c). Further comparison of global Ascl2 binding sites with Ascl2-regulated gene list showed that 145 among 4374 Ascl2-bound genes were transcriptionally regulated by Ascl2 (Fig. 3d).
As anticipated, analysis of Ascl2-binding peaks identified E-box protein binding site (5′-CANNTG-3′) as the consensus motif 5 (Fig. 3e). Ascl2 binding sites were identified in groups of gene loci including receptor genes (Cxcr5, Cxcr4, Ccr7, Selplg, Il2ra and Il2rb), inflammatory signature genes (Ifng, Tbx21, Il2, and Rorc) (Fig. 3f), but not in some of Tfh-related genes (Bcl6, Prdm1, pdcd1, sh2d1a, Icos, Il21, and Cd40lg) (data not shown).
Particularly, the Cxcr5 locus was found with multiple Ascl2 binding sites in the conserved non-coding sequence (CNS) regions (Fig. 3f–g). Moreover, these Ascl2 binding sites at the Cxcr5 locus were confirmed in in vivo-generated Tfh cells: two strong binding sites at intronic regions (CNS5 and CNS4-1), and three ones at distal promoter region (CNS1-1, CNS1, and CNS2) (Fig. 3f–h). Of note, the strongest peak of Ascl2 at CNS5 region was consistent with E47 (bHLH family member) binding site at the Cxcr5 locus 22 (Fig. 3f–h), implying the potentially redundant role of E2A in transcriptional regulation of the Cxcr5 gene 23.
To examine the functional significance of Ascl2 binding in regulating CXCR5 expression, we introduced Id3, an inhibitor of E-box protein 22, into Ascl2-overexpressing OT-II cells by retroviral infection, and observed a substantial reduction of Ascl2-regulated CXCR5 expression (Extended Data Fig. 4a), and this reduction was due to the inhibition of Ascl2 binding at the Cxcr5 locus revealed by ChIP assay (Extended Data Fig. 4b). Additionally, luciferase reporter assay defined that CNS5 and CNS1 were responsive to Ascl2 binding (Extended Data Fig. 4c–d). Together, these results offer evidences for Ascl2 in direct control of Tfh cell programming.
Extended Data Figure 4. CXCR5 expression is directly mediated by Ascl2.
Naïve CD4+ T cells were pre-activated and transduced with empty-RV-GFP, Id3-RV-GFP, Ascl2-RV-GFP, or Ascl2-RV-GFP together with Id3-RV-GFP retrovirus, respectively.
a. Flow cytometry analysis of CXCR5 expression in retroviral virus-infected T cells. Data are a representative of two independent experiments.
b. GFP+ cells were sorted from Ascl2-RV-GFP, or Ascl2-RV-GFP plus Id3-RV-GFP retrovirus-infected T cells, and subject to Ascl2 binding analysis on Cxcr5 gene locus via ChIP assay. Primer information was listed in Supplementary Table II. Data are a representative of two independent experiments.
c. Luciferase reporter assay of enhancer activity for Ascl2-bound region at Cxcr5 locus. Conserved noncoding sequence (CNS)-containing PGL3 plasmid was transfected with either empty-RV-GFP or Ascl2-RV-GFP into EL4 T cell line. Bar graph displayed as mean ± SD, n = 3, **P<0.01, two-tailed t-test.
d. Map of Cxcr5 gene locus and Ascl2 binding peaks at Cxcr5 locus. Arrow indicates the Ascl2-responsive CNS region.
As Maf 10, Batf 11,12 and IRF4 13 are required in Tfh cell differentiation, we compared genome-wide occupancy of these transcriptional factors, and found that Ascl2-bound genes hardly correlated with those by Maf (Extended Data Fig. 5a). For instance, IL-21 is directly regulated by Maf 24, but not Ascl2 (Extended Data Fig. 5b). Additionally, there was no binding site of Maf at Ascl2 locus (data not shown), or vice versa (Extended Data Fig. 5b), suggesting that Ascl2 and Maf are functionally independent in Tfh cells. A large proportion of Ascl2 occupancy co-localized with Batf/IRF4 binding sites (Extended Data Fig. 5c), including at CNS of gene loci including Cxcr5, Cxcr4, Ccr7, Selplg, Il2, Il2ra, and Il2rb (Extended Data Fig. 5d), but not at Bcl6, Prdm1, Maf (Extended Data Fig. 5b). Interestingly, Batf/IRF4 and Ascl2 peaks were found independently localized in Th1, Th2, and Th17 signature genes (Extended Data Fig. 5e–g), supporting that Batf/IRF4 acts downstream of TcR in activation of effector programs 25, whereas Ascl2 mediates suppression. These data strongly support Ascl2 as a specific regulator in Tfh cells.
Extended Data Figure 5. Coordinated function of Ascl2 and Batf/IRF4 in regulating Tfh-related genes.
(ChIP-Seq data of Maf, Batf, and IRF4 were derived from GSE40918 25)
a. Venn diagram of ChIP-Seq peaks from Ascl2 and Maf.
b. Distribution of ChIP-Seq peaks by Ascl2, Batf, and IRF4 on gene loci including Bcl6, Prmd1, Ascl2 and Maf.
c. Venn diagram of ChIP-Seq peaks from Ascl2, Batf and IRF4.
d. Distribution of ChIP-Seq peaks by Ascl2, Batf, and IRF4 on gene loci including Cxcr5, Cxcr4, Ccr7, Selplg1, Il2, Il2ra, and Il2rb. Blue frame represents the co-localization of peaks.
e. Distribution of ChIP-Seq peaks by Ascl2, Batf, and IRF4 on Th1-related Tbx21 and Ifng gene loci.
f. Distribution of ChIP-Seq peaks by Ascl2, Batf, and IRF4 on Th2-related Gata3, Il4, Il13 and Il5 gene loci.
g. Distribution of ChIP-Seq peaks by Ascl2, Batf, and IRF4 on Th17-related Rorc, Il17a, Il17f and Il21 gene loci.
ChIP-Seq assay of Ascl2 were performed on Ascl2-overexpressed T cell cultured under Th0 condition. ChIP-Seq assay of Maf, Batf and IRF4 were performed on Th0 cells by Dan R. Littman’s group, derived from GSE40918 25.
Tfh cells provide important help to B cells in the induction of efficient anti-virus antibody during viral infection 6,26. To address the functional roles of Ascl2, we generated Ascl2fl/fl/CD4-Cre mice, in which T cells were developmentally intact (data not shown), and assessed the requirement of Ascl2 in Tfh cell development in vivo with influenza virus infection. After intranasal (i.n.) infection, Ascl2fl/fl/CD4-Cre mice displayed gradual body weight loss from day 3 to day 9, while control littermate mice recovered after day 8 (Extended Data Fig. 6a). At day 9 post infection (dpi), the viral HA mRNA expression from lungs of Ascl2fl/fl/CD4-Cre mice was over 5-fold higher than those in control mice (Extended Data Fig. 6b), whereas CXCR5+Bcl6+ Tfh cells in lung dLNs were over 50 % decrease in percentages and a ~3–4 fold decrease in total cell numbers in the absence of Ascl2 (Fig. 4a), likely accounting for a severe reduction in GC B cell formation and antiviral IgG production (Extended Data Fig. 6c–d). Also, Tfh and GC B cell development in spleen was reduced in Ascl2fl/fl/CD4-Cre mice (Extended Data Fig. 6e–f). CD8+ T cell populations in lung, bronchoalveolar lavage fluid (BALF), spleen and dLNs were found no difference between Ascl2fl/fl/CD4-Cre mice and control mice (Extended Data Fig. 6g), although CD8+ T cell activation from dLNs of Ascl2fl/fl/CD4-Cre mice was slightly enhanced (Extended Data Fig. 6h). Moreover, we applied the same assay in tamoxifen-treated mixed bone marrow chimeric mice receiving both Ascl2fl/fl/ERT2-Cre and Ascl2+/+/ERT2-Cre bone marrow cells, and found that ~50% decrease of Tfh cells in Ascl2fl/fl/ERT2-Cre counterpart, while no defect in Ascl2-ablated CD8+ T cells (Extended Data Fig. 6i–j).
Extended Data Figure 6. Loss of Ascl2 in CD4+ T cells leads to impairment of germinal center responses during Influenza virus infection.
Control and Ascl2fl/fl/CD4-Cre mice were infected intranasally with influenza virus A/PR8.
a. Body weight of control and Ascl2fl/fl/CD4-Cre mice were monitored daily after infection.
b. Mice were sacrificed at day9 after infection, viral titer in the lungs were assessed by measurement of active Hemagglutinin (HA) gene expression with Quantitative RT-PCR.
c. Virus-specific total IgGs in the sera were measured by ELISA.
d. Flow cytometry analysis of germinal center B cells (GL-7hi Fashi) in lung dLNs from influenza-infected control and Ascl2fl/fl/CD4-Cre mice.
e. Frequencies of Tfh cells in spleens from influenza-infected control and Ascl2fl/fl/CD4-Cre mice;
f. Frequencies of GC B cells in spleens from influenza-infected control and Ascl2fl/fl/CD4-Cre mice;
g. After 9 day post infection (dpi), measurement of CD4+/CD8+ T cell ratio in lung, bronchoalveolar lavage fluid (BALF), spleen and dLNs from control and Ascl2fl/fl/CD4-Cre mice.
h. Flow cytometry analysis of Granzyme B and IFNγ production from both CD4+ and CD8+ T cell in dLNs.
(i–j). Mixed chimeric mice were reconstituted with both Ascl2+/+/ETR2-Cre and Ascl2fl/fl/ETR2-Cre bone marrow cells at a ratio of 1:1. Eight weeks later, chimeric mice were administered 200μl tamoxifen (5mg/ml) in sunflower seed oil i.p every other day for total 5 days, and then followed with influenza virus A/PR8 infection. At 9 dpi, measurement of CD4+/CD8+ T cell ratio and Tfh cell generation in dLNs (i) and spleens (j).
All the Data above are a representative of three independent experiments. Graph displayed as mean ± SD, n = 5 per group, *P<0.05, **P<0.01, (a, c) two way ANOVA, (b, e, f, and g–j) two-tailed t-test, N.S, no significance.
Figure 4. Loss of Ascl2 in peripheral CD4+ T cells inhibits Tfh differentiation.
a. Tfh cells (CXCR5hi Bcl6hi) in mediastinal LNs from Ascl2fl/fl/CD4-Cre and Ascl2+/+/CD4-Cre mice that received a sublethal dose of A/PR/8 (H1N1) virus intranasal infection nine days before, n = 5. b. Flow cytometry analysis of donor cells or host cells in mice receiving Cre-RV-GFP retroviral infected Ascl2fl/fl/OT-II or Ascl2+/+/OT-II GFP positive cells and subcutaneous 5-day- immunization of OVA/CFA, respectively, n = 3. c. Flow cytometry analysis of donor-derived Tfh cells in mice that received Id3-RV-GFP or control viral vector transduced OT-II cells and subcutaneous OVA/CFA immunization, n = 3. d. Flow cytometry analysis of donor-derived Tfh cells in Rag−/− mice receiving equal numbers of naïve Id3−/− and WT CD4+ T cells and subcutaneous KLH/CFA immunization for seven days, n = 5. e. Analysis and quantification of Tfh cells in chimeric mice reconstituted with equal number of Ascl2fl/fl/ERT2-Cre and Ascl2+/+/ERT2-Cre bone marrow cells after 5-day-tamoxifen treatment and 7-day- KLH/CFA immunization, n = 4. All above data are a representative of three independent experiments; Bar graph displays as mean ± SD, two-tailed t-test, N.S, no significance.
As Ascl2 can form heterodimers with other three relatives including E22, E47 and HEB in human cells 5,23, suggesting that the partial defect of Tfh cell differentiation in Ascl2fl/fl/CD4-Cre mice might be caused by compensation from other bHLH members. Indeed, Tfh cells and GC responses were normal in Ascl2fl/fl/CD4-Cre mice after immunization of KLH/CFA (data not shown). A substantial enhancement of E47 expression was noticed in naïve T, Tfh and even non-Tfh cells from Ascl2fl/fl/CD4-Cre mice, compared with control mice (Extended Data Fig. 7a), and expression of E47 also increased CXCR5 expression in CD4+ T cells (Extended Data Fig. 7b), consistent with a recent report that increased E47 activities in Id3−/− mice associated with increased Tfh-like cells 22. Furthermore, we examined T cell maturation and Tfh differentiation in chimeric mice receiving both Ascl2fl/fl/CD4-Cre and Ascl2+/+/CD4-Cre, or only Ascl2fl/fl/CD4-Cre bone marrow cells. As shown in Extended Data Fig. 7c–d, in chimeric mice, Ascl2 deficiency reduced TCRβhiCD69lo mature T cells, while mice receiving only Ascl2fl/fl/CD4-Cre cells had less defects in T cell maturation. Accordingly, after immunization with KLH/CFA, Tfh generation was inhibited in Ascl2fl/fl/CD4-Cre counterpart from mixed chimeric mice (Extended Data Fig. 7e–g). These data suggest that Ascl2 deletion at an early developmental stage induces compensatory mechanisms to allow T cell maturation.
Extended Data Figure 7. In the absence of Ascl2, bHLH family member E47 may play a redundant role in Tfh cell differentiation.
a. Quantitative RT-PCR measurement of E47 expression in naïve CD4+ T cell from Ascl2fl/fl/CD4-Cre and litter mate control mice; Tfh and non-Tfh were obtained from dLNs of Ascl2fl/fl/CD4-Cre and littermate control mice immunization with KLH in CFA, the expression of E47 was measured by real time RT-PCR.
b. Flow cytometry analysis of CXCR5 expression in T cell infected with E47-RV-CFP or control vector retrovirus.
(c–g). T cell-depleted bone marrow cells were obtained from Ascl2+/+/CD4-Cre (CD45.1+ CD45.2+) and Ascl2fl/fl/CD4-Cre (CD45.2+) mice and mixed at a ratio of 1:1 or 0:1 before transferred into irradiated Rag1−/− recipient mice (8×106 cell/mouse). Eight weeks later, mice were either utilized for measurement of thymic T cell maturation (c–d), or immunized with KLH in CFA for monitoring peripheral Tfh cell differentiation (e–g).
c. Flow cytometry analysis of T cells maturation in thymus of mixed chimeric mice containing both Ascl2+/+/CD4-Cre and Ascl2fl/fl/CD4-Cre bone marrow cells.
d. Flow cytometry analysis of T cells maturation in thymus of chimeric mice containing only Ascl2fl/fl/CD4-Cre bone marrow cells.
e. Seven days after immunization, Tfh cells in dLNs of mixed chimeric mice (Ascl2+/+/CD4-Cre and Ascl2fl/fl/CD4-Cre) were measured by flow cytometry.
f. Flow cytometry analysis of Tfh cells in dLNs of chimeric mice (Ascl2fl/fl/CD4-Cre).
g. The percentages of both CD4+ T cells and B220+ B cells in dLNs of mixed chimeric mice (Ascl2+/+/CD4-Cre and Ascl2fl/fl/CD4-Cre).
All the Data above are a representative of two independent experiments. Graph displayed as mean ± SD, n = 3 per group, two-tailed t-test, N.S, no significance.
To overcome the inducible compensation mechanism, we deleted Ascl2 gene acutely using a cre-expressing retrovirus and examined Tfh cell differentiation. As shown in Fig. 4b, Ascl2 deficiency in this case resulted in an absolute impairment in Tfh development in vivo. In contrast, Bcl6 deficient OT-II cell exhibited intact CXCR5 expression and homing ability at day 3 post immunization (Extended Data Fig. 8a–e). Moreover, Id3 overexpression impaired Tfh generation and GC responses (Fig. 4c and Extended Data Fig. 9a–c). Conversely, Id3 deficiency enhanced Tfh population (Fig. 4d). Together with the observation that Bcl6 overexpression could not rescue Id3-induced Tfh blockage (Extended Data Fig. 9d), Ascl2 appears to have an earlier function than Bcl6 in Tfh development.
Extended Data Figure 8. Loss of Bcl6 in CD4+ T cells does not affect early Tfh cell homing ability in vivo.
Equal amount of retrovirus Cre-RV-GFP-transduced WT/OT-II, Bcl6−/−/OT-II and Ascl2 fl/fl/OT-II cells were transferred into congenic mice, respectively, and followed with subcutaneous OVA/CFA immunization.
a. At day 3 post immunization, flow cytometry analysis of donor-derived Tfh cell generation with CXCR5 and PD1 staining.
b. Quantification of donor-derived CXCR5+ T cell.
c. At day 8 post immunization, examination of donor-derived Tfh cell generation with CXCR5 and PD1 staining.
d. Quantification of donor-derived CXCR5+ PD1+ Tfh cells.
e. At day 3 post immunization, dLNs were isolated and subject to histochemistry staining of B cell follicles and donor T cells. Green, GFP; Red, anti-B220; Scale bar, 100μm, n=8. All the Data above are a representative of two independent experiments. Graph displayed as mean ± SD, n = 4, two-tailed t-test, N.S, no significance.
Extended Data Figure 9. Ectopic expression of Id3 inhibits Tfh cell generation in vivo.
(a–c) Naïve OT-II CD4+ T cells were activated and transduced with Id3-RV-GFP or control viral vector (empty-RV-GFP) for three days. GFP+ T cell were then sorted and transferred into naïve congenic mice subsequently immunized with OVA/CFA.
a. At day3 post immunization, immunohistochemistry staining of section slide of draining lymph nodes. Red, B220+ B cells; Green, GFP+ donor derived OT-II cells. Data are a representative of two independent experiments, n = 6.
b. Quantification of GFP+ OT-II cell distributions in dLNs. Data are a representative of two independent experiments; Dot graph displayed as mean ± SD, n=17, two-tailed t-test.
c. OD values of OVA-specific antibodies in serum from mice day 7 after immunization, measured by 3 fold serial dilution in OVA (100μg/ml)-coated plates. Data are a representative of two independent experiments; Graph displayed as mean ± SD, n = 6, *P<0.05, **P<0.01, one way ANOVA.
d. Naïve CD4+ OT-II cells were pre-activated and co-transduced with Empty-RV-GFP/Empty-RV-hCD2, Empty-RV-GFP/Bcl6-RV-hCD2, Id3-RV-GFP/Bcl6-RV-hCD2, or Id3-RV-GFP/Empty-RV-hCD2, respectively. Sorted hCD2+GFP+ OT-II cells were transferred into congenic mice, followed with subcutaneous OVA/CFA immunization for seven days. Measurement of donor-derived Tfh cells with CXCR5 and PD1 staining. Data are representative of two independent experiments; Bar graph displayed as mean ± SD, n = 3, two-tailed t-test.
To confirm the above result, we examined Tfh differentiation in either mixed chimeric mice receiving equal numbers of Ascl2fl/fl/ETR2-Cre and Ascl2+/+/ETR2-Cre bone marrow cells (Fig. 4e), or intact Ascl2fl/fl/ETR2-Cre mice (data not shown), after tamoxifen treatments and KLH/CFA immunization, Ascl2 deletion inhibited Tfh cell differentiation and GC response in vivo (Fig. 4e and data not shown). Therefore, these data verify that Ascl2 is intrinsically necessary for Tfh differentiation.
In summary, we have identified a new player, the Ascl2 transcription factor that is crucial for Tfh development and function. On one hand, similar to Bcl6, Ascl2 acts as a novel suppressor of Th1, Th2 and Th17 cell differentiation. On the other hand, Ascl2 uniquely regulates Tfh cell migration and development via increasing CXCR5 and CXCR4 expression, and suppressing CCR7 and PSGL-1 expression, and IL-2 signaling. Our data suggest that Ascl2 and Bcl6 act in order to program Tfh generation. Activated T cells by antigen-presenting cells gain Ascl2 expression, which allows their migration towards B cell follicles. At the T-B border, the cognate B cells provide another signal for precursor Tfh cell to increase Bcl6 expression that further completes Tfh polarization and GC formation. Therefore, Id3 and Ascl2 may serve as an early checkpoint during Tfh cell development in promoting appropriate antibody response to infection while keeping autoimmune diseases in check (Extended Data Fig. 10). Further investigation into this axis may offer new ways to modulate antibody responses in infection and autoimmunity.
Extended Data Figure 10. The schematic model on the sequential roles of Ascl2/Id3 and Bcl6/Blimp1 during Tfh differentiation.
Ascl2 expression plus simultaneous Id3 reduction in activated CD4+ T cells orchestrates T cells to migrate toward B cell follicles and initiate Tfh program by inducing chemokine receptor CXCR5, CXCR4 expression, and suppressing CCR7, PSGL1, IL-2 signal pathway, as well as Th1, Th17 differentiation. Upon interacting with cognate B cells at T-B border, CXCR5+ T cells begin to increase Bcl6 expression, which eventually facilitates Tfh maturation in B follicles and GC formation.
Methods
Mice
Mice were housed in specific pathogen-free animal facility at MD Anderson Cancer Center and Tsinghua University, and animals were used according to the protocols approved by Institutional Animal Care and Use Committee. 6–8-week-old mice were used for all experiments, and were randomly allocated into treatment group. C57BL/6 mice were from National Cancer Institute. ETR2-Cre, CD4-Cre, OT-II, B6SJL, TCRβ−/−, Rag1−/−, and Batf−/− mice were from Jackson Laboratories. Stat5−/− 15, Id3−/− 29, Bcl6-RFP 4 and Bcl6−/− 8 mice were previously described.
The Ascl2fl/fl mice were generated previously and have been backcrossed with C57BL/6 for at least six generations5. Ascl2fl/fl mice were bred with CD4-Cre mice to generate Ascl2fl/fl/CD4-Cre and control mice Ascl2+/+/CD4-Cre. Ascl2fl/fl mice were crossed with ETR2-Cre mice to generate Ascl2fl/fl/ETR2-Cre and control mice Ascl2+/+/ETR2-Cre. Ascl2 deletion in Ascl2fl/fl/ETR2-Cre cells was achieved by administering 200μl tamoxifen (5mg/ml) in sunflower seed oil subcutaneously or intraperitoneally (i.p) every other day for total five days.
KLH, NP-KLH, OVA and NP-OVA Immunization
Mice and their wild-type controls (6–8 wk old; three per group) were immunized with antigen (0.5 mg/ml) emulsified in CFA (0.5 mg/ml) subcutaneously (100 μl each mouse,). After immunization, these mice were sacrificed and analyzed individually. Germinal center B cells were determined by staining with FITC-labeled anti-GL7, PE-labeled anti-FAS and PerCP-labeled anti-B220 mAb (Pharmingen). Tfh cells were determined by staining with PerCP-labeled anti-CD4 mAb and biotinylated anti-CXCR5 mAb (Pharmingen), followed by APC-labeled streptavidin (Jackson ImmunoResearch Laboratories, Inc.) and surface staining by PE-labeled anti-PD1 mAb or intracellular staining by PE-labeled anti-Bcl6 mAb (Pharmingen). Sera from immunized mice were collected, and antigen-specific IgM, IgA, IgG1, IgG3, IgG2a and Ig2b antibodies were measured by using ELISA. Briefly, isotype-specific antibodies to NP were measured in plate coated with NP4-BSA with SBA Clonotying System (Southern Biotech). Titers were presented as the maximum serum dilution exceeding 1.5 fold above the average background. KLH- or OVA-specific titers were measured in a 3-fold serial dilution onto plates pre-coated with 100 μg/ml KLH or OVA.
Influenza virus infection
Influenza virus A/Puerto Rico/8 (PR8, H1N1) was purchased from Charles River Laboratories. 6–8 -week-old Ascl2fl/fl/CD4-Cre mice (3–5 mice per group) were anesthetized by intraperitoneal injection with ketamine and infected intranasally with 0.5 LD50 of PR8 influenza virus 27. Mice were monitored daily, and weight loss was recorded. To analyze influenza virus-specific germinal center (GC) responses, Tfh and GC B cells from lung mediastinal lymph nodes and spleens were analyzed by flow cytometry. BAL fluid and sera were collected from virus-infected mice on 9 dpi. Virus-specific IgG antibodies were measured by using ELISA. Briefly, serum samples were added in a 3-fold serial dilution onto plates pre-coated with heat-inactivated virus. Bound antibodies were detected by the incubation of horseradish peroxidase-conjugated anti–mouse total IgG (1:10,000; Southern Biotech) antibodies. Lung viral titer were monitored by examining influenza virus hemagglutinin (HA) and neuraminidase (NA) gene expression using real time RT-PCR as previously described 27.
Retroviral transduction and T cell differentiation
Naïve CD4+CD25−CD44lowCD62Lhigh T cells from Ascl2fl/fl/OT-II, OT-II or C57BL6 mice were FACS-sorted and activated with plate-bound anti-CD3e (clone, 2C11) and anti-CD28 (clone, 37.51) under neutral conditions. 36 hours after activation, cells were infected by retroviruses Ascl2-RV-GFP, Bcl6-RV-GFP, Batf-RV-GFP, Maf-RV-GFP, Id3-RV- GFP, Cre-RV-GFP, E47-RV-CFP (a kind gift from Dr. Yuan Zhuang) or control empty vector (empty-RV-GFP or empty-RV-CFP). One day after infection, GFP cells were FACS sorted for adoptive transfer, or followed by re-stimulation with pre-coated anti-CD3e. Also, virus-infected cell were polarized under Th1 (IL-12, anti-IL-4), Th2 (IL-4, anti-IFNγ), Th17 (TGF-β, IL-6, IL-23, anti-IL-4, anti-IFN-γ) and Treg (TGF-β) conditions. Four days after culture, cells were re-stimulated with PMA and ionomycin in the presence of Golgi-stop for 4 h, after which IFNγ-, IL-4/IL-5/IL-13-, IL-17-, and Foxp3-expressing cells were analyzed using intracellular staining, respectively. Cytokines including IL-4, IL-5, and IL-13 from Ascl2-RV-GFP-infected Th2 cells were measured by ELISA. Cell transfer numbers for different cells were as follows: 1×105 for GFP+T cells; 8×106 for bone marrow cells.
Mixed chimeric bone marrow mice
To generate mixed bone marrow chimera, T cell-depleted bone marrow cells were obtained from Ascl2fl/fl/CD4-Cre or Ascl2fl/fl/ERT2-Cre and their respective congenic WT (CD45.1+ CD45.2+) mice, and mixed at ratio of 1:1 before being transferred into irradiated Rag1−/− (750 rad). Six to eight weeks later, the reconstituted mice were subject to immunization and analysis as above.
Microarray and ChIP-Seq
Total cellular RNA was extracted from cells transduced with vector or Ascl2-expressing retrovirus that were purified by their expression of GFP marker with TRIzol reagent (Invitrogen). DNA microarray labeling and analysis were performed by microarray core at the Institute for System Biology. Approximately 10ug RNA was labeled and hybridized to GeneChip Mouse Genome 430.2 arrays (Affymetrix) according to the manufacturer’s protocols. Expression values were defined with GeneChip Operating Software (GCOS) software.
ChIP-Seq was performed as described previously 28. Briefly, Sorted T cells were fixed by 1% paraformaldehyde, and followed with digestion with Mnase cocktail (Active motif). Chromatin from 5×106 cells was used for each ChIP experiment. Antibodies against H3K4me3 (Cat# 07-473, Millipore), H3K27me3 (Cat# 07-449, Millipore), and Ascl2 mAb (clone, 7E2, Millipore) were used. The pull-down DNA fragments were blunt-ended ligated with Solexa adaptors, and amplified for sequencing. The DNA fragments were sequenced with Illumina 1G Analyzer at the Institute for System Biology. Output of the Solexa Analysis Pipeline was converted to browser-extensible data (BED) files, and the data were viewed in the UCSC genome browser.
Immunohistochemistry
The protocol for immunohistochemical staining was as described previously30. Staining reagents included biotinylated PNA (Vector), AlexaFluor 647 anti-B220, biotinylated anti-IgD, AlexaFluor 488 anti-CD45.2 (Biolegend), and streptavidin AlexaFluor 568 (Invitrogen). All stained slides were mounted with ProlongGold antifade reagents (Invitrogen) and examined with an Olympus FV1000 confocal system.
Real-time RT-PCR analysis
Reactive tonsils were obtained from children undergoing elective tonsillectomy after informed consent on an institutional review board-approved protocol 20. Total RNA was extracted with Trizol reagent (Invitrogen). Oligo (dT), and MMLV reverse transcriptase (Invitrogen) were used to generate cDNA. Gene expressions were examined by iQ™ SYBR real-time kit (Bio-Rad Laboratories). The data were normalized to a reference gene beta-actin. The primer pairs for real time RT-PCR analysis of Bcl6, Batf, CXCR5, IL-21, IL-4, Gata3, IFN-γ, T-bet, Il-17A, and RORγ were previously described 4. Primer pair for detection of mouse Ascl2 is: forward, 5′-CGCTGCCCAGACTCATGCCC-3′; reverse, 5′-GCTTTACGCGGTTGCGCTCG -3′.
Statistics
Unless specifically indicated, otherwise, comparison between two different groups were done with unpaired two-tailed Student’s t-tests or two-way ANOVA. All P-values below 0.05 considered significant. Statistical analysis was performed with Graphpad Prism 6.
Supplementary Material
Acknowledgments
We thank Drs. Jeffery A. Whitsett and James P. Bridges at the University of Cincinnati for their provision of Ascl2 conditional knockout mice, Danielle Yi for help in ChIP-Seq and microarray analysis, Dr. Riccardo Dalla-Favera for Bcl6−/− mice, Dr. Hongbo Hu for histochemistry staining and the Dong lab members for their help. The work was supported in part by grant from NIH (AI106654 to C.D.), intramural research program (NIDCR to W.C. and H.N.), a NIH Lymphoma SPORE (to X.L.), Odyssey fellowship from MD Anderson Cancer Center (to X.L. and B.Z.), Chinese Ministry of Science and Technology “973” program grant (No. 2014CB542501 and 2012CB910402) and National Natural Science Foundation of China grant (No. 81361120397 to H.Q.).
Footnotes
Author contributions X.L. designed and performed the experiments, and wrote the manuscript; X.C., B.Z., A.W., X.W., R.I.N., F.C., S.S.N. and H.Q. contributed to the performance of the experiments; X.Y., P.C., Q.T. and L.W. performed microarray, ChIP product sequencing and data analysis; L.G.F., H.N., W.C. and H.C. provided important mouse strains; and C.D. designed and directed the research.
Microarray and ChIP-Seq data have been deposited at GenBank under accession codes ().
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Liu X, Nurieva RI, Dong C. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunological reviews. 2013;252:139–145. doi: 10.1111/imr.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Xu H, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. The Journal of experimental medicine. 2012;209:1841–1852. S1841–1824. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Flier LG, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 6.Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 7.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nature immunology. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. The Journal of experimental medicine. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature immunology. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon H, et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurieva RI, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. The Journal of biological chemistry. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nature medicine. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballesteros-Tato A, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SK, et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. The Journal of experimental medicine. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen CD, et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nature immunology. 2004;5:943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 20.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang HE, et al. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nature immunology. 2012;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki M, et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nature immunology. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murre C, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 24.Hiramatsu Y, et al. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. Journal of leukocyte biology. 2010;87:703–712. doi: 10.1189/jlb.0909639. [DOI] [PubMed] [Google Scholar]
- 25.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo JK, Fish EN, Braciale TJ. LAPCs promote follicular helper T cell differentiation of Ag-primed CD4+ T cells during respiratory virus infection. The Journal of experimental medicine. 2012;209:1853–1867. doi: 10.1084/jem.20112256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. MKP-1 is necessary for T cell activation and function. The Journal of biological chemistry. 2009;284:30815–30824. doi: 10.1074/jbc.M109.052472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama T, et al. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nature immunology. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.