Abstract
Herbal medicines and traditionally used medicinal plants present an untapped potential for novel molecular target discovery using systems science and OMICS biotechnology driven strategies. Since up to 40% of the world's poor people have no access to government health services, traditional and folk medicines are often the only therapeutics available to them. In this vein, North East (NE) India is recognized for its rich bioresources. As part of the Indo-Burma hotspot, it is regarded as an epicenter of biodiversity for several plants having myriad traditional uses, including medicinal use. However, the improvement of these valuable bioresources through molecular breeding strategies, for example, using genic microsatellites or Simple Sequence Repeats (SSRs) or Expressed Sequence Tags (ESTs)-derived SSRs has not been fully utilized in large scale to date. In this study, we identified a total of 47,700 microsatellites from 109,609 ESTs of 11 medicinal plants (pineapple, papaya, noyontara, bitter orange, bermuda brass, ratalu, barbados nut, mango, mulberry, lotus, and guduchi) having proven antidiabetic properties. A total of 58,159 primer pairs were designed for the non-redundant 8060 SSR-positive ESTs and putative functions were assigned to 4483 unique contigs. Among the identified microsatellites, excluding mononucleotide repeats, di-/trinucleotides are predominant, among which repeat motifs of AG/CT and AAG/CTT were most abundant. Similarity search of SSR containing ESTs and antidiabetic gene sequences revealed 11 microsatellites linked to antidiabetic genes in five plants. GO term enrichment analysis revealed a total of 80 enriched GO terms widely distributed in 53 biological processes, 17 molecular functions, and 10 cellular components associated with the 11 markers. The present study therefore provides concrete insights into the frequency and distribution of SSRs in important medicinal resources. The microsatellite markers reported here markedly add to the genetic stock for cross transferability in these plants and the literature on biomarkers and novel drug discovery for common chronic diseases such as diabetes.
Introduction
Nature has produced a significant number of medicinal plants and herbs that contain numerous active ingredients and complex molecules that have yet to be scientifically identified and analyzed. Medicinal plants have long been considered as a healthy source of life for all human beings. In developing countries, including India, over 80% of the population depends directly on plants for their medicinal and nutritional requirements (WHO, 2002). In fact, as up to 40% of the world's poor people have no access to government health services, under such circumstances traditional and folk medicine is the only medicine available to them.
The northeastern (NE) region of India is blessed with a wide range of physiographic and ecoclimatic conditions, and most importantly is the geographical gateway for much of India's endemic flora. In addition, the region represents a vital component of the Indo-Myanmar biodiversity hotspot, among the 25 global biodiversity hotspots recognized to date across the globe. Many precious medicinal plants are closely associated socially as well as culturally. In addition, these plants are widely used for nutritional purpose by the local inhabitants and indigenous communities of this region. Many plants with potential medicinal value also used for various domestic purposes in daily lives are not listed under the medicinal plant category. An extensive literature survey revealed 11 plants having proven antidiabetic properties that are largely distributed across the geographical niche of NE India. The plants include: Ananas comosus (pineapple), Carica papaya (papaya), Catharanthus roseus (noyontara), Citrus aurantium (bitter orange), Cynodon dactylon (bermuda grass), Dioscorea alata (ratalu), Jatropha curcas (barbados nut), Mangifera indica (mango), Morus indica (mulberry), Nelumbo nucifera (lotus), and Tinospora cordifolia (guduchi) (Aderibigbe et al., 1999; Xie et al., 2005; Huralikuppi et al., 2006; Jarald et al., 2008; Sharma et al., 2008; Mishra et al., 2010; Rasineni et al., 2010; Kumar et al., 2010; Maithili et al., 2011; Juarez-Rojop et al., 2012; Sangeetha et al., 2013). Although much progress have been made in recent years in many medicinal plants, the study of genetic potential and improvement through molecular breeding has not been attempted to date for the above mentioned plants.
The ability to investigate DNA sequences directly became available to population biologists only during the late 1970s. Molecular markers have been demonstrated as potential tools to detect genetic diversity and to aid the management of plant genetic resources (Ford-Lloyd et al., 1997; Virk et al., 2000; Song et al., 2003). Currently, a number of DNA-based techniques are widely used for analyzing the genetic diversity in natural populations. These include (i) restriction fragment length polymorphism (RFLP; Botstein et al., 1980), (ii) polymerase chain reaction (PCR; Mullis and Faloona, 1987), and its derivatives, termed as random amplified polymorphic DNA (RAPD; Williams et al., 1990); AP-PCR (Welsh and McClelland, 1990), and (iii) a hybrid of both the above techniques named amplification fragment length polymorphism (AFLP) (Vos et al., 1995). The microsatellite markers are the most powerful derivation of PCR technology that are being widely used in marker assisted breeding programmes. Abundantly dispersed in genome (i.e., both coding and noncoding regions of DNA sequences), SSRs (simple sequence repeats or microsatellites) are short repeat motifs (Toth et al., 2000; Katti et al., 2001; Gupta et al., 2007) that show a high level of length polymorphism due to insertion or deletion mutations in one or more repeats (Tautz and Renz, 1984). As compared to other DNA based markers, SSRs are more convenient, simple, stable, multiallelic, reproducible, and polymorphic, which make them the markers of choice in plant genetics and breeding.
Expressed sequence tags (ESTs) represent short, unedited, randomly selected single-pass sequence reads derived from cDNA libraries and serve as the main source for in silico identification of microsatellites. Because of the utility, speed with which ESTs are generated, and the low cost associated with the second generation sequencing technologies, ESTs became point of attention to many scientists. Importantly, most of the projects result in hundreds or thousands of ESTs that are released to the public domain for unrestricted use by the scientific community across the globe. ESTs are highly error prone and require several computational methods for pre-processing, clustering, assembly, and annotation to yield biological information.
Microsatellites developed from ESTs, popularly known as EST-SSRs or genic SSRs, which correspond to functional molecular markers, can be obtained from database searches and other in silico methodologies. With the advent of high-throughput next generation sequencing technologies in recent years, focus on functional genomics revolutionized in generation ESTs in large scale from model and nonmodel organisms, including important medicinal plants. In this scenario, evolving high throughput bioinformatics tools have complemented in mining microsatellites from large scale ESTs in a time and cost effective manner (Varshney et al., 2002). Because of the above mentioned advantages, genic SSRs have been identified and extensively studied in most of the model plants and major crop species, such as Arabidopsis, tobacco, rice, maize, wheat, poplar, pineapple, peach, and others (Varshney et al., 2005, Victoria et al., 2011, Duran et al., 2013).
The traditional methods of developing simple sequence repeat (SSR) markers are usually time consuming, and cost- and labor-intensive. SSR markers can be rapidly and cheaply identified through computational methods from various public domains. Not only is the in silico approach time- and cost effective, but also it allows for the discovery of SSRs from ESTs that represent only the coding region in the genome (Scott et al., 2000; Kantety et al., 2002; Varshney et al., 2002). Recent trends in marker development and studies in plants are more towards gene-specific markers rather than random DNA markers, and microsatellite markers are of great importance in identification of genes (Zhao et al., 2012). Additionally, bioinformatics tools also supplement existing approaches by automating the task of SSR identification from available DNA sequences.
The neglected and the under-utilized status of these locally important crops indicate a risk of disappearance of important plant material without knowing their exact genetic background. One of the important factors restricting their large-scale production and development of better varieties is that very little information is available about their genetic diversity, inter- and intraspecific variability, and genetic relationship among these species. And as such, availability of informative marker is also very scanty. Therefore, attempts to analyze possible untapped genetic diversity and development of marker become extremely essential for breeding and crop improvement. SSR marker reported in the present study shall act as a resource bank for analyzing genetic diversity in various accessions, and the SSR marker linked to the antidiabetic gene shall act as a benchmark for further improvement of the crop using marker assisted breeding program.
Materials and Methods
Data source and EST assembly
Sequence data were collected from the public domain ‘dbEST’ at NCBI (National Center for Biotechnology Information) website (http://www.ncbi.nlm.nih.gov) (Boguski et al., 1993). EST sequences were collected for 11 plants: 5941 for Ananas comosus, 77393 for Carica papaya, 20168 for Catharanthus roseus, 14584 for Citrus aurantium, 20497 for Cynodon dactylon, 44134 for Dioscorea alata, 46862 for Jatropha curcas, 1665 for Mangifera indica, 4526 for Morus indica, 2207 for Nelumbo nucifera, and 5498 for Tinospora cordifolia. For the pre-processing and assembly steps, an in-house software pipeline ESMP (Sarmah et al., 2012) and EGassembler (Masoudi-Nejad et al., 2006) were used. The updated vector sequences were downloaded from the ‘UniVec’ database (ftp://ftp.ncbi.nih.gov/pub/UniVec) of NCBI and checked against the downloaded ESTs. The sequences were cleaned to remove the vector contamination using cross_match with min-match and min-score value of 20. The poly-A trimming was done with the Trimest tool from EMBOSS. The value for minimum length and mismatch in case of Trimest was used as 4 and 1 respectively.
The high quality sequences obtained after pre-processing were assembled using CAP3, which resulted in contigs and singlets. The parameters used in assembly using CAP3 (Huang and Madan, 1999) are listed in Supplementary Material (supplementary material is available online at www.liebertpub.com. Raw data are available from the authors). The assembly in case of Carica papaya was done with the help of EGassembler due to the sequence size limitation in CAP3. EGassembler uses CAP3 in an iterative process so that it takes sequence size of more than 50000 bp. In this study the stand-alone processing option was selected in EGassembler, where we allotted 8 CPUs for the analysis.
SSR detection and primer designing
Microsatellite repeats were obtained using MISA (MIcroSAtellite identification tool). MISA is a freely available Perl script, which was downloaded from the Web (http://pgrc.ipk-gatersleben.de/misa/misa.html). Along with “misa.pl” another file viz., “misa.ini,” which contains the search parameters, was also downloaded. The following search parameters were employed in the detection of SSR in MISA: the maximum difference between two SSRs that interrupts to form a compound microsatellite was 100 and the minimum length parameter for the repeated units (unit size/minimum number of repeats): at least ten mononucleotides (1/10); at least six dinucleotides (2/6); at least five trinucleotides (3/5); and five tetranucleotides (4/5), pentanucleotides (5/5), and hexanucleotides (6/5). Primer pairs were designed for the SSRs containing singlets and contigs with the help of primer3 tool (Rozen and Skaletsky, 2000). Perl scripts available on the web (http://pgrc.ipk-gatersleben.de/misa/primer3.html) enabled us simultaneously to use Primer3 for designing primers only for SSR-ESTs and sorting out the results of primer pairs for further studies. For designing primer pairs to flank the EST-SSRs, primer-specificity was optimized for primers greater than 10 bp on either side of the identified SSR and maximum product size of 100–280 bp. The following parameters were employed to design primer pairs in Primer3: optimal size of primer was set to 18 bp with maximum up to 27, melting temperature of 55°C with a minimum of 50°C and maximum of 70°C, and a maximum GC content of 65%.
Functional annotation and GO term analysis
The EST contig sequences containing microsatellites were assigned to putative functions with the help of BLASTx. The nonredundant (nr) protein sequences were downloaded from ftp://ftp.ncbi.nlm.nih.gov/blast/db and configured on the local server with the following command: ‘formatdb −p T −i nr.fasta −n nr_db’ where ‘nrq.fasta’ represents the nonredundant protein sequences from nr database of NCBI. SSR containing contigs were given as input to run BLASTx with the following command: ‘blastx −query <input fasta sequence> -db nr_db −outfmt 6 −out <result file name>’, where all the other parameters were kept as default. InterProScan at EBI is an important tool which was used for analyzing the SSR-contigs to get the functional domain markers (FDMs) (Quevillon et al., 2005). GO terms were assigned to the SSR-contigs using QuickGo (http://www.ebi.ac.uk/QuickGO) at EBI. Custom in-house perl scripts (as listed in Supplementary Material) were used for preparation of the input files for all the above tools.
Prediction of amino acid content for SSR loci
The amino acid content for SSR loci was detected in all the SSR-EST sequences of 11 species using MEGA v. 5.2 and the mean value calculated for each amino acid was extensively studied incorporating Boxplot analysis in R.
Analysis of antidiabetic SSR markers
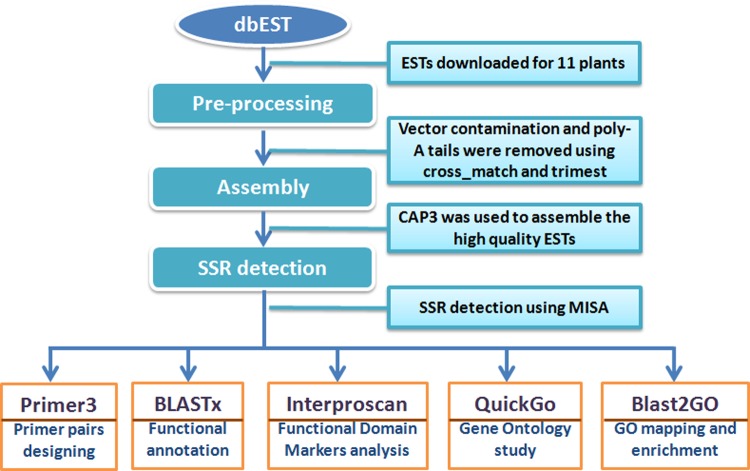
Antidiabetic compound information in several plants was available in a previous report (Perez et al., 1998). The gene sequences involved with diabetes were curated manually from GenBank of NCBI with reference to the antidiabetic compounds. Here, we have employed locally installed BLASTn program using the SSR containing EST sequences and the gene responsible for antidiabetic compounds. The antidiabetic genes were formatted to create a BLAST database, and the SSR-ESTs were used as query sequences. From the BLAST results, the SSR-EST sequences having significant matches with the genes responsible for antidiabetic compounds were collected and subjected to Blast2go V.2.7.0 to map gene ontology (GO) terms and enrichment analysis. Two datasets were prepared as test and reference for the SSR-ESTs having BLAST matches and the antidiabetic gene sequences, respectively. The enrichment analysis (Fisher's Exact Test) was performed between the two datasets at p<1.5. The GO terms were reduced to more specific terms using the respective option in Blast2GO. An interactive network graph was constructed for the enriched GO terms into three different functional categories [i.e., biological process (BP), molecular function (MF), and cellular component (CC)] using Gephi (https://gephi.org/). The overall workflow of present work is summarized in Figure 1.
FIG. 1.
Work flowchart for the EST-SSR mining. The description of total in silico analysis (i.e., SSR detection, functional annotation, gene ontology study, and GO enrichment analysis).
Results
In the present study, ESTs of 11 important plants having antidiabetic property were screened for identification of genic microsatellites and functional characterization. Initially the raw sequences obtained from public domains were preprocessed and subsequently subjected to sequence assembly programme CAP3. The number of SSRs obtained from assembled (i.e., contigs and siglets) sequences of all the eleven plants and related information has been summarized in Table 1.
Table 1.
Statistical Summary of the SSRs Identified from 11 Medicinal Plants of North-East India with Potent Antidiabetic Properties
| Plant common name | Plant botanical name | No. of sequences examined | No. of SSRs | SSR frequency | No. of SSR-ESTs | No. of cSSRs |
|---|---|---|---|---|---|---|
| Pineapple | Ananas comosus | 3647 | 1199 | 1 SSR/2.4 kb | 898 | 182 |
| Papaya | Carica papaya | 31554 | 19662 | 1 SSR/1.3 kb | 8536 | 8979 |
| Noyontara | Catharanthus roseus | 8559 | 1753 | 1 SSR/2.7 kb | 1401 | 229 |
| Bitter orange | Citrus aurantium | 11934 | 5924 | 1 SSR/1.6 kb | 3926 | 1149 |
| Bermuda brass | Cynodon dactylon | 11953 | 9310 | 1 SSR/0.8 kb | 5508 | 3046 |
| Ratalu | Dioscorea alata | 22583 | 4711 | 1 SSR/2.5 kb | 3608 | 730 |
| Barbados nut | Jatropha curcas | 11334 | 2783 | 1 SSR/2.6 kb | 2125 | 410 |
| Mango | Mangifera indica | 1241 | 125 | 1 SSR/5.3 kb | 100 | 10 |
| Mulberry | Morus indica | 2455 | 434 | 1 SSR/2.2 kb | 389 | 26 |
| Lotus | Nelumbo nucifera | 1634 | 1008 | 1 SSR/0.9 kb | 707 | 155 |
| Guduchi | Tinospora cordifolia | 2715 | 791 | 1 SSR/2.6 kb | 616 | 97 |
SSR detection and primer designing
The SSR survey detected a total of 47,700 numbers of SSRs in 109,609 numbers of ESTs from 11 plants. The density of SSRs in total was found to be 1 SSR per 1.54 kb, and 25.38 % of the total ESTs contain these SSRs. The SSR density ranged from 1 SSR/5.3 kb in Mangifera indica to 1 SSR/0.8 kb in Cynodon dactylon. A total of 15,013 numbers of SSRs were found in the compound formation. The highest numbers of compound microsatellites (cSSRs) were present in Carica papaya (i.e., 8979) and the lowest in Mangifera indica (i.e., 10) as shown in Table 1.
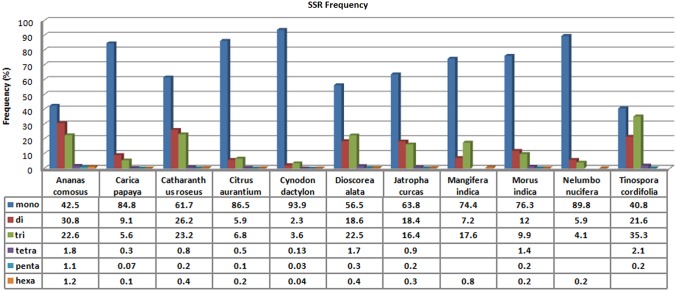
The frequency of SSRs across the EST sequences was analyzed very carefully. Generally, mononucleotides were found to be the abundant repeat type. Apart from mononucleotide repeats (MNR), dinucleotide repeats (DNR) and trinucleotide repeats (TNR) were also present abundantly. The density of DNRs was found to be 30.8% and 2.3% in Ananas comosus and Cynodon dactylon respectively and TNRs was 35.3% and 3.6% in Tinospora cordifolia and Cynodon dactylon, respectively. The hexanucleotide repeats were completely absent in Tinospora cordifolia and the tetra- and pentanucleotide repeats were completely absent in both Mangifera indica and Nelumbo nucifera (Fig. 2).
FIG. 2.
Frequency of the individual repeat types in the SSRs obtained from MISA analysis.
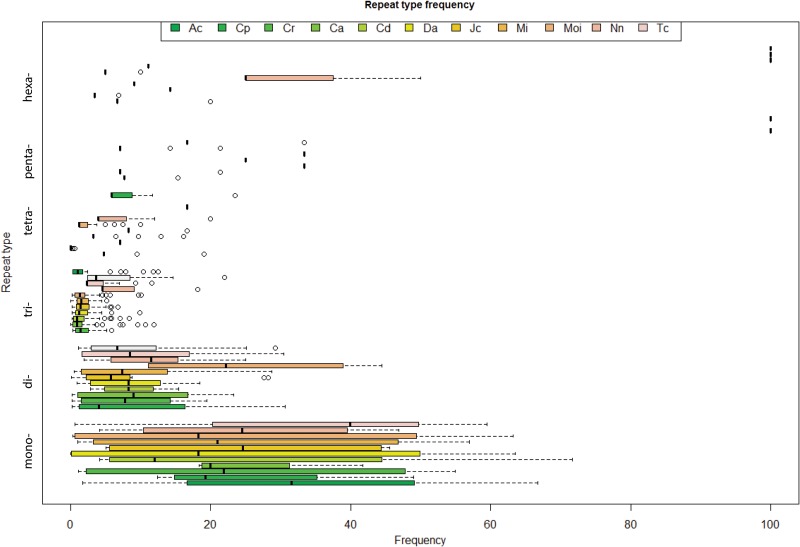
The most frequent classified repeat types were A/T in mononucleotides, AG/CT in dinucleotides, and AAG/CTT in trinucleotides. The exceptions were found in Dioscorea alata in case of dinucleotides, where AT/AT was the abundant type, and in case of trinucleotides, AGC/CTG and CCG/CGG were the exceptions found in Citrus aurantium and Cynodon dactylon, respectively. The frequency of A/T was found to be 60.9% and 99.7% in Citrus aurantium and Dioscorea alata, respectively, the frequency of AG/CT was 50% and 88.9% in Citrus aurantium and Mangifera indica, respectively, and the frequency of AAG/CTT was found to be 19.9% and 55.6% in Ananas comosus and Tinospora cordifolia, respectively (Table 2). A boxplot plotted using R and Bioconductor describes the distribution of the unit repeat type SSRs across the 11 plant species (Fig. 3).
Table 2.
Summary of the Each Classified SSR Motif Types with Their Frequency in 11 Medicinal Plants with Potent Antidiabetic Properties
| Plant name | Mononucleotides | Mononucleotide frequency | Dinucleotides | Dinucleotide frequency | Trinucleotides | Trinucleotide frequency |
|---|---|---|---|---|---|---|
| Ananas comosus | A/T | 98.2 | AG/CT | 84.6 | AAG/CTT | 19.9 |
| Carica papaya | A/T | 70.4 | AG/CT | 57.4 | AAG/CTT | 51.5 |
| Catharanthus roseus | A/T | 95.5 | AG/CT | 63.6 | AAG/CTT | 36.2 |
| Citrus aurantium | A/T | 60.9 | AG/CT | 50 | AGC/CTG | 24.2 |
| Cynodon dactylon | A/T | 88.9 | AG/CT | 60.2 | CCG/CGG | 27.3 |
| Dioscorea alata | A/T | 99.7 | AT/AT | 55.9 | AAG/CTT | 23 |
| Jatropha curcas | A/T | 88.7 | AG/CT | 79.4 | AAG/CTT | 39.8 |
| Mangifera indica | A/T | 93.5 | AG/CT | 88.9 | AAG/CTT | 45.5 |
| Morus indica | A/T | 98.8 | AG/CT | 62.2 | AAG/CTT | 37.2 |
| Nelumbo nucifera | A/T | 63.5 | AG/CT | 84.7 | AAG/CTT | 39.1 |
| Tinospora cordifolia | A/T | 99.4 | AG/CT | 67.3 | AAG/CTT | 55.6 |
FIG. 3.
Box plot to illustrate distribution of the unit repeat type SSRs across the 11 plant species.
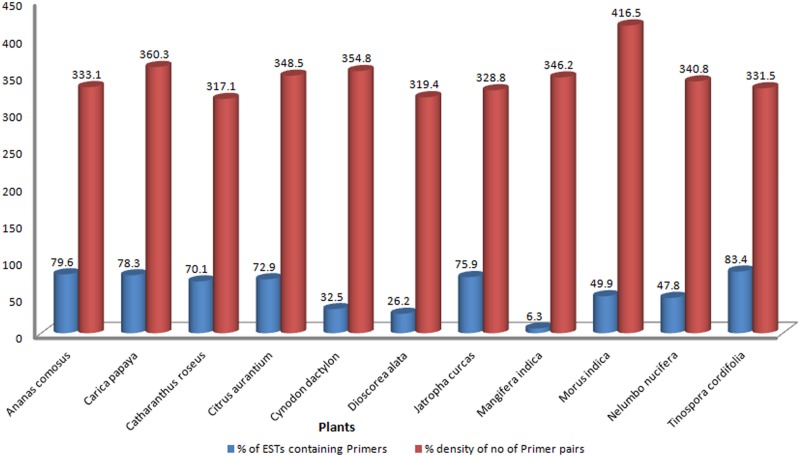
Primer pairs were not possible to design for all the SSR containing ESTs with the optimum parameter values. However, a total of 57957 numbers of primer pairs were designed from 16724 unique ESTs (as listed in Supplementary Table S1). Highest percentage (83.4%) was found in Tinospora cordifolia out of all the SSR-ESTs and the lowest was found in case of Mangifera indica with a percentage of only 6.3. The output of primer3 tool was analyzed, which provided the percentage density of number of primer pairs designed viz., the number of primer pairs with respect to 100 of unique SSR-ESTs for which primer designing was possible. The percentage ranged from 317.1% to 416.5% in Catharanthus roseus and Morus indica, respectively (Fig. 4).
FIG. 4.
Primer statistics of SSR containing ESTs.
Functional annotation
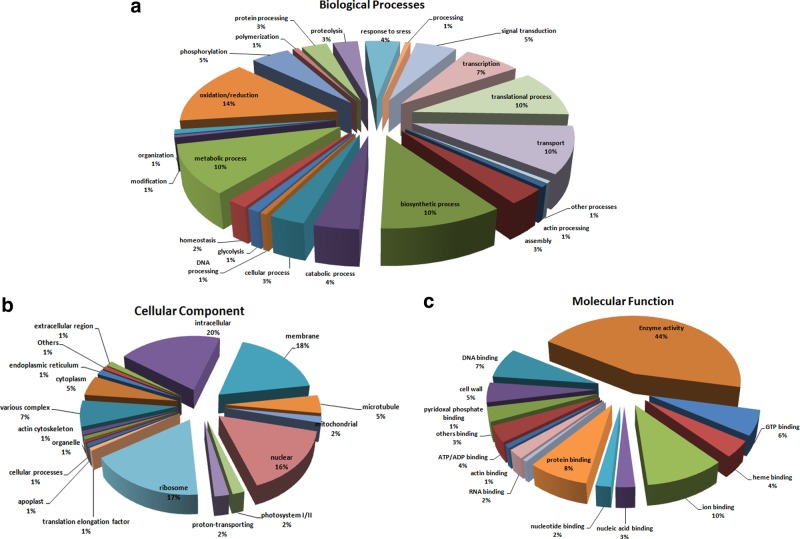
BLASTx identified significant matches for a total of 3736 SSR containing contigs in 11 plant species (as listed in Supplementary Table S2). The average range percentage (%) of sequence identity was 79.3% to 100%. Highest numbers of matches (1472) were found in case of Cynodon_dactylon, whereas in Mangifera indica, the least number of matches (i.e., 7) was found. The InterProScan results can be summarized with the obtained 21,568 numbers of functional domain markers (FDMs) for a total of 4728 unique contigs as listed in Supplementary Table S3. The gene ontology (GO) term annotation provides a common terminology for the functional description of transcripts comprised of three sub-ontologies: biological process (BP), molecular function (MF), and cellular component (CC). The GO term annotation revealed a total of 9634, 4740, and 18611 numbers of biological processes, cellular components, and molecular functions (Fig. 5). Several important processes related to oxidation, reduction, metabolic, biosynthesis, transport, translation etc. were assigned to the SSR containing contigs. In case of cellular components, the intracellular, membrane, ribosomal, and nuclear components were abundant, and so many others were found for all the SSR-contigs analyzed. Enzyme activity was found to be the most frequent with 44% of the total molecular functions detected. In addition, the other molecular functions were mostly related to binding functions such as ion binding and protein binding.
FIG. 5.
Statistics of GO annotation of the SSR-contigs; (a) biological processes, (b) cellular components, and (c) molecular functions.
Analysis of antidiabetic SSR markers
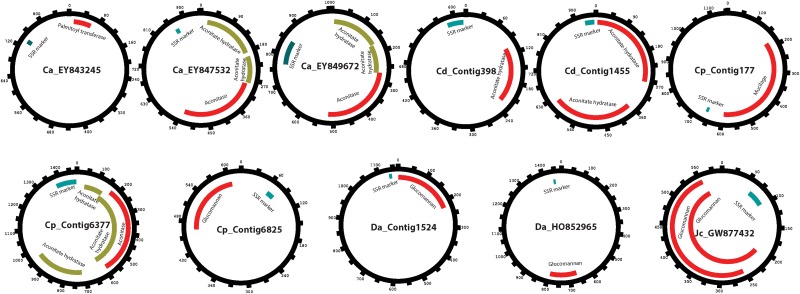
A total of 134 gene sequences involved in the synthesis of antidiabetic compounds were collected manually from available literature and GenBank. The BLASTn between SSR containing ESTs and antidiabetic genes identified a total of 11 unique SSR-ESTs in 5 out of all 11 plants examined, which matched significantly with 30 antidiabetic gene sequences. Figure 6 illustrates the positions of SSR markers, antidiabetic gene similarity region (the redundant regions were avoided in the image). Citrus aurantium, Carica papaya, Cynodon dactylon, Dioscorea alata, and Jatropha curcas were found to contain 3, 3, 2, 2, and 1 antidiabetic SSR-ESTs, respectively. Blast2GO analysis identified 126, 37, and 30 numbers of GO terms associated with several biological processes, molecular functions, and cellular components, respectively. The statistical significance of GO terms associated with the 11 SSR-ESTs (test datasets) having match with the 30 antidiabetic genes (reference datasets) was well explored with the help of enrichment analysis in Blast2go tool. The enrichment analysis at p<1.5 detected total of 80 specific GO terms distributed in to three sub-ontologies (i.e., 53 biological processes, 17 molecular functions and 10 cellular components respectively).
FIG. 6.
Figures showing the positions of SSR markers and regions similar to antidiabetic gene for all 11 SSR-ESTs.
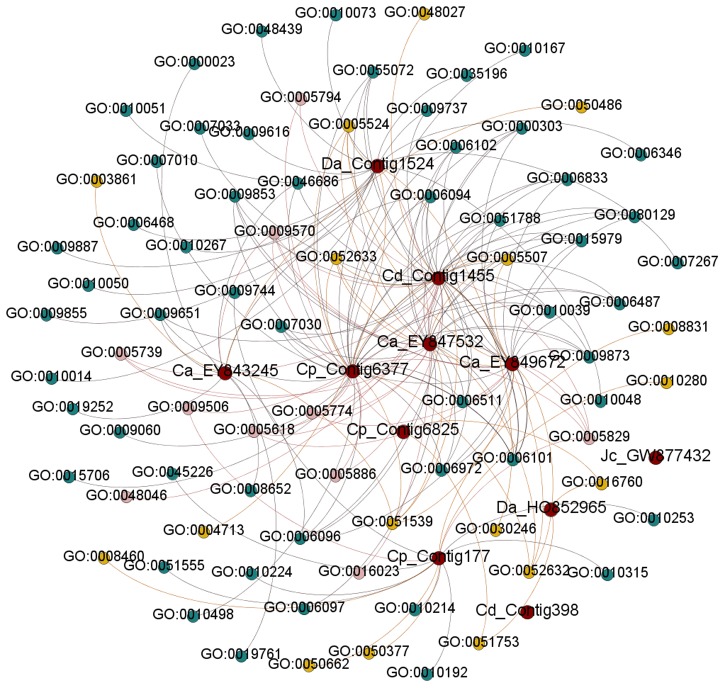
A modular architecture (Fig. 7) was constructed with the help of Gephi which clearly illustrates the networks between the antidiabetic SSR-ESTs and enriched GO terms associated with them. Both the Contig1455 and Contig6377 of Cynodon dactylon and Carica papaya were found to be connected with 37 enriched GO terms, whereas the Contig6825 of Carica papaya was connected with least numbers of GO terms (i.e., 3). The Contig398 of Cynodon dactylon and GW877432 of Jatropha curcas were found to have no association with the enriched GO terms. The most frequent enriched GO terms associated with biological processes were found to be GO: 0006096 and GO: 0006094, and each were mapped to five unique SSR-ESTs. In the case of molecular functions, there were four GO terms (i.e., GO: 0005829, GO: 0009570, GO: 0051539 and GO: 0005524) connected with maximum (i.e., five numbers of unique SSR-ESTs). GO: 0005829 and GO: 0009570 are the two enriched GO terms that belong to cellular component and mapped to five unique SSR-ESTs which is the highest.
FIG. 7.
Modular architecture of the GO terms associated with the anti-diabetic genes. Red, yellow, green, and blue color nodes represent the EST-IDs, biological processes, cellular components, and molecular functions, respectively.
Discussion
Over the last 2 decades, molecular techniques have been widely used for the genetic analysis of various crop plants due to their high efficiency (Ford-Lloyd et al., 1997; Virk et al., 2000; Song et al., 2003). The molecular markers, in particular, genic microsatellite (SSR) markers, are the markers of choice due to their high levels of cross-taxon portability, rapid and less expensive development. The applications of SSR markers have been reported in many crops to scrutinize DNA sequence variation(s). The multiallelic nature, reproducibility, high abundance, and extensive genome coverage make microsatellites a powerful tool to get into the genetic makeup of plants (Tautz and Renz, 1984; Gupta et al., 1996; Toth et al., 2000; Katti et al., 2001; Kantety et al., 2002; Varshney et al., 2002). The traditional methods of developing SSR markers are usually time consuming and labor-intensive processes involving genomic library construction, hybridization with the repeated units of nucleotides, and sequencing of the clones. The computational approach for developing SSR markers from ESTs provides a better platform than the conventional approach. Computational biology, along with high throughput bioinformatics tools (both standalone and web-based), have paved the way to screen publicly available EST and GSS data to design EST-SSR markers on a large scale. The publicly available computational tools and large set of EST data available on the web helps researchers to perform data mining rapidly with ease from their local system at a very low cost.
In this study, we present mining of EST sequences of 11 important plants bearing antidiabetic properties, and abundant in the Northeastern region of India, that include pineapple, papaya, noyontara, bitter orange, bermuda brass, ratalu, barbados nut, mango, mulberry, lotus, and guduchi. Though these plants are mainly known for their uses of fruits and flowers, they are also very effective against diabetes, as evident from literature surveys. But as of now, no efforts have been put into development of efficient molecular markers in these plants, so an attempt was made to derive EST-SSR markers in silico. Our in silico survey of microsatellite from the ESTs of these plants revealed a total 1199, 19662, 1753, 5924, 9310, 4711, 2783, 125, 434, 1008, and 791 numbers of SSRs markers. The densities of SSRs found to be 1 SSR per 2.4 kb, 1.3 kb, 2.7 kb, 1.6 kb, 0.8 kb, 2.5 kb, 2.6 kb, 5.3 kb, 2.2 kb, 0.9 kb, and 2.6 kb, respectively, for the above plants. Previous studies have shown the SSR densities in several other plants as 1 SSR per 2–10 kb (Morgante et al., 2002; Kantety et al., 2002; Varshney et al., 2002; Kumpatla and Mukhopadhyay, 2005; Poncet et al., 2006; Scaglione et al., 2009), which signifies that SSRs detected are more frequent in our study. A total of 58,159 number of primer pairs were possible to design for all the 11 plants (see Results section).
Mononucleotide repeats were the most frequent repeat type in all the plants. Apart from mononucleotide repeats, dinucleotide and trinucleotide-SSR motifs were predominant, whereas tetra-, penta-, and hexanucleotide motifs were detected in much smaller amounts. In Mangifera indica and Nelumbo nucifera, tetranucleotides and pentanucleotides were completely absent, whereas in Tinospora cordifolia, hexanucleotide was completely absent. The dinucleotide repeats (DNRs) are more frequent than the trinucleotide repeats (TNRs) in ESTs of Ananas comosus, Carica papaya, Catharanthus roseus, Jatropha curcas, Morus indica, and Nelumbo nucifera, similar to the previous reports in Actinidia (Fraser et al., 2004), Camellia (Sahu et al., 2012), and Picea species (Rungis et al., 2004). But the frequencies of TNRs were found to be more than DNRs in Citrus aurantium, Cynodon dactylon, Dioscorea alata, Mangifera indica, and Tinospora cordifolia, similar to most previous studies (Cordeiro et al., 2001; Kantety et al., 2002; Varshney et al., 2002; Thiel et al., 2003; Nicot et al., 2004). In most cases, the frequent classified repeat types are found to be A/T in mononucleotides, AG/CT in dinucleotides. The frequently classified repeat types observed in this study perfectly correlate with the earlier studies by Temnykh et al. (2000) and Kantety et al. (2002). In the case of trinucleotides, AAG/CTT is the most frequent classified repeat in nine plants, unlike the previous study by Morgante and co-workers (2002). In Dioscorea alata, AT/AT is the most frequent dinucleotide classified repeat type, which is the same as in nonvascular plants, as found for Physcomitrella patens (Victoria et al., 2011). AGC/CTG and CCG/CGG are the most frequent trinucleotide classified repeat type in Citrus aurantium and Cynodon dactylon, similar to Cymbidium spp. (Moe et al., 2012).
InterProScan helped functional analysis of translated nucleotides by classifying them into families and predicting domains and important sites. Most importantly, the GO annotations revealed a total of 2619 unique GO terms. Gene ontology analysis revealed that majority of SSR loci were involved in 1332 unique molecular functions, 938 biological processes, and 349 cellular components. Among the discrete biological processes inferred through gene ontology analysis, oxidation/reduction, which was the most abundant biological process, accounted for 14%. Other processes that were more frequently noted include metabolic, translational, transport, biosynthesis, and transcription processes, whereas aminoacylation, peptide processing, and RNA processing were found to be very negligible. Of cellular components, intracellular components had the highest frequency of 20%, followed by membrane at 18%, ribosome at 17%, and nuclear at 16%. The less frequent components were associated with cytoskeleton, chromosomes, and lipid storage bodies. A number of important molecular functions were predicted, among which the most abundant types were enzyme activity with a frequency of 44%. The other frequent functions were associated with ion binding, and protein binding, whereas phospholipid binding, ribosome, and transporter activity weree also detected with less frequency.
Previous studies revealed that the frequency of SSRs detected in a survey greatly depends upon the size of the sequence data, SSR parameter, search criteria and the mining tools used (Varshney et al., 2005). Compound microsatellites (cSSRs) have already been proven to be the most important despite their low density in the nucleotide sequences (Bull et al., 1999). In the present study, the percentage of cSSRs was found to be the highest in Carica papaya at 45.7%. In other plants, the percentage of detected cSSRs in the total SSRs were: Cynodon dactylon (32.7%), Citrus aurantium (19.4%), Dioscorea alata (15.5%), Nelumbo nucifera (15.4%), Ananas comosus (15.2%), Jatropha curcas (14.7%), Catharanthus roseus (13.1%), Tinospora cordifolia (12.3%), Mangifera indica (8%) and Morus indica (6%).
The frequencies of cSSRs in most of the plants in the present report are better than the previous studies where the frequencies of cSSRs were 11% in human, 4%–25% in seven fully sequenced species (Maccaca mulatta, Mus musculus, Rattus norvegicus, Ornithorhynchus anatinus, Gallus gallus, Danio rerio, and Drosophila melanogaster), and 1.75%–2.85% in complete Escherichia coli genomes (Weber, 1990; Kofler et al., 2008; Chen et al., 2011). In this study, we identified a total of 7164 cSSRs, where only 6856 unique cSSRs were retained after removal of redundant ones. Out of all the compound microsatellites, 213 were detected more than once. The 6 unique cSSRs: (A)11g(A)12, (A)15g(A)30, (A)22g(A)29, (A)35g(A)29, (A)39g(A)29, and (C)10gg(C)10 were found to have the highest frequency.
The microsatellite markers are the most preferable for studying genetic diversity, linkage map analysis, identification of new genes, and many other important fields. Mainly in plants these markers have shown quite impressive ability to explore the genetic information (Liu et al., 1996; Struss et al., 1998; Ramsay et al., 2000; Ritschel et al., 2004; Varshney et al., 2005; Tang et al., 2006) and to study the cross-transferability across related species (Cordeiro et al., 2001; Morgante et al., 2002; Saha et al., 2006; Wohrmann and Weising, 2011).
In the present study, 11 SSR markers were found to be associated with five unique genes: palmitoyl transferase (carnitine palmitoyltransferase), aconitase, aconitate hydratase, mucilage, and glucomannan (glucomannan 4-beta-mannosyltransferase), which were shown to be the key regulatory elements in curing diabetes. Carnitine palmitoyltransferases 1 and 2 (CPTs; EC: 2.3.1.21) were found to be the key enzymes in the import of long-chain fatty acids into mitochondria and recently have received considerable attention due to their catalytic activity and for the development of novel drugs against diabetes (Rufer et al., 2006). Aconitase is an alternative name for Aconitate hydratase, and both indicate the same enzyme that is associated with diabetes (Boquist et al., 1985). Though the exact connection is still not fully known, studies have shown that lower aconitase activity is responsible for influencing the rates of the activity of the tricarboxylic acid cycle, thereby impairing mitochondrial function and energy metabolism in diabetic hearts (Lin et al., 2009). The low cost, lack of toxicity, easy availability, soothing action, and nonirritant nature of mucilage makes them preferable for semi-synthetic and synthetic excipients (Malviya et al., 2011). The effect of fenugreek seed mucilage on disaccharide activities has been proven to be beneficial to increase specific activities of intestinal disaccharides significantly during diabetes; it is also found to be better than turmeric (Kumar et al., 2005). A direct association is reported of glycemia with glucomannan, a water-soluble polysaccharide used as a dietary fiber (Vuksan et al., 1999). Although at this point, the exact mechanisms underlying the link of antidiabetic properties and SSR could not be proposed, further studies involving microsatellite linked association with trait studies can shed more light into the microsatellite–gene alliance with medicinal importance in plants.
Conclusions
For the first time, we developed and characterized potential microsatellite markers as well as several efficient gene-based SSR markers for the identification of hypoglycemic agents in 11 traditional plants with potential antidiabetic property. The genic microsatellites developed from our study can be considered as an important repository for future improvement of the crop through marker assisted breeding and also for identification of new ideotypes with antidiabetic property. Moreover, the SSR markers have been proven to be cross transferable across species of the same family. Hence, the EST-SSR markers developed in this study would be useful tools for genetic diversity and conservation studies in several plant species.
Supplementary Material
Acknowledgments
The authors acknowldge Department of Electronics and Information Technology (DeitY), Ministry of Communications and Information Technology, Government of India, Bioinformatics Centre, Assam University, Silchar, Biotechnology Information System Network (BTISNet), and Department of Biotechnology (DBT), Government of India, for providing financial assitance and necessary facility to carry out the present research work. We also acknowledge Mr. Abhijit Mitra, Research Scholar, Assam University, Silchar for providing list of antidiabetic plants of North East India.
Author Disclosure Statement
The authors declare that there are no competing financial interests.
References
- Aderibigbe AO, Emudianughe TS, and Lawal BA. (1999). Antihyperglycaemic effect of Mangifera indica in rat. Phytotherapy Res 13, 504–507 [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, and Tolstoshev CM. (1993). dbEST–database for expressed sequence tags. Nature Genet 4, 332–333 [DOI] [PubMed] [Google Scholar]
- Boquist L, Ericsson I, Lorentzon R, and Nelson L. (1985). Alterations in mitochondrial aconitase activity and respiration, and in concentration of citrate in some organs of mice with experimental or genetic diabetes. FEBS Lett 183, 173–176 [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, and Davis RW. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Human Genet 32, 314–331 [PMC free article] [PubMed] [Google Scholar]
- Bull LN, Pabón-Peña CR, and Freimer NB. (1999). Compound microsatellite repeats: Practical and theoretical features. Genome Res 9, 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zeng G, Tan Z, et al. (2011). Compound microsatellites in complete Escherichia coli genomes. FEBS Lett 585, 1072–1076 [DOI] [PubMed] [Google Scholar]
- Cordeiro GM, Casu R, McIntyre CL, Manners JM, and Henry RJ. (2001). Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160, 1115–1123 [DOI] [PubMed] [Google Scholar]
- Duran C, Singhania R, Raman H, Batley J, and Edwards D. (2013). Predicting polymorphic EST-SSRs in silico. Mol Ecol Res 13, 538–545 [DOI] [PubMed] [Google Scholar]
- Ford-Lloyd BV, Jackson MT, and Newbury HJ. (1997). Molecular markers and the management of genetic resources in seed gene banks: A case study of rice. In: Biotechnology and Plant Genetic Resources. Conservation and Use. CAB International, Wallingford, UK, pp. 103–118 [Google Scholar]
- Fraser LG, Harvey CF, Crowhurst RN, and Silva HN. (2004). EST-derived microsatellites from Actinidia species and their potential for mapping. Theoret Appl Genet 108, 1010–1016 [DOI] [PubMed] [Google Scholar]
- Gupta S, Pandey-Rai S, Srivastava S, Naithani SC, Prasad M, and Kumar S. (2007). Construction of genetic linkage map of the medicinal and ornamental plant Catharanthus roseus. J Genet 86, 259–268 [DOI] [PubMed] [Google Scholar]
- Huang X, and Madan A. (1999). CAP3: A DNA sequence assembly program. Genome Res 9, 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huralikuppi JC, Christopher AB, and Stephen PM. (2006). Anti-diabetic effect of Nelumbo nucifera (Gaertn): Part I. Preliminary studies in rabbits. Phytother Res 5, 54–58 [Google Scholar]
- Jarald EE, Joshi SB, and Jain DC. (2008). Antidiabetic activity of aqueous extract and non polysaccharide fraction of Cynodon dactylon Pers. Ind J Exp Biol 46, 660–667 [PubMed] [Google Scholar]
- Juárez-Rojop IE, Díaz-Zagoya JC, Ble-Castillo JL, et al. (2012). Hypoglycemic effect of Carica papaya leaves in streptozotocin-induced diabetic rats. BMC Complement Alt Med 12, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantety RV, La Rota M, Matthews DE, and Sorrells ME. (2002). Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48, 501–510 [DOI] [PubMed] [Google Scholar]
- Katti MV, Ranjekar PK, and Gupta VS. (2001). Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18, 1161–1167 [DOI] [PubMed] [Google Scholar]
- Kofler R, Schlötterer C, Luschützky E, and Lelley T. (2008). Survey of microsatellite clustering in eight fully sequenced species sheds light on the origin of compound microsatellites. BMC Genom 9, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GS, Shetty AK, Sambaiah K, and Salimath PV. (2005). Antidiabetic property of fenugreek seed mucilage and spent turmeric in streptozotocin-induced diabetic rats. Plant Foods Human Nutrit 60, 87–91 [DOI] [PubMed] [Google Scholar]
- Kumar RP, Sujatha D, Saleem TSM, Chetty CM, and Ranganayakulu D. (2010). Potential antidiabetic and antioxidant activities of Morus indica and Asystasia gangetica in alloxan-induced diabetes mellitus. J Pharmacol Exp Therapeut 2, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpatla SP, and Mukhopadhyay S. (2005). Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48, 985–998 [DOI] [PubMed] [Google Scholar]
- Lin G, Brownsey RW, and MacLeod KM. (2009). Regulation of mitochondrial aconitase by phosphorylation in diabetic rat heart. Cell Mol Life Sci 66, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZW, Biyashev RM, and Maroof MA. (1996). Development of simple sequence repeat markers and their integration into a barley linkage map. Theoret Appl Genet 93, 869–876 [DOI] [PubMed] [Google Scholar]
- Maithili V, Dhanabal SP, Mahendran S, and Vadivelan R. (2011). Antidiabetic activity of ethanolic extract of tubers of Dioscorea alata in alloxan induced diabetic rats. Ind J Pharmacol 43, 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malviya R, Srivastava P, and Kulkarni GT. (2011). Applications of mucilages in drug delivery; A review. Adv Biol Res 5, 01–07 [Google Scholar]
- Masoudi-Nejad A, Tonomura K, Kawashima S, et al. (2006). EGassembler: Online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Res 34, W459–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SB, Vijayakumar M, Ojha SK, and Verma A. (2010). Antidiabetic effect of Jatropha curcas L. leaves extract in normal and alloxan-induced diabetic rats. Intl J Pharmaceut Sci 2, 482–487 [Google Scholar]
- Moe KT, Hong WJ, Kwon SW, and Park YJ. (2012). Development of cDNA-derived SSR markers and their efficiency in diversity assessment of Cymbidium accessions. Electron J Biotechnol 15, 1–23 [Google Scholar]
- Morgante M, Hanafey , and Powell W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature Genet 30, 194–200 [DOI] [PubMed] [Google Scholar]
- Mullis KB, and Faloona FA. (1987). Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 155, 335–350 [DOI] [PubMed] [Google Scholar]
- Nicot N, Chiquet V, Gandon B, et al. (2004). Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs). Theoret Appl Genet 109, 800–805 [DOI] [PubMed] [Google Scholar]
- Perez RMG, Zavala MAS, Perez SG, and Perez CG. (1998). Antidiabetic effect of compounds isolated from plants. Phytomedicine 5, 55–75 [DOI] [PubMed] [Google Scholar]
- Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, and Hamon P. (2006). SSR mining in coffee tree EST databases: Potential use of EST-SSRs as markers for the Coffea genus. Mol Genet Genom 276, 436–449 [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, et al. (2005). InterProScan: Protein domains identifier. Nucleic Acids Res 33, W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, degli Ivanissevich S, et al. (2000). A simple sequence repeat-based linkage map of barley. Genetics 156, 1997–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasineni K, Bellamkonda R, Singareddy SR, and Desireddy S. (2010). Antihyperglycemic activity of Catharanthus roseus leaf powder in streptozotocin-induced diabetic rats. Pharmacognosy Res 2, 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritschel PS, Lins TC, Tristan RL, Buso GS, Buso JA, and Ferreira ME. (2004). Development of microsatellite markers from an enriched genomic library for genetic analysis of melon (Cucumis melo L.). BMC Plant Biol 4, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, and Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132, 365–386 [DOI] [PubMed] [Google Scholar]
- Rufer AC, Thoma R, Benz J, et al. (2006). The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment. Structure 14, 713–723 [DOI] [PubMed] [Google Scholar]
- Rungis D, Berube Y, Zhang J, et al. (2004). Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theoret Appl Genet 109, 1283–1294 [DOI] [PubMed] [Google Scholar]
- Saha MC, Cooper JD, Mian MAR, Chekhovskiy K, and May GD. (2006). Tall fescue genomic SSR markers: Development and transferability across multiple grass species. Theoret Appl Genet 113, 1449–1458 [DOI] [PubMed] [Google Scholar]
- Sahu J, Sarmah R, Dehury B, et al. (2012). Mining for SSRs and FDMs from expressed sequence tags of Camellia sinensis. Bioinformation 8, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangeetha MK, Priya CD, and Vasanthi HR. (2013). Anti-diabetic property of Tinospora cordifolia and its active compound is mediated through the expression of Glut-4 in L6 myotubes. Phytomedicine 20, 246–248 [DOI] [PubMed] [Google Scholar]
- Scaglione D, Acquadro A, Portis E, Taylor CA, Lanteri S, and Knapp SJ. (2009). Ontology and diversity of transcript-associated microsatellites mined from a globe artichoke EST database. BMC Genom 10, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti I, Magni F, Fink R, Powell W, Binelli G, and Hedley PE. (2000). Microsatellite repeats are not randomly distributed within Norway spruce (Picea abies K.) expressed sequences. Genome 43, 41–46 [PubMed] [Google Scholar]
- Sharma M, Fernandes J, Ahirwar D, and Jain R. (2008). Hypoglycemic and hypolipidimic activity of alcoholic extract of citrus aurantium in normal and alloxan-induced diabetic rats. Pharmacology online 3, 161–171 [Google Scholar]
- Song ZP, Xu X, Wang B, Chen JK. and Lu BR. (2003). Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theoret Appl Genet 107, 1492–1499 [DOI] [PubMed] [Google Scholar]
- Struss D, and Plieske J. (1998). The use of microsatellite markers for detection of genetic diversity in barley populations. Theoret Appl Genet 97, 308–315 [Google Scholar]
- Tang JH, Fu ZY, Hu YM, Li JS, Sun LL, and Ji HQ. (2006). Genetic analyses and mapping of a new thermo-sensitive genic male sterile gene in maize. Theoret Appl Genet 113, 11–15 [DOI] [PubMed] [Google Scholar]
- Tautz D, and Renz M. (1984). Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12, 4127–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S, Park WD, Ayers N, et al. (2000). Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.) Theoret Appl Genet 100, 697–712 [Google Scholar]
- Thiel T, Michalek W, Varshney RK, and Graner A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoret Appli Genet 106, 411–422 [DOI] [PubMed] [Google Scholar]
- Toth G, Gaspari Z, and Jurka J. (2000). Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res 10, 967–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Thiel T, Stein N, Langridge P, and Graner A. (2002). In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7, 537–546 [PubMed] [Google Scholar]
- Varshney VK, Dayal R, Bhandari RS, Jyoti KN, Prasuna AL, Prasad AR, and Yadav JS. (2005). Behavioral response of the borer beetle Hoplocerambyx spinicornis to volatile compounds of the tree Shorea robusta. Chem Biodiversity 2, 785–791 [DOI] [PubMed] [Google Scholar]
- Victoria FC, da Maia LC, and de Oliveira AC. (2011). In silico comparative analysis of SSR markers in plants. BMC Plant Biology 11, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk PS, Newbury JH, Bryan GJ, Jackson MT, and Ford-Lloyd BV. (2000). Are mapped or anonymous markers more useful for assessing genetic diversity? Theoret Appl Genet 100, 607–613 [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res 23, 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuksan V, Jenkins DJ, Spadafora P, et al. (1999). Konjac-mannan (glucomannan) improves glycemia and other associated risk factors for coronary heart disease in type 2 diabetes. A randomized controlled metabolic trial. Diabetes Care 22, 913–919 [DOI] [PubMed] [Google Scholar]
- Weber JL. (1990). Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics 7, 524–530 [DOI] [PubMed] [Google Scholar]
- Welsh J, and McClelland M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18, 7213–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2002). World Health Organization, WHO Traditional Medicine Strategy 2002–2005. World Health Organization, Geneva: WHO/EDM/TRM/2002.1 [Google Scholar]
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, and Tingey SV. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18, 6531–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohrmann T, and Weising K. (2011). In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae. Theoret Appl Genet 123, 635–647 [DOI] [PubMed] [Google Scholar]
- Xie W, Xing D, Sun H, Wang W, Ding Y, and Du L. (2005). The effects of Ananas comosus L. leaves on diabetic-dyslipidemic rats induced by alloxan and a high-fat/high-cholesterol diet. Am J Chinese Med 33, 95–105 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Williams R, Prakash CS, and He G. (2012). Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol 12, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.