The great planetary nitrogen cycle, which includes the cyclic conversion of nitrogen gas (N2) into “fixed” nitrogen that can be used by plants, is in large part mediated by metalloenzymes that catalyze the elementary chemical reactions. On page 1666 of this issue, Hino et al. (1) take an important step toward understanding the chemical function and evolution of one of these enzymes. They describe the structure of a nitric oxide reductase (NOR) from a common bacterium which plays an important role in the nitrogen cycle—and in human disease.
Today, human activities such as the widespread use of agricultural fertilizers and the burning of fossil fuels are influencing Earth’s nitrogen cycle (2). Of considerable interest is the increasing release into the atmosphere of nitrous oxide (N2O), a greenhouse gas (3) about 300 times as powerful, on an equimolar basis, as CO2 (2). The vast majority of N2O originates from microbes that break down nitrogen compounds in soil and water. Approximately equal quantities of N2O come from two processes: nitrification (which converts ammonia into nitrite), in which N2O is an unintended by-product of the oxidation of hydroxylamine (NH2OH), and anaerobic denitrification, in which nitric oxide (NO) and N2O are formed as diffusible intermediates (2, 4).
Hino et al. studied cytochrome c–dependent nitric oxide reductase (cNOR), in which the cytochrome acts as the electron donor in an enzyme that catalyzes the reduction of NO in Pseudomonas aeruginosa, a ubiquitous, denitrifying bacterium. It is also a nasty pathogen, particularly among burn victims and patients with chronic infections of their airways (5, 6). Similar infectious bacteria, such as Neisseria meningitides and N. gonorrhoeae, depend on NOR activity to withstand the defenses of host cells (7). Hino et al. describe the x-ray structure of cNOR at 2.7 Å resolution. In addition to offering insight into the detailed chemical mechanisms that affect the nitrogen cycle, the structure offers evidence for the existence of a common ancestor connecting the NORs to the heme-copper oxidase (HCO) superfamily of enzymes, and raises intriguing questions about how one diverged from the other. Finally, the structure provides the basis for an atomic-level understanding of the chemistry of both NOR and HCO enzymes.
Both HCOs and NORs are integral membrane proteins. The HCO superfamily contains at least three types that share a common core structure but vary in the number of peripheral subunits, heme types, physiological electron donors, and proton pathways that can generate the heme-copper cluster (8). The NOR family is also composed of several types (9). P. aeruginosa cNOR consists of a small (NorC) and a large (NorB) subunit. Although NorB and the large subunits of HCOs are clearly homologs, both families show diversity in the smaller subunits (especially in the associated electron donor). It is hard to see how the predominantly β-sheet cupredoxin fold of subunit II containing CuA (see the figure, left panel) and the predominantly helical cytochrome c fold of NorC (right) could be linked evolutionarily via a series of mutations. Rather, it seems more likely that the smaller subunits evolve partly by “jumping” from one type of large subunit to another.
Figure. Compare and contrast.
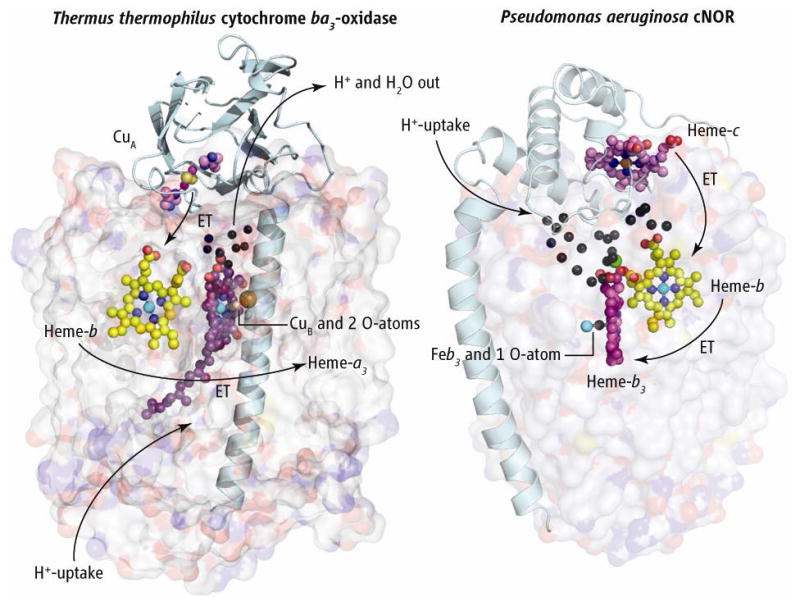
The structures of Thermus thermophilus (Tt) cytochrome ba3 oxidase (left) and cNOR from Pseudomonas aeruginosa (Pa) (right), with large subunits shown as transparent surfaces. The two structures are aligned vertically at the level of the b-hemes and the cNOR structure is rotated approximately 180° about its vertical axis to bring the b-hemes into the same plane. Electrons flow from the subunit II of ba3 that anchors the dinuclear copper cluster CuA, and from the cytochrome c in cNOR. These two redox centers and globular domains exemplify the variability in redox partners in the oxidases and NO reductases, as evolution may allow for occasional interchange of large and small subunits. Electrons flow into the b-hemes before passing to the binuclear active sites. Presumed proton flow in ba3 oxidase and its water exit are represented by arrows. Black spheres represent crystallographically defined water molecules. The single Ca2+ ion is shown as a green sphere. In cNOR, protons appear to flow from the outside into the extensive water cluster, and from there into the binuclear heme-b3/FeB active site.
Hino et al. compare their structure to the simplest known HCO, the B-type cytochrome ba3-oxidase from Thermus thermophilus (8, 10), and a comparison to the C-type cbb3 oxidase from P. stutzeri (11) is also informative. Although the cbb3 enzyme appears to be closer to cNOR than does ba3, similarities in the amino acid sequences of NorB and subunit I of cbb3 are low (<40%). Regardless, the two proteins exhibit highly similar three dimensional structures. Specifically, 12 central transmembrane helices are conserved that share a topology of tightly packed helices arranged around a low-spin heme and a binuclear active site (i.e., a heme-b3/FeB or a heme-a3/CuB). In ba3, the active-site heme-a3/CuB center is buried in the hydrophobic core of the enzyme, requiring that both polar (H+, e−, and H2O) and lipophilic (O2) reactants move along structurally specified pathways to their intended destinations. In cNOR, the active-site heme-b3/FeB is found essentially at the same location in the hydrophobic core. The three histidine side chains that coordinate the CuB in HCO are conserved in cNOR and are supplemented with a glutamyl carboxylate group, providing a favorable coordination shell for an iron ion at the FeB site.
The structure of cNOR also shows that transmembrane pathways (D- and K-paths) for proton uptake are absent; this supports previous findings that cNOR does not pump protons (12) and sheds light on how heme-Cu oxidases acts as a proton pump. Hino et al. also propose pathways for electron transfer from the cytochrome c via the low-spin heme and proton entries from the periplasmic side to the binuclear center. As in cbb3, a Ca2+ ion bridges and may stabilize an efficient electron transfer route between the two hemes in NorB. Another remarkable feature of the cNOR structure is the presence of a Y-shaped hydrophobic channel similar to that reported by Luna et al. (13) in ba3 and by Buschmann et al. (11) in cbb3. In cNOR, this channel must serve to carry NO from the lipid bilayer to the active site.
The authors do not assign the exit route of the N2O product, but the factor of 10 increase in water solubility of N2O relative to O2 or NO may allow this small molecule to diffuse through both hydrophobic and hydrophilic channels. A question worthy of future investigation is whether the exit route of N2O from cNOR influences its capture by the periplasmic, copper-containing N2O reductase, and its escape into the atmosphere. The structure also leaves two other questions unanswered: (i) How do the two NO molecules bind at the diiron site? and (ii) How do the N-N bond formation and N-O bond cleavage occur? A distal glutamic acid within 5 Å of the iron cluster that appears conserved in NorB sequences may provide a proton to a putative hyponitrite anion (N2O22−) and facilitate N-O bond cleavage. Although most investigators might agree that the first NO binds to the ferrous heme b3, the mode of addition of a second NO to this mononitrosyl complex remains to be defined, as do the respective roles of the two Fe ions (14). Bioengineering models of the heme b3/FeB site in the simpler protein scaffold of myoglobin will be valuable tools for conducting these mechanistic investigations (15). Structural characterization of other transmembrane NORs in parallel with HCOs will further clarify evolutionary variation in FeB/CuB coordination spheres, electron and proton transfer pathways, and hydrophobic channels to the catalytic dinuclear center.
References and Notes
- 1.Hino T, et al. Science. 2010;330:1666. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 2.Canfield DE, Glazer AN, Falkowski PG. Science. 2010;330:192. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 3.Lacis AA, Schmidt GA, Rind D, Ruedy RA. Science. 2010;330:356. doi: 10.1126/science.1190653. [DOI] [PubMed] [Google Scholar]
- 4.Mosier A, et al. Nutr Cycl Agroecosyst. 1998;52:225. [Google Scholar]
- 5.Barraud N, et al. J Bacteriol. 2006;188:7344. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romeo T. J Bacteriol. 2006;188:7325. doi: 10.1128/JB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevanin TM, Laver JR, Poole RK, Moir JWB, Read RC. Microbes Infect. 2007;9:981. doi: 10.1016/j.micinf.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Pereira MM, Santana M, Teixeira M. Biochim Biophys Acta. 2001;1505:185. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 9.de Vries S, Schroder I. Biochem Soc Trans. 2002;30:662. doi: 10.1042/bst0300662. [DOI] [PubMed] [Google Scholar]
- 10.Hunsicker-Wang LM, Pacoma RL, Chen Y, Fee JA, Stout CD. Acta Crystallogr D Biol Crystallogr. 2005;61:340. doi: 10.1107/S0907444904033906. [DOI] [PubMed] [Google Scholar]
- 11.Buschmann S, et al. Science. 2010;329:327. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 12.Flock U, et al. J Biol Chem. 2008;238:3839. doi: 10.1074/jbc.M704615200. [DOI] [PubMed] [Google Scholar]
- 13.Luna VMM, Chen Y, Fee JA, Stout CD. Biochemistry. 2008;47:4657. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 14.Moënne-Loccoz P. Nat Prod Rep. 2007;24:610. doi: 10.1039/b604194a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung N, et al. Nature. 2009;462:1079. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]


