Abstract
Phosphatidylethanolamine N-methyltransferase (PEMT) plays a critical role in breast cancer progression. However, the epigenetic mechanism regulating PEMT transcription remains largely unknown. Here, we show that the first promoter-specific transcript 1 is the major PEMT mRNA species, and methylation of the -132 site is a key regulatory element for the PEMT gene in BRCA1-mutated breast cancer. Mechanistically, hypermethylated -132 site-mediated loss of active histone marks H3K9ac and increase of repressive histone marks H3K9me enrichment synergistically inhibited PEMT transcription. Clinicopathological data indicated that a hypermethylated -132 site was associated with histological grade (P = 0.031) and estrogen receptor status (P = 0.004); univariate survival and multivariate analyses demonstrated that lymph node metastasis was an independent and reliable prognostic factor for BRCA1-mutated breast cancer patients. Our findings imply that genetic (e.g., BRCA1 mutation) and epigenetic mechanisms (e.g., DNA methylation and histone modifications) are jointly involved in the malignant progression of PEMT-related breast cancer.
Keywords: PEMT, DNA methylation, Histone modifications, BRCA1, Breast cancer
INTRODUCTION
Breast cancer is the most common malignancy and a major cause of mortality in women worldwide [1]. Accumulating evidence indicates that an increased risk for breast cancer is associated with dietary factors [2] and a few significant-risk genetic components (e.g., BRCA1) [3]. BRCA1 is a tumor suppressor gene which plays a key role in numerous cellular processes, including transcription regulation, DNA damage repair and protein ubiquitination.4 Recent research has confirmed that BRCA1 is an important transcriptional regulator, and BRCA1-depleted breast cancer cells shows changes to approximately 7% of the mRNAs expressed [4]. Moreover, our recent study also indicated that angiotensin II type 1 receptor and epidermal growth factor receptor displayed different expression patterns in BRCA1-defective cancer cells [5,6], and confirmed that differential epigenetic regulation of transcription exist along with BRCA1 inactivation [7,8]. Therefore, one can speculate that there are wide ranges of gene expression and regulation differences between BRCA1 dysfunction and the basal phenotype. To date, choline is among the well-studied essential nutrients that are involved in breast cancer; for example: (i) choline-containing compounds are significantly changed in breast cancer [9,10]; (ii) choline intake is inversely correlated with breast cancer risk [11-13]; and (iii) aberrant choline metabolism is often associated with malignant transformation, invasion, and metastasis of breast cancer [14-16]. Phosphatidylethanolamine N-methyltransferase (PEMT) is a small integral membrane protein that catalyzes the de novo synthesis of choline using S-adenosylmethionine as a methyl donor [17]. The human PEMT gene is located on chromosome 17p11.2 and consists of nine exons and eight introns, and differential promoter usage generates multiple transcripts [18]. It is interesting to note that functional polymorphisms of the PEMT gene play an important role in breast cancer development [12,19-21]. However, the expression patterns and transcriptional regulation of individual transcripts remain to be elucidated. Therefore, the present study was undertaken to investigate PEMT transcriptional regulation from genetic (BRCA1 mutation or not) and epigenetic (promoter methylation and histone modifications) aspects, and to provide novel insight into epigenetic change-mediated abnormal PEMT expression in BRCA1-mutated breast cancer progression.
RESULTS
BRCA1-mutated breast cancer displayed a hypermethylated -132 site and promoter region
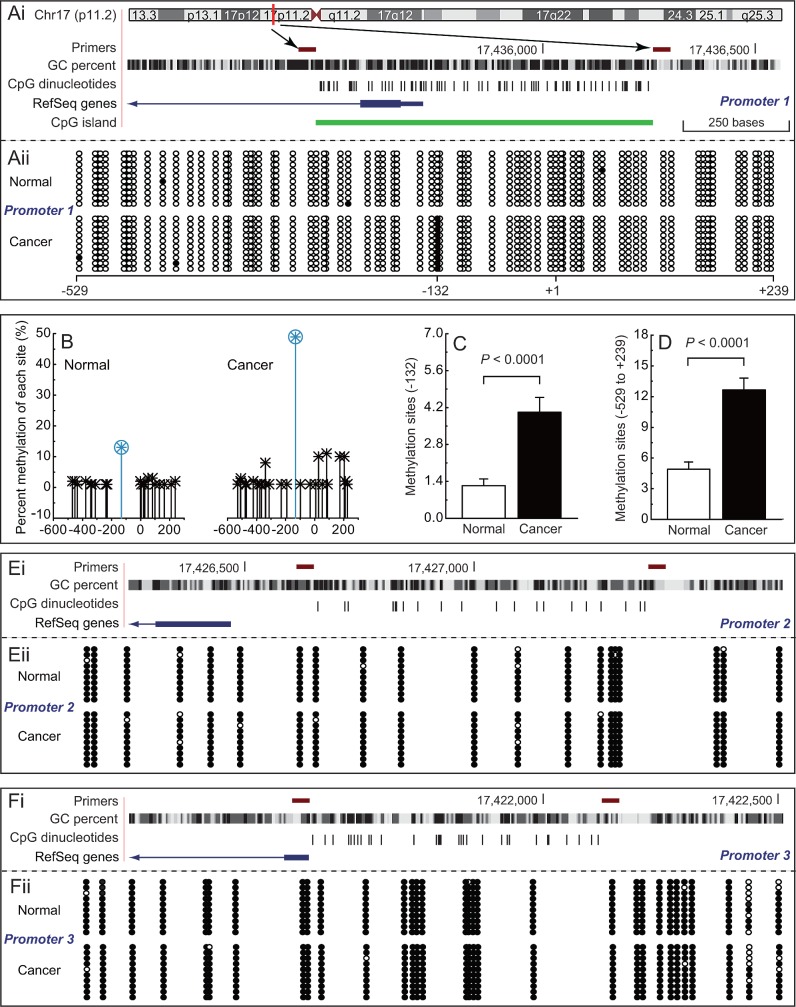
To investigate the epigenetic regulation of PEMT, we compared the DNA methylation patterns of three promoters between BRCA1-mutated or non-mutated breast cancer and their adjacent normal breast tissues. As shown in Fig. 1Aii and B, BRCA1-mutated breast cancer exhibited hypermethylation of the first promoter (P < 0.001; Fig. 1D), especially around the -132 site (P < 0.001; Fig. 1C). However, no significant methylation differences were observed in the second and third promoters in BRCA1-mutated breast cancer (Figs. 1E and F), and in all three promoters in non-mutated breast cancer (Supplementary Fig. S1A), compared with normal breast tissues.
Fig 1. Methylation patterns of differential PEMT promoter in BRCA1-mutated breast cancer.
Ai, location of CpG sites in the first promoter of PEMT. Genomic coordinates are shown, along with the primer-amplified fragments, GC percentage, location of individual CpG dinucleotides (dashes), the PEMT RefSeq gene (exon 1 shown as a blue box and intron shown as an arrowed line), and CpG island (green bar). The arrow indicates the transcriptional direction, +1 is the transcription initiation site. Aii, comparative analysis of methylation patterns in the first promoter of PEMT in BRCA1-mutated breast cancer, and their adjacent normal breast tissues (each group, n = 68). The circles correspond to CpG sites denoted by the black dashes in Fig. 1Ai. Closed circles, methylation; open circles, unmethylation. Ten individual clones were sequenced for each sample. B, summary of the methylation patterns of the first PEMT promoter in Fig. 1Aii. The y-axis shows the mean methylation sites. C and D, overall methylation percentage of position -132 and the PEMT core promoter region (−529 to +239). Bar graphs show mean ± SD. E and F, comparative analysis of methylation patterns in the second and third promoters of PEMT, respectively (each group, n = 68)
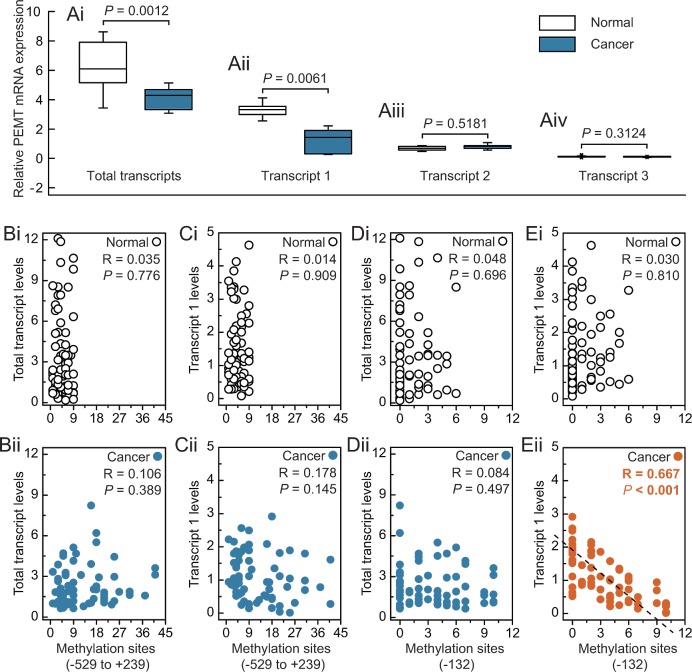
Low PEMT transcript 1 levels showed a significant inverse correlation with hypermethylation of the -132 site in BRCA1-mutated breast cancer
Multiple promoters of the human PEMT gene generate at least three transcripts and exhibit tissue-specific expression; for example, transcript 1, transcript 2, and transcript 3 are mainly initiated from the first, second, and third promoter, respectively. Our results indicated that transcript 1 was the major PEMT mRNA species in human breast tissues, and reduced PEMT mRNA may be dependent on reduced levels of the first-promoter-specific transcript 1 in human breast tissues (Fig. 2A and Supplementary Fig. S1B). In addition, expression levels of PEMT were decreased in BRCA1-mutated breast cancer compared to their adjacent normal breast tissues. However, no expression differences for different transcripts between non-BRCA1-mutated breast cancer and their adjacent normal breast tissues were observed (Supplementary Fig. S1B). In addition, we analyzed the correlation between total transcript levels or transcript 1 levels, and the number of methylated sites at -132 site or around the PEMT core promoter region (−529 to +239) in BRCA1-mutated breast cancer and normal breast tissues (Figs. 2B–E). It is interesting to note that a significant inverse correlation was only observed between levels of PEMT transcript 1 and numbers of methylated -132 sites (R = 0.667, P < 0.001; Fig. 2Eii).
Fig 2. Expression features of PEMT in BRCA1-mutated breast cancer.
A, relative PEMT mRNA levels of differential promoter utilization (each group, n = 68). B and C, correlation between the methylated sites (−592 to +239), and total transcript or transcript 1 levels in BRCA1-mutated breast cancer and adjacent normal breast tissues, respectively. D and E, correlation between the methylated sites (-132), and total transcript or transcript 1 levels in BRCA1-mutated breast cancer and adjacent normal breast tissues, respectively. Open circles, normal breast tissues. Closed circles, breast cancer tissues (each group, n = 68).
Hypermethylated -132 site is a key regulatory mechanism for PEMT transcription in BRCA1-mutated breast cancer
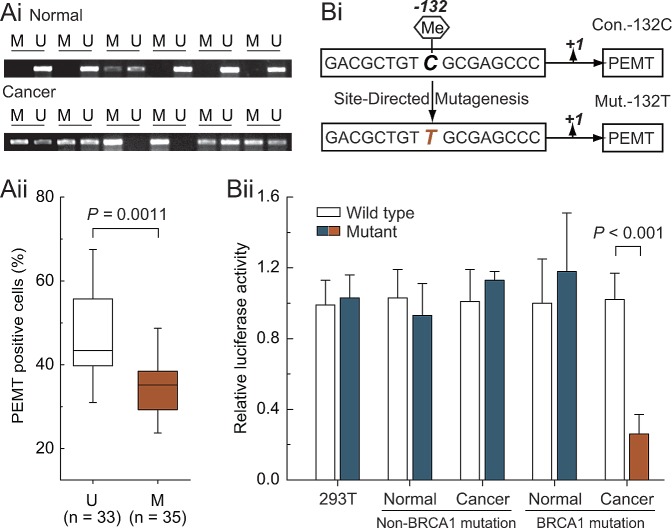
Based on the results above, we further explored the relationship between -132 site methylation and PEMT expression in BRCA1-mutated breast cancer specimens. BRCA1-mutated cancer specimens were classified into unmethylated and methylated groups for comparing their protein expression of PEMT. The methylated group showed a significantly lower expression of PEMT compared with the unmethylated group (P = 0.0011; Fig. 3Ai and Aii). To further confirm the role of the cytosine located at the -132 site, a point mutation of cytosine to thymine was constructed (Fig. 3Bi). Notably, -132 was the critical site for PEMT transcription only in primary BRCA1-mutated breast cancer cells (Fig. 3Bii). Moreover, methylated -132 site can also significantly inhibit the transcription of PEMT (Supplementary Fig. S2A and B).
Fig 3. Methylated -132 site and PEMT transcriptional activity.
Ai, comparative analysis of -132 site methylation between BRCA1-mutated breast cancer and normal breast tissues using methylation-specific PCR. Aii, relationship between PEMT protein expression and promoter methylation in BRCA1-mutated breast cancer (U, unmethylated group, n = 33; M, methylated group, n = 35). Bi, the schematic represents the nucleotide sequence around the -132 site (Con.-132C) that was point mutated at position -132 (C to T) to generate the Mut.-132T. Bii, 293T cells (repeated 12 times), and primary non-mutated (n = 75) and BRCA1-mutated breast cancer (n = 68) and their normal breast cells were transfected with Con.-132C and Mut.-132T. At 24 hours after transfection, whole-cell extracts were analyzed for luciferase activity. Bar graphs show mean ± SD.
Loss of H3K9ac and increase of H3K9me enrichment around the methylated -132 site in BRCA1-mutated breast cancer
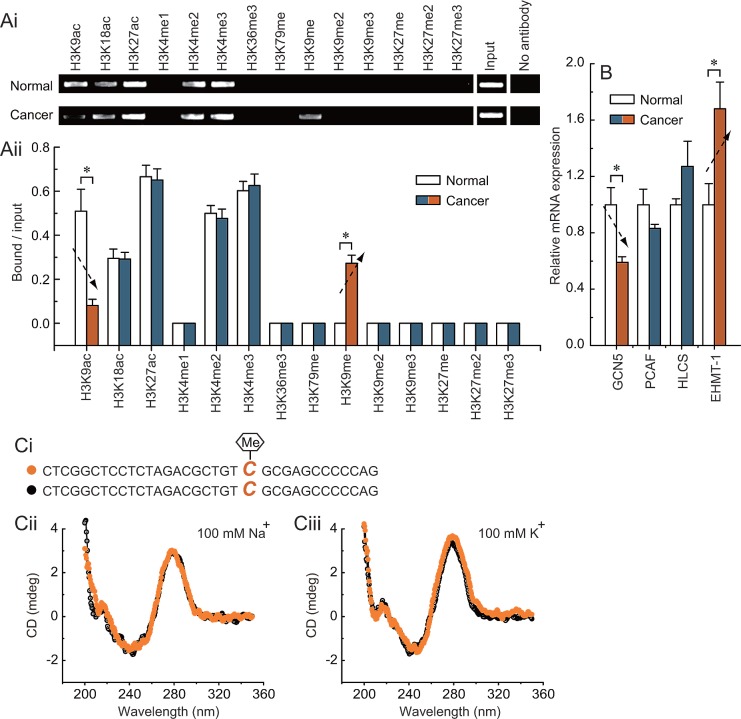
To obtain further understanding of the regulatory potential of the crosstalk between DNA methylation and histone modification in controlling PEMT transcription, we examined the active histone markers H3K9ac, H3K18ac, H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3, and H3K79me, and the repressive histone markers H3K9me, H3K9me2, H3K9me3, H3K27me, H3K27me2, and H3K27me3 in the first promoter of PEMT, especially around the -132 site. Chromatin immunoprecipitation analysis indicated that the levels of H3K9ac were reduced, and levels of H3K9me were increased around the methylated -132 site in BRCA1-mutated breast cancer (Fig. 4A). Chromatin-modifying enzymes that facilitate the creation of H3K9ac and H3K9me were also analyzed: GCN5 and PCAF are capable of creating H3K9ac; HLCS and EHMT-1 are capable of creating H3K9me. Although there was no significant change in the expression of PCAF and HLCS, the expression level of GCN5 was reduced, and EHMT-1 was increased in BRCA1-mutated breast cancer (Fig. 4B; P < 0.05). In addition, there was little difference in histone markers around the -132 site between BRCA1 wild type and mutant tissue, such as the lack of specific H3K9me in BRCA1 wild type tissue. However, no differences were found for histone markers around the -132 site between normal and cancer tissues in BRCA1 wild type cases (Supplementary Fig. S3).These results, together with the methylation data in Figs. 1, 2, and 3, suggest that PEMT transcription may be associated with changes in epigenetic features, including reduced GCN5-related H3K9ac enrichment and increased EHMT-1-related H3K9me enrichment around the hypermethylated -132 site in BRCA1-mutated breast cancer. Recently, a substantial body of evidence suggests that most of the known genes contain specific motifs in their promoter regions, which can modulate gene transcription by affecting the binding of histone [22]. Therefore, CD spectra were used to gain information on whether methylation of the -132 site associated abnormal H3K9ac and H3K9me enrichment, which may involve changes in PEMT promoter structure. However, our results showed that -132 site methylation may not affect the structure of the first promoter of PEMT (Fig. 4C).
Fig 4. Characteristic histone modification pattern of -132 site methylation in BRCA1-mutated breast cancer.
Ai, chromatin immunoprecipitation was performed using antibodies to H3K9ac, H3K18ac, H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K79me, H3K9me, H3K9me2, H3K9me3, H3K27me, H3K27me2, and H3K27me3. PCR was performed for regions around the -132 site. A negative control without antibodies is included for comparison. Aii, representative results of primary BRCA1-mutated breast cancer and their normal breast tissues are shown (each group, n = 68). Bar graphs show mean ± SD. * P < 0.05 vs. normal. B, expression levels of the chromatin-modifying enzymes GCN5, PCAF, HLCS, and EHMT-1 in BRCA1-mutated breast cancer and normal breast tissues (each group, n = 68). Bar graphs show mean ± SD. * P < 0.05 vs. normal. Ci, the schematic represents the selected nucleotide sequence with or without a methyl group at the fifth position of the cytosine pyrimidine ring at position -132. Cii and Ciii, the CD spectra of the selected nucleotide sequence in the presence of 100 mM Na+ or 100 mM K+ are shown
H3K9ac and H3K9me present at the -132 site are responsible for transcriptional regulation of PEMT in BRCA1-mutated breast cancer
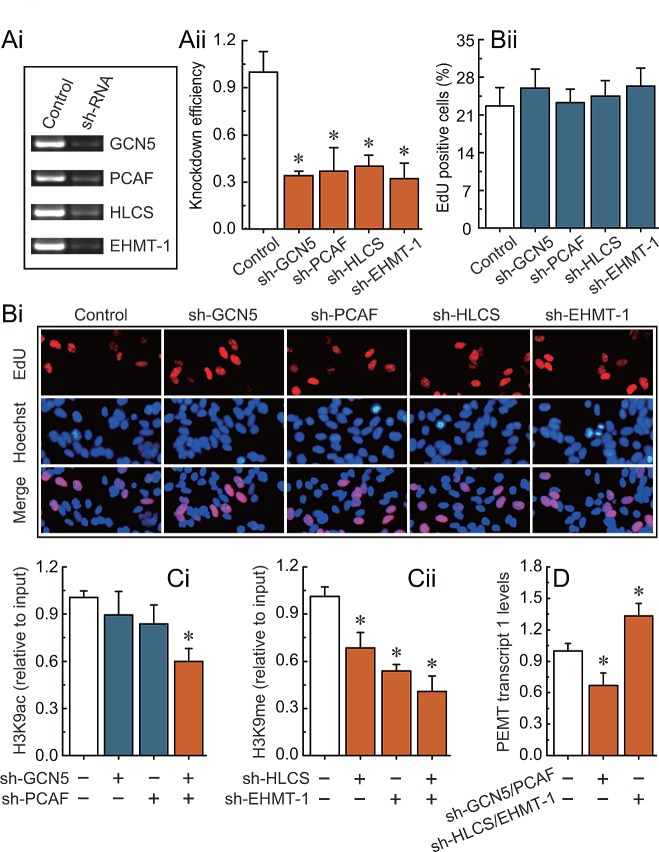
We observed that the knockdown of GCN5, PCAF, HLCS, and EHMT-1 (Fig. 5A) had no detectable effect on cell morphology and proliferation (Fig. 5B). Combined knockdown of GCN5 and PCAF, or HLCS and EHMT-1 specifically induced a decrease in H3K9ac or H3K9me enrichment around the -132 site, respectively (Figs. 5Ci and Cii). Notably, after the deletion of H3K9ac or H3K9me, the transcription of PEMT was significantly down-regulated or up-regulated, respectively (Fig. 5D). The phenomenon was further confirmed by other specific siRNAs (Supplementary Fig. S4).
Fig 5. H3K9ac and H3K9me-mediated transcriptional regulation of PEMT in BRCA1-mutated breast cancer cells.
Ai, RT-PCR showing GCN5, PCAF, HLCS, and EHMT-1 levels before and after knockdown by shRNAs, and normalized to β-actin expression. Aii, the results from three independent experiments are represented as mean ± SD. Bi, EdU labeling showing the proliferation of GCN5-, PCAF-, HLCS-, and EHMT-1-silenced in primary BRCA1-mutated breast cancer cells (each group, n = 68). Blue, Hoechst 33342 labeling of cell nuclei; red, EdU labeling of nuclei of proliferative cells. Bii, the EdU incorporation rate was expressed as the ratio of EdU-positive cells to total Hoechst 33342-positive cells. Ci, analysis of histone modification H3K9ac enrichment around the -132 site after the deletion of GCN5 and PCAF in primary BRCA1-mutated breast cancer cells (each group, n = 68). Cii, analysis of histone modification H3K9me enrichment around the -132 site after the deletion of HLCS and EHMT-1 in primary BRCA1-mutated breast cancer cells (each group, n = 68). D, PEMT transcript 1 levels after the deletion of H3K9ac and H3K9me around the -132 site in primary BRCA1-mutated breast cancer cells (each group, n = 68). Bar graphs show mean ± SD. * P < 0.05 vs. control.
Correlation of -132 site methylation with clinicopathological characteristics in BRCA1-mutated breast cancer
The correlation between -132 site methylation and clinicopathological parameters was analyzed using Fisher's exact test. As shown in Table 1, PEMT methylation was associated with histological grade (P = 0.031) and estrogen receptor status (P = 0.004). No significant associations were observed between PEMT methylation and age at diagnosis, menstrual status, tumor size, lymph node, progesterone receptor, c-erbB-2, p53, Ki67, or E-cadherin status.
Table 1. Association between PEMT promoter methylation and clinicopathological features of BRCA1-mutated breast cancer.
| n | M | (%) | UM | (%) | P | |
|---|---|---|---|---|---|---|
| Age at diagnosis | 0.799 | |||||
| ≤ 50 y | 23 | 11 | 31.43 | 12 | 36.36 | |
| > 50 y | 45 | 24 | 68.57 | 21 | 63.64 | |
| Menstrual status | 1.000 | |||||
| Premenopausal | 28 | 14 | 40.00 | 14 | 42.42 | |
| Postmenopausal | 40 | 21 | 60.00 | 19 | 57.58 | |
| Tumor size | 0.137 | |||||
| ≤ 5 cm | 43 | 19 | 54.29 | 24 | 72.73 | |
| > 5 cm | 25 | 16 | 45.71 | 9 | 27.27 | |
| Histological grade | 0.031 | |||||
| I-II | 49 | 21 | 60.00 | 28 | 84.85 | |
| III | 19 | 14 | 40.00 | 5 | 15.15 | |
| LN status | 0.809 | |||||
| Positive | 28 | 15 | 42.86 | 13 | 39.39 | |
| Negative | 40 | 20 | 57.14 | 20 | 60.61 | |
| ER status | 0.004 | |||||
| Positive | 37 | 13 | 37.14 | 24 | 72.73 | |
| Negative | 31 | 22 | 62.86 | 9 | 27.27 | |
| PR status | 0.144 | |||||
| Positive | 41 | 18 | 51.43 | 23 | 69.70 | |
| Negative | 27 | 17 | 48.57 | 10 | 30.30 | |
| c-erbB-2 status | 1.000 | |||||
| Positive | 29 | 15 | 42.86 | 14 | 42.42 | |
| Negative | 39 | 20 | 57.14 | 19 | 57.58 | |
| p53 status | 0.300 | |||||
| Positive | 21 | 13 | 37.14 | 8 | 24.24 | |
| Negative | 47 | 22 | 62.86 | 25 | 75.76 | |
| Ki67 status | 0.406 | |||||
| Positive | 51 | 28 | 80.00 | 23 | 69.70 | |
| Negative | 17 | 7 | 20.00 | 10 | 30.30 | |
| E-cadherin status | 0.767 | |||||
| Positive | 54 | 27 | 77.14 | 27 | 81.82 | |
| Negative | 14 | 8 | 22.86 | 6 | 18.18 | |
M, methylated; UM, unmethylated; LN, lymph node; ER, estrogen receptor; PR, progesterone receptor.
Multivariate and univariate analysis of overall survival for patients with BRCA1-mutated breast cancer
We analyzed overall survival to assess the prognostic significance of clinicopathological parameters. Multivariate Cox regression analysis indicated that lymph node metastasis (P = 0.090), estrogen receptor (P = 0.149), progesterone receptor (P = 0.157), and p53 status (P = 0.136) showed a trend for an independent prognostic factor for predicting the overall survival of BRCA1-mutated breast cancer patients (Supplementary Table S1). We also performed the Kaplan-Meier analysis and log-rank tests for overall survival in defining prognostic subgroups. The results revealed that lymph node metastasis (Supplementary Fig. S5E, P < 0.05) and E-cadherin status (Supplementary Fig. S5K, P < 0.05) were significant prognostic factors. Moreover, patients with high histological grade (Supplementary Fig. S5D, P = 0.248) or estrogen receptor-negative (Supplementary Fig. S5F, P = 0.278), c-erbB-2-positive (Supplementary Fig. S5H, P = 0.244), p53-negative (Supplementary Fig. S5I, P = 0.072), or Ki67-positive (Supplementary Fig. S5J, P = 0.145) breast cancer showed a trend for poor overall survival, although not statistically significant. No significant difference in overall survival was found among patients with different age at diagnosis, premenopausal, tumor size, progesterone receptor status, or PEMT methylation (Supplementary Fig. S5A, B, C, G, and L).
DISCUSSION
Promoter methylation, with concurrent changes in histone modifications, is an epigenetic phenomenon that can affect the conformation of chromatin and tissue-specific gene expression[23,24]. In this study, we report that the first-promoter-specific transcript 1 is the major PEMT mRNA species in human breast tissues, and that methylation of the -132 site is a key regulatory element for PEMT transcription in BRCA1-mutated breast cancer. The molecular mechanism may involve the hypermethylated -132 site-mediated loss of active histone marker H3K9ac and an increase of the repressive histone marker H3K9me enrichment, which synergistically inhibit the transcription of PEMT. Interestingly, the synergistic inhibitory effect of hypermethylated -132 site and H3K9ac and H3K9me histone modification were only observed primarily in cells originating from BRCA1-mutated breast cancer; the transformed cell line 293T and non-mutated breast cancer were insensitive to the loss of H3K9ac and H3K9me enrichment around the -132 site in the first promoter of PEMT. Accordingly, specific regulatory mechanisms may exist, and PEMT expression is likely to be the result of a complex interaction of multiple factors in BRCA1-mutated breast cancer cells, such as changes in chromatin-modifying enzymes that reduce GCN5 levels and increase EHMT-1 levels. Remarkably, as shown in Supplementary Fig. S6, BRCA1-mutated breast cancer tissues exhibited lower levels of choline compared with normal breast tissues and non-mutated cancer tissues. Choline is known to be an effective methyl donor that is involved in DNA methylation. This may help to explain why global DNA methylation was further reduced in BRCA1-mutated breast cancer (Supplementary Fig. S7). Moreover, DNA methyltransferase 1 (DNMT1) plays an important role in maintaining DNA methylation patterns [25,26], Shukla showed that DNMT1 was a transcriptional target of BRCA1 [27]. Our recent study confirmed that DNMT1 mRNA and protein were decreased in BRCA1-mutated breast cancer.7 Therefore, abnormal BRCA1 gene-mediated reduced levels of DNMT1 may also be involved in the progression of genomic DNA hypomethylation in breast cancer. In addition, Xu observed that low choline levels were associated with more frequent BRCA1 promoter methylation-mediated expression inhibition [28]. It can be speculated that dynamic crosstalk may exist between PEMT-related choline synthesis and choline-related BRCA1 inactivity. Meanwhile, choline plays a critical role in the methionine cycle [29]. Beetstra confirmed that defects in methionine metabolism may be related to breast cancer risk in BRCA carriers [30]. These results, together with our observations, suggest that low levels of PEMT may be involved in the development of BRCA-mutated breast cancer through choline deficiency.
Promoter hypermethylation is often associated with adverse clinical events [31]. In line with this, clinicopathological data indicated that hypermethylation of the -132 site may be an effective indicator for histological grade and estrogen receptor status in BRCA1-mutated breast cancer tissues (Table 1). Moreover, univariate survival and multivariate analyses indicated that lymph node metastasis was an independent and reliable prognostic factor which is associated with worse outcomes for BRCA1-mutated breast cancer patients.
This study provides new insights into the causes and prognosis of PEMT inactivation in BRCA1-mutated breast cancer. The mechanism involves the synergistic effects of promoter methylation and histone modification. Therefore, a more specific epigenetic therapy could be developed for BRCA1-mutated breast cancer.
METHODS
Ethics Statement
Investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and according to national and international guidelines and has been approved by the authors' institutional review board.
Patients and tissue collection
Sixty-eight invasive ductal carcinomas from BRCA1 mutation carriers and 75 invasive ductal carcinomas from non-BRCA1 mutation carriers were enrolled between 2007 and 2009, and all patients gave informed consent. Fresh breast cancer and adjacent normal breast tissues were obtained at the time of primary surgery before any chemotherapy or radiotherapy. Hematoxylin and eosin staining of the samples for histopathological diagnosis and grading were performed by three staff pathologists using the Nottingham Combined Histologic Grade. The tumor stages were classified according to the National Comprehensive Cancer Network guidelines. Their characteristics are given in Supplementary Table S2.
Cell culture, lentiviral infection, and cell proliferation assay
Detailed isolation and cultivation protocols were established as previously described [32]. Briefly, tissues were washed, minced, and digested in 0.1% collagenase type III (Sigma, CA, USA) overnight at 37°C. The suspension was filtered through a 100-μm nylon mesh to remove the remaining clumps. Following gentle centrifugation at 100 g for 5 min, the epithelial and stromal fractions were cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (Invitrogen, CA USA) for 24 h to promote cell attachment. Breast epithelial cells were maintained in CnT-27 mammary epithelium medium (CELLnTEC, Bern, Switzerland), and used in all experiments were passage 2 to passage 5. Human 293T cells were maintained in DMEM with 10% fetal bovine serum (Invitrogen), and have been tested and authenticated at Jun 1th, 2012. The short hairpin RNAs (shRNAs) lentiviral particles of histone acetyltransferase (GCN5), p300/ CBP-associated factor (PCAF), Holocarboxylase synthetase (HLCS) and Euchromatic histone-lysine N-methyltransferase 1 (EHMT-1) were purchased from Santa Cruz Biotechnology (CA, USA), see details in Supplementary Table S3. Transfections were performed using polybrene and enhanced infection solution (Genechem, Shanghai, China) according to the manufacturer's recommended protocol. The knowdown effiency was confirmed by RT-PCR and western blotting (Supplementary Fig. S8). After 48-hour infection, cell proliferation was determined using the Cell-Light™ EdU Apollo®643 In Vitro Imaging Kit (Ribobio, Guangzhou, China) following the manufacturer's instructions.
DNA methylation analysis
Genomic DNA from the breast cancer and adjacent normal breast tissues was extracted using the TIANamp Genomic DNA kit (Tiangen Biotech, Beijing, China). Sodium bisulfite conversion, PCR amplification, and general experimental procedures are described in the Supplementary Methods. The specific primer sequences for bisulfite sequencing and methylation-specific PCR are shown in Supplementary Table S4. Genomic DNA methylation assay are described in the Supplementary Methods.
Real-time PCR, immunohistochemistry and western blotting analysis
Real-time PCR, immunohistochemistry and western blotting analysis are described in the Supplementary Methods. The specific primer sequences for real-time PCR are shown in Supplementary Table S4. The primary antibody for immunohistochemistry and western blotting analysis are shown in Supplementary Table S3.
Chromatin immunoprecipitation (ChIP), site-directed mutagenesis, transfection, and dual-luciferase reporter assay
ChIP, site-directed mutagenesis, transfection, and dual-luciferase reporter assay are described in the Supplementary Methods. The specific primer sequences for site-directed mutagenesis and ChIP are shown in Supplementary Table S4. The specific antibodies for ChIP are shown in Supplementary Table S3.
Circular dichroism (CD) spectra
CD spectra were obtained using a Jasco J-810 spectropolarimeter at 25°C using a 0.1 cm path length cell. Data were collected with a 2 nm slit width from 350 to 200 nm at 0.5 nm intervals and averaged over three scans. CD experiments were carried out on DNA samples (5 μM) using a buffer containing 0.2 M phosphate buffer (pH 7.0) in the presence of 100 mM Na+ or K+. The DNA samples were annealed by heating to 95°C for 5 min followed by cooling to room temperature over 10 h before analysis. The DNA sequence was as follows: 5-CTCGGCTCCTCTAGACGCTGT “C (CH3 or non-CH3)” GCGAGCCCCCAG, and synthesized by Sangon Biotech Ltd (Shanghai, China).
Measurement of choline levels
Tissue choline content was measured using the Choline/Acetylcholine Quantification kit (Biovision, Mountain View, CA, USA) following the manufacturer's instructions. The absorbance of each plate was measured at 570 nm using a Bio-Rad model 550 microplate reader (Hercules, CA, USA).
Statistical analysis
Regression analysis was used to examine the possible relationship between PEMT mRNA and status of promoter methylation. The association between clinicopathological features and PEMT promoter methylation was determined using Fisher's exact test. Univariate analysis of survival was performed using the Kaplan-Meier method. Multivariate Cox regression analysis was performed to identify independent prognostic factors for overall survival. The data are presented as mean ± standard deviation (SD). Statistical differences in the data were evaluated by Student's t test or one-way analysis of variance (ANOVA) as appropriate, and were considered significant at P < 0.05.
SUPPLEMENTARY FIGURES TABLES AND METHODS
Acknowledgments
This work was supported by the 973 Program of China (No. 2011CB933504), Natural Science Foundation of China (No. 81071072) and the Higher Specialized Research Fund for Doctoral Program of Ministry of Education of China (No. 20122104110027).
Editorialnote
This paper has been accepted based in part on peer-review conducted by another journal and the authors' response and revisions as well as expedited peer-review in Oncotarget.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Slattery ML, John EM, Torres-Mejia G, Lundgreen A, Lewinger JP, Stern MC, Hines L, Baumgartner KB, Giuliano AR, Wolff RK. Angiogenesis genes, dietary oxidative balance and breast cancer risk and progression: The breast cancer health disparities study. Int J Cancer. 2013;134:629–644. doi: 10.1002/ijc.28377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevens KN, Vachon CM, Couch FJ. Genetic susceptibility to triple-negative breast cancer. Cancer Res. 2013;73:2025–2030. doi: 10.1158/0008-5472.CAN-12-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dacheux E, Vincent A, Nazaret N, Combet C, Wierinckx A, Mazoyer S, Diaz JJ, Lachuer J, Venezia ND. BRCA1-Dependent Translational Regulation in Breast Cancer Cells. PLoS One. 2013;8:e67313. doi: 10.1371/journal.pone.0067313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi FF, Li D, Cao C, Li CY, Yang Q. Regulation of angiotensin II type 1 receptor expression in ovarian cancer: a potential role for BRCA1. J Ovarian Res. 2013;6:89. doi: 10.1186/1757-2215-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Bi FF, Cao JM, Cao C, Li CY, Yang Q. Effect of BRCA1 on epidermal growth factor receptor in ovarian cancer. J Exp Clin Cancer Res. 2013;32:102. doi: 10.1186/1756-9966-32-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Bi FF, Cao JM, Cao C, Liu B, Yang Q. Regulation of DNA methyltransferase 1 transcription in BRCA1-mutated breast cancer: A novel crosstalk between E2F1 motif hypermethylation and loss of histone H3 lysine 9 acetylation. Mol Cancer. 2014;13:26. doi: 10.1186/1476-4598-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Bi FF, Cao JM, Cao C, Li CY, Liu B, Yang Q. Poly (ADP-ribose) polymerase 1 transcriptional regulation: A novel crosstalk between histone modification H3K9ac and ETS1 motif hypomethylation in BRCA1-mutated ovarian cancer. Oncotarget. 2013;5:1. doi: 10.18632/oncotarget.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Song Y, Cho N, Chang JM, Koo HR, Yi A, Kim H, Park S, Moon WK. An HR-MAS MR metabolomics study on breast tissues obtained with core needle biopsy. PLoS One. 2011;6:e25563. doi: 10.1371/journal.pone.0025563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin HJ, Baek HM, Cha JH, Kim HH. Evaluation of breast cancer using proton MR spectroscopy: total choline peak integral and signal-to-noise ratio as prognostic indicators. AJR Am J Roentgenol. 2012;198:W488–W497. doi: 10.2214/AJR.11.7292. [DOI] [PubMed] [Google Scholar]
- 11.Cho K, Mabasa L, Bae S, Walters MW, Park CS. Maternal high-methyl diet suppresses mammary carcinogenesis in female rat offspring. Carcinogenesis. 2012;33:1106–1112. doi: 10.1093/carcin/bgs125. [DOI] [PubMed] [Google Scholar]
- 12.Xu X, Gammon MD, Zeisel SH, Lee YL, Wetmur JG, Teitelbaum SL, Bradshaw PT, Neugut AI, Santella RM, Chen J. Choline metabolism and risk of breast cancer in a population-based study. FASEB J. 2008;22:2045–2052. doi: 10.1096/fj.07-101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CX, Pan MX, Li B, Wang L, Mo XF, Chen YM, Lin FY, Ho SC. Choline and betaine intake is inversely associated with breast cancer risk: a two-stage case-control study in China. Cancer Sci. 2013;104:250–258. doi: 10.1111/cas.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 15.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 16.Salvi F, Gadda G. Human choline dehydrogenase: medical promises and biochemical challenges. Arch Biochem Biophys. 2013;537:243–252. doi: 10.1016/j.abb.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vance DE. Physiological roles of phosphatidylethanolamine N-methyltransferase. Biochim Biophys Acta. 2013;1831:626–632. doi: 10.1016/j.bbalip.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Morita SY, Takeuchi A, Kitagawa S. Functional analysis of two isoforms of phosphatidylethanolamine N-methyltransferase. Biochem J. 2010;432:387–398. doi: 10.1042/BJ20100490. [DOI] [PubMed] [Google Scholar]
- 19.Dória ML, Cotrim CZ, Simões C, Macedo B, Domingues P, Domingues MR, Helguero LA. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J Cell Physiol. 2013;228:457–468. doi: 10.1002/jcp.24152. [DOI] [PubMed] [Google Scholar]
- 20.Fischer LM, da Costa KA, Galanko J, Sha W, Stephenson B, Vick J, Zeisel SH. Choline intake and genetic polymorphisms influence choline metabolite concentrations in human breast milk and plasma. Am J Clin Nutr. 2010;92:336–346. doi: 10.3945/ajcn.2010.29459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Gammon MD, Zeisel SH, Bradshaw PT, Wetmur JG, Teitelbaum SL, Neugut AI, Santella RM, Chen J. High intakes of choline and betaine reduce breast cancer mortality in a population-based study. FASEB J. 2009;23:4022–4028. doi: 10.1096/fj.09-136507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristensen LS, Raynor MP, Candiloro I, Dobrovic A. Methylation profiling of normal individuals reveals mosaic promoter methylation of cancer-associated genes. Oncotarget. 2012;3:450–461. doi: 10.18632/oncotarget.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hossain MZ, Healey MA, Lee C, Poh W, Yerram SR, Patel K, Azad NS, Herman JG, Kern SE. DNA-intercalators causing rapid re-expression of methylated and silenced genes in cancer cells. Oncotarget. 2013;4:298–309. doi: 10.18632/oncotarget.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amodio N, Leotta M, Bellizzi D, Di Martino MT, D'Aquila P, Lionetti M, Fabiani F, Leone E, Gullà AM, Passarino G, Caraglia M, Negrini M, Neri A, et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget. 2012;3:1246–1258. doi: 10.18632/oncotarget.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla V, Coumoul X, Lahusen T, Wang RH, Xu X, Vassilopoulos A, Xiao C, Lee MH, Man YG, Ouchi M, Ouchi T, Deng CX. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 2010;20:1201–1215. doi: 10.1038/cr.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Gammon MD, Zhang Y, Bestor TH, Zeisel SH, Wetmur JG, Wallenstein S, Bradshaw PT, Garbowski G, Teitelbaum SL, Neugut AI, Santella RM, Chen J. BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat. 2009;115:397–404. doi: 10.1007/s10549-008-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imbard A, Smulders YM, Barto R, Smith DE, Kok RM, Jakobs C, Blom HJ. Plasma choline and betaine correlate with serum folate, plasma S-adenosyl-methionine and S-adenosyl-homocysteine in healthy volunteers. Clin Chem Lab Med. 2013;51:683–692. doi: 10.1515/cclm-2012-0302. [DOI] [PubMed] [Google Scholar]
- 30.Beetstra S, Suthers G, Dhillon V, Salisbury C, Turner J, Altree M, McKinnon R, Fenech M. Methionine-dependence phenotype in the de novo pathway in BRCA1 and BRCA2 mutation carriers with and without breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2565–2571. doi: 10.1158/1055-9965.EPI-08-0140. [DOI] [PubMed] [Google Scholar]
- 31.Cao J, Song Y, Bi N, Shen J, Liu W, Fan J, Sun G, Tong T, He J, Shi Y, Zhang X, Lu N, He Y, et al. DNA methylation-mediated repression of miR-886-3p predicts poor outcome of human small cell lung cancer. Cancer Res. 2013;73:3326–3335. doi: 10.1158/0008-5472.CAN-12-3055. [DOI] [PubMed] [Google Scholar]
- 32.Speirs V, Green AR, Walton DS, Kerin MJ, Fox JN, Carleton PJ, Desai SB, Atkin SL. Short-term primary culture of epithelial cells derived from human breast tumours. Br J Cancer. 1998;78:1421–1429. doi: 10.1038/bjc.1998.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.