Abstract
Osterix (Osx) is a zinc-finger-containing transcription factor that is highly specific to osteoblasts in vivo. Because Osx homozygous null mutants die in the immediate perinatal period showing a complete absence of bone formation, it is impossible determine the role that Osx plays in bones that have already formed after birth. To determine whether Osx is essential for bone maintenance and homeostasis, we conditionally inactivated the Osx gene in adult bone using the Cre/loxP recombination system. In previous reports, 2.3-kb Col1a1-CreERT2 mice that expressed a Cre recombinase that is transiently inducible by 4-hydroxytamoxifen (4-OHT) were intercrossed with Rosa26R (R26R) reporter mice, which resulted in the production of Cre-expressing osteoblasts that were detected upon X-gal staining. In the present study, inducible Col1a1-CreERT2 transgenic mice and conditional Osx mice (Osxflox/+) were used to generate Osxflox/−; Col1a1-CreERT2 mice. The Osx gene in Osxflox/−; Col1a1-CreERT2 mice was inactivated in the osteoblasts of already formed bones by active Cre recombinase after the administration of 4-OHT. The bones from 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice and oil-treated control mice were analyzed by radiography, histology, and histomorphometry. Even though no significant difference was observed in the radiographic images of the whole mouse skeletons, the mineralized trabecular bone volume and number in lumbar vertebrae were remarkably reduced in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. In addition, the rate of bone formation and area of mineralized surface were also reduced in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. Osx inactivation in already formed bones during the postnatal period caused a functional defect in osteoblasts that was followed by a reduction of bone formation, even though there were no apparent differences in osteoblast proliferation and osteoclast formation. Taken together, these results indicate that Osx is required to maintain osteoblast function following adult bone maintenance.
Keywords: Osterix, Col1a1-CreERT2, Postnatal, Osteoblasts, Bone maintenance
Introduction
Osterix (Osx) is a major effector of the osteoblast genetic program. Osx-null mice have a normal cartilage skeleton but completely defective endochondral and intramembranous bone formation [1]. In Osx-null mutants, preosteoblasts are arrested in their differentiation and unable to express a wide variety of genes characteristic of the osteoblast phenotype, though the expression of Runx2, another transcription factor needed for osteoblast differentiation [2–5], is normal [1]. As a result, these mutant mice die at birth. Perinatal lethality prevents study of the role of Osx in postnatal bone development and physiology. To avoid perinatal lethality, gene-targeting using site-and time-specific recombination based on the Cre/loxP system has been used to delete genes in specific tissues and stages of development before birth or during growth after birth [6–9].
The Cre recombinase system is the most efficient strategy used to bypass perinatal lethality and achieve conditional gene knockouts [10–12]. The conditional knockout mouse lines with silent genetic alterations are activated by Cre-mediated excision using this system. Therefore, the genomic alterations can be generally or tissue- and stage-specifically generated depending on the Cre expression [10]. The use of inducible Cre recombinase has been developed to delicately regulate the onset of Cre expression [13–16]. To render the Cre recombinase inducible, Cre recombinase fuses to the mutated ligand-binding domain (LBD) of the estrogen receptor (ER). The CreER fusion protein becomes active only after administration of the synthetic ligand estrogen antagonist, tamoxifen or 4-hydroxytamoxifen (4-OHT), after which nuclear localization from the cytoplasm and recombination at the target DNA flanked by loxP occurs [13,14,17–19].
Type I collagen is synthesized by various types of fibroblasts, as well as by mesenchymal cells [20]. In previous studies, transgenic mice harboring a 2.3-kb Col1a1 promoter were identified and found to exhibit a high activity of the transgene in osteoblasts and odontoblasts [21–23]. Recently, we generated 2.3-kb Col1a1-Cre transgenic mice and demonstrated that bone transcription factor Osx positively regulates adult bone formation [24]. We also reported that inducible CreER transgenic mice with the same promoter are good models for the spatio-temporal expression of Cre recombinase [25].
Here, we evaluated osteoblast-specific transcription factor Osx to determine whether it is essential for maintenance of the osteoblast function and for bone homeostasis in already formed bones after birth. To address this question, we generated Osxflox/−; Col1a1-CreERT2 mice that contain one Osx-null allele [1] and one conditional Osxflox allele [26], as well as an inducible Col1a1-CreERT2 transgene [25]. Osx exon 2 flanked by loxP sites in the Osxflox allele of these mice was excised by active Cre recombinase in the osteoblasts of all bones after the administration of 4-OHT at 2 weeks of age. Even though radiographic imaging revealed no differences between 4-OHT- and oil-treated Osxflox/−; Col1a1-CreERT2 mice, Osx deficiency in osteoblasts by 4-OHT treatment after birth resulted in reduced bone formation that was evident upon histological and histomorphometric analysis. This was due to the reduction in osteoblast function by Osx, not due to osteoclasts. These results demonstrate that Osx plays an essential role in the maintenance of osteoblast function and bone formation in postnatal bones and should help define the role of Osx in adult bone physiology and diseases.
Materials and methods
Animals
Conditional Osx floxed (Osxflox/+) mice [26] were used to generate OsxΔex/+ and OsxΔex/− mice or Osxflox/−; Col1a1-CreERT2 mice. Briefly, to generate mice carrying the deleted Osx exon 2 (OsxΔex) allele in which the Osx exon 2 flanked by loxP was excised, the conditional Osxflox/+ heterozygous mice were crossed with Protamine I-Cre (PrmI-Cre) transgenic mice [27,28]. OsxΔex/− null mutant mice were prepared by crossing OsxΔex/+ with Osx+/− heterozygous mice with a LacZ knock-in in the Osx locus [1]. Osxflox/−; Col1a1-CreERT2 mice were then generated by crossing conditional Osxflox/+ mice with Osx+/− mice and a Col1a1-CreERT2 transgenic line [25]. To delete Osx, Osxflox/−; Col1a1-CreERT2 mice were intraperitoneally injected with the synthetic estrogen antagonist, 4-hydroxytamoxifen (4-OHT, Sigma-Aldrich). Mice from the same litter of the same gender and age that had a similar size and body weight and contained the floxed allele, the LacZ allele, and the Col1a1-CreERT2 transgene were utilized in the experiment. All procedures concerning animal experiments were conducted with the approval of Kyungpook National University.
PCR genotyping
PCR genotyping of the offspring was conducted using 100 ng of genomic DNA from the yolk sac or tail to detect flox, LacZ, and Cre from the floxed allele, LacZ allele, and Cre transgene, respectively. The primers and reactions used for PCR amplification of the flox, LacZ, and Cre genes have been described previously [24,25]. To detect the OsxΔex allele after the administration of 4-OHT, PCR was conducted using the Δex-specific primers, 5′-CTTGGGAACACTGAAGCTGT-3′ and 5′-GCACACCGGCCTTATTCC-3′.
Skeletal preparation and evaluation of GFP expression by confocal microscopy
Skeletons from embryos at embryonic day 18.5 (E18.5) were prepared and stained with alcian blue for cartilage and alizarin red S for bone. Briefly, the skin and viscera were removed, after which the embryos were fixed in 95% ethanol overnight and then stained in 150 mg/l alcian blue solution (Sigma) with 20% acetic acid and 80% ethanol overnight. The embryos were then rinsed in 95% ethanol for at least 3 h, after which they were treated with 2% KOH for 24 h to clarify soft tissues. Next, the bones were stained in 50 mg/l alizarin red solution with 1% KOH. Finally, the skeletons were cleared in 1% KOH with 20% glycerol for at least 2 days. To detect the EGFP expression that recapitulates the Osx expression after deletion of the Osx gene, the bones that had the skin removed were observed using a confocal microscope (Leica, Germany).
Radiographic imaging, histology, and histomorphometry in bones
To observe the skeletal deformations, radiographic images of the total skeletons were taken using a small animal X-ray at 36 kV for 1 min. The mice were injected with 30 mg/kg of calcein at 6 and 2 days prior to sacrifice. The bone tissues were then fixed in 4% paraformaldehyde (PFA) at 4 °C overnight. Next, the undecalcified lumbar vertebrae were embedded in destabilized methyl methacrylate resin [29], after which 5-μm sectioned lumbar vertebrae were stained with van Gieson and von Kossa reagents. Static and dynamic histomorphometric analysis was conducted on the lumbar vertebrae using the Bioquant program (Bio-Quant. Inc., San Diego) [30]. Statistical differences were assessed by t-test. Decalcified long bones were embedded in paraffin and then sectioned in 6 μm. The sections were subjected to hematoxylin and eosin (H and E), alcian blue, and tartrate-resistant acidic phosphatase (TRAP) staining. BrdU incorporation in mice that had been intraperitoneally injected with 100 μg BrdU per gram of body weight for 3 h before sacrifice was detected using anti-BrdU antibody (Zymed).
Immunohistochemistry
Immunodetection of Osx was performed using anti-mouse Osx antibody (Abcam, Massachusetts, USA) with a standard protocol. Briefly, 6-μm sections from 4% PFA-fixed paraffin-embedded blocks were blocked with 3% bovine serum albumin (BSA) at room temperature for 1 h. To minimize the non-specific staining, avidin/biotin blocking was performed (Vector Laboratories, Burlingame, USA). The sections were incubated with anti-mouse Osx antibody diluted 1:500 in 0.03% phosphate-buffered saline (PBS) overnight at 4 °C. After incubation with primary antibody, the slides were washed in PBS and then incubated with a biotinylated anti-goat IgG antibody (VectaStain ABC Kit, Vector Laboratories) at room temperature for 1 h. Endogenous peroxidase activity was quenched in 0.3% H2O2 in methanol for 10 min, washed in PBS, and then treated with the ABC reagents (Vector Laboratories). Signal for antibody binding was visualized with diaminobenzidine (DAB) substrate (Zymed Laboratories, California, USA). Counterstaining was performed with methyl green.
Quantitative real-time RT-PCR analysis
Total RNA was isolated from long bones in oil-and 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice at 4 weeks of age using TRI reagent (Sigma-Aldrich). RNA was subjected to quantitative real-time RT-PCR as described [24]. The following primers for marker genes of osteoblast differentiation were used: alkaline phosphatase (ALP), 5′-AACCCAGACACAAGCATTCC-3′ and 5′-GCCTTTGAGGTTTTTGGTCA-3′; bone sialoprotein (BSP), 5′-ACCCCAAGCACAGACTTTTGA-3′ and 5′-CTTTCTGCATCTCCAGCCTTCT-3′; Col1a1, 5′-CCTGAGTCAGCAGATTGAGAACA-3′ and 5′-CCAGTACTCTCCGCTCTTCCA-3′; osteocalcin (OCN), 5′-GCGCTCTGTCTCTCTGACCT-3′ and 5′-ACCTTATTGCCCTCCTGCTT-3′. The quantified individual RNA expression was normalized to GAPDH and depicted as relative RNA expression levels with the corresponding oil-treated control mice set to 1.0.
Results
Generation of Osx-inactivated mice with EGFP expression in bones
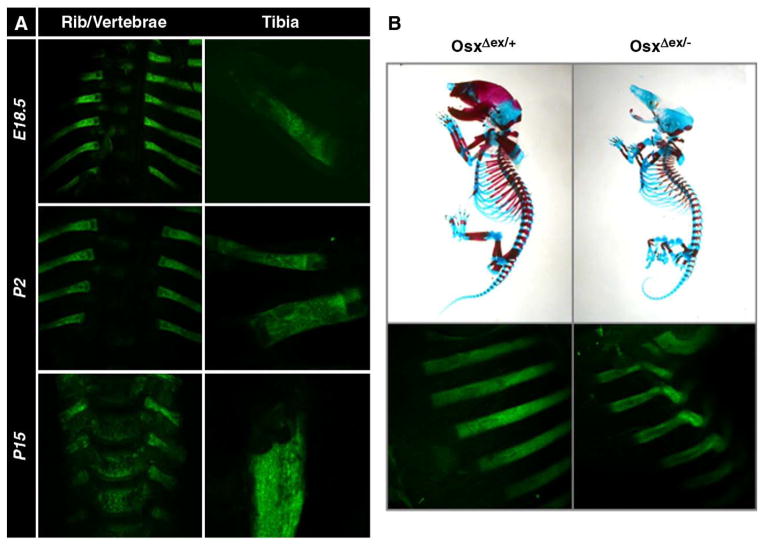
Osx heterozygous mice are normal and fertile but Osx homozygous null mutant mice die in the immediate perinatal period due to an inability to breathe [1]. In null mutants, osteoblast differentiation is arrested. In a previous report, conditional Osxflox/+ mice were generated in which inactivation of Osx is able to be induced in the osteoblasts using the Cre/loxP system [26]. The conditional Osxflox/+ mice were crossed with PrmI-Cre transgenic mice [27,28] to generate mice carrying the OsxΔex/+ allele, in which the Osx exon 2 flanked by loxP is excised by Cre-mediated recombination to inactive Osx function. After deletion of the Osx gene by PrmI-Cre, EGFP expression was linked under control of the regulatory elements of the Osx gene instead of the deleted Osx using a targeting vector strategy described in a previous study [26]. During development and growth, heterozygous embryos and pups carrying the OsxΔex allele expressed EGFP which recapitulated the expression of Osx in all bones (Fig. 1A). Under confocal microscopy, EGFP expression was easily observed in all bones from E15.5 without using any staining methods. Bone deformity in response to EGFP expression was examined in OsxΔex/− null mutants, which were prepared by crossing OsxΔex/+ mice with Osx+/− mice that contained a LacZ knock-in in the Osx locus [1]. Skeletal preparation of OsxΔex/+ heterozygous and OsxΔex/− null mutant embryos revealed that they had the same phenotype as Osx+/− and Osx−/− embryos (Fig. 1B, upper). Furthermore, bending of the ribs which was observed in EGFP expression was clearly evident in OsxΔex/− null mutant embryos compared to OsxΔex/+ heterozygotes (Fig. 1B, lower), indicating that bone deformity was easily observed by EGFP expression without any staining methods. No EGFP expression was detected in the bones of conditional Osxflox/+ mice prior to Cre-mediated recombination (data not shown).
Fig. 1.
EGFP expression that recapitulates the Osx expression in bones after Cre-mediated Osx excision. (A) EGFP expression in OsxΔex/+ heterozygous embryo and pups at embryonic day 18.5 (E18.5) and postnatal days 2 and 15 (P2 and P15). Heterozygotes carrying the OsxΔex allele expressed EGFP to recapitulate Osx expression in all bones from embryos and pups. (B) Skeletons of OsxΔex/+ and OsxΔex/− mice were stained with alcian blue and alizarin red at E18.5, indicating an absence of mineralization in OsxΔex/− null mutants. Bone deformity was remarkably observed in OsxΔex/− null mutants by EGFP expression.
Generation of mice in which Osx inactivation in adult bones was inducible
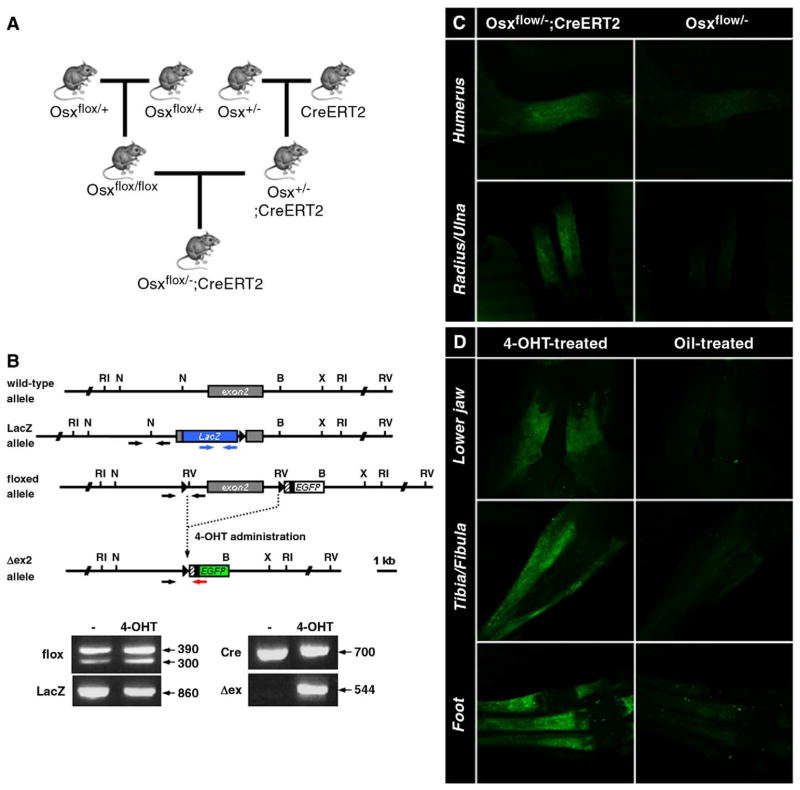
To examine the role of Osx in adult bones, it was necessary to inactivate Osx in the bones of postnatal mice using the inducible Cre system. Inactivation of Osx was conducted using Col1a1-CreERT2 transgenic mice in which a 2.3-kb mouse Col1a1 osteoblast-specific promoter drives Cre recombinase fused to a mutant ligand binding domain (LBD) of the estrogen receptor (ER). In these mice, the CreER fusion polypeptide becomes active only after administration of the synthetic estrogen antagonist, 4-OHT [25]. Here, Osxflox/−; Col1a1-CreERT2 mice were generated by crossing Osx+/− mice with inducible Col1a1-CreERT2 transgenic mice and then with conditional Osxflox/+ mice (Fig. 2A). In Osxflox/−; Col1a1-CreERT2 mice, PCR genotyping for the flox allele, LacZ allele, and Cre transgene was conducted using genomic DNAs extracted from embryos or pups (Fig. 2B). Osxflox/−; Col1a1-CreERT2 mice harboring these alleles matured normally, as did Osx heterozygous mice. Following the administration of 4-OHT, the Osx exon 2, which was flanked by loxP sites in a floxed Osx allele, was excised by active Cre recombinase in all bones. Precise excision of the Osx exon 2 was validated by PCR genotyping as shown in Δex (Fig. 2B). To determine the ability of Cre to induce a specific excision in bone, 4-OHT was administrated intraperitoneally into pregnant dams in which Osxflox/−; Col1a1-CreERT2 embryos were generated or into Osxflox/−; Col1a1-CreERT2 pups after birth. Osxflox/−; Col1a1-CreERT2 embryos from dams treated with 4-OHT showed EGFP expression in bone, whereas treatment of Osxflox/− embryos induced no EGFP expression (Fig. 2C). After intraperitoneal injection with 4-OHT for 5 consecutive days starting at 12 days of age, EGFP expression was detected in all bones (Fig. 2D). Strong EGFP expression was observed in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 pups compared to oil-treated controls. No EGFP expression was detected in the bones of Osxflox/−; Col1a1-CreERT2 pups before Cre-mediated excision was induced by the administration of 4-OHT (data not shown).
Fig. 2.
Generation of the Osxflox/−; Col1a1-CreERT2 mice. (A) Breeding scheme to generate Osxflox/−; Col1a1-CreERT2 mice by crossing conditional Osxflox/+ mice with Osx+/− mice with a LacZ knock-in in the Osx locus and a Col1a1-CreERT2 transgenic line. Inducible Col1a1-CreERT2 transgenic mice were used to excise the Osx gene from osteoblasts by the administration of 4-OHT. (B) PCR genotyping to detect the flox, LacZ, Cre allele, and deleted Osx exon 2 (Δex) before and after the administration of 4-OHT. Primers for each PCR genotyping were indicated in structure of the genomic Osx locus: black arrows for the floxed allele, blue arrows for the LacZ allele, and black and red arrows for the Δex allele. In PCR with primers of black arrows, the wild-type and the floxed alleles were amplified to generate a 300-bp and 390-bp fragments, respectively. (C, D) 4-OHT-induced expression of EGFP in Osxflox/−; Col1a1-CreERT2 embryos and pups. (C) Pregnant females from the breeding scheme shown in (A) were injected with 4-OHT. Only Osxflox/−; Col1a1-CreERT2 embryos from females expressed EGFP in bone. (D) Osxflox/−; Col1a1-CreERT2 pups were injected with 1 mg of 4-OHT for 5 consecutive days starting at postnatal day 12, while control pups were injected with oil. EGFP expression was observed in bone, including the jaw, digits, and tibia/fibula. No EGFP expression was detected in oil-injected control animals.
Reduced bone formation in mice which the Osx gene has been inactivated postnatally
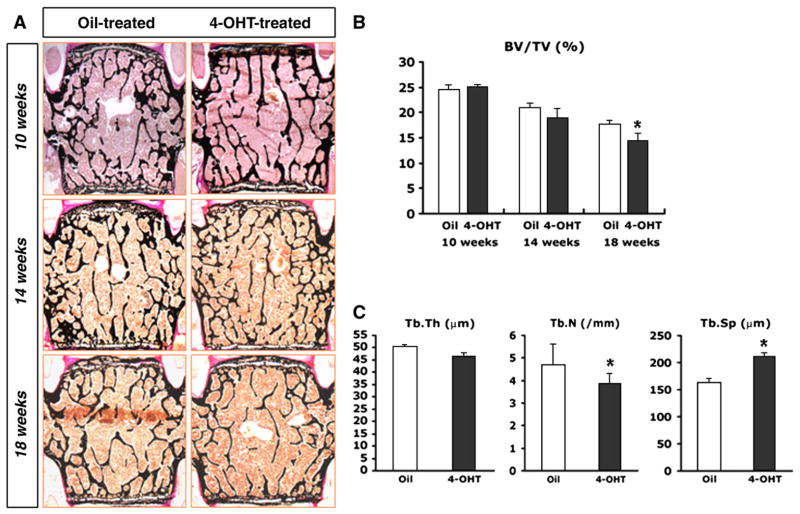
To evaluate the possible role of Osx in bones that had already formed, Osxflox/−; Col1a1-CreERT2 mice were intraperitoneally injected with 1 mg of 4-OHT or oil for 5 consecutive days, two times as shown in the study scheme (Fig. 3A). The injected mice were then sacrificed at 4, 8 or 12 weeks after the final injection, after which their bones were analyzed. Radiographic imaging of the whole mouse skeleton using an X-ray did not reveal any significant differences in the bone density of 4-OHT-and oil-treated Osxflox/−; Col1a1-CreERT2 mice at 12 weeks after the final injection (Fig. 3B). Histological analysis of the bones was conducted by staining undecalcified lumbar vertebrae from 4-OHT-and oil-treated Osxflox/−; Col1a1-CreERT2 mice with von Kossa method (Fig. 4A). Although there was almost no difference observed between groups at 10 and 14 weeks of age, the mineralized trabecular bone volume was reduced in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice at 18 weeks of age (Fig. 4A, B). Additionally, the trabecular bone mass was low in all 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice, regardless of gender (data not shown). Histomorphometric analysis of the lumbar vertebra at 18 weeks of age revealed a significant reduction in trabecular number (Tb.N) and an increase in trabecular separation (Tb.Sp) in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice compared to oil-treated mice (Fig. 4C).
Fig. 3.
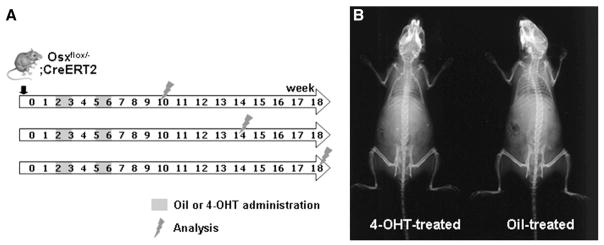
Induction of recombination in the bones of Osxflox/−; Col1a1-CreERT2 mice. (A) Scheme for 4-OHT administration to inactivate Osx. Mice were intraperitoneally injected with 1 mg of 4-OHT or oil for 5 consecutive days, twice. The treated mice were sacrificed 4, 8, or 12 weeks after the final injection and their bones were then analyzed. (B) X-ray radiography of whole skeletons of 4-OHT or oil-treated Osxflox/−; Col1a1-CreERT2 mice at 18 weeks of age. No difference in the lucency of the entire skeleton was observed between 4-OHT-treated mice and the oil-treated controls.
Fig. 4.
Reduced bone mass in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. (A) Histological analysis with von Kossa staining of the lumbar vertebrae from Osxflox/−; Col1a1-CreERT2 mice. Osx was inactivated in the osteoblasts of intact bones after birth by the administration of 4-OHT using an inducible Cre system. Decreased bone mass was observed in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice compared to oil-treated controls at 18 weeks of age. (B, C) Histomorphometric analysis in Osxflox/−; Col1a1-CreERT2 mice. At 18 weeks of age, the significant decrease of bone mass and trabecular numbers was observed in 4-OHT-treated (black bar) mice compared to oil-treated Osxflox/−; Col1a1-CreERT2 mice (white bar), whereas trabecular separation was increased in 4-OHT-treated mice. BV/TV, bone volume per tissue volume; Tb.Th, trabecular thickness; Tb.N, trabecular number; Tb.Sp, trabecular separation; *p<0.05.
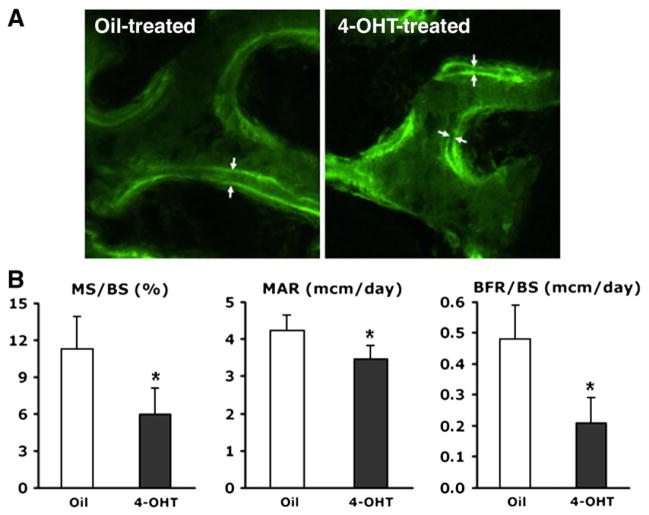
To determine whether Osx has an effect on osteoblast function during the postnatal period, the bone forming rate (BFR) was examined in 4-OHT-and oil-treated Osxflox/−; Col1a1-CreERT2 mice using calcein double labeling. Calcein double labeling was observed in the surfaces of the trabecular bone of Osxflox/−; Col1a1-CreERT2 mice (Fig. 5A). The surface of calcein labeling and the distances between the double labeling were clearly reduced in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice compared to oil-treated mice. In 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice, histomorphometric analysis revealed a significant reduction in the mineralized surface (MS), mineral apposition rate (MAR), and BFR (Fig. 5B). As a result, 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice that lacked the Osx gene in the osteoblasts of already formed bones showed a reduced ability to maintain bones.
Fig. 5.
Decreased bone formation in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice at 18 weeks of age. (A) Fluorescent micrographs of calcein double labeling with the distance indicating osteoblast functional activity. A reduced distance between the two labels was observed in 4-OHT-treated mice compared to oil-treated Osxflox/−; Col1a1-CreERT2 mice. (B) Histomorphometric analysis of calcein-labeled lumbar vertebrae in oil-and 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. MS, MAR, and BFR were reduced remarkably in the bones of 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice (black bar). MS/BS, mineralized surface per bone surface; MAR, mineral apposition rate; BFR/BS, bone forming rate per bone surface; *p<0.05.
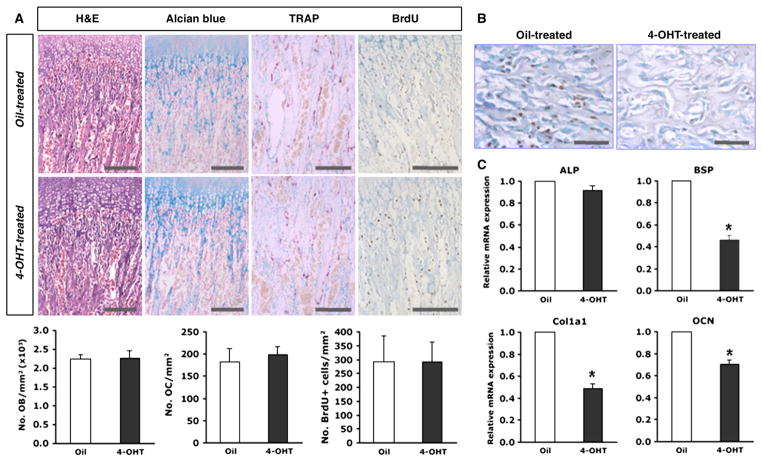
Histological analysis of the tibial bones of 4-OHT-and oil-treated Osxflox/−; Col1a1-CreERT2 mice was conducted (Fig. 6A). Alterations in the histological morphology of bone and differentiating chondrocytes were not observed in 4-OHT-treated Osxflox/−; Col1a1-Cre mice that were analyzed by H and E and alcian blue staining, respectively. The number of osteoblasts per trabecular bone area was also identical in both mice (Fig. 6A). To evaluate the osteoclastic cell function, the trabecular regions of the tibia were compared in TRAP-positive cells. Multinucleated giant cells representing functional osteoclasts were identical in both 4-OHT- and oil-treated Osxflox/−; Col1a1-CreERT2 mice (Fig. 6A). These results indicated that Osx inactivation in the osteoblasts of already formed bones did not affect osteoclast differentiation and function, and that the reduced bone mass in these Osx-inactivated mice was not due to bone resorption. Due to the altered osteoblast function, the BrdU incorporation of proliferating cells was analyzed to determine whether osteoblast proliferation was affected by Osx inactivation. The number of BrdU-labeled osteoblasts per trabecular bone area made no difference between 4-OHT- and in oil-treated Osxflox/−; Col1a1-CreERT2 mice, indicating no significant distinction in osteoblast proliferation between both groups (Fig. 6A). The frequency of the deletion of the Osx exon 2 was investigated in previous report which was more than 60% of the osteoblasts by Cre recombinase after 4-OHT treatment [25]. To confirm this frequency, immunohistochemistry using anti-Osx antibody was performed in 4-OHT-treated Osxflox/−; Col1a1-CreERT2. Consequently, Osx expression was almost not detected in bones of 4-OHT-treated Osxflox/−; Col1a1-CreERT2 compared to oil-treated control mice (Fig. 6B), indicating that the frequency of the deletion of the Osx exon 2 was more than 90% of the osteoblasts in Osxflox/−; Col1a1-CreERT2 by Cre recombinase after the administration of 4-OHT. To evaluate the function of osteoblasts, the expression of osteoblast differentiation marker genes was examined in Osxflox/−; Col1a1-CreERT2 after the administration of 4-OHT. Marker genes of osteogenic differentiation, bone sialoprotein (BSP) and type I collagen (Col1a1) were decreased in these mice, whereas early marker genes, alkaline phosphatase (ALP) and osteopontin were not affected (Fig. 6C and data not shown). Osteocalcin, a late marker gene of osteoblast differentiation, was also significantly reduced in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 (Fig. 6C). Finally, these results indicated that reduced bone formation in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice was due to altered osteoblast function in response to Osx inactivation, but that this inactivation did not cause any defects in osteoblastic cell proliferation or bone resorption by osteoclasts.
Fig. 6.
Osteoblast differentiation in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. (A) No overt histological phenotype in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. Longitudinal sections of tibiae were subjected to H and E, alcian blue, and TRAP staining. No morphological differences were observed in H and E staining. In differentiating chondrocytes and mature osteoclasts by alcian blue and TRAP staining, respectively, no significant differences were observed between 4-OHT-treated and oil-treated Osxflox/−; Col1a1-CreERT2 mice. In vivo cell proliferation was analyzed based on the incorporation of BrdU into the mice tibia. BrdU-positive cells were not altered in the tibia of 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice compared to the controls. The numbers of osteoblasts, osteoclasts, and BrdU-positive cells were quantified in both mice. No significant differences were observed. Representative images and analysis were shown in mouse bones at 10 weeks of age. Scale bar=200 μm. (B) Immunohistochemical analysis using anti-Osx antibody. Osteoblasts with Osx expression were detected in black. No signal was observed in osteoblasts of 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. Representative images were shown in mouse bones at 10 weeks of age. Scale bar=50 μm. (C) Expression of marker genes related to osteoblastic cell differentiation by quantitative real-time RT-PCR analysis. The expression of osteogenic markers, BSP and Col1a1, and a late marker of osteoblast differentiation, OCN, were obviously reduced, whereas the expression of an early marker gene, ALP, was not significantly changed in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 compared to oil-treated controls. *p<0.05.
Discussion
Bone remodeling occurs throughout life and involves breakdown of the bone matrix through resorption by osteoclasts and subsequent rebuilding through new bone formation by osteoblasts [31,32]. Usually, these two processes balance each other and a stable level of bone mass is maintained. In young adults, the amount of new bone growth is equal to the amount of bone resorption. As people age, however, more resorption than formation occurs. Osteoblasts are bone-forming cells that are derived from mesenchymal precursor cells. Osteoblasts synthesize a large number of proteins that represent the genetic program of these cells. In particular, type I collagen comprises more than 90% of the organic matrix of bone [20]. To evaluate the age-dependent functions of specific genes in the skeleton, it is important to disrupt these genes at different ages in an osteoblast-specific manner. Based on the characteristics of osteoblasts that change as they mature [33,34], specific genes have been disrupted in bone under the control of type I collagen promoter. For example, 3.6-kb and 3.2-kb Col1a1 promoters are expressed early in osteoblastogenesis, pre-osteoblasts [35,36]. A 2.3-kb Col1a1 promoter is highly active in maturing osteoblasts, while osteocalcin promoter is active very late in mouse embryogenesis and highly restricted to differentiated osteoblasts [23,35]. An inducible Cre system with these promoters more effectively controls the onset of the gene disruption in osteoblasts at any age in response to the administration of 4-OHT [25,36]. Therefore, this study was conducted by employing a CreER system under the control of a 2.3-kb Col1a1 promoter, which is very useful for evaluation of the role of osteoblast-specific genes in bones that have already formed bones after birth.
There are many factors that regulate osteoblast function during the formation and mineralization of bones. One of the primary regulators, Osx, is an osteoblast-specific transcription factor in vivo [1]. Before birth, Osx is expressed strongly in cells that are associated with bone trabeculae and bone collar formation. After birth, expression remains strong in bone trabeculae and in secondary ossification centers. Osx-null mutant embryos show a complete absence of bone formation. Indeed, in such embryos, bone trabeculae are completely absent and no mineralization occurs in the skeletal elements [1]. In a previous study using 2.3-kb Col1a1-Cre, Osx was inactivated in osteoblasts after bone collar formation at mouse embryonic day 14.5 [24]. Conditional Osx inactivation in osteoblasts under the control of 2.3-kb Col1a1-Cre resulted in osteopenia in adult bone during growth. Interestingly, even though Osx inactivation was started at embryonic day, severe difference was observed in bones of Osxflox/−; Col1a1-Cre from 6 weeks of age while no alteration of bone phenotype was observed in embryonic day or newborns. This result may be explained that the differentiation and function of osteoblasts by Osx inactivation was reduced first, followed by the visual decreases in bone mass. Like this report, even though Osx was inactivated in Osxflox/−; Col1a1-CreERT2 mice after 4-OHT administration at 2 weeks of age, the reduction of Col1a1 and BSP expression was drastic at around 4 weeks of age. Sequentially, bone loss was visually detected in several weeks later due to reduced differentiation and function of osteoblasts and finally the mild reduction of bone mass was observed. This result indicated that a series of sequential process and sufficient time may be needed to exhibit the reduced bone morphology by Osx gene which was inactivated in already formed intact bones of postnatal stage. Eventually, these findings designate that Osx is needed for preosteo-blasts to differentiate into fully functioning osteoblasts or osteoblasts to maintain their function, and for subsequent bone formation and mineralization to occur. In this study, inducible Col1a1-CreERT2 mice were used to inactivate the Osx gene in osteoblasts via the administration of 4-OHT during postnatal periods to identify critical evidence regarding the possible role of Osx in adult bones. Whereas no severe alterations were observed in histological analysis, reduced bone formation was observed in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice due to a functional defect in osteoblasts as a result of Osx inactivation. These findings demonstrate the importance of Osx in postnatal bone formation and maintenance.
Processes of osteoblastogenesis and osteoclastogenesis are tightly coupled. It has been studied that failure of osteoblast differentiation affects osteoclast maturation and function [31,32]. However, previous reports have shown that the delayed osteoblast differentiation by Osx inactivation did not alter osteoclast maturation and function [1,24]. In the study with Osx null mutants [1], multinucleated osteoclastic cells with MMP-9 and cathepsin K expression were present in bones in absence of Osx, indicating that Osx null mutants have functional osteoclasts. The comparable result was also demonstrated in Osx-inactivated mice with Col1a1-Cre [24]. Even though Osx was inactivated in osteoblasts with 2.3-kb Col1a1-Cre, no difference in osteoclast activity for bone resorption was found by the number of TRAP-positive osteoclasts and the measurement of urinary deoxypyridinoline crosslinks. With the increase of immature osteoblasts in Osx-inactivated mice with Col1a1-Cre, RANKL and OPG, which are important for osteoclast differentiation and activity, were also increased. However, the relative ratio of RANKL/OPG was not significantly changed and hence, bone resorption was not altered in this mice. Similar to these reports, the osteoclast function was not changed by the reduced osteoblast function in 4-OHT-treated Osxflox/−; Col1a1-CreERT2 mice. Finally, this suggests that Osx inactivation was affected to osteoblast function not to osteoclast function or activity in bones. Taken together, the characterization of mice in which Osx is inactivated after birth should help define the role that Osx plays in bone physiology. If Osx is indeed essential for the maintenance of osteoblast function and bone homeostasis after birth, then this transcription factor should be a potential target for drugs designed to correct osteopenia, osteoporosis, and eventually other bone diseases.
Acknowledgments
This work was supported by the Korea Research Foundation Grant funded by Korea Government (MOEHRD, Basic Research Promotion Fund) (KRF-2005-204-E00006), the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MOST) (R01-2007-000-20005-0), and the Brain Korea 21 Project in 2009.
Footnotes
Conflict of interest statement
The authors state that they have no conflicts of interest.
References
- 1.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G. Role of Cbfa1 in osteoblast differentiation and function. Semin Cell Dev Biol. 2000;11:343–6. doi: 10.1006/scdb.2000.0188. [DOI] [PubMed] [Google Scholar]
- 3.Komori T. Runx2, a multifunctional transcription factor in skeletal development. J Cell Biochem. 2002;87:1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- 4.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 5.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 6.Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J, et al. Conditional gene targeting. J Clin Invest. 1996;98:600–3. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobe CG, Nagy A. Conditional genome alteration in mice. Bioessays. 1998;20:200–8. doi: 10.1002/(SICI)1521-1878(199803)20:3<200::AID-BIES3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 8.Rossant J, McMahon A. “Cre”-ating mouse mutants—a meeting review on conditional mouse genetics. Genes Dev. 1999;13:142–5. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 9.Le Y, Sauer B. Conditional gene knockout using Cre recombinase. Methods Mol Biol. 2000;136:477–85. doi: 10.1385/1-59259-065-9:477. [DOI] [PubMed] [Google Scholar]
- 10.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 11.Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–38. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–60. [PubMed] [Google Scholar]
- 13.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci USA. 1996;93:10887–90. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–5. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennighausen L, Furth PA. The right time and place for molecular scissors. Nat Biotechnol. 1999;17:1062–3. doi: 10.1038/15046. [DOI] [PubMed] [Google Scholar]
- 16.Utomo AR, Nikitin AY, Lee WH. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat Biotechnol. 1999;17:1091–6. doi: 10.1038/15073. [DOI] [PubMed] [Google Scholar]
- 17.Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch JL, Chambon P, et al. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc Natl Acad Sci USA. 1997;94:14559–63. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71–80. doi: 10.1006/meth.2001.1159. [DOI] [PubMed] [Google Scholar]
- 19.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–7. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 20.Rossert J, de Crombrugghe B. Type I collagen: structures, synthesis, and regulation. In: Bilezikian J, Raisz L, Rodan G, editors. Principles of Bone Biology. San Diego: Academic Press; 1996. pp. 127–42. [Google Scholar]
- 21.Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-alpha-1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–32. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossert JA, Chen SS, Eberspaecher H, Smith CN, de Crombrugghe B. Identification of a minimal sequence of the mouse pro-alpha-1(I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc Natl Acad Sci USA. 1996;93:1027–31. doi: 10.1073/pnas.93.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha-1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–51. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 24.Baek WY, Lee MA, Jung JW, Kim SY, Akiyama H, de Crombrugghe B, et al. Positive regulation of adult bone formation by osteoblast-specific transcription factor Osterix. J Bone Miner Res. 2009;24:1055–65. doi: 10.1359/jbmr.081248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JE, Nakashima K, de Crombrugghe B. Transgenic mice expressing a ligand-inducible Cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth. Am J Pathol. 2004;165:1875–82. doi: 10.1016/S0002-9440(10)63240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA. 2005;102:14665–70. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peschon JJ, Behringer RR, Brinster RL, Palmiter RD. Spermatid-specific expression of protamine 1 in transgenic mice. Proc Natl Acad Sci USA. 1987;84:5316–9. doi: 10.1073/pnas.84.15.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–7. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erben RG. Embedding of bone samples in methyl methacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. J Histochem Cytochem. 1997;45:307–13. doi: 10.1177/002215549704500215. [DOI] [PubMed] [Google Scholar]
- 30.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 31.Lemaire V, Tobin FL, Greller LD, Cho CR, Suva LJ. Modeling the interactions between osteoblast and osteoclast activities in bone remodeling. J Theor Biol. 2004;229:293–309. doi: 10.1016/j.jtbi.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Blair HC, Zaidi M, Schlesinger PH. Mechanisms balancing skeletal matrix synthesis and degradation. Biochem J. 2002;364:329–41. doi: 10.1042/BJ20020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubin JE. Advances in the osteoblast lineage. Biochem Cell Biol. 1998;76:899–910. [PubMed] [Google Scholar]
- 34.Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–55. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- 35.Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–53. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 36.Kamiya N, Ye L, Kobayashi T, Lucas DJ, Mochida Y, Yamauchi M, et al. Disruption of BMP signaling in osteoblasts through type IA receptor (BMPRIA) increases bone mass. J Bone Miner Res. 2008;23:2007–17. doi: 10.1359/JBMR.080809. [DOI] [PMC free article] [PubMed] [Google Scholar]