The innate immune system is critical for the early detection of invading pathogens and dangers. In the last few years, key cellular sensors responsible for sensing pathogen-derived RNA and triggering innate immune signaling have been discovered. However, it is less clear how the cell triggers innate immune signaling in response to cytoplasmic DNA species originating from human pathogenic microbes.
Recently, numerous cytosolic DNA sensors have been described, including DNA-dependent activator of IFN-regulatory factors (DAI, also known as Z-DNA-binding protein or ZBP-1), RNA polymerase III, leucine-rich repeat (LRR)-containing protein 1 (LRRFIP1), aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box helicase (DHX) 9, DHX36, Ku70, IFN-γ-inducible protein 16 (IFI16) and (DExD/H)-box polypeptide 41 (DDX41).1,2,3,4,5,6,7,8 These sensors are able to bind DNA directly and engage signaling pathways converging on NF-κB and IRF3 activation, leading to type I IFN induction. They display some selectivity in terms of cell type, the pathogens sensed and the exact nature of the DNA ligand tested. Research on DNA sensors is now of great interest in not only determining the mechanisms underlying how the pathogens are sensed by DNA sensors, but also understanding how DNA sensors elicit immune responses, including the induction of type I IFN.
DDX41 is a newly identified DNA sensor responsible for the recognition of cytosolic double-stranded DNA (dsDNA) and can recruit the stimulator of interferon genes (STING, also known as MITA, MPYS and ERIS) to activate IRF3 and NF-κB.9,10,11,12 Knockdown of DDX41 expression through short-hairpin RNA in both mouse and human cell lines blocked induction of the gene encoding IFN-β and other genes of the innate immune system in response to synthetic DNA sequences and DNA viruses. DDX41 is a member of the DEXDc family of helicases, which has two conserved domains: a DEAD domain and a helicase domain. DDX41 binds dsDNA or CDNs (Cyclic dinucleotides) and interacts with STING-dependent on its DEAD domain. However, how the binding of dsDNA or CDNs to the DEAD domain triggers the recruitment of signaling adaptors (or STING directly) and thereby stimulate downstream signaling is unknown. Another interesting question that needs to be resolved is the regulation of the DNA sensor-induced signaling pathway.
Zhiqiang et al.13 identified Trim21 as a DDX41-interacting protein through immunoprecipitation with an antibody to DDX41 in the mouse mDC line D2SC and protein sequencing by liquid chromatography-mass spectrometry. Trim21, also known as Ro52, is an interesting protein that belongs to the Trim family. Studies have demonstrated that many Trim family members play important roles in regulating the innate immune response. Trim21 itself has been reported to be a regulator of type I IFN production and the antiviral response. However, there is much debate about the function of Trim21. Higgs et al.14 provided evidence that Trim21 targeted IRF3 for polyubiquitin-mediated degradation and thus turned off the production of IFN-β. Subsequently, Yang and his co-workers15 identified Trim21 as an IRF3-interacting protein but with an opposite function. They reported that Trim21 enhanced IFN-β production by preventing IRF3 ubiquitination and degradation. The issue was further complicated when another group revealed that Trim21−/− elongation factors or immune cells exhibited no defect in type I IFN induction in response to Newcastle disease virus and the Toll-like receptor ligands poly(I:C), CpG and lipopolysaccharide, even though ubiquitylation of IRF3 was significantly reduced.16
Here, Zhiqiang et al.13 describe in their articles that Trim21 deletion led to only approximately 20% more IFN-β production in bone marrow-derived dendritic cells (BMDCs) treated with 5′-triphosphate RNA, lipopolysaccharide, poly(I:C) or reovirus, and Trim21 might have a much more important effect on the innate immune response to intracellular double-stranded DNA, as 1.5- to 4-fold more IFN-β was observed in Trim21-deficient BMDCs in response to poly(dA:dT), vaccinia virus DNA, adenovirus and herpes simplex virus type 1 (HSV-1).
To confirm their observations, they investigated the antiviral role of Trim21 in vivo. Trim21-deficient mice showed more than 90% lower viral titers than wild-type mice infected by HSV-1, with two-fold more type I IFN and IL-6 production, whereas only a small effect on type I IFN production was detected in Trim21-deficient mice in response to vesicular stomatitis virus, which is an RNA virus. Furthermore, similar results were obtained in Trim21 knockdown human THP-1 monocytes infected with intracellular DNA or HSV-1.
Trim21 bound to the DEADc domain of DDX41 through its SPRY–PRY domain. This interaction occurred in resting cells and diminished after 4 h of stimulation with intracellular DNA. Subcellular localization analysis showed that Trim21 and DDX41 localized together in the cytosol under resting conditions and that TRIM21 dissociated from DDX41 after stimulation with dsDNA. The facts that Trim21 had no effect in DDX41-silenced THP-1 cells in response to intracellular DNA or HSV-1 and that Trim21 interacted with DDX41 suggest that DDX41 might be the regulation target of Trim21.
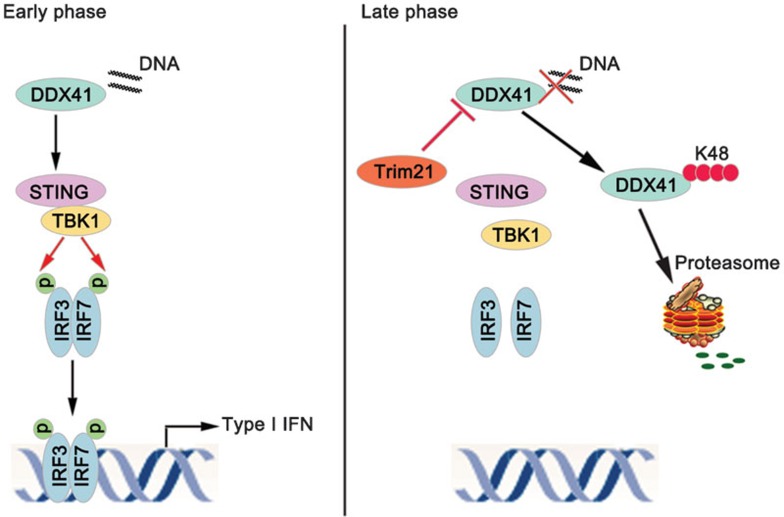
DDX41 was characterized as a protein that undergoes robust ubiquitination and degradation (Figure 1). The ubiquitin–proteasome pathway regulates the expression of many proteins involved in diverse cellular processes. Proteins targeted for degradation by the proteasome must first be tagged with polyubiquitin chains, linked through lysine at position 48 of ubiquitin (Lys48). Generally, ubiquitin tagging of proteins through an enzymatic cascade involves three enzymes: E1, E2 and E3.17
Figure 1.
Trim21 negatively regulates the DDX41-mediated signaling pathway. In the early phase, DNA from diverse microbes is sensed by DDX41 in the cytosol of infected cells as a danger signal. STING recruits TBK1 to its C-terminal tail, and TBK1 then interacts with and phosphorylates IRF3/7. Phosphorylated IRF3/7 dimerizes and translocates to the nucleus, thereby promoting the expression of immune and inflammatory genes. In the late phase, Trim21 interacts with DDX41 and causes the K48-linked ubiquitination of DDX41. After ubiquitination, DDX41 is degraded by the proteasome, thus preventing ‘downstream' cytokine induction.
Trim21 overexpression enhanced the K48-linked ubiquitination of DDX41 dependent on its RING domain. Moreover, K48-linked ubiquitination was detected in wild-type BMDCs, but not Trim21-deficient BMDCs. In vitro ubiquitination research showed that Trim21 directed modified DDX21 in the presence of E1, ubiquitin, ATP and E2 enzyme. Finally, Lys9 and Lys115 were identified as the ubiquitination sites of DDX41.
Many critical questions are raised by this study. It is notable that Trim21 expression was induced after stimulation with poly(dA:dT). However, Trim21 interacted with DDX41 under resting conditions and then dissociated from DDX41 after stimulation with dsDNA. Therefore, Trim21 regulated DDX41 at the steady state and the early stage of stimulation with dsDNA. It is puzzling that if the regulatory role of Trim21 ends at the early stage of stimulation, why would the host cells need to upregulate Trim21 expression? Does Trim21 target another protein after it dissociates from DDX41? IRF3 has been reported to interact with Trim21, but it appears that Trim21 has no effect on IRF3 expression. Then, does Trim21 have an effect on STING and TBK1, which signals downstream of DDX41? The answers to these questions may help us to better understand the role of Trim21 in the host cell response to intracellular dsDNA.
Another interesting question is how Trim21 deficiency affects the interferon response of dendritic cells to Listeria monocytogenes, a bacterial pathogen that induces interferon via the secretion of c-di-AMP.18 There are different opinions on the main sensor of cyclic dinucleotides. One theory is that DDX41 is the main direct sensor of cyclic dinucleotides,19 and the other recognizes STING in this role.20 If Trim21 has no effect on the STING-triggered signaling pathway, Trim21-deficient mice could be used to solve this problem. If Trim21 deficiency has no effect on interferon in response to L. monocytogenes, STING is more likely to be the sensor of cyclic dinucleotides. If Trim21-deficient mice have an impaired response to L. monocytogenes, DDX41 plays an important role in sensing the cyclic dinucleotides. Regardless of the actual results, they will help us to learn about DNA sensing.
Innate immune signaling plays an important role in the elimination of invading pathogens. However, the uncontrolled production of type I IFN induced by DNA and RNA may lead to autoimmune or auto-inflammatory diseases, such as systemic lupus erythematosus.21 Auto-inflammatory patients often have high levels of inflammatory cytokines, which are produced by the activation of innate immunity. Autoantibodies against the Trim21 protein can be detected in several different autoimmune disease patients and sometimes in healthy individuals. In systemic lupus erythematosus as well as systemic sclerosis and autoimmune myositis patients, anti-Trim21 is detected in approximately one-third of patients. The identification of Trim21 as a negative regulator of sensors of intracellular dsDNA will have important implications for understanding not only antiviral innate immunity, but also the pathogenesis of human autoimmune diseases, such as systemic lupus erythematosus.
References
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14:172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Shi HX, Liu XY, Shan YF, Wei B, Chen S, et al. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol. 2009;182:3782–3792. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- Yoshimi R, Chang TH, Wang H, Atsumi T, Morse HC 3 rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]