Abstract
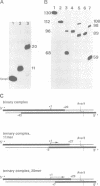
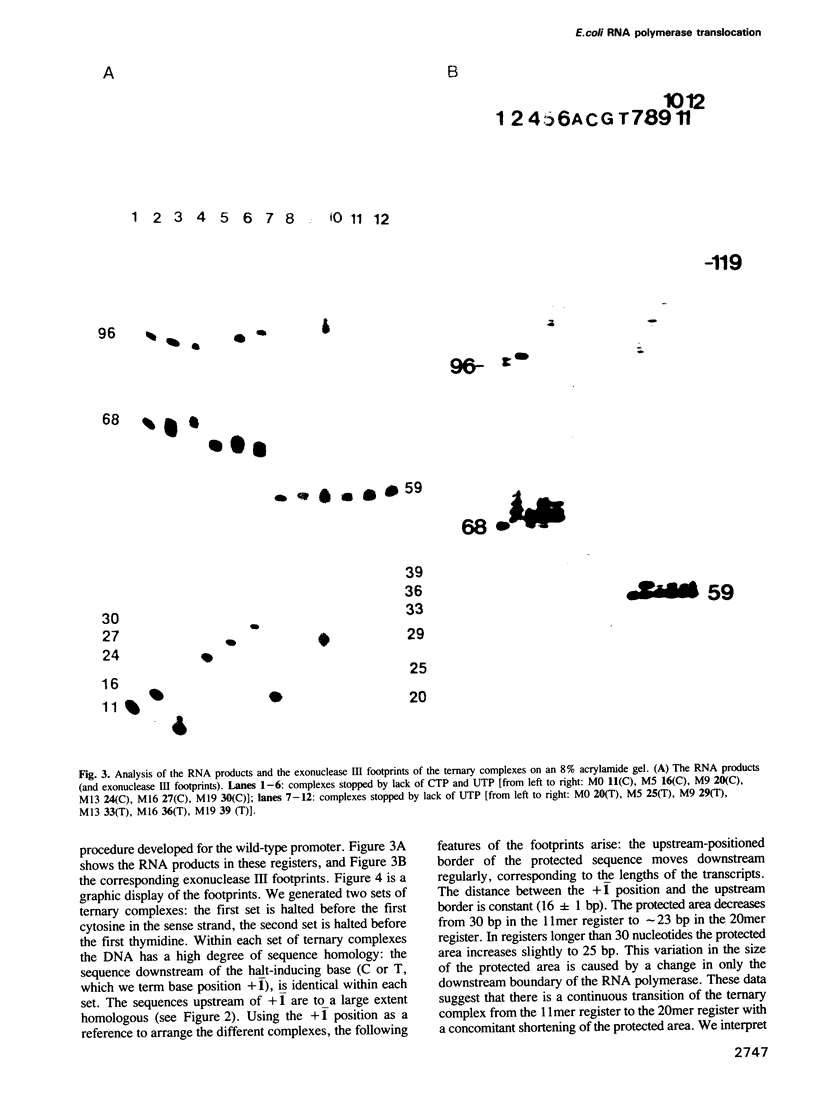
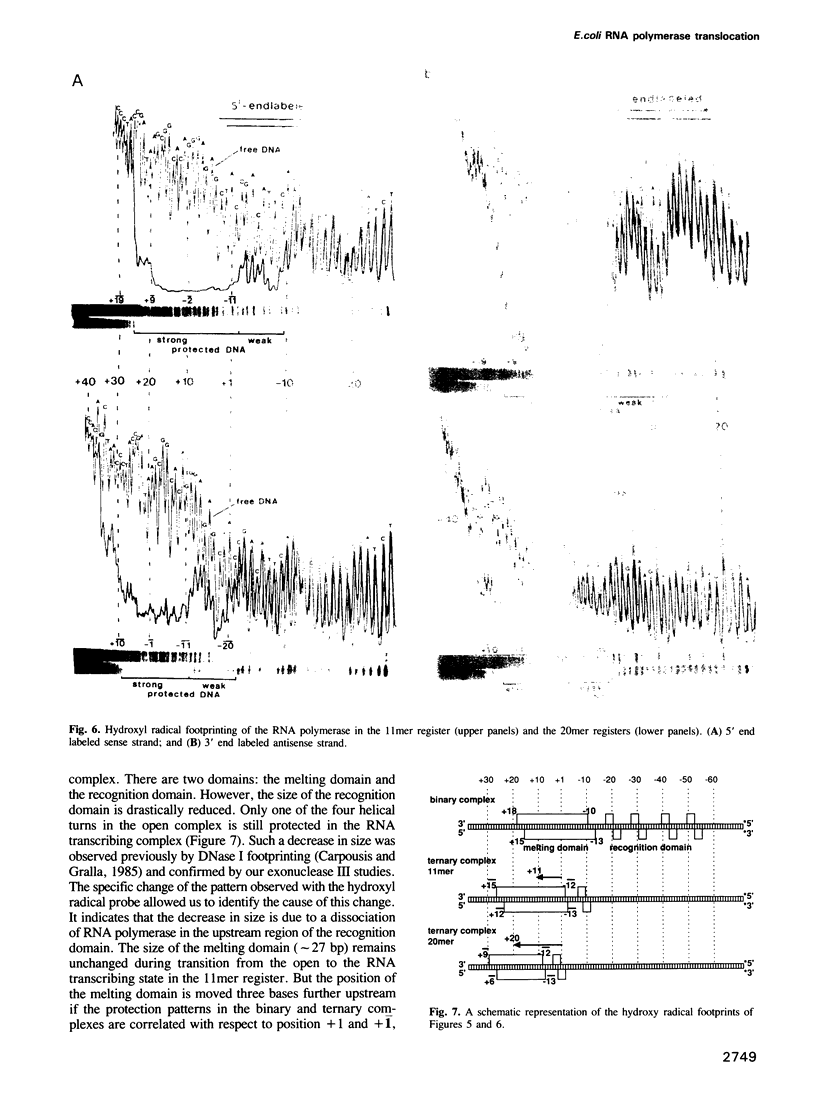
A series of RNA synthesizing transcription complexes, initiated at the T7 A1 promoter and halted at specific base positions ranging from +12 to +40, were analyzed by footprinting techniques; exonuclease III was used to determine the position of the bound RNA polymerase on the DNA and hydroxyl radicals were used to visualize the protein--DNA contact sites within the protected areas. In the binding (open) complex without RNA there are two DNA-domains, differing in their protection pattern. The first, extending from position +18 to -13, termed 'melting domain', is fully protected, whereas the second, extending from -14 to -55, termed 'recognition domain', shows only partial protection. At this domain, RNA polymerase is attached to one side of the DNA only, as indicated by the 10-bp periodicity of the protection pattern. Our data show that the formation of a mature RNA transcribing complex is characterized by dissociation of the RNA polymerase from the recognition domain, whereby the size of the melting domain remains constant. This process is accomplished if the nascent RNA has reached a length of 11 bases. As the RNA reaches a length of 20 bases, the size of the melting domain decreases from approximately 30 to 23 bp. Further RNA synthesis leaves the protection pattern essentially unchanged. These data demonstrate that the formation of a mature RNA transcribing complex can be described by at least two transitions.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Gralla J. D. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980 Jul 8;19(14):3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Gralla J. D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J Mol Biol. 1985 May 25;183(2):165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- Dennis D., Sylvester J. E. RNA polymerase: a model for rotational translocation. FEBS Lett. 1981 Feb 23;124(2):135–139. doi: 10.1016/0014-5793(81)80121-4. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982 May;29(1):81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Overexpression and purification of the sigma subunit of Escherichia coli RNA polymerase. Gene. 1983 Dec;26(2-3):109–118. doi: 10.1016/0378-1119(83)90180-4. [DOI] [PubMed] [Google Scholar]
- Hanna M. M., Meares C. F. Topography of transcription: path of the leading end of nascent RNA through the Escherichia coli transcription complex. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4238–4242. doi: 10.1073/pnas.80.14.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U. M., McClure W. R. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. II. Release of sigma from ternary complexes. J Biol Chem. 1980 Oct 25;255(20):9564–9570. [PubMed] [Google Scholar]
- Heumann H., Lederer H., Baer G., May R. P., Kjems J. K., Crespi H. L. Spatial arrangement of DNA-dependent RNA polymerase of Escherichia coli and DNA in the specific complex. A neutron small angle scattering study. J Mol Biol. 1988 May 5;201(1):115–125. doi: 10.1016/0022-2836(88)90443-3. [DOI] [PubMed] [Google Scholar]
- Heumann H., Lederer H., Kammerer W., Palm P., Metzger W., Baer G. Large-scale preparation of a DNA fragment containing the strong promoter A1 of the phage T7. Biochim Biophys Acta. 1987 Jul 14;909(2):126–132. doi: 10.1016/0167-4781(87)90034-0. [DOI] [PubMed] [Google Scholar]
- Heumann H., Metzger W., Niehörster M. Visualization of intermediary transcription states in the complex between Escherichia coli DNA-dependent RNA polymerases and a promoter-carrying DNA fragment using the gel retardation method. Eur J Biochem. 1986 Aug 1;158(3):575–579. doi: 10.1111/j.1432-1033.1986.tb09793.x. [DOI] [PubMed] [Google Scholar]
- Heumann H., Ricchetti M., Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988 Dec 20;7(13):4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer W., Deuschle U., Gentz R., Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 1986 Nov;5(11):2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa H., Mabuchi K., Futai M. Nucleotide sequence of the promoter region of the gene cluster for proton-translocating ATPase from Escherichia coli and identification of the active promotor. Biochem Biophys Res Commun. 1982 Jul 30;107(2):568–575. doi: 10.1016/0006-291x(82)91529-7. [DOI] [PubMed] [Google Scholar]
- Kinsella L., Hsu C. Y., Schulz W., Dennis D. RNA polymerase: correlation between transcript length, abortive product synthesis, and formation of a stable ternary complex. Biochemistry. 1982 May 25;21(11):2719–2723. doi: 10.1021/bi00540a022. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. R., Chamberlin M. J. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987 Jul 5;196(1):61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- McClure W. R., Cech C. L., Johnston D. E. A steady state assay for the RNA polymerase initiation reaction. J Biol Chem. 1978 Dec 25;253(24):8941–8948. [PubMed] [Google Scholar]
- Ricchetti M., Metzger W., Heumann H. One-dimensional diffusion of Escherichia coli DNA-dependent RNA polymerase: a mechanism to facilitate promoter location. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4610–4614. doi: 10.1073/pnas.85.13.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovskaya T. A., Chenchik A. A., Beabealashvilli R. Sh. Processive pyrophosphorolysis of RNA by Escherichia coli RNA polymerase. FEBS Lett. 1982 Jan 11;137(1):100–104. doi: 10.1016/0014-5793(82)80323-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalloway D., Kleinberger T., Livingston D. M. Mapping of SV40 DNA replication origin region binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980 Jun;20(2):411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Van Houten B., Hearst J. E. Interaction of Escherichia coli RNA polymerase with DNA in an elongation complex arrested at a specific psoralen crosslink site. J Mol Biol. 1988 Jan 20;199(2):277–293. doi: 10.1016/0022-2836(88)90314-2. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Spassky A. Visualization of the movement of the Escherichia coli RNA polymerase along the lac UV5 promoter during the initiation of the transcription. J Mol Biol. 1986 Mar 5;188(1):99–103. doi: 10.1016/0022-2836(86)90484-5. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987 Jan 20;193(2):267–278. doi: 10.1016/0022-2836(87)90218-x. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Comparison of the open complexes formed by RNA polymerase at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987 Jan 20;193(2):279–292. doi: 10.1016/0022-2836(87)90219-1. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A., Lamond A. I., Mace H. A., Berman M. L. RNA polymerase interactions with the upstream region of the E. coli tyrT promoter. Cell. 1983 Nov;35(1):265–273. doi: 10.1016/0092-8674(83)90229-5. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Zechel K., Halbwachs H. J. A new method of large scale preparation of highly purified DNA-dependent RNA-polymerase from E. coli. Hoppe Seylers Z Physiol Chem. 1970 Feb;351(2):221–224. doi: 10.1515/bchm2.1970.351.1.221. [DOI] [PubMed] [Google Scholar]